Abstract
The flexible tubulin C-terminal tails (CTTs) have recently been implicated in the walking mechanism of dynein and kinesin. To address their role in the case of conventional kinesin, we examined the structure of kinesin–microtubule (MT) complexes before and after CTT cleavage by subtilisin. Our results show that the CTTs directly modulate the motor–tubulin interface and the binding properties of motors. CTT cleavage increases motor binding stability, and kinesin appears to adopt a binding conformation close to the nucleotide-free configuration under most nucleotide conditions. Moreover, C-terminal cleavage results in trapping a transient motor–ADP–MT intermediate. Using SH3-tagged dimeric and monomeric constructs, we could also show that the position of the kinesin neck is not affected by the C-terminal segments of tubulin. Overall, our study reveals that the tubulin C-termini define the stability of the MT–kinesin complex in a nucleotide-dependent manner, and highlights the involvement of tubulin in the regulation of weak and strong kinesin binding states.
Keywords: cryo-electron microscopy, kinesin, kinesin neck-linker, SH3 domain, tubulin C-terminus
Introduction
Microtubules (MTs) provide the cell with molecular tracks traveled by dynein and kinesin motor proteins. These polar cylindrical structures, composed of αβ-tubulin dimers, are fundamental for many cellular functions such as organelle trafficking, cell division, localization of developmental morphogens, and signal transduction pathways (Goldstein and Philp, 1999). To date, most studies on kinesins have been carried out on conventional kinesin (reviewed in Vale and Milligan, 2000; Vale, 2003), a dimer of two identical heavy chains producing processive movement that is mediated by alternating head catalysis of adenosine triphosphate (ATP) (Hackney, 1994; Ma and Taylor, 1997; Gilbert et al, 1998).
Binding of the kinesin core on MTs is dominated by ionic interactions. Kinesin contributes mainly positive charges to the tubulin binding interface, clustered in loops L7/L8, L11, and the switch II cluster α4/L12/α5 (Woehlke et al, 1997). Dimeric kinesin dimerizes through an extended coiled-coil region separated into neck and stalk (reviewed in Vale, 2003). The neck connects to the core motor domain via the so-called neck-linker (β9–β10). The Absence of ATP in the catalytic domain renders the neck-linker highly flexible, while AMP-PNP locks the neck-linker to the motor core (Rice et al, 1999; Skiniotis et al, 2003).
The C-terminal domains of both α- and β-tubulin consist of helices H11 and H12 (Nogales et al, 1998) at the outer surface of MTs (Li et al, 2002). The last 10 and 18 C-terminal residues in the α-and β-tubulin, respectively, are highly disordered and not visible in the crystal structure. These flexible C-terminal tails (referred to here as CTTs; according to Sackett et al, 1985), along with the C-terminal helices, especially H12, contribute a large amount of negative charges to the kinesin–tubulin interface (Hoenger et al, 2000; Kikkawa et al, 2001). The binding site for conventional kinesin has been established by a variety of methods to be predominantly on the outer surface of β-tubulin, with a partial overlap with α-tubulin (Hoenger et al, 2000).
CTTs can be removed from MTs using subtilisin, which cleaves ∼10 amino acids from the α- and ∼20 amino acids from β-tubulin C-termini (Serrano et al, 1984a; Sackett et al, 1985). According to limited proteolysis studies, CTTs provide binding sites for tau and MAP2 proteins (Serrano et al, 1984b; Marya et al, 1994; Kar et al, 2003), and they are also required for the MT depolymerizing activity of KIN I kinesins (Moores et al, 2002; Niederstrasser et al, 2002). Conventional kinesin and dynein still bind MTs in the absence of the C-terminal peptides, but their processivity is significantly reduced (Thorn et al, 2000; Wang and Sheetz, 2000).
To address the role of the tubulin tails in the case of conventional kinesin, we generated helical 3D reconstructions from cryo-electron microscopy (cryo-EM) data to compare the structure of kinesin–MT complexes in the presence or absence of the CTTs. We were able to identify distinct differences in the motor conformation according to the nucleotide conditions, before and after subtilisin cleavage of the CTTs. Our interpretations are supported by steady-state and pre-steady-state kinetics as well as equilibrium binding experiments. We also examined the location of the neck in relation to the presence and absence of CTTs using an SH3 density label as recently described in Skiniotis et al (2003).
Results
Kinesin in the presence of AMP-PNP assumes a more mobile configuration than in a nucleotide-free state
In a first set of experiments, we investigated the structure and the binding characteristics of a monomeric rat kinesin construct (rK354: Sack et al, 1997) on native MTs, in the presence of AMP-PNP or adenosine diphosphate (ADP), and in the absence of nucleotides. Due to technical limitations with helical 3D reconstructions (Beuron and Hoenger, 2001), we focused exclusively on monomeric motor constructs. Statistically significant density differences between groups of helically reconstructed 3D maps were identified using t-test analysis (Milligan and Flicker, 1987). Difference volumes include an area reflecting a certainty of >99% for a real difference in density.
Figure 1 compares 3D reconstructions of rK354 on native MTs in a nucleotide-free versus an AMP-PNP state (Figure 1A and B). With the underlying MT remaining unchanged, the motor domains exhibit different shapes. In the presence of AMP-PNP, the head forms a slightly narrower nose towards the plus-end. Docking of the X-ray structure into the EM-derived envelopes reveals an approximately 12° anticlockwise rotation from a nucleotide-free state (Figure 1C) to an AMP-PNP state (Figure 1D). Additionally, t-test analysis between nucleotide-free and AMP-PNP-bound motor–MT complexes revealed a lower density close to the motor–tubulin interface in the AMP-PNP state (Figure 2A).
Figure 1.
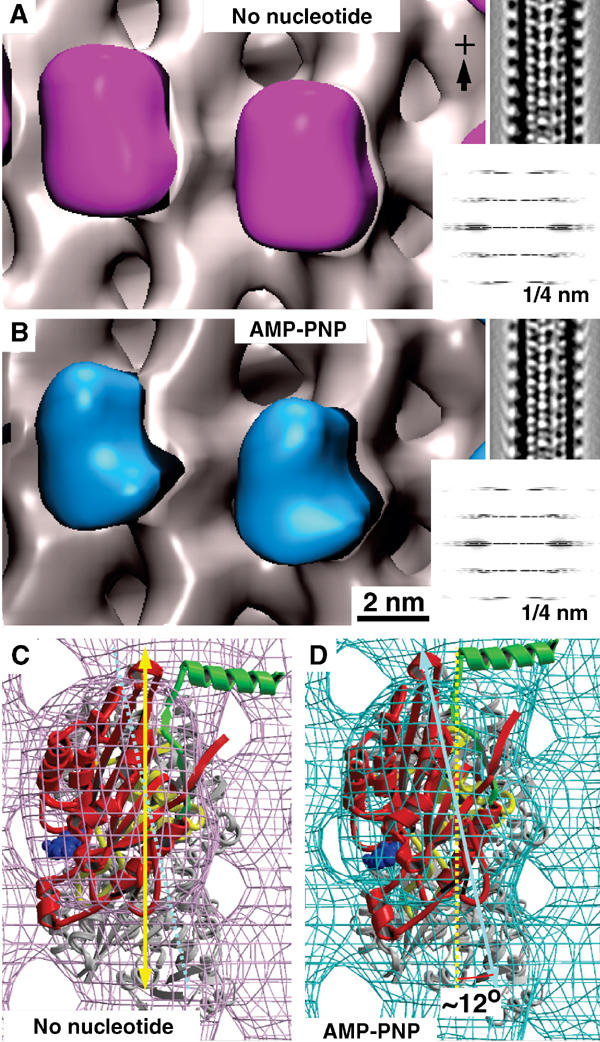
Nucleotide-dependent structural changes in monomeric kinesin motor domains. (A) 3D reconstruction of MTs decorated with monomeric rat kinesin rK354 in the absence of nucleotide (magenta), and (B) in the presence of AMP-PNP (cyan). Corresponding lateral projections and diffraction patterns are shown in the insets. In both reconstructions, decoration of tubulin dimers by motor domains was complete (see Figure 3). Docking of the X-ray structure into the EM-derived envelopes reveals an approximately 12° anticlockwise head rotation after transition from a nucleotide-free state (C) to an AMP-PNP state (D).
