Abstract
An animal's ability to detect and avoid toxic compounds in the environment is crucial for survival. We show that the nematode Caenorhabditis elegans avoids many water-soluble substances that are toxic and that taste bitter to humans. We have used laser ablation and a genetic cell rescue strategy to identify sensory neurons involved in the avoidance of the bitter substance quinine, and found that ASH, a polymodal nociceptive neuron that senses many aversive stimuli, is the principal player in this response. Two G protein α subunits GPA-3 and ODR-3, expressed in ASH and in different, nonoverlapping sets of sensory neurons, are necessary for the response to quinine, although the effect of odr-3 can only be appreciated in the absence of gpa-3. We identified and cloned a new gene, qui-1, necessary for quinine and SDS avoidance. qui-1 codes for a novel protein with WD-40 domains and which is expressed in the avoidance sensory neurons ASH and ADL.
Keywords: avoidance, bitter taste, chemosensory neurons, quinine, WD-40 domain
Introduction
The detection of water-soluble substances is commonly defined as taste or gustation. A bitter sensation in mammals is mediated by a family of G-protein-coupled taste receptors expressed in the tongue, the T2Rs (Adler et al, 2000; Chandrashekar et al, 2000; Matsunami et al, 2000), which are coexpressed with the taste-specific G protein Gustducin. The molecular bases of bitter sensation in other animals are not known.
Caenorhabditis elegans is endowed with both olfaction and taste. Its major chemosensory organ, the amphid, functions in olfaction, taste, thermosensation and mechanosensation (reviewed in Bargmann and Mori, 1997). Of the 12 sensory neurons in each amphid, 11 are chemosensory and are associated with the amphid channel that opens through the cuticle to the outside world (Ward et al, 1975; Ware et al, 1975; White, 1986) (Figure 1). Genetic and laser microsurgery studies have been used to define functions for all amphid sensory neurons. Each neuron has a characteristic function that is reproducible between animals (reviewed in Bargmann and Mori, 1997). ASH, ADL, ASK and ASE sensory neurons have been shown to be responsible for the detection of some chemical repellents (Bargmann et al, 1990; Sambongi et al, 1999; Hilliard et al, 2002). ASH plays a major role in avoidance, whereas ADL, ASK and ASE play minor roles that are only evident when ASH is missing (Bargmann et al, 1990; Sambongi et al, 1999; Hilliard et al, 2002). In addition to its role in the avoidance of water-soluble repellents, the polymodal ASH sensory neuron also mediates avoidance of volatile repellents (Troemel et al, 1997) and avoidance of a mechanical stimulus such as touch on the nose (Kaplan and Horvitz, 1993).
Figure 1.
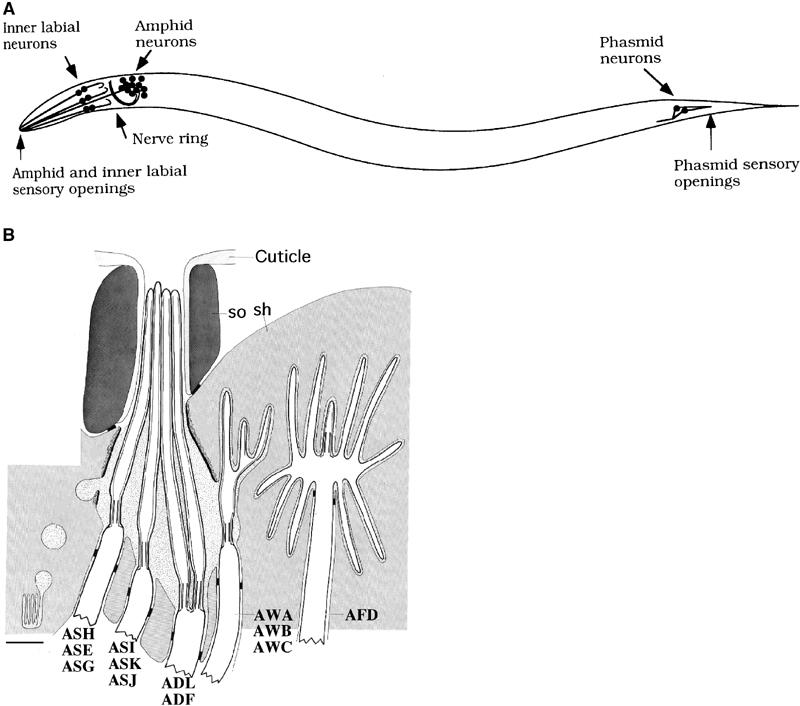
C. elegans chemosensory organs. Adapted from Ward et al (1975), Hedgecock et al (1985), Perkins et al (1986) and Bargmann and Mori (1997). (A) Scheme of the position of main sensilla in the animal. Dendritic processes of the sensory neurons extend to the external environment. The sensory cilia are the terminal part of the dendrites. (B) Details of amphid structure. The cilia of the AWA, AWB, AWC and AFD neurons are embedded in the sheath cell (sh). The cilia of the remaining eight chemosensory neurons reach the outside world through the amphid channel constituted by the socket cell (so). Bar is 1 μm.
Many chemosensory defective mutants that cannot avoid high osmotic strength (osm for OSMotic avoidance defective) have been isolated (Dusenbery, 1973; Ward, 1973; Lewis and Hodgkin, 1977; Culotti and Russell, 1978; Perkins et al, 1986; Bargmann et al, 1990, 1993; Starich et al, 1995; Wicks et al, 2000). Some of the osm mutants have defects in the structure of sensory cilia. Others have more specific defects and intact structure of the cilia. The molecular identification of these genes has revealed interesting new molecules involved in the avoidance of toxic stimuli. Osmotic avoidance requires the Gi-like G-α protein ODR-3, the TRP-related channel OSM-9 and the novel cytoplasmic protein OSM-10 (Colbert et al, 1997; Roayaie et al, 1998; Hart et al, 1999; Jansen et al, 1999).
In this paper, we identify 11 new chemicals that act in C. elegans as water-soluble repellents, and show that they largely overlap with molecules that are toxic and taste bitter to humans. We then focus on avoidance of the bitter substance quinine and, using laser ablations, show that ASH is the main sensory neuron responsible for quinine detection and that ASK plays a minor role. A genetic cell rescue strategy performed in a mutant background with shortened sensory cilia revealed that three cells, ASH, ADL and ASK, are sufficient for quinine avoidance. As for the molecules in sensory neurons necessary for quinine avoidance, we show that the G protein α subunit, GPA-3, is required and that the Gi-like G-α protein, ODR-3, plays a role that is revealed only when GPA-3 is missing. Finally, in a forward genetic screen, we identified and cloned qui-1, a new gene necessary for quinine and SDS (Sodium Dodecyl Sulfate) avoidance. QUI-1 is a novel large protein of 1592 aa, containing WD-40 repeats. qui-1∷GFP fusion gene is expressed in a few neurons, including the avoidance sensory neurons ASH and ADL.
