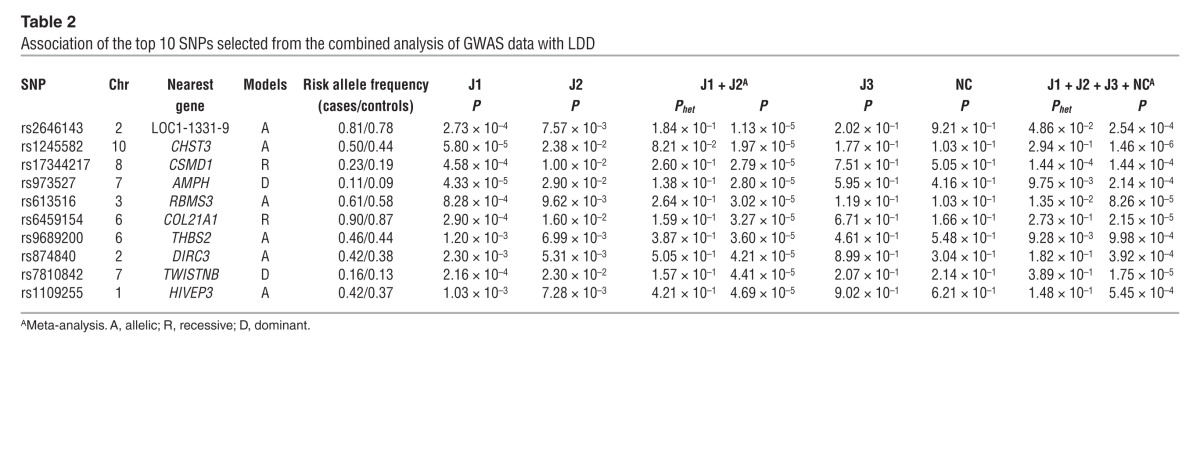
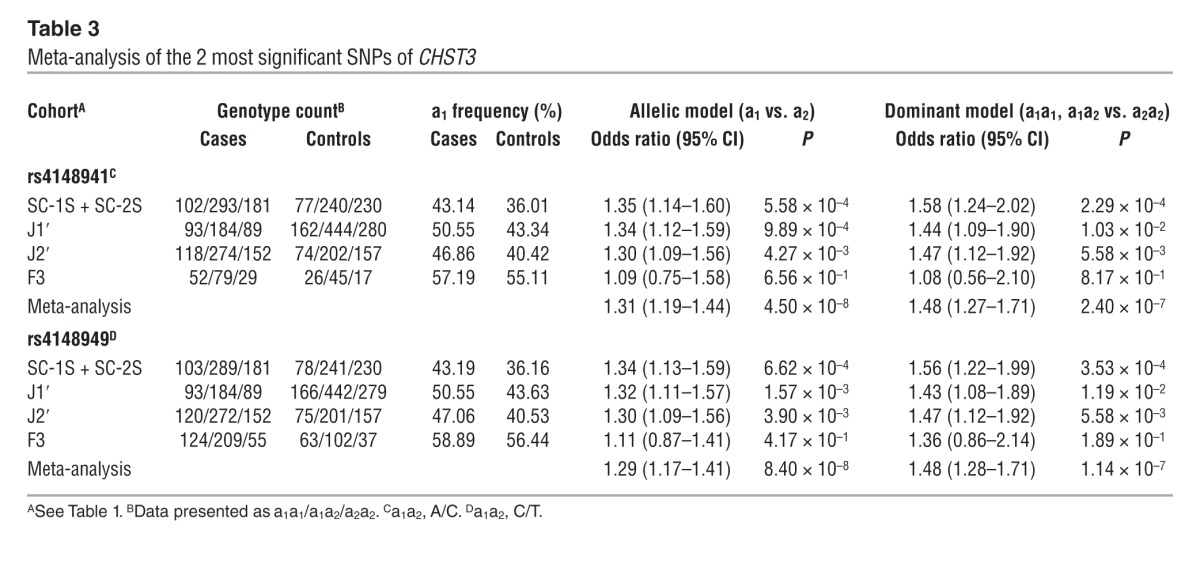
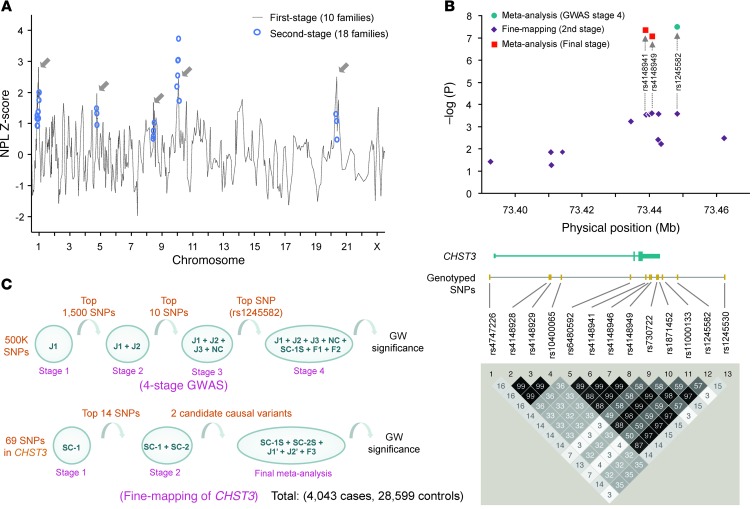
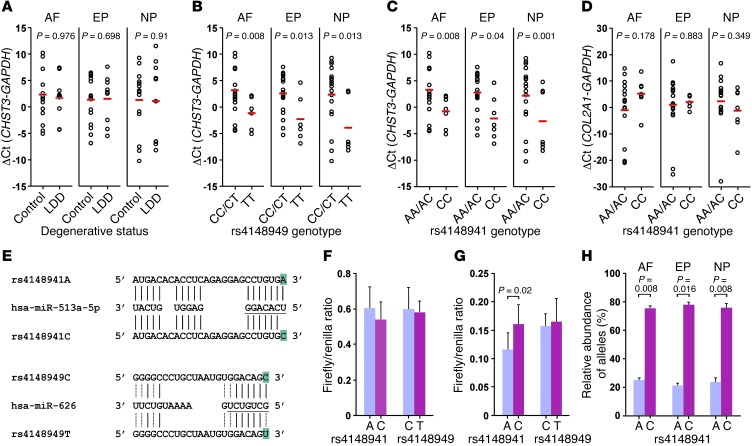
You-Qiang Song
You-Qiang Song
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
1,
Tatsuki Karasugi
Tatsuki Karasugi
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
2,3,
Kenneth MC Cheung
Kenneth MC Cheung
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
4,
Kazuhiro Chiba
Kazuhiro Chiba
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
5,
Daniel WH Ho
Daniel WH Ho
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
1,
Atsushi Miyake
Atsushi Miyake
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
2,5,
Patrick YP Kao
Patrick YP Kao
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
1,
Kit Ling Sze
Kit Ling Sze
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
1,
Anita Yee
Anita Yee
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
1,
Atsushi Takahashi
Atsushi Takahashi
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
6,
Yoshiharu Kawaguchi
Yoshiharu Kawaguchi
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
7,
Yasuo Mikami
Yasuo Mikami
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
8,
Morio Matsumoto
Morio Matsumoto
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
5,
Daisuke Togawa
Daisuke Togawa
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
9,
Masahiro Kanayama
Masahiro Kanayama
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
9,
Dongquan Shi
Dongquan Shi
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
10,11,
Jin Dai
Jin Dai
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
10,11,
Qing Jiang
Qing Jiang
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
10,11,
Chengai Wu
Chengai Wu
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
12,
Wei Tian
Wei Tian
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
13,
Na Wang
Na Wang
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
13,
John CY Leong
John CY Leong
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
14,
Keith DK Luk
Keith DK Luk
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
4,
Shea-ping Yip
Shea-ping Yip
