Abstract
As the only imaging method available, Imaging Mass Spectrometry (IMS) can determine both the identity and the distribution of hundreds of molecules on tissue sections, all in one single run. IMS is becoming an established research technology, and due to recent technical and methodological improvements the interest in this technology is increasing steadily and within a wide range of scientific fields. Of the different IMS methods available, matrix-assisted laser desorption/ionization (MALDI) IMS is the most commonly employed. The course at IMSC 2012 in Kyoto covered the fundamental principles and techniques of MALDI-IMS, assuming no previous experience in IMS. This mini review summarizes the content of the one-day course and describes some of the most recent work performed within this research field.
Keywords: Imaging Mass Spectrometry, sample preparation, heat stabilization, MALDI IMS
INTRODUCTION
A detailed understanding of molecular distribution and co-localization in tissue is vital for the progress in drug development and basic biological research. Imaging mass spectrometry is a label-free method that is suitable for this need, increasingly employed for the analysis of biological tissue. Lipids, proteins, peptides, drugs and their metabolites can be analyzed for their distribution and relative concentration, at spatial resolutions down to cellular levels and for sample sizes up to whole body model animals. Tissue origins are ranging from plant tissue1) to model animals, human samples or even inorganic material such as coronary stents.2) The common trait of the existing imaging mass spectrometry technologies is the ionization of molecules in a raster over a tissue with a subsequent ion detection step, where an individual mass spectrum is recorded for each pixel. The signal intensities for a specific m/z value are subsequently visualized in a colored or grey scale over the full tissue area. As long as the molecules are ionizable, no labeling or other derivatization methods are required and hence, molecular distribution can be assessed in an unbiased manner. A number of ionization methods have been developed, the most commonly employed to date being matrix-assisted laser/desorption ionization (MALDI). A textbook written by members of our lab describes the MALDI imaging process in detail,3) and a number of recent review articles provide up-to-date tips for IMS analyses.1,4–7) For this article we have chosen to give a brief description of important sample preparation steps, and also review some very recent developments that we believe will be helpful for the reader in terms of successful sample preparation and/or data analyses.
SAMPLE PREPARATION
Throughout the sample preparation procedure, special care should be taken not to degrade or de-localize the molecule(s) of interest. For the earliest IMS applications, proteins and peptides were in focus, and hence many sample preparation strategies are formulated with these molecules in mind. Beginners in IMS should take extra care to find or develop sample preparation strategies that suit their tissues or analytes of interest. General recommendations for successful imaging experiments are given below.
Tissue stabilization
There are useful strategies for hampering the post mortal changes of biological samples. Studies have shown rapidly occurring increases or decreases in abundance for a number of molecules when the samples are kept at room temperature and at normal room humidity.8–10) Stabilization is thus highly recommended and can be performed through heat stabilization, microwave irradiation, formalin fixation or simply through flash freezing. For freezing, the use of powdered dry ice keeps the samples from cracking. Powdered dry ice is easily manufactured by breaking up dry ice with a hammer and separating the resulting powder from the larger pieces of ice through a common sieve.3) For the analysis of rapidly degrading neurotransmitters such as acetylcholine, in situ freezing (ISF) has been shown to ameliorate detectability. For ISF, the model animal is deeply anesthetized and sacrificed by carefully dipping the tip of the head into liquid nitrogen.10) Heat stabilization using the Denator system, where samples are rapidly heated to 95°C, has been proven effective for a number of proteins and peptides.8,11) Preliminary data suggests that heat stabilization is useful also for the analysis of lipids (data not shown). The heat-stabilized tissue may become somewhat fragile when frozen. Resulting difficulties in cryosectioning may be circumvented by utilizing double-adhesive carbon tape, as recently described in a paper by Goodwin et al.9)
Formalin fixation paraffin embedding (FFPE) is a preservation method with a long history in pathology, with millions of clinical samples available in biobanks around the world. Fortunately, sample preparation protocols have been developed that enable the analysis of proteins and peptides in FFPE tissue.12,13) For the analysis of lipids, some species may be analyzed with no special pre-treatment of the samples.14) One should remember that heat stabilization, formalin fixation and microwave irradiation permanently protects samples from enzymatic changes, whereas freezing only preserves the sample state until the temperature is risen and enzymatic processes re-occur.