Results
Bitter substances and alkaloids are sensed as repellents
To identify stimuli that act as repellents, 73 chemicals (see Supplementary Material 1) were tested with the drop test (Hilliard et al, 2002; see Materials and methods) on populations of wild-type N2 animals. Of these chemicals, 16 were found to induce clear avoidance responses. Figure 2 reports the responses obtained with these 16 stimuli at different concentrations. Five of the stimuli reported in Figure 2 had previously been identified as repellents for C. elegans: Cu2+, garlic extract, low pH, SDS and high osmotic strength. Two general features of the 11 new repellents stand out. First, many are plant alkaloids or derivatives (quinine, amodiaquine, chloroquine, primaquine and veratridine); a sixth repellent, shikimic acid, is an important precursor in the biosynthetic pathway of plant alkaloids. Second, most of the chemicals that trigger the avoidance reflex in C. elegans are sensed as bitter by humans or are discarded in double choice assays by other vertebrates. One of them, denatonium benzoate, is commonly considered as one of the bitterest natural compounds.
Figure 2.
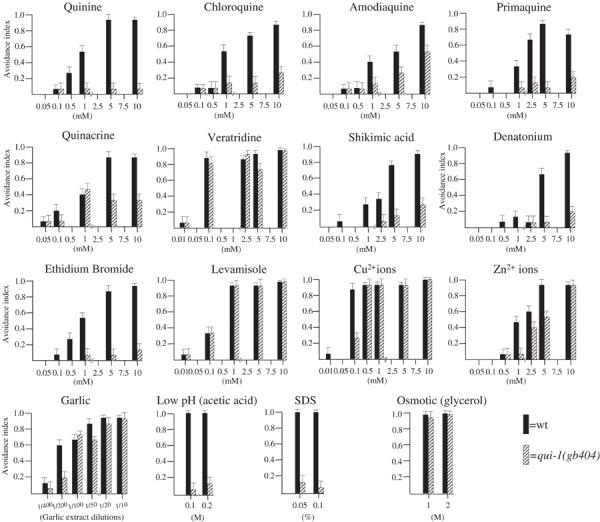
Eleven new water-soluble repellents. Each data set represents the response curves of the wild-type and qui-1(gb404) mutants to a defined repellent. The identified bitter substances are in the two top rows. For the other substances, no indication of their bitterness was found. Each black bar represents the avoidance index of a population of 100 wild-type (wt) animals, each tested with a single drop of repellent. The striped bars are the qui-1(gb404) responses. Bars indicate standard error of proportion values.
The active concentration capable of inducing an avoidance response in more than 50% of the trials was between 1 and 10 mM for most of the substances. In every case, lowering the concentration of the repellent produced a reduction in the avoidance response.
ASH is the main sensory neuron for the detection of quinine
We used laser ablation to identify sensory neurons necessary for quinine avoidance (Figure 3). Each operated animal was tested with 10–70 drops to give a quantitative index for its avoidance. Animals in which the ASH neurons (left and right) were killed showed strongly reduced responses to quinine hydrochloride (avoidance index=0.45; Figure 3), indicating that this cell is a central player in quinine detection. The ASH neurons have previously been implicated in osmotic, copper ions and SDS avoidance as well as in volatile and nose touch avoidance (Bargmann et al, 1990; Kaplan and Horvitz, 1993; Troemel et al, 1997; Hart et al, 1999; Sambongi et al, 1999; Hilliard et al, 2002). The residual avoidance response observed in ASH-ablated animals suggested that other sensory neurons play a role in quinine detection. To confirm that this residual response was not due to residual function of ASH processes, we tested these ablated animals for their avoidance response to the high osmotic strength stimulus, which is mediated only by ASH. Operated animals had an osmotic avoidance index of 0.1, suggesting that ASH function was mostly abolished.
Figure 3.
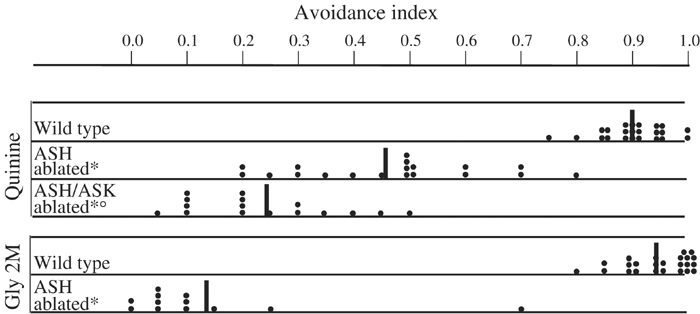
ASH and ASK are involved in avoidance. Scatter diagram plotting the avoidance index of single operated animals (each black dot represents one animal tested in a minimum of 10 drop tests). For each ablation, a minimum of 10 animals was generated and tested. Some animals were tested in the same way more than once. ASH-ablated animals showed a strong reduction in the response to quinine and an almost completely abolished response to high osmotic strength (glycerol). Ablation of ASH and ASK reduced the response to quinine compared to killing only ASH (P<0.01). * Indicates a statistically significant difference with the wild-type strain. ○ Indicates a statistically significant difference with the ASH-ablated animals. Black vertical bars indicate the mean response.
The ADL and ASK neurons have been implicated in the detection of other water-soluble repellents (Bargmann et al, 1990; Sambongi et al, 1999; Hilliard et al, 2002), and are thus good candidates for the supplementary cells mediating quinine avoidance. Ablation of the ASK sensory neuron alone did not reduce the response to quinine (data not shown). However, animals in which the ASH and ASK neurons had both been killed showed responses to quinine that were significantly lower than those of animals in which only the ASH neurons had been killed (Figure 3). This result indicates that ASK has a minor role in quinine avoidance, which is only revealed when ASH is killed. Ablation of ADL alone or in combination with ASH did not produce a significant reduction of quinine avoidance compared to wild-type and ASH-ablated animals, respectively (data not shown).
ASH, ASK and ADL are sufficient for quinine avoidance
To ask wether the three avoidance sensory cells ASH, ADL and ASK were sufficient for avoidance of quinine, we performed genetic cell rescue in the osm-6 mutant strain. In osm-6 mutants, most of the ciliated neurons have significantly shortened cilia that are not exposed to the environment (Lewis and Hodgkin, 1977; Perkins et al, 1986; Collet et al, 1998). As a result, osm-6 mutants are unable to detect many chemicals and show reduced avoidance responses to repellents, including quinine (Figure 4B). Because of these defective cilia, the sensory neurons of osm-6 animals do not take up the vital fluorescent dye DiO (Perkins et al, 1986; Starich et al, 1995). The osm-6 gene encodes a 472-residue protein that is a component of the intraflagellar transport machinery (Cole et al, 1998; Collet et al, 1998; Orozco et al, 1999; Qin et al, 2001). Mosaic analysis has shown that it acts cell-autonomously with respect to the DiO staining phenotype (Collet et al, 1998).
Figure 4.