Figure 2.
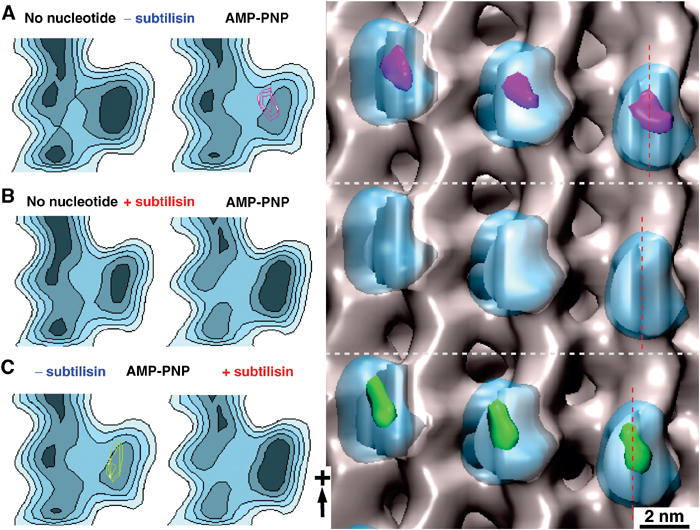
Statistical 3D difference mapping between monomeric kinesins in the presence or absence of the tubulin C-terminus. Difference volumes include areas with a significance of >99%. The left panels show sections through the reconstructions along the red lines indicated on the right. (A) On untreated MTs, comparison between nucleotide-free and AMP-PNP states revealed a significant density difference (magenta) at the motor–tubulin interface (for atomic details, see Figure 5). (B) The differences detected under native conditions shown in (A) disappear when subtilisin-treated MTs are used. (C) Comparing subtilisin-treated versus untreated kinesin–MT complexes in the presence of AMP-PNP reveals a similar difference volume (green) as found in (A), indicating that the AMP-PNP-bound state of kinesin approximates a no-nucleotide state once the CTTs are cleaved off.
Removal of CTTs decreases the binding flexibility of kinesin to MTs in the presence of AMP-PNP
The effects of the CTTs in kinesin–MT interaction were investigated by cleaving CTTs with subtilisin on intact MTs. The CTT cleavage was monitored by SDS–PAGE (Figure 3), and an electrophoretic shift indicative of proteolysis in the CTTs of α- and β-tubulin was observed (Figure 3D; see also Wang and Sheetz, 2000). The different extent of cleavage between α- and β-tubulin is revealed in the two bands obtained after subtilisin treatment. Previous studies have identified the lower band as β-tubulin, and the upper band as the α-tubulin (Bhattacharyya et al, 1985; Sackett et al, 1985).
Figure 3.

Interaction of monomeric kinesin motor domains with subtilisin-treated MTs. MT pelleting assays and SDS–PAGE of subtilisin-treated (+subt.) and native (−subt.) MTs incubated with monomeric rK354 (A) in the absence of nucleotide and (B) the presence of AMP-PNP, or (C) excess ADP (s=supernatant, p=pellet). Subtilisin-treated MTs reveal the two characteristic lower tubulin bands as indicated. Kinesin motor domains in the absence of nucleotides (A) and in the presence of AMP-PNP (B) bind to both native and subtilisin-treated MTs at comparable amounts, indicating full decoration of the MT surface. (C) In the presence of excess ADP (20 mM), subtilisin treatment clearly increases the amount of kinesin motor domains in the pellet. (D) SDS–PAGE, comparing native and subtilisin-treated tubulin under strongly separating conditions, reveals the molecular mass shifts for both α- and β-tubulin (assignments of α- and β-tubulin are according to Bhattacharyya et al, 1985).
After subtilisin treatment of MTs, density differences between nucleotide-free and AMP-PNP-bound motor–MT complexes are no longer detected (Figure 2B). Both nucleotide-free and AMP-PNP states approached the nucleotide-free binding configuration on native MTs. AMP-PNP maps before and after subtilisin treatment revealed a very similar density difference as between AMP-PNP and nucleotide-free states on native MTs (Figure 2C).
Docking of the crystal structures of rK354 (Sack et al, 1997) and tubulin (Nogales et al, 1998) in the EM density locates loops L8, L11, L12, and the switch II helix α4 at the tubulin interface (Figure 5; see also Hoenger et al, 2000; Kikkawa et al, 2001; Li et al, 2002). The difference densities obtained between AMP-PNP and nucleotide-free states reside directly above the switch II cluster region.
Figure 5.
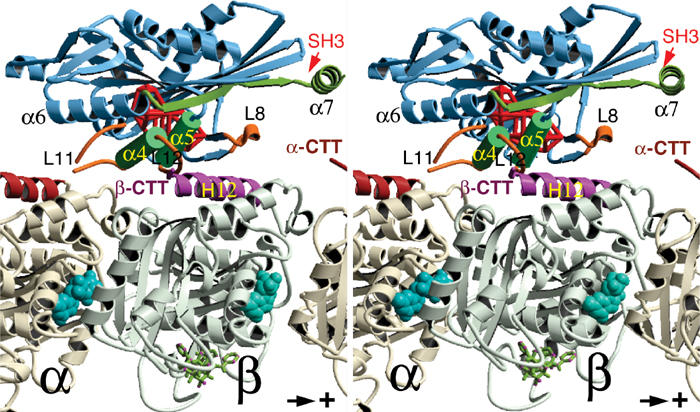
The C-terminal tail of β-tubulin locates at the vicinity of the switch II cluster. Stereo image of the kinesin–tubulin complex as derived from molecular docking of the atomic-resolution structures into cryo-EM density envelopes of MT–kinesin complexes. The statistically relevant (>99%) density difference found between AMP-PNP and nucleotide-free states is shown as a red wire frame (located behind α4 and α5). The same location was found for a density difference between MT–kinesin complexes in the AMP-PNP state, before and after subtilisin treatment (see Figure 2). The C-terminus of β-tubulin locates in close proximity to loop 11, α4, loop 12 and α5 of kinesin. ‘SH3' marks the insertion sites of SH3 density labels shown in Figure 6.
CTT cleavage strongly increases the binding affinity of kinesin motor domains in the presence of excess ADP
The most remarkable effect related to CTTs was found under conditions using excess ADP (20 mM). Under these conditions, kinesin motor domain decoration on native MTs is only very sparse. This is reflected in the almost complete absence of an 8 nm layer line cluster in diffraction images (Figure 4A) and is very distinct on MT pelleting assays (Figure 3C). Cryo-EM images do not reveal any motor-related mass on the MT surface (Figure 4A, insets). However, cleavage of CTTs changes the situation entirely: under otherwise identical conditions as used above, complete MT decoration by motor domains was observed (Figure 4B; green volumes). Micrographs now show an axially repeating pattern of motor-related densities at the outer MT surface, and diffraction patterns exhibit a strong 8 nm layer line (Figure 4B).
Figure 4.
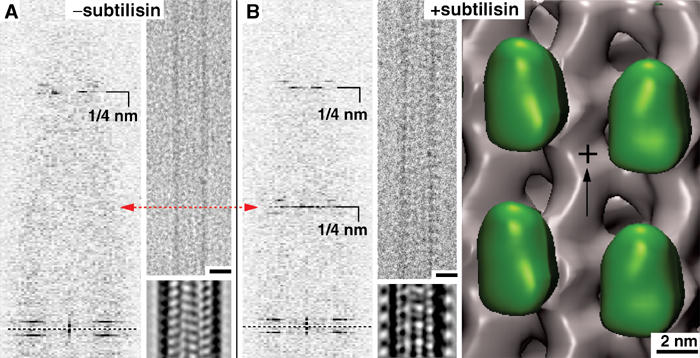
Monomeric kinesin binding in the presence of excess ADP on native or subtilisin-treated MTs. (A) The presence of excess ADP (20 mM) strongly reduces the MT decoration efficiency of kinesin motor domains. Cryo-EM images of MTs under these conditions do not show any signs of motor decoration (inset: averaged projections), which is reflected in the absence of a strong 1/8 nm layer line, corresponding to the axial tubulin dimer repeat. (B) After cleavage of CTTs, ADP–rK354 decorates MTs very efficiently. Cryo-EM images reveal a much rougher outer MT surface, giving rise to a strong 1/8 nm reflection (one motor head per αβ-tubulin dimer). In 3D –reconstructions, the motor heads (green) are clearly visible on the MT portion. Scale bars on micrographs=20 nm.