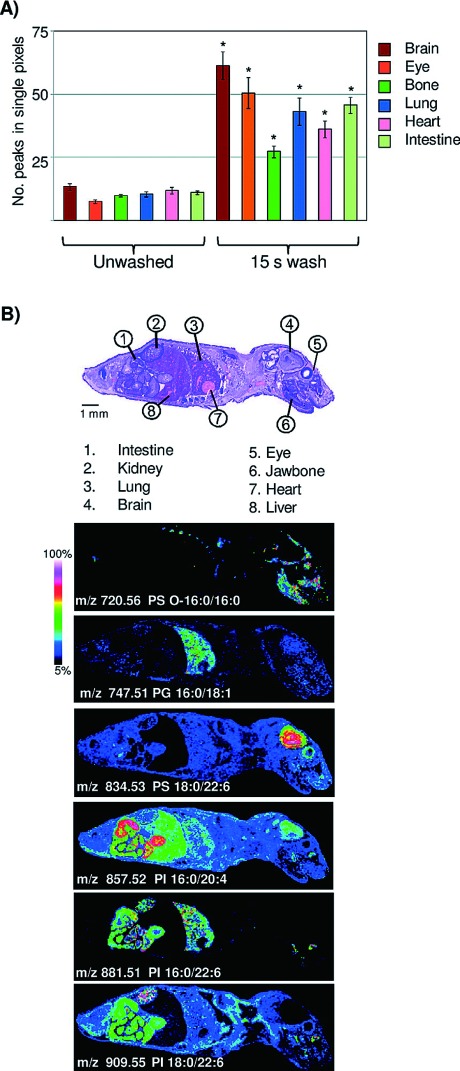
Fig. 1. Aqueous washing illustrated on a whole body mouse pup by MALDI TOF. (A) Comparison of peak number between unwashed and washed tissue. ∗=Student’s t test p-value <2.0×10−4. (B) Examples of images obtained across a whole body mouse pup, illustrating organ specific lipid expression in the negative ion mode. Figure reprint by permission, Angel et al. Anal. Chem. 7(84): 1557–1564, 2012.
Cryosectioning
Preparation of tissue samples starts with tissue extraction and is followed by tissue embedding, sectioning, and mounting. Post mounting, the samples can either be directly prepared for analysis, or stored at −80°C. Analysis preparation steps for the tissue sections include washing, drying and coating with a carefully selected MALDI matrix. Our most important tips for the cryosectioning are the following; i) Due to the rapid metabolic turnover, work rapidly and in a reproducible manner; ii) Extracted tissue should be frozen or stabilized immediately; iii) Embedding should be performed using a material that does not interfere with mass spectrometry (MS) analysis. We recommend a semi-liquid gel of 2% sodium carboxymethylcellulose (CMC)15) or even a small pool of water. Optimal cutting temperature (OCT) compound or other polymeric materials should be avoided; iv) A tissue thickness of <20 μm improves spectrum quality.3)
Washing
Washing procedures should be chosen with the analyte(s) of interest. For peptide or protein analysis, salts and lipids should be removed in order to avoid analytically disturbing ion suppression and hence, the washing solutions will in general contain organic solvents.16,17) These washing solutions are evidently unsuitable for lipid analysis. In addition, organic solvents for tissue washing are often not suitable for the analysis of drug compounds, since drug solubility leads to displacement or even total loss from the tissue. Shariatgorji et al. developed a protocol where the pHs of the washing solutions were adjusted to levels where the drug would have low solubility. Mouse brain tissue treated with one of three different drug compounds (cimetidine, imipramine or “compound c”) had their sections washed with ammonium acetate buffer at pH 10, significantly increasing signal intensities as compared to the results from using acidic or neutral buffers.18) For the analysis of lipids, washing steps are usually avoided. However, Angel et al. developed a washing protocol for the analysis of lipids in negative mode utilizing ammonium formate at pH 6.4 or ammonium acetate at pH 6.7, which significantly increased the number of detectable analytes along with their signal intensities.19)
A third example where washing procedures were specifically developed to fit the analysis is a washing protocol for fragile tissue sections; van Hove et al. developed a method where, instead of immersing the tissue in washing solution, a fiber-free paper tissue was pre-wetted in washing solution and placed on top of the tissue section for 30 to 60 s. If desirable, smaller sections of the tissue can be specifically targeted for this washing procedure.20) Before moving to the matrix application step, the tissue sections should be somewhat dried, either under a swift air of nitrogen gas or in a desiccator for 10–30 min. This process enhances the stability of tissue adhesiveness to the slides in the mass spectrometer.