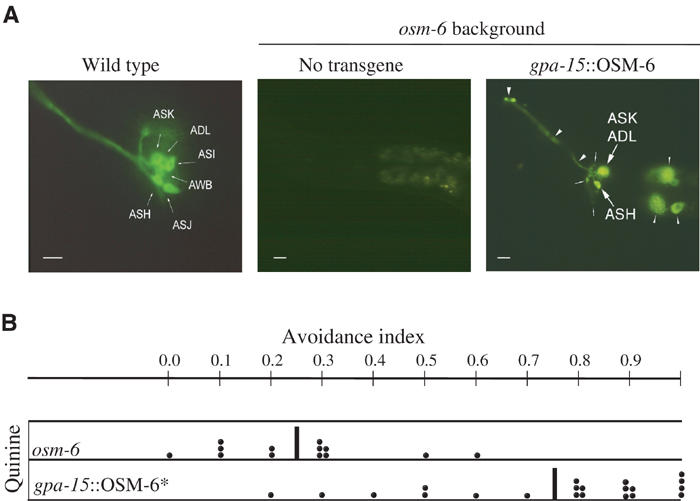
Anatomical and functional rescue of osm-6 mutants. (A) DiO staining of wild-type, osm-6 and gpa-15∷OSM-6 transgenic rescued strain. Animals were stained with DiO, which enters through functional cilia, and viewed with epifluorescence at high resolution. Thick arrows point to neuronal cell bodies, thin arrows point to axons; thick arrowheads point to dendrites and thin arrowheads point to intestinal cells expressing GFP from the elt-2∷GFP transformation marker. A control wild-type DiO staining shows the six cells on each side of the animal that take up the dye. An osm-6(p811) animal (no transgene) shows no staining of any of the amphidial neurons. In a gpa-15∷OSM-6 transformed animal, the cell bodies, axons and dendrites of the rescued neurons ASH, ASK and ADL are visible upon DiO staining. (B) Avoidance responses to quinine of osm-6 animals and of gpa-15∷OSM-6 rescued strain. The scatter diagram is obtained by plotting the avoidance index of single operated animals tested with a minimum of 10 drops (each black dot). Each row represents a minimum of 10 animals. Some of the animals were tested in the same way more than once. Behavioral assays were conducted prior to DiO staining. Only animals showing bright bilateral fluorescence with DiO were included in the figure. * Indicates a statistically significant difference from osm-6 (P<0.01). Black vertical bars indicate the mean.
A cell-specific promoter from the regulatory region of gpa-15 was fused in-frame to the wild-type osm-6 cDNA so that OSM-6 expression was restricted to ASH, ASK and ADL chemosensory cells. The fusion gene was injected into osm-6 animals and several transformant lines were obtained. In transformed animals, ASH, ASK and ADL sensory cilia extended the entire length of the amphid channel and these sensory neurons recovered the capacity to stain with DiO (Figure 4A). These rescued neurons also recovered their function in avoidance. Animals in which ASH, ASK and ADL staining with DiO was restored recovered their responses to quinine, high osmotic strength, SDS and copper (Figure 4B and data not shown). These rescue experiments represent an independent confirmation of the laser ablation experiments, and demonstrate that ASH, ASK and ADL sensory neurons are sufficient to restore avoidance of quinine.
GPA-3 and ODR-3 G-α subunits mediate quinine detection
In vertebrates, the taste-specific G protein α subunit Gustducin has been shown to mediate bitter transduction, and its expression is associated with that of bitter specific 7-TM receptors (Wong et al, 1996; Adler et al, 2000; Chandrashekar et al, 2000). In C. elegans, the complete family of genes encoding G proteins has been described and the expression pattern of each gene determined (Zwaal et al, 1997; Roayaie et al, 1998; Jansen et al, 1999). We asked if the G protein α subunits expressed in the C. elegans avoidance sensory neuron ASH could be responsible for quinine avoidance. ASH expresses several G-α genes, including gpa-3, gpa-13, gpa-15 and odr-3. The quinine avoidance response is significantly reduced in gpa-3 mutants but not in gpa-13, gpa-15 or odr-3 animals (Figure 5A). gpa-3 is expressed in ASH, ASK and ADL avoidance sensory neurons as well as in other sensory neurons, and the GPA-3 protein is predominantly localized at the tip of the sensory cilia, suggesting a function immediately downstream of the receptors (Zwaal et al, 1997; Jansen et al, 1999). The effect of the gpa-3 mutation on quinine detection is not due to a general loss of ASH function since, unlike ASH-ablated animals, gpa-3 animals are still capable of responding to high osmotic strength (Figure 5B, Zwaal et al, 1997; Jansen et al, 1999). A simple interpretation of this result suggests that GPA-3 function in the sensory neurons as the main signal transduction molecule downstream of the quinine receptor.
Figure 5.
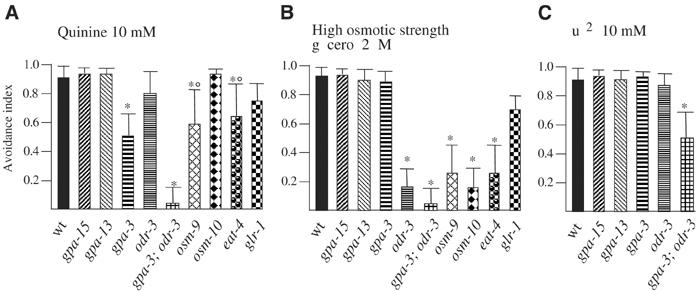
Avoidance responses of mutant animals. Avoidance index of (A) quinine, (B) high osmotic strength and (C) Cu2+ in different mutant animals. Each data point was obtained on populations of ⩾10 animals tested each with a minimum of 10 drops. The bars indicate the s.e.m. values. * Indicates a statistically significant difference with the wild-type strain, P<0.01. ○ Significantly different from the osm response (P<0.05).
Although reduced, the quinine response in gpa-3 mutant animals was not completely abolished. The residual response observed could be due to the effects of other G protein α subunits or to different unknown signal transduction molecules. We found that in the double mutants gpa-3;odr-3 the response to quinine was completely abolished (Figure 5A). This strong synthetic effect of odr-3 and gpa-3 revealed that ODR-3 is the other G protein α subunit mediating quinine avoidance, and that its function is masked when GPA-3 is present. Since the gpa-3;odr-3 double mutants are still capable of responding to copper ions (Figure 5C), it is likely that other G-α subunits, besides GPA-3 and ODR-3 or different signalling molecules, are involved in the avoidance response to other repellents. ODR-3 is expressed in five pairs of sensory neurons: AWA, AWB, AWC, ASH and ADF. Thus, ASH and ADF are the only neurons in which both proteins, GPA-3 and ODR-3, are expressed and, since ASH is the only one to which a function in water-soluble avoidance has been assigned, it is likely that ODR-3 exerts its function in quinine avoidance in ASH.
To discover other molecules involved in the pathway of the quinine response, we tested animals mutated in candidate chemoreception genes expressed in ASH (Figure 5A and B and Supplementary Material 2).
OSM-10 is a cytosolic protein expressed in ASH and a few other neurons (ASI, PHA and PHB), and necessary for high osmotic strength avoidance (Hart et al, 1999). Quinine avoidance in osm-10 mutants was at a level comparable to wild type (Figure 5A), indicating that this molecule is not used for quinine response. Conversely, osm-10 animals presented a strong reduced response to high osmotic strength (Figure 5B), confirming the result previously obtained by others using a different chemoreception assay (Bargmann et al, 1990; Hart et al, 1999).
OSM-9 is a TRPV-related, cation selective channel expressed in ASH as well as in ASK, ADL and ASE (Colbert et al, 1997; Tobin et al, 2002). Quinine avoidance was significantly reduced in osm-9 animals, indicating the participation of OSM-9 in response to quinine (Figure 5A). In addition, the presence of a significant residual response also suggests that other pathways not using OSM-9 contribute to the quinine avoidance (Figure 5A).