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
15,
Stacey S Cherny
Stacey S Cherny
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
16,
Junwen Wang
Junwen Wang
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
1,
Stefan Mundlos
Stefan Mundlos
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
17,
Anthi Kelempisioti
Anthi Kelempisioti
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
18,
Pasi J Eskola
Pasi J Eskola
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
18,
Minna Männikkö
Minna Männikkö
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
18,
Pirkka Mäkelä
Pirkka Mäkelä
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
19,
Jaro Karppinen
Jaro Karppinen
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
20,21,
Marjo-Riitta Järvelin
Marjo-Riitta Järvelin
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
22,23,
Paul F O’Reilly
Paul F O’Reilly
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
23,
Michiaki Kubo
Michiaki Kubo
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
24,
Tomoatsu Kimura
Tomoatsu Kimura
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
7,
Toshikazu Kubo
Toshikazu Kubo
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
8,
Yoshiaki Toyama
Yoshiaki Toyama
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
5,
Hiroshi Mizuta
Hiroshi Mizuta
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
3,
Kathryn SE Cheah
Kathryn SE Cheah
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
1,
Tatsuhiko Tsunoda
Tatsuhiko Tsunoda
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
25,
Pak-Chung Sham
Pak-Chung Sham
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
16,
Shiro Ikegawa
Shiro Ikegawa
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
2,
Danny Chan
Danny Chan
1Department of Biochemistry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
2Laboratory for Bone and Joint Diseases, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
3Department of Orthopaedic and Neuro-Musculoskeletal Surgery, Faculty of Medical and Pharmaceutical Sciences, Kumamoto University, Kumamoto, Japan.
4Department of Orthopaedics and Traumatology, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