The location of the kinesin neck region is not influenced by CTT cleavage
In Skiniotis et al (2003), we introduced a clonable density marker (SH3 domain) inserted at the transition between neck-linker and neck coiled-coil, to localize the neck region on monomeric and dimeric kinesins. In the presence of AMP-PNP, the neck region was found locked towards the MT plus-end, but there were striking variations between monomeric and dimeric constructs (compare yellow and cyan volumes in Figure 6). In that work, we could show that subtilisin treatment did not affect the position of the neck in dimeric constructs (Figure 6; compare cyan and magenta volumes). In the present work, in order to exclude the fact that steric constraints between the two motor heads of a dimer might prohibit the detection of the neck–CTT interaction, we further investigated the neck positions in monomers. Again, the position of the marker remained unchanged upon subtilisin treatment (Figure 6; compare yellow and green volumes).
Figure 6.
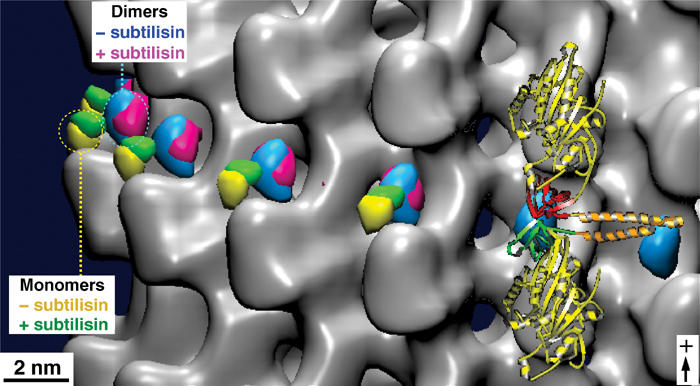
Tubulin C-terminal cleavage does not affect the positions of the neck region in monomeric and dimeric kinesins. The kinesin neck region position was probed by monomeric and dimeric constructs carrying an SH3 domain at the transitions between the neck-linker and the neck coiled-coil helix α7. In the presence of AMP-PNP, the kinesin neck is found in a stable position towards the plus-end in both complexes with native and subtilisin-treated MTs. However, its position in dimers (blue, before treatment; magenta, after treatment) locates closer to the tubulin surface than in monomers (yellow, before treatment; green, after treatment). Hence, for both monomeric and dimeric constructs, their positions were not affected by the presence or absence of the β-tubulin C-terminus. The atomic model of the docked dimeric rK379–SH3 construct is shown on the right (see also Skiniotis et al, 2003).
Biochemical characterization of the MT–kinesin complexes
We evaluated the effect of CTT cleavage on the mechanochemistry of the MT–rK354 complexes to provide an insight into the structural conformations observed (Figures 1, 2, 4 and 6). Although there was no apparent difference in the steady-state kcat (Figure 7A, B and G), the subtilisin-treated MT–rK354 complexes appeared to bind the MT lattice more tightly (K1/2,MT=0.25 μM) than observed with the native MT–rK354 complexes (K1/2,MT=0.6 μM). However, the equilibrium dissociation constant determined for both complexes in the absence of added nucleotide was comparable (Kd,MT ∼0.4 μM, Figure 7C). These results suggested that the tighter affinity for subtilisin-treated MTs based on the steady-state K1/2,MT is the result of one or more nucleotide intermediate states during ATP turnover and not simply reflecting a tighter binding of the motor to subtilisin-treated MTs.
Figure 7.
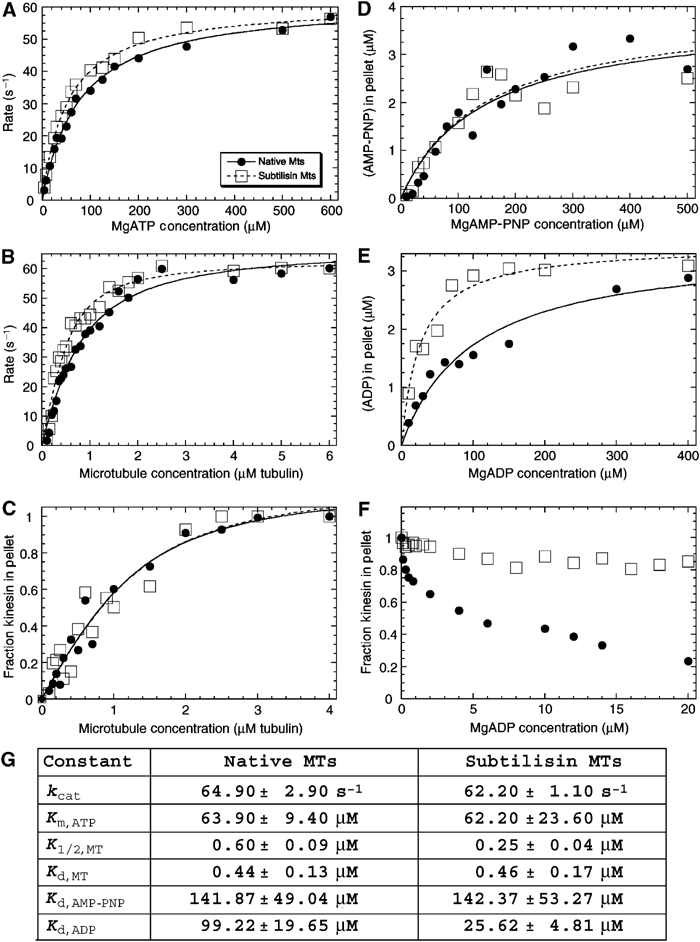
Characterization of the MT–kinesin complexes. (A) Native MT–rK354 and subtilisin-treated MT–rK354 complexes (0.1 μM rK354 plus taxol-stabilized MTs at 8 μM tubulin) were preformed and incubated with [α32P]ATP. The steady-state ATPase is shown as a function of MgATP concentration: native MT, kcat=61.9±1.2 s−1 and Km,ATP=77.4±4.2 μM; subtilisin-treated MT, kcat=61.2±0.9 s−1 and Km,ATP=52.8±2.5 μM. (B) Native MT–rK354 and subtilisin-treated MT–rK354 complexes were preformed with 0.25 μM rK354 plus taxol-stabilized MTs (0−6 μM tubulin) and reacted with 1 mM [α32P]ATP. The steady-state ATPase parameters were determined for native MTs (kcat=67.8±2.3 s−1 and K1/2,MT=0.51±0.06 μM) and subtilisin-treated MTs (kcat=63.3±1.9 s−1 and K1/2,MT=0.21±0.03 μM). (C) Equilibrium binding of 2 μM rK354 with varying concentrations of native and subtilisin-treated MTs (0−4 μM tubulin) in the absence of nucleotide. The fraction of rK354 in the MT pellet was plotted as a function of the total MT concentration. For native MTs, Kd,MT=0.44±0.13 μM with maximum fractional binding at 1.1±0.1. For subtilisin-treated MTs, Kd,MT=0.46±0.17 μM with maximum fractional binding at 1.2±0.1. (D) Equilibrium binding of 4 μM rK354, varying concentrations of [α32P]AMP-PNP (0−600 μM), and native or subtilisin-treated MTs (5 μM tubulin). The concentration of [α32P]AMP-PNP that partitioned with the MT–rK354 complexes was plotted as a function of MgAMP-PNP concentration. Kd,AMP−PNP was 142±49 μM for native MT–rK354 complexes with the maximal AMP-PNP binding at 3.83±0.52 μM sites. For subtilisin-treated MTs, Kd,AMP-PNP=142±53 μM with maximal AMP-PNP binding at 3.93±0.58 μM sites. (E) Equilibrium binding of 3 μM rK354, varying concentrations of [α32P]ADP (0−400 μM), and native or subtilisin-treated MTs (4 μM tubulin). The concentration of [α32P]ADP that partitioned with the MT–rK354 complex was plotted as a function of MgADP concentration. Kd,ADP=99±20 μM for the native MT–rK354 complex with maximal ADP binding at 100% sites. For the subtilisin-treated MT–rK354 complex, Kd,ADP=26±5 μM with maximal ADP binding at 100% sites. (F) Partitioning of 2 μM rK354 with 3 μM MTs as a function of MgADP concentration. (G) Summary of the MT–kinesin constants based on 3–7 independent experiments for each. The conditions were 80 mM PIPES (pH 6.8) with NaOH, 1 mM MgCl2, 1 mM EGTA at 25°C.