Finally, we tested mutants in eat-4 and glr-1, which are believed to function in avoidance to relay the signals from the sensory neurons to the movement commanding interneurons (Hart et al, 1995; Maricq et al, 1995; Lee et al, 1999; Mellem et al, 2002). EAT-4 may function as a glutamate vesicular transporter acting also in the sensory neurons ASH and ASK; GLR-1 is a glutamate receptor expressed in motor neurons and interneurons, including those that are synaptic targets of the ASH sensory neuron (Hart et al, 1995). The response to quinine of eat-4 mutants is significantly reduced compared to that of the wild type, while the response observed in glr-1 mutants, although slightly reduced, was not significantly different from that of wild type (Figure 5A). These results show that the response to quinine is, at least partially, mediated by glutamate and that glutamate receptors other than GLR-1 might be necessary for the avoidance response.
The avoidance gene qui-1 encodes a novel protein with WD-40 domains and is expressed in the avoidance sensory neuron ASH
To discover new genes that play a role in the avoidance of bitter substances, we used the drop test to screen for mutants that have lost the ability to avoid quinine. We analyzed 7500 haploid genomes after ethylmethanesulfonate (EMS) mutagenesis (see Materials and methods) and found 14 qui mutants (QUInine nonavoider mutants). Five mutants presented an unaltered structure of the sensory cilia when stained with the fluorescent dye DiO and a wild-type response to light touch, indicating relatively specific defects. One of these five mutants, qui-1(gb404), showed a very low avoidance of quinine as well as of SDS and low pH. Avoidance responses remained instead apparently unchanged to high osmotic strength and to some other repellents, while a smaller reduction of the response was observed for heavy metals when tested at intermediate concentrations (Figure 2). We cloned qui-1 using an SNP mapping strategy (Wicks et al, 2001). Initially, we positioned qui-1 in a small interval of about 120 kb on LG IV, between the two SNPs Y45F10A and C08F11A. We then rescued the quinine avoidance mutant phenotype by injecting long wild-type DNA as PCR products into the qui-1(gb404) strain (Figure 6 and Supplementary Material 3). The avoidance index for quinine was rescued to over 0.9 in qui-1(gb404) animals harboring the 20 kb fragment Y45F10B (1–21). For this region of the C. elegans genome, GENEFINDER predicted only a single gene, Y45F10B.10, containing 10 exons and spanning the last 6937 bp of the rescuing fragment. We determined the structure of qui-1 by analyzing cDNAs obtained from the C. elegans EST project (yk76c2, a gift from Yuji Kohara) as well as by RT–PCR and RACE experiments (Supplementary Material 3). The qui-1 coding sequence includes nine additional exons at the 5′ end of Y45F10B.10 as predicted by GENEFINDER, and is thus made up of 19 exons (Figure 6). The complete coding sequence (from ATG to TAA) spans 12 469 bp and is entirely contained in Y45F10B (1–21). The mature mRNA begins at position 6822 of the deposited Y45F10B sequence and is transpliced to SL1, 11 nt before the presumptive ATG starting codon. The mRNA is 4868 nt long with an ORF for a 1592 aa protein and a 3′ UTR 78 nt long. The TAA termination codon is at position 19301 of Y45F10B. Extensive RACE and RT–PCR analyses failed to detect alternative or shorter transcripts. We determined the sequence of Y45F10B.10 from two mutant strains that genetically define qui-1: the original gb404 mutant and gb681, a gb404 noncomplementing mutant isolated in a successive genetic screen. In qui-1(gb404), a CAA to TAA transition generates a stop codon in the fifth exon of Y45F10B.10, at aa 250 of QUI-1. In qui-1(gb681), whose phenotype is indistinguishable from that of gb404, a TGG to TGA transition generates a stop codon in the 13th exon at aa 1022 of QUI-1. The sequences of the two mutant alleles and the results obtained in the rescue experiments indicate that qui-1 corresponds to the revised version of Y45F10B.10.
Figure 6.
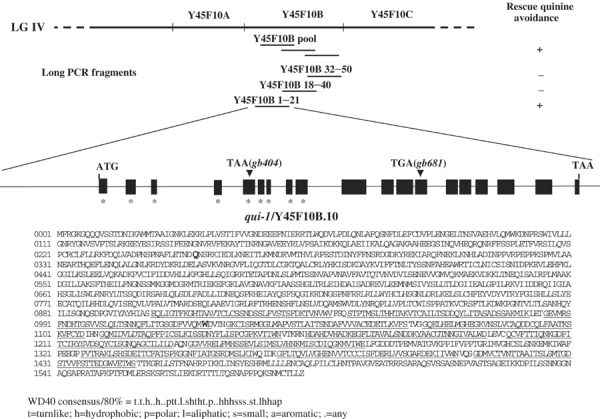
Rescue of the mutant phenotype and structure of qui-1. The PCR amplification products of the Y45f10B genomic sequence were injected in the qui-1(gb404) mutant strain, and the quinine avoidance behavior observed was indicated as + (rescued) or − (nonrescued). The structure of qui-1 with 19 exons is reported and the nine additional exons not predicted in the WormBase released sequence (GeneFinder) are indicated with an *. Exons are indicated as boxes and introns as lines, and both are not strictly to scale. The position and the nature of the mutation in the two alleles isolated, gb404 and gb681, are indicated with a triangle. In the protein sequence, the two amino-acid residues mutated to the stop codon in the two alleles gb404 and gb681 are written in bold and larger font. They are, respectively, in position 250 and 1022 aa. The WD-40 domains as determined using the SMART web program (Simple Modular Architecture Research Tool) are underlined. The WD-40 consensus is also shown. The revised version of the Y45F10B.10/qui-1 sequence has been submitted to WormBase, http://www.wormbase.org/.
Analysis of the predicted amino-acid sequence of QUI-1 reveals that it is a novel protein. The C. elegans genome codes for another protein, T05C3.2, whose primary sequence is weakly but significantly related to QUI-1 and whose function is unknown. QUI-1 appears to have been conserved through evolution since proteins with sequence similarity encompassing the whole length of QUI-1 are predicted in insects (Drosophila melanogaster NP_611338.1 and NP_650521.1) and mammals (Homo sapiens XP_211460.2 and XP_049078.3; mouse NP_795914.1 and XP_132047.3). In QUI-1 the region from aa 900 to aa 1447 contains 12 putative WD-40 domains, functional modules for protein–protein interactions present in a variety of proteins from many organisms. WD-40 repeats are present in a similar position also in T05C3.2 and in the homologs from insects and mammals. QUI-1 does not appear to have a protein secretion signal sequence or transmembrane segments, and thus it is likely to function within the cell (Figure 7). However, its cellular function cannot, at present, be inferred from its amino-acid sequence.
Figure 7.