Matrices and matrix application strategies
All sample preparation steps are important for high quality imaging results. However, it could be argued that the single most crucial step for successful IMS is the matrix application—both in regards to the choice of matrix, and the tissue coating procedure. There is a plethora of matrices available, all with their individual compound compatibilities, which means that the matrix should be chosen with each analyte at hand.3) New matrices and novel methods for their coating are continuously tested.21–24) There are some practical parameters to keep in mind when choosing matrices. For example, the sublimation rate of your matrix in the vacuum environment of the MS instrument should be investigated; the thickness of the matrix layer should be consistent throughout the analysis, or else the signal intensity is at risk of decreasing over time. In some cases, additives can be used to prolong matrix lifetime or to reduce the effect of alkali adducts.21) In addition, if your matrix peaks overlap with the m/z of your analyte(s), it might be possible to induce a matrix peak mass shift by deuterating the matrix. Thereby, the masking problem is avoided.25) In cases where a non-biased omics approach is employed, a matrix with a low number of matrix peaks is beneficial, to minimize the risk of masking potential biomarkers. Regarding application strategies; there are a number of choices, ranging from manual sprayers to spotting instruments. Any method is satisfactory, as long as it results in homogenous coating and small crystal sizes. Wetting of the tissue should be avoided, since the matrix solvent might de-localize the analytes of interest. When applying matrix through TLC sprayers or air-brushes, the first rounds of spraying should be short in order to create a first layer of seeding crystals. Make sure to wait long enough between the spraying rounds for the tissue to fully dry. Dry matrix can be applied for the analysis of lipids or drugs,26–28) however the results for peptides and proteins have not yet been satisfactory. A large number of publications focus on the different matrices and matrix application strategies,3) and we strongly recommend the beginner in IMS to read in to this particular part of the experiments.
Compound identification
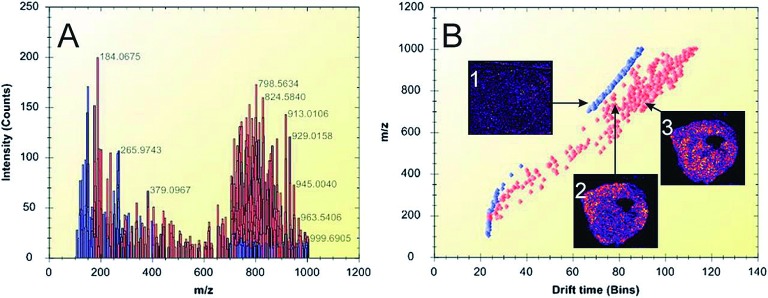
A challenge for the IMS technology compared to other MS technologies is the lack of upstream separation technologies. Due to the extraordinarily large number of molecules present, in combination with the matrix-inherent ion signals, there is an obvious risk of detecting molecules with overlapping m/z values. This in turn leads to a difficulty in ensuring non-ambiguous molecular identification. Fortunately, there are several possible solutions to this problem. For example, the combination of the imaging platform with high-resolution mass analyzers such as orbitrap is one route that has been successfully tested.29) Also, ion mobility is a strategy that offers a post-ionization separation step where isobaric molecules may be distinguished based on their unique molecular cross section area. This approach was successfully used to separate lipid molecules of m/z 746.5 and 746.6, respectively, and illustrate their differing molecular distribution in xenograft breast cancer tissue30) (Fig. 2). A third approach is the use of imaging MS/MS where the distribution images for daughter ions are matched to those of the precursor ion, thereby adding an extra dimension to the confidence in the molecular identification process.10)
Fig. 2. Ion mobility separation of biomolecular ions detected from thin tumor tissue sections. A) Representative spectrum of peak-picked data acquired during an IMS imaging experiment. The lipid peaks are highlighted in red, while matrix-related ions are shown in blue. B) Drift plot of the separated ions. Ion image 1 shows the distribution of a background matrix ion. Ion images 2 (m/z 746.5) and 3 (m/z 746.6) show the distribution of different lipids in the tumor tissue. Lipid-related ions (highlighted in red) were separated from background ions (blue). Figure reprint by permission, Chughtai et al. J. Lip. Res. 54(2): 333–344, 2013.