To determine the nucleotide state that was binding subtilisin-treated MTs with higher affinity than native MTs, we pursued a series of equilibrium binding experiments. Figure 7D shows the experiment in which we measured directly the concentration of [α32P] AMP-PNP that partitions with the MT–rK354 pellet. The data for subtilisin-treated and native MTs were similar, and the Kd,AMP-PNP for each was the same at 142 μM. However, when the experiment was repeated with [α32P]ADP, there was a significant difference (Figure 7E–G). At 100 μM [α32P]ADP, all motors partitioned to the subtilisin-treated MT–rK354 pellet, and the motor affinity for ADP was tighter, Kd,ADP=26 μM. In contrast, for native MT–rK354, the Kd,ADP was 99 μM. These equilibrium binding experiments were critical because they also indicate that the MT–rK354 complexes used for the cryo-EM do have AMP-PNP or ADP at the active site. The equilibrium binding experiments presented in Figure 7D and E were designed to determine the Kd,AXP, thus requiring low concentrations of AMP-PNP or ADP. At these conditions, the motor may still be bound to native MTs. To evaluate the partitioning of rK354 as a function of MgADP concentration, we designed an experiment similar to the cryo-EM conditions. Figure 7F shows that as a function of MgADP, the motor partitions off the native MTs, yet even at 20 mM MgADP (cryo-EM conditions) the motor remains bound to subtilisin-treated MTs. These studies reinforce our structural interpretations that with the subtilisin-treated MT–rK354 complexes, we have increased the conformational stability of the AMP-PNP state. Moreover, we have trapped an ADP state that is otherwise too transient to visualize on native MTs.
CTT cleavage does not affect the rate of ADP release but alters the pre-steady-state kinetics of MT association and detachment
The structural studies (Figures 1, 2, 4 and 6) and initial biochemical characterization (Figure 7) indicated that the CTTs of αβ-tubulin modulate MT interactions with kinesin during the ATPase cycle. To determine the specific nucleotide-dependent step affected, we measured the pre-steady-state kinetics of 2′(3′)-O-(N-methylanthraniloyl)-adenosine 5′ diphosphate (mantADP) release, MT association with rK354, and the ATP-promoted dissociation kinetics for motor detachment from the MT. Figure 8A shows that the rate of mantADP release increased as a function of MT concentration, with the maximum rate constant for both native and subtilisin-treated MTs at ∼340 s−1. However, the concentration of MTs required to reach one-half the maximal rate was somewhat tighter for subtilisin MTs with K1/2,MT=54 μM as compared to 70 μM for native MTs.
Figure 8.
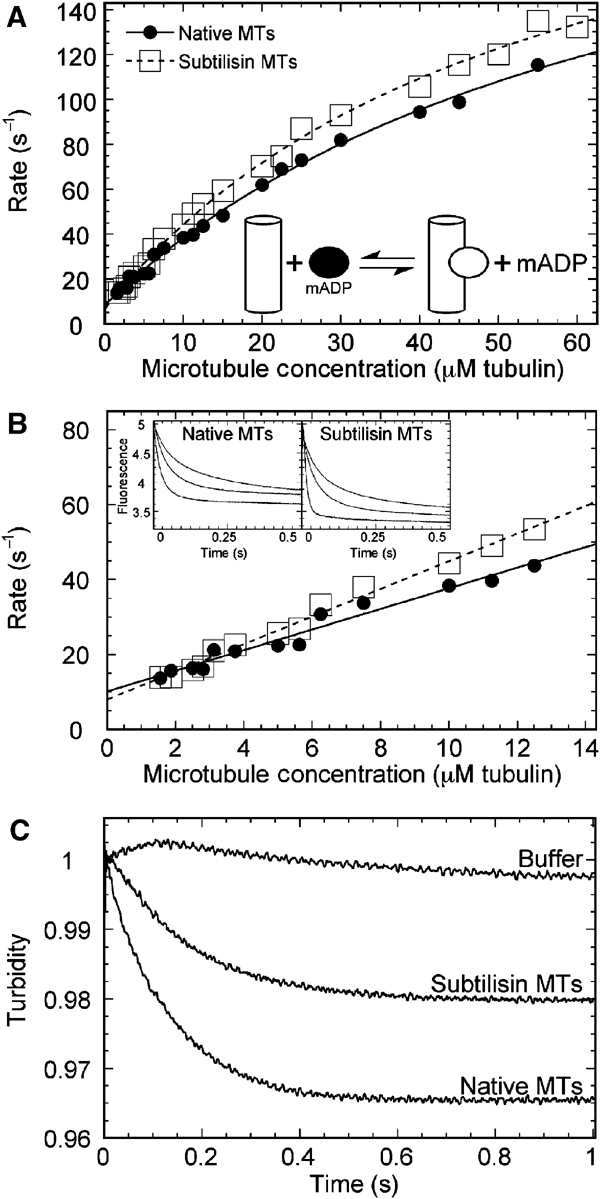
Pre-steady-state kinetics of the MT–rK354 complexes. (A) MantADP release kinetics. The rK354–mantADP complex was preformed and rapidly mixed in the stopped-flow instrument with varying concentrations of native and subtilisin-treated MTs+1 mM MgATP (final concentrations after mixing, 1.5 μM rK354+3 μM mantADP for MTs from 1.56–2.8 μM and 3 μM rK354+6 μM mantADP for MTs from 3.125–60 μM). Representative transients are shown in the inset of (B) (3.125, 6.25, 12.5 μM MTs from top to bottom transient). Each transient was fit to an exponential function, and the rate of the fluorescence decrease was plotted as a function of MT concentration. The fit of the data to hyperbolae provided the maximum rate constant for mantADP release for native MTs (kmax=239±18 s−1, K1/2,MT=69.7±9.1 μM) and subtilisin MTs (kmax=242±11 s−1, K1/2,MT=54.1±5.3 μM). (B) The data at low MT concentrations shown in (A) were re-plotted here to determine the second-order rate constant for MT association. The slope predicts the minimum estimate for MT association. Native MTs: 2.75±0.16 μM−1 s−1; subtilisin MTs: 3.69±0.11 μM−1 s−1. (C) MT–rK354 complexes were preformed and rapidly mixed with MgATP+KCl (final concentrations after mixing: 4 μM rK354, 5 μM MTs, 1 mM MgATP, 50 mM KCl in BRB80 buffer). The transients were each normalized to begin at 1 V to compare amplitudes. Note that the buffer control with 50 mM KCl exhibited no significant change in turbidity. Native MTs: kobs=7.9±0.02 s−1; subtilisin MTs: kobs=5.9±0.02 s−1.