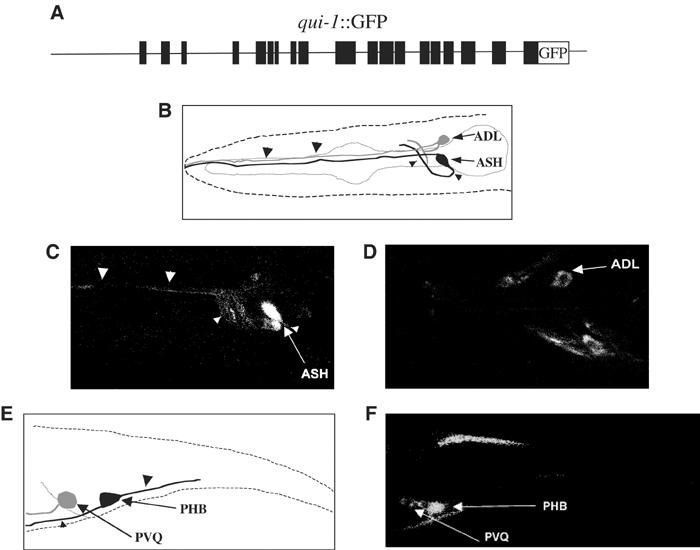
qui-1∷GFP expression and localization. (A) Scheme of the injected construct containing 6 kb of 5′ upstream sequences and the full-length gene fused to GFP. (B) Schematic diagram of the animal's head with the position and shape of ASH and ADL sensory neurons. The anterior is at the left and the ventral side is down in all the panels. (C, D) Fluorescent confocal image of an L1 animal carrying the transgene qui-1∷GFP. The GFP expression is visible in ASH (C) and the cell is uniformly stained in all cell compartments, cytosol, nucleus, dendrite and axon. In ADL (D), GFP is weaker, less diffused and absent from the nucleus. (E) Schematic diagram of the animal tail with highlighted PHB and PVQ neurons. (F) Fluorescent confocal image of an L1 animal where PHB (brighter) and PVQ (weaker) are visible. Arrows point to cell bodies, large arrowheads point to dendrites and small arrowheads point to axons.
The expression of qui-1 was examined using fusions of the gene to the reporter protein GFP. The coding sequence of GFP was fused to qui-1 immediately before its terminal codon within a 21 kb fragment, qui-1∷GFP (Figure 7A) (see Supplementary Material 3), which is the same as the one used for the rescue experiment except that it did not include the 761 bp that follow the QUI-1 stop codon. GFP was visible in the processes of the nerve ring and in several neurons of the lateral and ventral ganglia, including the avoidance sensory neurons ASH and ADL (Figure 7B–D). We did not detect GFP expression in ASK sensory neurons. Four additional neurons in the retrovesicular ganglion and two neurons in the lumbar ganglion, PVQ and the sensory neuron PHB, also express GFP (Figure 7E and F). This expression pattern partially overlaps but is not completely coincident with that obtained with a much shorter construct in which GFP was fused to the second exon, and the construct was thus missing part of exon 2 and the last 17 introns and exons of the gene (not shown). Expression from this shorter construct included neither ASH nor PHB, indicating that important regulatory elements are present within the coding region of qui-1. In the ASH sensory neurons, the reporter protein uniformly stained the cell body, nucleus, dendrite and axon. The staining was too faint to unambiguously determine whether QUI-1∷GFP localized also in the cilia. In most of the other neurons, expression of GFP was lower in all cellular compartments and apparently absent from the nucleus. The expression of QUI-1 in ASH and ADL suggests that, for its role in quinine avoidance, it functions in these cells.
Discussion
Water-soluble avoidance and bitter taste
Many toxic compounds are tasted as bitter by humans, and elicit aversive reflexes such as tongue retraction, retarded swallowing and nausea. The same bitter substances are tasted as unpalatable by many other vertebrate and invertebrate species (Glendinning, 1994). C. elegans lives in the soil at the air–water interface and is exquisitely sensitive to chemical cues. Its survival requires it to avoid predators and poisonous compounds in the environment. We have shown that, of the chemicals that C. elegans avoids, an important group is constituted by plant alkaloids, including quinine. These are tasted as bitter by humans and are toxic for most animals. We speculate that the avoidance reflex of C. elegans mirrors the aversive behavior observed in vertebrates and suggests a central evolutionary drive, maintained across different phyla, to avoid dangerous molecules. In mammals, bitter taste is mediated by G-protein-coupled receptors of the T2R family (Adler et al, 2000; Chandrashekar et al, 2000; Matsunami et al, 2000). One mammalian denatonium receptor has been identified (Chandrashekar et al, 2000) but, although C. elegans senses denatonium as a repellent and possesses a huge family of 7-TM candidate chemoreceptors, no close homolog of the mammalian gene can be identified in the nematode genome. The lack of conservation of T2Rs in nematode suggests that different primary receptors might underlie a common behavioral strategy. It is interesting to note that many substances tasted and smelled by humans and lower animals are produced by Angiosperms, the flowering plants. These plants originated only about 150 million years ago, long after the separation of the animal phyla 800–1000 million years ago. The relatively recent evolution of novel tastes and odors in fruits, flowers and other plant parts may have driven the evolution of chemosensory receptors in different animal phyla starting from the same basic molecular structure of seven transmembrane proteins.
Avoidance sensory neurons
Our laser ablation results show that the amphid polymodal avoidance neuron ASH is the main sensory neuron necessary for proper response to quinine. This result attributes yet another function to this neuron, which appears to mediate a wide diversity of aversive behavioral responses.
A second cell, ASK, has a minor role in the avoidance of quinine. The role of ASK is evident only in animals where ASH is missing. ASK was previously shown to have a minor role in chemotaxis of attractant stimuli (Bargmann and Horvitz, 1991). It is intriguing that this cell contributes to both attractant and repulsive behaviors; the mechanisms underlying these opposite roles remain to be elucidated.
The results of the cell-specific osm-6 rescue show that the three avoidance sensory neurons ASH, ASK and ADL are sufficient among ciliated neurons for the response to quinine. Sensory functions that are redundant in many sensory neurons or encoded in complex networks would be misinterpreted in experiments relying entirely on laser ablation. The cell-specific genetic rescue is a valuable complementary approach to identify sensory functions. Since osm-6 mutant animals are defective in most of the behaviors mediated by sensory cilia, this approach will open new possibilities in the study of sensory cell function.
The results presented in this paper are in agreement with a model in which receptors for various repellents, in this case quinine, are present on more than one sensory neuron and that each avoidance neuron can detect more than one repellent. A similar arrangement of overlapping functions with one major sensory cell has been described for the organization underlying chemotaxis to attractants in C. elegans, and a similar strategy is used for sensory control of entry into and exit from the Dauer developmental pathway (reviewed in Bargmann and Mori, 1997).
Genes involved in avoidance of water-soluble repellents
In vertebrates, bitter taste uses T2Rs 7-TM receptors and Gustducin as the intracellular signalling molecule (Wong et al, 1996; Adler et al, 2000; Chandrashekar et al, 2000; Matsunami et al, 2000). Although a specific bitter chemoreceptor protein in C. elegans has not yet been identified, our results show that two G protein α subunits, GPA-3 and ODR-3, expressed and localized on the cilia of the avoidance sensory neurons, are necessary for quinine detection. However, while GPA-3 shows a predominant role in detecting quinine, the function of ODR-3 is revealed only when GPA-3 is missing. Considering the central role played in quinine avoidance by ASH sensory neurons, it is reasonable to think that GPA-3 and ODR-3 are both specifically required in this cell to relay the quinine signal, while GPA-3 will be acting also into the ASK (and/or ADL) sensory neuron. In fact, the completely abolished response observed in the double mutants gpa-3;odr-3 resembles that observed in the ASH/ASK-ablated animals, suggesting that the two signalling proteins are key components of the sensory machinery for quinine detection in these neurons. Further studies might reveal a possible specific role that each of these two G proteins play in the sensory neurons where they are expressed.
We used the same candidate molecules approach to identify other genes that are part of the quinine response pathway.