Data analysis
All IMS data are comprised of x- and y-coordinates, m/z values and ion signal intensities, and theses four dimensions can be illustrated through individual or combined ion intensity images or as spectra. Many leading edge data analyses have been introduced to IMS data analysis; Principal component analysis degenerates data dimensions and enhances the difference among data.31,32) Hierarchical cluster analysis reveals relationships among molecules and classifies them.31,33) Multivariate curve resolution can reveal mixed spectra from observed single spectrum.34) Markov chain Monte Carlo analysis infers true spectra from incomplete ones, such as in MS/MS.35) Such analyses are helpful for the extraction of the nature of IMS data; however, they are usually too complicated and over engineered. Through extensive research, we found that a simple extension of traditional image processing is the most useful approach.35) Conventional region of interest (ROI) analysis has long been used for the analysis of labeled targets using staining or fluorescent dyes, where localization or distribution of target molecules have been evaluated and ROI-to-ROI are compared. Since mass spectrometry allows for non-targeted studies, IMS enables us to reveal molecules that characterize a specific ROI. Therefore, ROI analysis of IMS data provides an efficient means for screening of biomarkers or key molecules in specific phenomena of interest. Ratio imaging is also popular in conventional image processing, e.g. fluorescence resonance energy transfers (FRET). As already described, IMS can reveal information about huge amount of molecules, which enable to calculate various quantity based on theoretical ideas and mathematical models. An example of such extended ratio analysis is visualization of energy charge, which was introduced to characterize metabolic energy state using number of phosphorylation sites calculated from abundance of ATP, ADP, and AMP.36,37) In the near future, various visualizations will be performed, based on more complex theoretical ideas.
A lot of effort is now being put into finding the best strategies for the extraction of useful information through processes such as baseline correction, denoising and normalization. A strong, collective effort is also put into finding common data formats such as imzML to enable a straightforward sharing of data sets between research groups.38)
Quantification and data normalization
For quantitation, it is important that the ion signal intensities truly reflect the actual on-tissue concentrations of the analyte in question. One should therefore take into account i) the ionization yield of the respective analytes, ii) matrix deposition homogeneity and iii) the varying tissue- or compartment specific properties that might affect the ionization.