In contrast, both the association and dissociation kinetics were affected by the loss of the CTTs. As mantADP release is rapid for kinesin, the initial linear part of the curves in Figure 8A predicts the second-order rate constant for motor association with the MTs. These data are shown in Figure 8B. The second-order rate constant was 3.7 μM−1 s−1 for subtilisin-treated MTs and 2.7 μM−1 s−1 for native MTs. As the fluorescence signal of mantADP release was used to obtain the association kinetics, the second-order rate constant determined also reflects the structural transitions required for mantADP release from the active site of the motor. In Figure 8C, the ATP-promoted dissociation kinetics are shown. For this experiment, an MT–rK354 complex was preformed and then rapidly mixed with MgATP+additional salt (final concentrations 4 μM rK354, 10 μM MTs, 1 mM MgATP, 50 mM KCl). The observed decrease in turbidity reflects the kinesin motors detaching from the MT. The additional salt is required to facilitate detachment of monomeric kinesin from MTs (Moyer et al, 1998). Note that the rate as well as the amplitude of the dissociation kinetics were both affected by removal of the CTTs from αβ-tubulin. For subtilisin MTs, kinesin detachment was observed at 5.9 s−1, whereas for native MTs this rate was 7.9 s−1. The amplitude of the subtilisin-treated MT transient was ∼50% of the native MT transient. These results indicate that the CTTs of αβ-tubulin play an important role in the structural transitions required for kinesin motor detachment from the MT after ATP hydrolysis.
Discussion
The function of kinesin is tightly coupled to an alternating nucleotide hydrolysis in its head domains (Hackney, 1994; Ma and Taylor, 1997; Gilbert et al, 1998). ADP-kinesin binds to MTs through immediate nucleotide release. ATP uptake in the leading head mediates the forward motion of the trailing motor through a conformational change in the neck region (Rice et al, 1999; Skiniotis et al, 2003), and ATP hydrolysis releases the motor again from the MT surface. If the MT on time is longer than the off time and hydrolysis occurs in an alternating fashion, long processive walks are possible. An important question is how the tubulin portion itself participates in the regulation of the kinesin walking mechanism apart from providing the structural track. Here, we have investigated the conformational states of kinesin motor domains complexed to MTs under different nucleotide conditions and the effect of the highly acidic αβ-tubulin C-termini on these binding conformations.
Nucleotide-dependent conformational changes in kinesin–MT complexes
This work revealed significant structural differences within kinesin motor domains in nucleotide-free versus AMP-PNP-bound states complexed to native MTs (Figures 1 and 2). The statistically most significant changes in density occur at the motor–tubulin interface, close to the switch II region of kinesin (Figure 5). Thereby, the nucleotide-free state exhibits a higher density at that location than the AMP-PNP state. The shape of the motor domain appears wider and rotated by about 12° (Figure 1C and D). Hence, the nucleotide-free state displays a more rigid interaction, despite its dislocated neck region (Rice et al, 1999; Skiniotis et al, 2003), while the AMP-PNP state appears more mobile. Due to the statistical procedure used here, density differences at the plus-end directed nose are not detected at the 99% significance level. Mobile elements produce increased variances and therefore density differences at such locations are statistically less significant.
The structural differences in binding states between nucleotide-free and AMP-PNP states, observed here, are consistent with the large conformational changes reported in the switch II helix during the ATPase cycle (Kikkawa et al, 2001; Kull and Endow, 2002). The switch II helix is supposed to be one of the sensors that react conformationally to the presence of the γ-phosphate in ATP (Vale and Milligan, 2000). Hence, we can assume that the AMP-PNP state loses part of the binding forces from loops 8 and 12, rendering its conformation more mobile (see also Figure 9). This could reflect a conformational state that prepares the motor for releasing from the MT surface after ATP hydrolysis. The nucleotide-free state, on the other hand, represents a clear rigor state depending on ATP to proceed. These results provide the first structural confirmation of biochemical and biophysical studies, which indicated that, while the ADP state of a motor displays the weakest, dissociation-connected binding (Figure 4), the nucleotide-free state induces the strongest binding affinity (Crevel et al, 1996; Xing et al, 2000; Kawaguchi and Ishiwata, 2001; Uemura et al, 2002).
Figure 9.
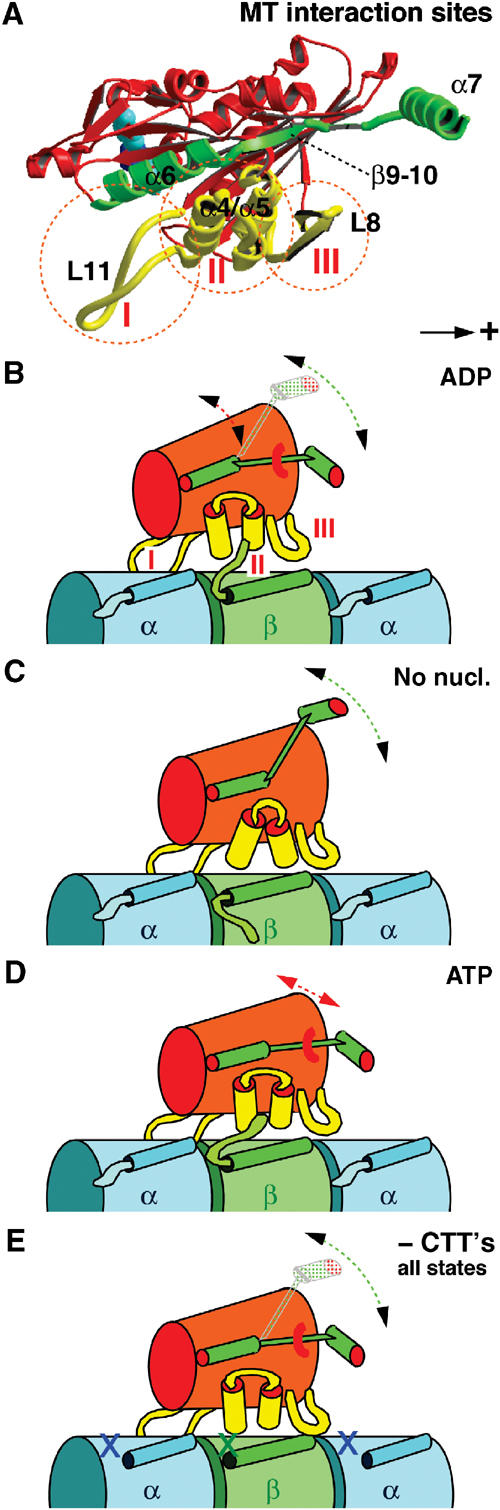
Schematic model for kinesin interaction with MTs at different nucleotide states and in dependence of the CTTs. (A) The kinesin–MT interface consists of three different clusters, loop 11 (I), α4-loop12-α5 (II), and loop 8 (III). (B) In the presence of ADP, the affinity of kinesin to MTs is rather weak and probably only mediated through loop 11 (Asenjo et al, 2003), which does not seem to be influenced by the CTTs. (C) Upon ADP release, binding sites II and III become more effective through rearrangements in the switch-II region. (D) Upon ATP uptake, the neck-linker (β9–10, α7) locks in its plus-end pointing position and binding sites II and III experience a stronger interference from the β-tubulin C-terminus. It may be possible that α7 interferes with the α-tubulin C-terminus, rendering the head more mobile. (E) Absence of CTTs renders all binding regions equally rigid through all nucleotide states.
Effects of CTTs on kinesin–MT complexes
We have incubated MTs with subtilisin under conditions that removed the CTTs from both α- and β-tubulin (Figure 3D; see also Bhattacharyya et al, 1985; Sackett et al, 1985). The structural data are illustrated in Figures 2 and 4, and the mechanistic analysis is summarized in Figures 7 and 8. Our t-test analysis suggests that C-terminal cleavage renders the binding configuration of AMP-PNP-bound motor heads close to that of a nucleotide-free head. The comparison of AMP-PNP states before and after subtilisin treatment produces a density difference (Figure 2C), which is very similar to the one detected between AMP-PNP and nucleotide-free states on native MTs (Figure 2A). As demonstrated in Figure 7D, on treated MTs 98% of AMP-PNP remains bound to the kinesin pocket, and therefore the observed conformational differences are not connected to a loss of nucleotide from the active site. This should be expected, since we found the neck-linker locked forward as with native MTs (Figure 6). Nucleotide-free kinesin heads bind equally tight to native or treated MTs (Figure 7C). The Kd,MT is 0.46 μM for subtilisin-treated MT and 0.44 μM for native MT. Therefore, the tighter affinity for subtilisin-treated MTs, based on the steady-state K1/2,MT (0.25 μM (+subt.) and 0.6 μM (−subt.); Figure 7B), is a result of one or more nucleotide intermediate states during ATP turnover, and does not reflect a tighter kinesin–MT binding once the CTTs are cleaved.