The TRPV-related cation selective channel OSM-9 is thought to be involved in the signalling and depolarization of the ASH sensory neuron (Colbert et al, 1997; Tobin et al, 2002). We found that this molecule is necessary for a proper quinine response, although its function might be redundant with other molecules since a residual response is still observed. Considering that all the ASH responses tested so far in osm-9, by us and by other researchers, seem to be totally or partially compromised, it is possible that in addition to some specific effects, this cation channel is also regulating more in general ASH function.
Another important aspect that emerged from our experiments is that the quinine avoidance response is achieved, at least partially, using glutamate as the neurotransmitter. eat-4 animals, defective in the glutamate vesicular transporter, showed a reduced avoidance index when tested with quinine. eat-4 is expressed in several neurons, including ASH and ASK sensory neurons, and it is reasonable to believe that its function in one or both of these cells might be necessary for a proper quinine response. The substantial residual response observed indicates that other neurotransmitters or different glutamate transporters must however also be involved. Our results also indicate that the glutamate receptor GLR-1 either has no role or has a minor one in relaying the quinine signal to the interneurons that control movement.
QUI-1, a new player in avoidance
We have identified a novel WD-40 domain containing protein QUI-1, which is required for quinine avoidance. qui-1, described in this paper, represents a new component of the machinery for sensing quinine, SDS and some other repellents, which is expressed in ASH, ADL and other neurons. Homologs of QUI-1 are predicted to exist in other organisms, including insects and mammals, but at the moment the relationship between QUI-1 and these proteins with respect to their function is still not clear. The sequence of QUI-1 reveals many WD-40 motifs, which are generally involved in protein–protein interactions but which can also have more specialized functions. QUI-1 is very similar in its WD-40 motifs to the Podospora anserina heterokaryon incompatibility gene het-e-1, a WD-40-containing protein that interacts genetically with het-c, a predicted glycolipid transfer protein (GLTP) (Espagne et al, 2002). Podospora heterokaryon instability results in cell death following inappropriate cell fusions, through signalling pathways that are not well understood. Mammalian GLTPs bind several glycosphingolipids and glyceroglycolipids and allow their transfer between donor and acceptor liposomes; if QUI-1 has a similar partner protein, it might be involved in lipid signalling in the ciliated sensory neurons. The WD-40 repeats in QUI-1 are also related to repeats in the mammalian apoptosis protease activating factor APAF-1. In APAF-1, the WD-40 repeats inhibit the apoptotic activity of the protein, and deleting them results in constitutive activation of apoptosis.
The behavioral defects shown by qui-1 mutants and the apparent lack of specific membrane localization of QUI-1∷GFP fusion protein suggest that it is not the receptor for quinine itself. Although it could be involved in sensory signal transduction, other proteins implicated in chemosensory signal transduction such as GPA-3 and ODR-3 are, like the receptors, enriched in the sensory cilia. A role for WD-40 proteins in sensory cilia biosynthesis has been revealed by CHE-2, a WD-40 domain protein necessary for proper cilia formation (Fujiwara et al, 1999). In che-2 mutant animals, all sensory cilia are shortened and do not reach the external environment; thus, CHE-2 may have an essential role in the process of biosynthesis of all cilia. qui-1 has a different expression pattern and appears to be more specific in its function since its chemosensory defects are less general and the structure of the cilia is apparently normal in the mutant. The function of the main avoidance neuron is not completely compromised in qui-1 mutants, which can still sense the high osmotic strength stimulus and the mechanical stimulus ‘nose touch', both mediated mostly by ASH. These results implicate qui-1 in a more specific aspect of ASH and ADL sensory function.
Expression of qui-1 in ADL, although consistent with the result of the cell rescue experiment, is in apparent contrast with the fact that laser ablation of ADL, alone or in combination with the ablation of ASH, did not result in a reduction of quinine avoidance. We do not know the reason for this apparent discrepancy. One possibility is that ADL is a very minor player in quinine avoidance and that QUI-1 has in ADL also a different function, not related to quinine avoidance.
Responses to various aversive stimuli are mediated in C. elegans by only a few sensory neurons, with ASH being the main one. Our results on quinine avoidance of mutants in various chemoreceptor genes, including the newly discovered qui-1, indicate that the proteins required to transduce the signals for the different stimuli downstream of the receptors overlap only partially. This supports the hypothesis that combinatorial strategies at the cellular and at the molecular level may be at work in the sensory system of this simple organism, and may underlie the capacity of worms to discriminate between different repellents even with the limited number of sensory neurons dedicated to the detection of repellent stimuli.
Materials and methods
Strains
Wild-type animals were C. elegans variety Bristol, strain N2. Wild-type Hawaiian strain CB4856 was used for SNP mapping strategy. Mutant strains included: PR808 osm-1(p808) X; CB1124 che-3(e1124) (aka osm-2) I; PR802 osm-3(p802) IV; CB1387 daf-10(e1387) (aka osm-4) IV; PR813 osm-5(p813) X; PR811 osm-6(p811) V; MT3564 osm-7(n1515) III; CX10 osm-9(ky10) IV; MT3641 osm-10(n1602) III; MT3643 osm-11(n1604) X; CX2937 tax-4(p678) III; KP4 glr-1(n2461) III; NL2330 gpa-13(pk1270) V; NL797 gpa-15(pk477) I; NL335 gpa-3(pk35) V; CX2205 odr-3(n2150) V; MT6318 eat-4(n2474) III. NA404 qui-1(gb404) IV and NA681 qui-1(gb681) IV were generated in the course of this work. Animals were grown under uncrowded conditions at 20°C on NGM agar plates seeded with Escherichia coli strain OP50 (Brenner, 1974).
Single-worm assay
A detailed protocol of the ‘drop test' assay is described in Hilliard et al (2002). In brief, a drop of a solution is delivered near the tail of a moving animal. When the substance is a repellent, the animal immediately ceases forward movement and reverses; if buffer alone is delivered, the animal will continue moving forward. The assay is conducted always on unseeded plates.
The response of a single animal to each drop delivered is recorded as either a positive or negative response. Responses are considered positive when the avoidance reflex is observed within 4 s after the animal encounters the substance. An inter stimuli interval (ISI) of at least 2 min is used between successive drops to the same animal. Each animal is tested with no more than 20 successive drops; the animal is then transferred onto a new seeded plate and allowed to recover for 1 or 2 h before starting a new set of experiments. No more than three sets of 20 drops each were conducted per day on each animal.
Population assays
Two methods were used to define the responses of populations of worms.
A population of well-fed adult animals was washed three times with M13 buffer (Tris 30 mM, 100 mM NaCl, 10 mM KCl) and then placed on an unseeded NGM agar plate (or an M13 plate for the experiment in Figure 3). The animals were allowed to rest for 15 min, and then at least two series of 50 animals of the same population were challenged with a single drop of the chosen substance and each response was recorded as either positive or negative.
In all, 10–50 well-fed adult animals of the same population were placed each on an NGM unseeded agar plate and allowed to rest for 5–10 min. Each animal was then tested with several successive drops (5–60) as described in the single-worm assay. The results of all animals belonging to the same population were then combined.
The results of either single-worm or population assays are represented as an avoidance index, which is defined as the number of positive responses divided by the total number of trials (drops delivered).