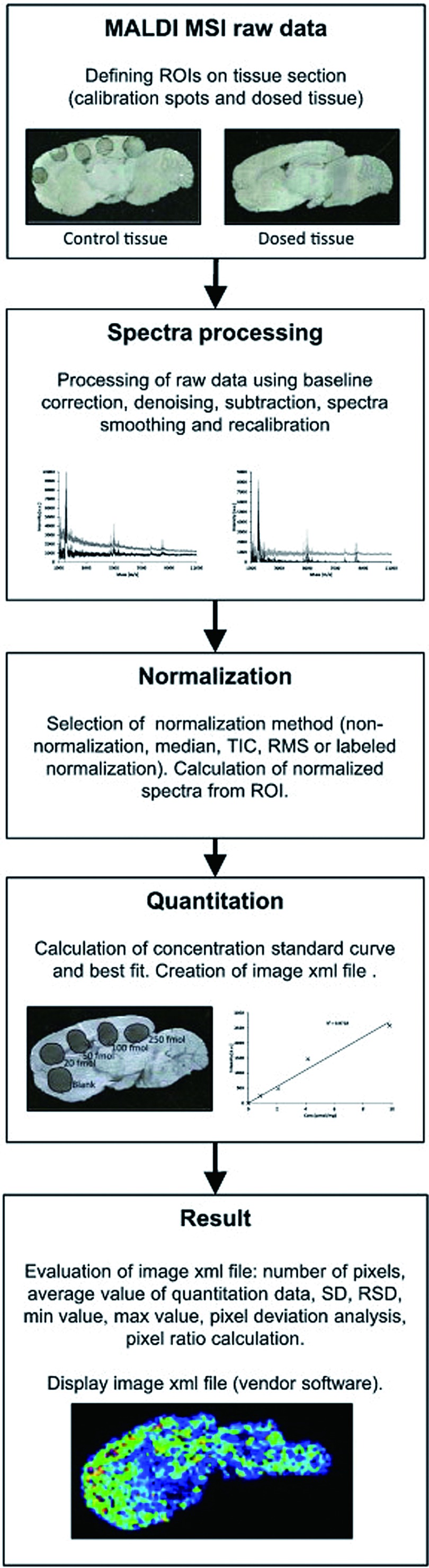
The first applications described for MALDI IMS were developed for protein or peptide analysis, followed by lipid analysis applications. Recently, interest in drug imaging has increased.5,39,40) Imaging of drug distribution is a crucial step in assessing drug function, and currently used methods such as positron emission tomography (PET) or whole body autoradiography (WBA) depend on labeling the drug compound with a radioactive tag. Not only is labeling a costly and time consuming procedure, but also, the subsequent detection process is targeted to the label rather than the drug compound itself, hampering the possibility to find and identify drug metabolites. In contrast, IMS offers the detection of drug molecules as well as their metabolites. Moreover, there is a possibility for un-biased detection of treatment related molecular alterations or reactions in the same tissue sections. Recently, the Andrén group presented a quantification method that is useful for the analysis of a pre-selected target molecule.18) Standard solutions of the target compound are spotted at known concentrations onto control tissue. The spots (5–8 in total) then serve as references for a calibration curve. An analogue to the target, e.g. a deuterated analogue, is added to the matrix solution to correct for ionization biases that the molecule of interest displays. Using the analysis strategy described in Fig. 3, Kallback et al. reached therapeutic levels (pmol) in their detection. Normalizing the data to the deuterated molecule to compensate for local ionization effects generated the best fit, with a correlation coefficient R2=0.985.
Fig. 3. Quantitation: structural overview of the quantitation software design. Figure reprint by permission, Källback et al. J. Proteomics 75(16): 4941–4951, 2012.
A similar approach for the analysis of drug distribution and quantitation in tissue was recently presented by Hamm et al. where they added a “pseudo internal standard,” i.e. the molecule of interest, at known concentrations to the matrix solution and spraying this mix onto control tissue sections. By collecting ion intensity data over a whole body section, drug specific as well as tissue specific ion suppression effects could be analyzed and summarized in a normalization factor denoted the Tissue Extinction Coefficient, TEC. As a consequence, the amount of “drug per area of tissue” could be determined (which in theory should be convertible to “drug per gram of tissue”). Using this method, the drugs propranolol and olanzapine were analyzed at limits of detection (LODs) down to 0.006 pmol/mm2 and 0.3 pmol/mm2, respectively. The IMS data correlated well with results using analytical techniques such as Quantitative Whole Body Autoradiography (QWBA) or LC-MSMS.41)
CONCLUSION
MALDI IMS has left its infancy and is becoming an established technique in various fields of biological research, especially for the analysis of lipids and drugs where alternative methods are scarce. The instrumentation and sample preparation methods have been refined, leaving data analysis and software design to be the current bottlenecks and primary challenges, although all aspects of MALDI IMS are under continuous improvement. We believe that MALDI IMS is an excellent choice for the un-biased assessment of molecular distribution in tissue, and we are observing a recent explosive increase in attention for this technique within a myriad of research areas. The future is exciting.