Removal of the CTTs has a very striking effect on the binding properties of kinesin motor domains in the presence of excess ADP, increasing kinesin affinity dramatically (Figure 4). Equilibrium binding showed that ADP remains in the motor nucleotide pocket and Kd,ADP drops from ∼99 μM on native MTs to ∼26 μM on subtilisin-treated MTs (Figure 7E and F). These data suggest that the removal of the acidic C-terminus traps the ADP–kinesin–MT complex in an otherwise very transient intermediate state, shortly before ADP release and transition into the nucleotide-free conformation. These observations at equilibrium conditions were reinforced by the pre-steady-state kinetics in which ATP-promoted motor dissociation from the subtilisin MTs was aberrant (Figure 8C). We also suggest that this effect may be the major reason for the lower processivity reported for kinesin by Wang and Sheetz (2000), as a strongly bound ADP state probably stalls the motor. The MT–kinesin association kinetics (Figure 8C) indicates that removal of the CTTs results in more rapid MT–rK354 complex formation that leads to ADP release. In dimeric kinesin, this could also mean a loss of cooperativity and possibly a destructive interference with the alternating head catalysis process.
Effects of subtilisin treatment on the conformation of the neck region
We further tested the effects of CTT removal on the configurations of the neck region in monomeric and dimeric motor constructs, using mutants with an engineered SH3 domain at the transition from neck-linker to the neck helix α7 (Skiniotis et al, 2003). Our results clearly show that in the presence of AMP-PNP, the neck-linker of monomeric and dimeric kinesins attaches to the kinesin motor core domain at essentially identical locations, irrespective of CTT presence or absence (Figure 6). Hence, we conclude that the configuration of the neck region itself is not affected by CTTs. In addition, subtilisin treatment does not seem to affect the flexible nature of the neck-linker in nucleotide-free (see also Skiniotis et al, 2003) and, presumably, ADP conditions. Hence, our results do not reflect the proposal made by Thorn et al (2000).
The C-terminal tail of β-tubulin locates at the vicinity of the switch II cluster
Molecular docking shows that the switch II cluster (α4/L12/α5) locates above the end of helix H12 from β-tubulin, in very close proximity to the disordered CTT (Figure 5). The calculated difference density related to the nucleotide state (Figure 5; red wireframe) locates in close proximity to this cluster. Hence, we assume that the obtained differences are connected with structural rearrangements of the switch II cluster in response to the presence of both the nucleotide in kinesin and the presence of the CTT from tubulin. Besides the switch II cluster (marked as interaction site II in Figure 9), loops L11 and L8 also reside at the tubulin interface (sites I and III in Figure 9), but both loops seem to be too distant from either of the tubulin C-termini to assume a strong interaction with them. The α-tubulin C-terminus, however, could interfere with kinesin binding via the neck helix (helix 7) of kinesin.
According to our data, removal of the CTTs allows kinesin to form a strong complex with MTs, irrespective of any nucleotide present in the active site. Hence, overall the CTTs appear to weaken, rather than strengthen, the interaction of the motor domain with the MT surface. This suggests that the conformational changes of module α4/L12/α5 from a so-called ‘open state' (nucleotide-free) to a ‘closed state' (ATP present; Kull and Endow, 2002) result in different levels of interference from the tubulin C-termini, thereby regulating MT binding and release. The state that appears to be the least affected by the CTTs is the nucleotide-free state. Thus, in contrast to the closed (retracted) state of the α4/L12 module in the presence of ATP, the extended state of the switch II cluster may reduce CTT interference (Figure 9). Solution studies showed that the helical propensity of β-tubulin CTT can be extended for at least nine more residues (Jimenez et al, 1999) compared to the structure reported in the crystallographic model (Nogales et al, 1998). Therefore, one may speculate that a helix ⇌ coil transition in the β-tubulin C-terminus could play a regulatory role in kinesin–tubulin interaction (Jimenez et al, 1999).
As discussed above, in the absence of CTTs, kinesin–ADP binds MTs very stably. This phenotype resembles that of a monomeric kinesin mutant, reported by Okada and Hirokawa (2000), which has the highly positively charged K-loop of KIF1A engineered into the position of loop L12. This mutant showed nucleotide-insensitive binding to MTs and its Kd values in the ADP state were very close to those with AMP-PNP or nucleotide-free states. In KIF1A, the CTTs appear to interact with the K-loop to keep the monomer on track, which might explain why KIF1A can also be a processive monomer (Okada and Hirokawa, 2000; Okada et al, 2003). For conventional kinesin however, the CTTs seem to interfere with motor binding in a way that facilitates a stepping mechanism with a high frequency of binding and releasing events. This is achieved by weakening the motor–MT interaction in two critical phases, thereby (a) preventing the second head from binding before the first (MT-bound) head exchanged ADP for ATP, and (b) mediating the release from the MT surface after ATP hydrolysis. Hence, the interaction of motor domains with the CTTs appears to be an important element for fine-tuning the processive walk of kinesin on MTs (Thorn et al, 2000; Wang and Sheetz, 2000).
Here, we showed that the interference from tubulin CTTs, in response to the nucleotide state of the motor, regulates weak and strong kinesin binding states. Thus, the CTTs play a crucial role in promoting processive walking of dimeric kinesin by facilitating motor release from the MT surface. The interactions between structural MAPs and CTTs (Serrano et al, 1984a; Marya et al, 1994; Al-Bassam et al, 2002) further suggest a functional relationship between kinesin and CTTs. One could speculate that MAPs, by saturating the charged CTTs, may impose a similar rigor state on kinesin as caused by their removal, thereby stalling the processive walk (Ebneth et al, 1998; Seitz et al, 2002), but still allow kinesin motors to decorate MTs fully (unpublished results). Similarly, the facts that the major part of tubulin heterogeneity is located at its C-terminus (Redeker et al, 1992), and that CTTs undergo post-translational modifications which all modify their charge (Luduena, 1998; Rosenbaum, 2000), indicate their central role in the modulation of MT-based motility and underline the necessity to further investigate kinesin binding properties to individual tubulin isoforms.
Experimental procedures
MT preparation
Bovine brain tubulin (Cytoskeleton, Inc.) was polymerized for 20 min at 37°C in 80 mM PIPES (pH 6.8), 2 mM MgCl2 at a concentration of 5 mg ml−1 and in the presence of 7.5% (v/v) DMSO, 2 mM GTP and 20 μM Taxol. In order to remove the C-terminal ends of α- and β-tubulin, MTs were treated with 200 μg ml−1 subtilisin for 1 h at 30°C. Proteolysis was stopped with the addition of 10 mM PMSF (Roche Molecular Biochemicals). Treated MTs were spun for 10 min at 16 000 rcf and were resuspended in BRB80 including 0.8% Taxol. All chemicals used were of analytical grade.
Cloning, expression, and purification of kinesin constructs
We cloned, expressed, and purified all kinesin constructs as described in Hoenger et al (2000) and in Skiniotis et al (2003). Briefly, non-SH3-tagged monomeric and dimeric constructs (rK354, rK379) were expressed from a pET-3a derivative vector. For kinesin–SH3 chimeras, we amplified by polymerase chain reaction the corresponding fragments and cloned them in vector pET-22b, including a stop codon to avoid expression of the His-tag. All constructs were expressed in Escherichia coli BL21 (DE3) by induction with 0.5 mM IPTG for 16 h at 24°C. Cells were resuspended in 20 mM PIPES (pH 6.9), 1 mM EGTA, 1 mM DTT, 1 mM MgCl2, 50 mM NaCl, 100 μM MgATP, and lysed with a French press (Aminco). Purification was carried out by two ion exchange columns (SP sepharose, monoQ) followed by a gel filtration run on G200 Hiload 16/60 (Pharmacia) in 20 mM PIPES (pH 7.2), 50 mM NaCl, 5 mM MgCl2.