Laser killing of neurons
Individual neurons were identified using Nomarski optics based on position and morphological cues as previously described (Bargmann and Horvitz, 1991). Cells were killed in the first larval stage (L1) using a laser microbeam focused through the 100× objective of a Zeiss Axioskop microscope, as described by Avery and Horvitz (1987, 1989). Animals were tested 48 h after the operation when they were young adults. Only animals whose growth rate was similar to that of unoperated ones were used for avoidance testing. After the behavioral tests had been completed, animals were individually stained with DiO or DiI to confirm that the expected cells had been ablated and that the integrity of the amphid structure was preserved. The results reported refer only to animals meeting these criteria.
osm-6 expression
All general molecular biology manipulations were performed using standard methods (Sambrook et al, 1989). gpa-15∷OSM-6 fusion gene was generated using an osm-6 cDNA (1476 nt) cloned in pBluescript (kindly given by Jocelyn Shaw, Minnesota).
The promoter sequences of gpa-15 (Jansen et al, 1999) were obtained by PCR amplification using the plasmid gpa-15∷GFP as a template (kindly given as a gift by Gert Jansen, Rotterdam). The amplified fragment contains the initiator ATG and was cloned in-frame with the osm-6 cDNA using appropriate restriction sites whose sequences had been added to the amplification primers. Details of the fusion gene and the gene itself are available on request.
The translational fusion gene, together with a selectable marker, was injected in osm-6 mutant animals following the standard C. elegans transformation procedure (Mello and Fire, 1995). Both transgene and marker were injected at 50 ng/μl. As a cotransfection marker, we used elt-2∷GFP, which is expressed in intestinal cells (a gift from J McGhee, Calgary). F2 stable lines were selected by scoring for GFP expression from the marker under a stereoscope equipped with epifluorescence. To confirm the anatomical rescue of the sensory cilia, transgenic animals were stained following the standard protocol with the fluorescent dye DiO (Starich et al, 1995). For the experiment in Figure 4, adult animals from transformed lines were chosen randomly and tested for avoidance. After testing, animals were individually scored for the presence of the GFP marker and stained with DiO to determine, in each animal, which cells had been rescued anatomically. In Figure 4B, we report only the results obtained on animals whose DiO staining pattern indicated the complete rescue of ASH, ASK and ADL.
Isolation of quinine nonavoider mutants
Wild-type animals were mutagenized with EMS as described by Brenner (1974), and allowed to self-fertilize for two generations. The F2 progeny was screened on NGM plates using the drop test with a solution of 10 mM quinine hydrochloride. For screening, a population of about 300 animals was washed three times with M13 buffer and then placed on the agar plate. The animals were tested individually with quinine drops to identify nonavoiders. Nonresponders were placed on individual plates, fed and tested again several times to confirm their quinine avoidance defect. The progeny of candidate mutants was assayed again to confirm that the phenotype bred true. Mutants were outcrossed at least twice with the wild-type strain N2. NA404(gb404) is one of a set of mutants isolated using this procedure. gb404 animals responded normally to light touch and the cilia of their exposed sensory neurons stained, with the fluorescent dye DiO, like those of wild-type animals. gb404 animals are somewhat dumpish and male tail abnormalities are often observed. Despite this defect, animals were able to mate efficiently. Mutants were tested for the nose touch response and only a minor reduction of the avoidance response was observed (avoidance index 0.76 versus wild type 0.9). gb404 animals are partially defective in the avoidance response to the volatile repellent octanol (time to respond 11.6±2 s versus 2.3±0.5 s for the wild type; Hana Sugimoto and Anne Hart, personal communication).
A second, noncomplementing allele of qui-1, gb681 was isolated in a successive screen. qui-1(gb681) mutant animals have normal sensory cilia and their behavioral characteristics are indistinguishable from those of qui-1(gb404).
Microscopy
Worms were viewed using a Zeiss Axioskop equipped with epifluorescence and DIC, and images were collected with an Axiocam digital camera. The expression of GFP in animals transformed with the qui-1∷GFP reporter was analyzed using a Leica TCS SP2 confocal microscope. Cell identification was under DIC optics and was aided, for the sensory neurons, by staining with DiI.
Statistical analysis
t-test, mean values, and s.d. and s.e.m. values were calculated using Primers of Biostatistics software (Stanton Glantz).
Supplementary Material
SUPPLEMENTARY MATERIAL 1
SUPPLEMENTARY MATERIAL 2
SUPPLEMENTARY MATERIAL 3
Acknowledgments
We thank Cori Bargmann for her crucial help with the laser ablations, for suggesting the osm-6 rescue experiments and for her invaluable contribution to the preparation of this manuscript; Gert Jansen for very generously providing gpa-15, gpa-13 and gpa-3 mutants and the plasmids with the gpa-15 promoter; Jocelyn Shaw for providing osm-6 cDNA; Jim McGhee for the elt-2∷GFP plasmid; Yuji Kohara for qui-1 cDNA; Steve Wicks for his kind assistance in the identification of SNPs; Alan Coulson and John Sulston for sending YACs and cosmids and the C. elegans Sequencing Project for the released sequences of Y45F10B.10. We thank Emma Lewis, who contributed greatly during the first stages of this work to the development of the drop test assay. We are grateful to Anna Sollo for excellent technical support and Umberto di Porzio, Franco Graziani, Carla Perrone Capano Elia di Schiavi, Mario de Bono and Maria Rosaria Sapio for advice during the course of this work and for reading the manuscript. Most of the strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by the National Institute of Health, National Center for Research Resources. This work was supported by grants to PB (Human Frontiers Science Program, MURST-Cluster 02, Telethon-Italy grant GP1122Y00, FIRB Neuroscienze RBNE01WY7P). MAH was partially supported by an EMBO short-term fellowship.