Acknowledgments
The authors are grateful to the committee of IMSC 2012 for all their kind help and effort in organizing the above described short course. Cecilia Eriksson’s work is financed by VINNMER International Qualification Programme, Vinnova, Sweden and by Japan Society for the Promotion of Science (JSPS), Japan.
References
- 1) Y. J. Lee, D. C. Perdian, Z. H. Song, E. S. Yeung, B. J. Nikolau. Use of mass spectrometry for imaging metabolites in plants. Plant J. 70: 81–95, 2012 [DOI] [PubMed] [Google Scholar]
- 2) J. T. Huang, L. Hannah-Qiuhua, R. Szyszka, V. Veselov, G. Reed, X. Wang, S. Price, L. Alquier, G. Vas. Molecular imaging of drug-eluting coronary stents: Method development, optimization and selected applications. J. Mass Spectrom. 47: 155–162, 2012 [DOI] [PubMed] [Google Scholar]
- 3) M. Setou. Imaging Mass Spectrometry: Protocols for Mass Microscopy, Springer, 2010
- 4) R. J. Goodwin. Sample preparation for mass spectrometry imaging: Small mistakes can lead to big consequences. J. Proteomics 75: 4893–4911, 2012 [DOI] [PubMed] [Google Scholar]
- 5) B. Prideaux, M. Stoeckli. Mass spectrometry imaging for drug distribution studies. J. Proteomics 75: 4999–5013, 2012 [DOI] [PubMed] [Google Scholar]
- 6) Y. Saito, M. Waki, S. Hameed, T. Hayasaka, M. Setou. Development of imaging mass spectrometry. Biol. Pharm. Bull. 35: 1417–1424, 2012 [DOI] [PubMed] [Google Scholar]
- 7) H. Wang, Z. Zhao, Y. Guo. Chemical and biochemical applications of MALDI TOF-MS based on analyzing the small organic compounds. Top. Curr. Chem.331: 165–192, 2013 [DOI] [PubMed] [Google Scholar]
- 8) R. J. Goodwin, J. C. Dungworth, S. R. Cobb, A. R. Pitt. Time-dependent evolution of tissue markers by MALDI-MS imaging. Proteomics 8: 3801–3808, 2008 [DOI] [PubMed] [Google Scholar]
- 9) R. J. A. Goodwin, S. L. Iverson, P. E. Andren. The significance of ambient-temperature on pharmaceutical and endogenous compound abundance and distribution in tissues sections when analyzed by matrix-assisted laser desorption/ionization mass spectrometry imaging. Rapid Commun. Mass Spectrom. 26: 494–498, 2012 [DOI] [PubMed] [Google Scholar]
- 10) Y. Sugiura, N. Zaima, M. Setou, S. Ito, I. Yao. Visualization of acetylcholine distribution in central nervous system tissue sections by tandem imaging mass spectrometry. Anal. Bioanal. Chem. 403: 1851–1861, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11) R. J. A. Goodwin, A. M. Lang, H. Allingham, M. Borén, A. R. Pitt. Stopping the clock on proteomic degradation by heat treatment at the point of tissue excision. Proteomics 10: 1751–1761, 2010 [DOI] [PubMed] [Google Scholar]
- 12) M. R. Groseclose, P. P. Massion, P. Chaurand, R. M. Caprioli. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 8: 3715–3724, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13) R. Casadonte, R. M. Caprioli. Proteomic analysis of formalin-fixed paraffin-embedded tissue by MALDI imaging mass spectrometry. Nat. Protoc. 6: 1695–1709, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14) C. L. Carter, C. W. McLeod, J. Bunch. Imaging of phospholipids in formalin fixed rat brain sections by matrix assisted laser desorption/ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 22: 1991–1998, 2011 [DOI] [PubMed] [Google Scholar]
- 15) M. Stoeckli, D. Staab, A. Schweitzer. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Int. J. Mass Spectrom. 260: 195–202, 2007 [Google Scholar]