Specimen preparation for cryo-EM
Decoration of MTs with kinesin constructs was carried out at a final concentration of tubulin at 0.5 mg ml−1. The different nucleotide states were generated by treatment of the motor with 5 mM apyrase (nucleotide-free), 5 mM AMP-PNP (ATP-like), or 20 mM ADP. For dimeric motors, we used 1.5 times motor per binding site for AMP-PNP decoration, and 2.5–3 times motor per binding site for nucleotide-free conditions. Motors with MTs were incubated for 2 min, followed by absorption of the sample on holey carbon grids and quick freezing in liquid ethane (Dubochet et al, 1988). Cryo-EM was performed on a Philips CM200-FEG microscope, using a GATAN-626 cryo-holder. Images were recorded on Kodak SO-163 electron microscopy film, at × 38.000 magnification, and at a defocus range between –1.5 and –2.0 μm.
Image processing and 3D reconstruction
For 3D reconstructions, we screened for 15-protofilament/2-start helical MTs (see Beuron and Hoenger, 2001). Micrographs were digitized using a Zeiss-SCAI scanner operated at 21 μm on the negative, which corresponds to 0.5526 nm on the sample. Suitable MTs were helically reconstructed by using the program suite PHOELIX (Whittaker et al, 1995). 3D maps were visualized using SUPRIM (Schroeter and Bretaudiere, 1996) and VolVis (SUNY Stonybrook). Significant differences between the 3D maps were calculated by a t-test within the PHOELIX suite, comparing all the data sets used for each reconstruction. In our figures, difference maps are displayed at volumes enclosing a certainty level higher than 99% for a significant difference in mass.
Molecular docking and modeling
Using the modeling program ‘O' (Jones et al, 1991), the atomic resolution structures of rK354 (Sack et al, 1997 (2KIN)) and tubulin (Nogales et al, 1998 (1TUB)) were manually docked in the EM envelopes obtained for rK354 complexed to MTs in the absence of nucleotide.
Steady-state ATPase kinetics
All biochemical assays were performed at 25°C in BRB80 buffer (80 mM PIPES (pH 6.8) with NaOH, 1 mM MgCl2, 1 mM EGTA). MTs were assembled from cold-depolymerized tubulin and stabilized with taxol. All concentrations reported are final concentrations after mixing.
The steady-state ATPase activity was determined by following [α32P]ATP hydrolysis (Gilbert and Mackey, 2000). Each data set in Figure 7A was fit to the Michaelis–Menten equation; the data in Figure 7B required quadratic Equation (1).
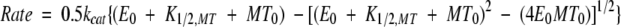 |
1 |
where Rate is the amount of hydrolysis product formed per second per active site, kcat is the maximum rate constant of product formation at saturating substrate, E0 is the rK354 concentration, K1/2,MT is an apparent dissociation constant defined by the MT concentration needed to provide one-half the maximal velocity, and MT0 is the tubulin concentration as taxol-stabilized MTs.
MT equilibrium binding assays
These experiments were performed as described previously (Klumpp et al, 2003). In the absence of added nucleotide (Figure 7C), rK354 at 2 μM was incubated with varying concentrations of native MTs or subtilisin MTs (0–4 μM tubulin) for 30 min, followed by centrifugation. The MT pellet was resuspended in BRB80 buffer to equal the volume of the supernatant, and samples of each were analyzed by SDS–PAGE. The Coomassie blue stained gels were scanned and quantified by NIH Image v1.62 to determine the concentration of rK354 in the supernatant and pellet at each MT concentration. The ratio of rK354 in the pellet to total rK354 (supernatant+pellet) was plotted as a function of MT concentration. The data were fit to quadratic Equation (2):
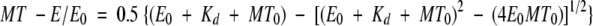 |
2 |
where MT-E/E0 is the fraction of rK354 partitioning with MTs, E0 is the total rK354 concentration, Kd is the dissociation constant, and MT0 is the total tubulin concentration.
Equilibrium binding of MT–rK354 as a function of [α32P]AMP-PNP or [α32P]ADP
Kinesin rK354 was incubated with varying concentrations of radiolabeled MgAMP-PNP or MgADP for 30 min, and then native or subtilisin MTs were added and incubated for 30 min. The reaction mixture was centrifuged, the supernatant was removed, and the unwashed pellet was resuspended in 4 N NaOH. The volume of the pellet was adjusted with buffer to equal the volume of the supernatant. Control reactions with MTs in the absence of rK354 were performed at each MT concentration to determine the concentration of [α32P]AMP-PNP or [α32P]ADP that was trapped in the MT pellet in the absence of motor. The concentration of [α32P]AXP for each MT–rK354 reaction was corrected for [α32P]AXP that sedimented with MTs in the absence of motor. Figure 7D and E show the normalized data, and each data set was fit to quadratic Equation (3).
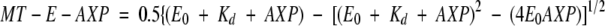 |
3 |
where MT-E-AXP is the concentration of [α32P]AMP-PNP or [α32P]ADP partitioning with the MT pellet, E0 is the total rK354, AXP is the total AMP-PNP or ADP present, and Kd is the dissociation constant.
Pre-steady-state kinetics
The kinetics of MT-activated ADP release were measured using the fluorescent ADP analogue, mantADP. For the mantADP release experiments, the excitation wavelength was 360 nm with emitted light measured through a 400 nm cutoff filter (mant λemm=450 nm). The rK354 motor was incubated with mantADP at a ratio of 1:2 for 60 min to allow exchange of ADP at the active site with mantADP. The rate constants for mantADP release at low MT concentrations (Figure 8B) were used to detect motor association with the MT as described previously (Moyer et al, 1998). This approach is valid because mantADP release is rapid for kinesin. The pre-steady-state kinetics of ATP-promoted dissociation was measured using the change in turbidity monitored at 340 nm. MantADP release, motor association with MTs, and ATP-promoted motor dissociation were determined using the SF-2003 KinTek stopped-flow instrument in BRB80 buffer at 25°C.
Acknowledgments
We thank Luis Serrano (EMBL Heidelberg) for the SH3 plasmid pBAT4.