References
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS (2000) A novel family of mammalian taste receptors. Cell 100: 693–702 [DOI] [PubMed] [Google Scholar]
- Avery L, Horvitz HR (1987) A cell that dies during wild-type C. elegans development can function as a neuron in a ced-3 mutant. Cell 51: 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery L, Horvitz HR (1989) Pharyngeal pumping continues after laser killing of the pharyngeal nervous system of C. elegans. Neuron 3: 473–485 [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Hartwieg E, Horvitz HR (1993) Odorant-selective genes and neurons mediate olfaction in C. elegans. Cell 74: 515–527 [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Horvitz HR (1991) Chemosensory neurons with overlapping functions direct chemotaxis to multiple chemicals in C. elegans. Neuron 7: 729–742 [DOI] [PubMed] [Google Scholar]
- Bargmann CI, Mori I (1997) Chemotaxis and thermotaxis. In C. elegans II, Riddle DL, Bluementhal T, Mayer BJ, Priess JR (eds) pp 717–737. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [PubMed] [Google Scholar]
- Bargmann CI, Thomas JH, Horvitz HR (1990) Chemosensory cell function in the behavior and development of Caenorhabditis elegans. Cold Spring Harbor Symp Quant Biol 55: 529–538 [DOI] [PubMed] [Google Scholar]
- Brenner S (1974) The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ (2000) T2Rs function as bitter taste receptors. Cell 100: 703–711 [DOI] [PubMed] [Google Scholar]
- Colbert HA, Smith TL, Bargmann CI (1997) OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci 17: 8259–8269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole DG, Diener DR, Himelblau AL, Beech PL, Fuster JC, Rosenbaum JL (1998) Chlamydomonas kinesin-II-dependent intraflagellar transport (IFT): IFT particles contain proteins required for ciliary assembly in Caenorhabditis elegans sensory neurons. J Cell Biol 141: 993–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet J, Spike CA, Lundquist EA, Shaw JE, Herman RK (1998) Analysis of osm-6, a gene that affects sensory cilium structure and sensory neuron function in Caenorhabditis elegans. Genetics 148: 187–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culotti JG, Russell RL (1978) Osmotic avoidance defective mutants of the nematode Caenorhabditis elegans. Genetics 90: 243–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dusenbery DB (1973) Countercurrent separation: a new method for studying behavior of small aquatic organisms. Proc Natl Acad Sci USA 70: 1349–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espagne E, Balhadere P, Penin ML, Barreau C, Turcq B (2002) HET-E and HET-D belong to a new subfamily of WD40 proteins involved in vegetative incompatibility specificity in the fungus Podospora anserina. Genetics 161: 71–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara M, Ishihara T, Katsura I (1999) A novel WD40 protein, CHE-2, acts cell-autonomously in the formation of C. elegans sensory cilia. Development 126: 4839–4848 [DOI] [PubMed] [Google Scholar]
- Glendinning JI (1994) Is the bitter rejection response always adaptive? Physiol Behav 56: 1217–1227 [DOI] [PubMed] [Google Scholar]
- Hart AC, Kass J, Shapiro JE, Kaplan JM (1999) Distinct signaling pathways mediate touch and osmosensory responses in a polymodal sensory neuron. J Neurosci 19: 1952–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart AC, Sims S, Kaplan JM (1995) Synaptic code for sensory modalities revealed by C. elegans GLR-1 glutamate receptor. Nature 378: 82–85 [DOI] [PubMed] [Google Scholar]
- Hedgecock EM, Culotti JG, Thomson JN, Perkins LA (1985) Axonal guidance mutants of Caenorhabditis elegans identified by filling sensory neurons with fluorescein dyes. Dev Biol 111: 158–170 [DOI] [PubMed] [Google Scholar]
- Hilliard MA, Bargmann CI, Bazzicalupo P (2002) C. elegans responds to chemical repellents by integrating sensory inputs from the head and the tail. Curr Biol 12: 730–734 [DOI] [PubMed] [Google Scholar]
- Jansen G, Thijssen KL, Werner P, van der Horst M, Hazendonk E, Plasterk RH (1999) The complete family of genes encoding G proteins of Caenorhabditis elegans. Nat Genet 21: 414–419 [DOI] [PubMed] [Google Scholar]
- Kaplan JM, Horvitz HR (1993) A dual mechanosensory and chemosensory neuron in Caenorhabditis elegans. Proc Natl Acad Sci USA 90: 2227–2231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R, Sawin ER, Chalfie M, Horvitz HR, Avery L (1999) EAT-4, a homolog of a mammalian sodium-dependent inorganic phosphate cotransporter, is necessary for glutamatergic neurotransmission in Caenorhabditis elegans. J Neurosci 19: 159–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JA, Hodgkin JA (1977) Specific neuroanatomical changes in chemosensory mutants of the nematode Caenorhabditis elegans. J Comp Neurol 172: 489–510 [DOI] [PubMed] [Google Scholar]
- Maricq AV, Peckol E, Driscoll M, Bargmann CI (1995) Mechanosensory signaling in C. elegans mediated by the GLR-1 glutamate receptor. Nature 378: 78–81 [DOI] [PubMed] [Google Scholar]
- Matsunami H, Montmayeur JP, Buck LB (2000) A family of candidate taste receptors in human and mouse. Nature 404: 601–604 [DOI] [PubMed] [Google Scholar]
- Mellem JE, Brockie PJ, Zheng Y, Madsen DM, Maricq AV (2002) Decoding of polymodal sensory stimuli by postsynaptic glutamate receptors in C. elegans. Neuron 36: 933–944 [DOI] [PubMed] [Google Scholar]
- Mello C, Fire A (1995) DNA transformation. Methods Cell Biol 48: 451–482 [PubMed] [Google Scholar]
- Orozco JT, Wedaman KP, Signor D, Brown H, Rose L, Scholey JM (1999) Movement of motor and cargo along cilia. Nature 398: 674. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG (1986) Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol 117: 456–487 [DOI] [PubMed] [Google Scholar]
- Qin H, Rosenbaum JL, Barr MM (2001) An autosomal recessive polycystic kidney disease gene homolog is involved in intraflagellar transport in C. elegans ciliated sensory neurons. Curr Biol 11: 457–461 [DOI] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI (1998) The G αprotein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron 20: 55–67 [DOI] [PubMed] [Google Scholar]
- Sambongi Y, Nagae T, Liu Y, Yoshimizu T, Takeda K, Wada Y, Futai M (1999) Sensing of cadmium and copper ions by externally exposed ADL, ASE, and ASH neurons elicits avoidance response in Caenorhabditis elegans. Neuroreport 10: 753–757 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Maniatis T, Fritsch EF (1989) Molecular Cloning: A Laboratory Manual, 2nd edn, Cold Spring Harbor, NY: Cold Spring Harbor Laboratory [Google Scholar]
- Starich TA, Herman RK, Kari CK, Yeh WH, Schackwitz WS, Schuyler MW, Collet J, Thomas JH, Riddle DL (1995) Mutations affecting the chemosensory neurons of Caenorhabditis elegans. Genetics 139: 171–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin D, Madsen D, Kahn-Kirby A, Peckol E, Moulder G, Barstead R, Maricq A, Bargmann C (2002) Combinatorial expression of TRPV channel proteins defines their sensory functions and subcellular localization in C. elegans neurons. Neuron 35: 307–318 [DOI] [PubMed] [Google Scholar]
- Troemel ER, Kimmel BE, Bargmann CI (1997) Reprogramming chemotaxis responses: sensory neurons define olfactory preferences in C. elegans. Cell 91: 161–169 [DOI] [PubMed] [Google Scholar]
- Ward S (1973) Chemotaxis by the nematode Caenorhabditis elegans: identification of attractants and analysis of the response by use of mutants. Proc Natl Acad Sci USA 70: 817–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S (1975) Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans. J Comp Neurol 160: 313–337 [DOI] [PubMed] [Google Scholar]
- Ware RW, Clark D, Crossland K, Russell RL (1975) The nerve ring of the nematode Caenorhabditis elegans: sensory input and motor output. J Comp Neurol 162: 71–110 [Google Scholar]
- White JG (1986) The structure of the nervous system of the nematode C. elegans. Philos Trans R Soc Lond Biol Sci 314: 1–340 [DOI] [PubMed] [Google Scholar]
- Wicks SR, de Vries CJ, van Luenen HG, Plasterk RH (2000) CHE-3, a cytosolic dynein heavy chain, is required for sensory cilia structure and function in Caenorhabditis elegans. Dev Biol 221: 295–307 [DOI] [PubMed] [Google Scholar]
- Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH (2001) Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat Genet 28: 160–164 [DOI] [PubMed] [Google Scholar]
- Wong GT, Gannon KS, Margolskee RF (1996) Transduction of bitter and sweet taste by gustducin. Nature 381: 796–800 [DOI] [PubMed] [Google Scholar]
- Zwaal RR, Mendel JE, Sternberg PW, Plasterk RH (1997) Two neuronal G proteins are involved in chemosensation of the Caenorhabditis elegans Dauer-inducing pheromone. Genetics 145: 715–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SUPPLEMENTARY MATERIAL 1
SUPPLEMENTARY MATERIAL 2
SUPPLEMENTARY MATERIAL 3