- 16) E. H. Seeley, S. R. Oppenheimer, D. Mi, P. Chaurand, R. M. Caprioli. Enhancement of protein sensitivity for MALDI imaging mass spectrometry after chemical treatment of tissue sections. J. Am. Soc. Mass Spectrom. 19: 1069–1077, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17) J. H. Yang, R. M. Caprioli. Matrix sublimation/recrystallization for imaging proteins by mass spectrometry at high spatial resolution. Anal. Chem. 83: 5728–5734, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18) P. Källback, M. Shariatgorji, A. Nilsson, P. E. Andrén. Novel mass spectrometry imaging software assisting labeled normalization and quantitation of drugs and neuropeptides directly in tissue sections. J. Proteomics 75: 4941–4951, 2012 [DOI] [PubMed] [Google Scholar]
- 19) P. M. Angel, J. M. Spraggins, H. S. Baldwin, R. Caprioli. Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal. Chem. 84: 1557–1564, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20) E. R. van Hove, D. F. Smith, L. Fornai, K. Glunde, R. M. Heeren. An alternative paper based tissue washing method for mass spectrometry imaging: Localized washing and fragile tissue analysis. J. Am. Soc. Mass Spectrom. 22: 1885–1890, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21) Y. Sugiura, M. Setou. Selective imaging of positively charged polar and nonpolar lipids by optimizing matrix solution composition. Rapid Commun. Mass Spectrom. 23: 3269–3278, 2009 [DOI] [PubMed] [Google Scholar]
- 22) D. Bonnel, J. Franck, C. Meriaux, M. Salzet, I. Fournier. Ionic matrices pre-spotted MALDI plates for patients markers following, drugs titration and MALDI MSI. Anal. Biochem., 2012 [DOI] [PubMed] [Google Scholar]
- 23) C. H. Le, J. Han, C. H. Borchers. Dithranol as a MALDI matrix for tissue imaging of lipids by Fourier transform ion cyclotron resonance mass spectrometry. Anal. Chem. 84: 8391–8398, 2012 [DOI] [PubMed] [Google Scholar]
- 24) Y. Fukuyama, R. Tanimura, K. Maeda, M. Watanabe, S. I. Kawabata, S. Iwamoto, S. Izumi, K. Tanaka. Alkylated dihydroxybenzoic acid as a MALDI matrix additive for hydrophobic peptide analysis. Anal. Chem. 84: 4237–4243, 2012 [DOI] [PubMed] [Google Scholar]
- 25) M. Shariatgorji, A. Nilsson, R. J. A. Goodwin, P. Svenningsson, N. Schintu, Z. Banka, L. Kladni, T. Hasko, A. Szabo, P. E. Andren. Deuterated matrix-assisted laser desorption ionization matrix uncovers masked mass spectrometry imaging signals of small molecules. Anal. Chem. 84: 7152–7157, 2012 [DOI] [PubMed] [Google Scholar]
- 26) S. M. Puolitaival, K. E. Burnum, D. S. Cornett, R. M. Caprioli. Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J. Am. Soc. Mass Spectrom. 19: 882–886, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27) R. J. Goodwin, L. Macintyre, D. G. Watson, S. P. Scullion, A. R. Pitt. A solvent-free matrix application method for matrix-assisted laser desorption/ionization imaging of small molecules. Rapid Commun. Mass Spectrom. 24: 1682–1686, 2010 [DOI] [PubMed] [Google Scholar]
- 28) R. J. A. Goodwin, C. L. Mackay, A. Nilsson, D. J. Harrison, L. Farde, P. E. Andren, S. L. Iverson. Qualitative and quantitative MALDI imaging of the positron emission tomography ligands raclopride (a D2 dopamine antagonist) and SCH 23390 (a D1 dopamine antagonist) in rat brain tissue sections using a solvent-free dry matrix application method. Anal. Chem. 83: 9694–9701, 2011 [DOI] [PubMed] [Google Scholar]