References
- Al-Bassam J, Ozer RS, Safer D, Halpain S, Milligan RA (2002) MAP2 and tau bind longitudinally along the outer ridges of microtubule protofilaments. J Cell Biol 157: 1187–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asenjo AB, Krohn N, Sosa H (2003) Configuration of the two kinesin motor domains during ATP hydrolysis. Nat Struct Biol 10: 836–842 [DOI] [PubMed] [Google Scholar]
- Beuron F, Hoenger A (2001) Structural analysis of the microtubule–kinesin complex by Cryo-electron microscopy. In Kinesin Protocols, Vernos I (ed) Totowa, NJ: Humana Press Inc. pp 235–254 [DOI] [PubMed] [Google Scholar]
- Bhattacharyya B, Sackett DL, Wolff J (1985) Tubulin, hybrid dimers, and tubulin S. Stepwise charge reduction and polymerization. J Biol Chem 260: 10208–10216 [PubMed] [Google Scholar]
- Crevel IM, Lockhart A, Cross RA (1996) Weak and strong states of kinesin and ncd. J Mol Biol 257: 66–76 [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P (1988) Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21: 129–228 [DOI] [PubMed] [Google Scholar]
- Ebneth A, Godemann R, Stamer K, Illenberger S, Trinczek B, Mandelkow E (1998) Overexpression of tau protein inhibits kinesin-dependent trafficking of vesicles, mitochondria, and endoplasmic reticulum: implications for Alzheimer's disease. J Cell Biol 143: 777–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert SP, Mackey AT (2000) Kinetics: a tool to study molecular motors. Methods 22: 337–354 [DOI] [PubMed] [Google Scholar]
- Gilbert SP, Moyer ML, Johnson KA (1998) Alternating site mechanism of the kinesin ATPase. Biochemistry 37: 792–799 [DOI] [PubMed] [Google Scholar]
- Goldstein LS, Philp AV (1999) The road less travelled: emerging principles of kinesin motor utilization. Annu Rev Cell Dev Biol 15: 141–183 [DOI] [PubMed] [Google Scholar]
- Hackney DD (1994) Evidence for alternating head catalysis by kinesin during microtubule-stimulated ATP hydrolysis. Proc Natl Acad Sci USA 91: 6865–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenger A, Thormählen M, Diaz-Avalos R, Doerhoefer M, Goldie KN, Müller J, Mandelkow E (2000) A new look at the microtubule binding patterns of dimeric kinesins. J Mol Biol 297: 1087–1103 [DOI] [PubMed] [Google Scholar]
- Jimenez MA, Evangelio JA, Aranda C, Lopez-Brauet A, Andreu D, Rico M, Lagos R, Andreu JM, Monasterio O (1999) Helicity of alpha(404–451) and beta(394–445) tubulin C-terminal recombinant peptides. Protein Sci 8: 788–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kar S, Fan J, Smith MJ, Goedert M, Amos LA (2003) Repeat motifs of tau bind to the insides of microtubules in the absence of taxol. EMBO J 22: 70–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K, Ishiwata S (2001) Nucleotide-dependent single- to double-headed binding of kinesin. Science 291: 667–669 [DOI] [PubMed] [Google Scholar]
- Kikkawa M, Sablin EP, Okada Y, Yajima H, Fletterick RJ, Hirokawa N (2001) Switch-based mechanism of kinesin motors. Nature 411: 439–445 [DOI] [PubMed] [Google Scholar]
- Klumpp LM, Brendza KM, Rosenberg JM, Hoenger A, Gilbert SP (2003) Motor domain mutation traps kinesin as a microtubule rigor complex. Biochemistry 42: 2595–2606 [DOI] [PubMed] [Google Scholar]
- Kull FJ, Endow SA (2002) Kinesin: switch I & II and the motor mechanism. J Cell Sci 115: 15–23 [DOI] [PubMed] [Google Scholar]
- Li H, DeRosier DJ, Nicholson WV, Nogales E, Downing KH (2002) Microtubule structure at 8 Å resolution. Structure 10: 1317–1328 [DOI] [PubMed] [Google Scholar]
- Luduena RF (1998) Multiple forms of tubulin: different gene products and covalent modifications. Int Rev Cytol 178: 207–275 [DOI] [PubMed] [Google Scholar]
- Ma YZ, Taylor EW (1997) Interacting head mechanism of microtubule–kinesin ATPase. J Biol Chem 272: 724–730 [DOI] [PubMed] [Google Scholar]
- Marya PK, Syed Z, Fraylich PE, Eagles PA (1994) Kinesin and tau bind to distinct sites on microtubules. J Cell Sci 107: 339–344 [DOI] [PubMed] [Google Scholar]
- Milligan RA, Flicker PF (1987) Structural relationship of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol 105: 29–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moores CA, Yu M, Guo J, Beraud C, Sakowicz R, Milligan RA (2002) A mechanism for microtubule depolymerization by KinI kinesins. Mol Cell 9: 903–909 [DOI] [PubMed] [Google Scholar]
- Moyer ML, Gilbert SP, Johnson KA (1998) Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry 37: 800–813 [DOI] [PubMed] [Google Scholar]
- Niederstrasser H, Salehi-Had H, Gan EC, Walczak C, Nogales E (2002) XKCM1 acts on a single protofilament and requires the C terminus of tubulin. J Mol Biol 316: 817–828 [DOI] [PubMed] [Google Scholar]
- Nogales E, Wolf SG, Downing KH (1998) Structure of the alpha beta tubulin dimer by electron crystallography. Nature 391: 199–203 [DOI] [PubMed] [Google Scholar]
- Okada Y, Higuchi H, Hirokawa N (2003) Processivity of the single-headed kinesin KIF1A through biased binding to tubulin. Nature 424: 574–577 [DOI] [PubMed] [Google Scholar]
- Okada Y, Hirokawa N (2000) Mechanism of the single-headed processivity: diffusional anchoring between the K-loop of kinesin and the C terminus of tubulin. Proc Natl Acad Sci USA 97: 640–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redeker V, Melki R, Prome D, Le Caer JP, Rossier J (1992) Structure of tubulin C-terminal domain obtained by subtilisin treatment. The major alpha and beta tubulin isotypes from pig brain are glutamylated. FEBS Lett 313: 185–192 [DOI] [PubMed] [Google Scholar]
- Rice S, Lin AW, Safer D, Hart CL, Naber N, Carragher BO, Cain SM, Pechatnikova E, Wilson-Kubalek EM, Whittaker M, Pate E, Cooke R, Taylor EW, Milligan RA, Vale RD (1999) A structural change in the kinesin motor protein that drives motility. Nature 402: 778–784 [DOI] [PubMed] [Google Scholar]
- Rosenbaum J (2000) Cytoskeleton: functions for tubulin modifications at last. Curr Biol 10: R801–R803 [DOI] [PubMed] [Google Scholar]
- Sack S, Muller J, Marx A, Thormählen M, Mandelkow EM, Brady ST, Mandelkow E (1997) X-ray structure of motor and neck domains from rat brain kinesin. Biochemistry 36: 16155–16165 [DOI] [PubMed] [Google Scholar]
- Sackett DL, Bhattacharyya B, Wolff J (1985) Tubulin subunit carboxyl termini determine polymerization efficiency. J Biol Chem 260: 43–45 [PubMed] [Google Scholar]
- Schroeter JP, Bretaudiere JP (1996) SUPRIM: easily modified image processing software. J Struct Biol 116: 131–137 [DOI] [PubMed] [Google Scholar]
- Seitz A, Kojima H, Oiwa K, Mandelkow E-M, Song Y-H, Mandelkow E (2002) Single-molecule investigation of the interference between kinesin, tau and MAP2c. EMBO J 21: 4896–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano L, Avila J, Maccioni RB (1984a) Limited proteolysis of tubulin and the localization of the binding site for colchicine. J Biol Chem 259: 6607–6611 [PubMed] [Google Scholar]
- Serrano L, Avila J, Maccioni RB (1984b) Controlled proteolysis of tubulin by subtilisin: localization of the site for MAP2 interaction. Biochemistry 23: 4675–4681 [DOI] [PubMed] [Google Scholar]
- Skiniotis G, Surrey T, Altmann S, Gross H, Song YH, Mandelkow E, Hoenger A (2003) Nucleotide-induced conformations in the neck region of dimeric kinesin. EMBO J 22: 1518–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn KS, Ubersax JA, Vale RD (2000) Engineering the processive run length of the kinesin motor. J Cell Biol 151: 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uemura S, Kawaguchi K, Yajima J, Edamatsu M, Toyoshima YY, Ishiwata S (2002) Kinesin–microtubule binding depends on both nucleotide state and loading direction. Proc Natl Acad Sci USA 99: 5977–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112: 467–480 [DOI] [PubMed] [Google Scholar]
- Vale RD, Milligan RA (2000) The way things move: looking under the hood of molecular motor proteins. Science 288: 88–95 [DOI] [PubMed] [Google Scholar]
- Wang Z, Sheetz MP (2000) The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophysical J 78: 1955–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker M, Carragher BO, Milligan RA (1995) PHOELIX: a package for semi-automated helical reconstruction. Ultramicroscopy 58: 245–259 [DOI] [PubMed] [Google Scholar]
- Woehlke G, Ruby AK, Hart CL, Ly B, Hom-Booher N, Vale RD (1997) Microtubule interaction site of the kinesin motor. Cell 90: 207–216 [DOI] [PubMed] [Google Scholar]
- Xing J, Wriggers W, Jefferson GM, Stein R, Cheung HC, Rosenfeld SS (2000) Kinesin has three nucleotide-dependent conformations. Implications for strain-dependent release. J Biol Chem 275: 35413–35423 [DOI] [PubMed] [Google Scholar]