5Department of Orthopaedic Surgery, School of Medicine, Keio University, Tokyo, Japan.
6Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
7Department of Orthopaedic Surgery, Faculty of Medicine, University of Toyama, Toyama, Japan.
8Department of Orthopaedics, Graduate School of Medical Science, Kyoto Prefectural University of Medicine, Kyoto, Japan.
9Spine Center, Hakodate Central General Hospital, Hakodate, Japan.
10The Center of Diagnosis and Treatment for Joint Disease, Drum Tower Hospital Affiliated to Medical School of Nanjing University, Nanjing, People’s Republic of China.
11Laboratory for Bone and Joint Diseases, Model Animal Research Center, Nanjing University, Nanjing, People’s Republic of China.
12Department of Orthopedics, Beijing Jishuitan Hospital, Beijing, People’s Republic of China.
13Department of Spinal Surgery, Beijing Jishuitan Hospital, The 4th Clinical Medical College of Peking University, Beijing, People’s Republic of China.
14Open University of Hong Kong, Hong Kong, People’s Republic of China.
15Department of Health Technology and Informatics, The Hong Kong Polytechnic University, Hong Kong, People’s Republic of China.
16Department of Psychiatry, The University of Hong Kong, Pokfulam, Hong Kong, People’s Republic of China.
17Institute for Medical Genetics, Charité-Universitätsmedizin Berlin, Berlin, Germany.
18Oulu Center for Cell-Matrix Research, Biocenter and Department of Medical Biochemistry and Molecular Biology of Oulu, Oulu, Finland.
19Department of Surgery, University Hospital of Oulu, Oulu, Finland.
20Finnish Institute of Occupational Health, Health and Work Ability, and Disability Prevention Centre, Oulu, Finland.
21Institute of Clinical Medicine, Department of Physical and Rehabilitation Medicine, and
22Institute of Health Sciences, Public Health and General Practice, University of Oulu, Oulu, Finland.
23Department of Epidemiology and Biostatistics, School of Public Health, Imperial College, Faculty of Medicine, London, United Kingdom.
24Laboratory for Statistical Analysis, Center for Genomic Medicine, RIKEN, Tokyo, Japan.
25Laboratory for Medical Informatics, Center for Genomic Medicine, RIKEN, Yokohama, Japan.
1