- 29) J. H. Jun, Z. H. Song, Z. J. Liu, B. J. Nikolau, E. S. Yeung, Y. J. Lee. High-spatial and high-mass resolution imaging of surface metabolites of Arabidopsis thaliana by laser desorption-ionization mass spectrometry using colloidal silver. Anal. Chem. 82: 3255–3265, 2010 [DOI] [PubMed] [Google Scholar]
- 30) K. Chughtai, L. Jiang, T. R. Greenwood, K. Glunde, R. M. Heeren. Mass spectrometry images acylcarnitines, phosphatidylcholines and sphingomyelin in MDA-MB-231 breast tumor models. J. Lipid Res., 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31) G. McCombie, D. Staab, M. Stoeckli, R. Knochenmuss. Spatial and spectral correlations in MALDI mass spectrometry images by clustering and multivariate analysis. Anal. Chem. 77: 6118–6124, 2005 [DOI] [PubMed] [Google Scholar]
- 32) I. Yao, Y. Sugiura, M. Matsumoto, M. Setou. In situ proteomics with imaging mass spectrometry and principal component analysis in the Scrapper-knockout mouse brain. Proteomics 8: 3692–3701, 2008 [DOI] [PubMed] [Google Scholar]
- 33) S. O. Deininger, M. P. Ebert, A. Fütterer, M. Gerhard, C. Röcken. MALDI imaging combined with hierarchical clustering as a new tool for the interpretation of complex human cancers. J. Proteome Res. 7: 5230–5236, 2008 [DOI] [PubMed] [Google Scholar]
- 34) J. A. Ohlhausen, M. R. Keenan, P. G. Kotula, D. E. Peebles. Multivariate statistical analysis of time-of-flight secondary ion mass spectrometry images using AXSIA. Appl. Surf. Sci. 231-232: 230–234, 2004 [Google Scholar]
- 35) S. R. Jefferys, M. C. Giddings. Baking a mass-spectrometry data PIE with McMC and simulated annealing: Predicting protein post-translational modifications from integrated top-down and bottom-up data. Bioinformatics 27: 844–852, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36) Y. Sugiura, R. Taguchi, M. Setou. Visualization of spatiotemporal energy dynamics of hippocampal neurons by mass spectrometry during a kainate-induced seizure. PLoS ONE 6: e17952, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37) N. Goto-Inoue, Y. Manabe, S. Miyatake, S. Ogino, A. Morishita, T. Hayasaka, N. Masaki, M. Setou, N. L. Fujii. Visualization of dynamic change in contraction-induced lipid composition in mouse skeletal muscle by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 403: 1863–1871, 2012 [DOI] [PubMed] [Google Scholar]
- 38) M. Stoeckli. MSImaging. 2011. http://www.maldi-msi.org/
- 39) K. Kuwayama, K. Tsujikawa, H. Miyaguchi, T. Kanamori, Y. T. Iwata, H. Inoue. Distribution measurements of 3,4-methylenedioxymethamphetamine and its metabolites in organs by matrix-assisted laser desorption/ionization imaging mass spectrometry using an automatic matrix spraying system with an air brush and a turntable. Anal. Bioanal. Chem. 404: 1823–1830, 2012 [DOI] [PubMed] [Google Scholar]
- 40) S. K. Shahidi-Latham, S. M. Dutta, M. C. Prieto Conaway, P. J. Rudewicz. Evaluation of an accurate mass approach for the simultaneous detection of drug and metabolite distributions via whole-body mass spectrometric imaging. Anal. Chem. 84: 7158–7165, 2012 [DOI] [PubMed] [Google Scholar]
- 41) G. Hamm, D. Bonnel, R. Legouffe, F. Pamelard, J. M. Delbos, F. Bouzom, J. Stauber. Quantitative mass spectrometry imaging of propranolol and olanzapine using tissue extinction calculation as normalization factor. J. Proteomics 75: 4952–4961, 2012 [DOI] [PubMed] [Google Scholar]