Abstract
Inadequate functional β cell mass underlies both type 1 and type 2 diabetes. β Cell growth and regeneration also decrease with age through mechanisms that are not fully understood. Age-dependent loss of enhancer of zeste homolog 2 (EZH2) prevents adult β cell replication through derepression of the gene encoding cyclin-dependent kinase inhibitor 2a (INK4a). We investigated whether replenishing EZH2 could reverse the age-dependent increase of Ink4a transcription. We generated an inducible pancreatic β cell–specific Ezh2 transgenic mouse model and showed that transgene expression of Ezh2 was sufficient to increase β cell replication and regeneration in young adult mice. In mice older than 8 months, induction of Ezh2 was unable to repress Ink4a. Older mice had an enrichment of a trithorax group (TrxG) protein complex at the Ink4a locus. Knockdown of TrxG complex components, in conjunction with expression of Ezh2, resulted in Ink4a repression and increased replication of β cells in aged mice. These results indicate that combined modulation of polycomb group proteins, such as EZH2, along with TrxG proteins to repress Ink4a can rejuvenate the replication capacity of aged β cells. This study provides potential therapeutic targets for expansion of adult β cell mass.
Introduction
Patients with type 1 or type 2 diabetes have inadequate functional β cell mass, a clinical need framing worldwide interest in the mechanisms controlling β cell growth and regeneration (1, 2). β Cell mass expands in response to increased metabolic demands associated with physiological growth, pregnancy, obesity, and insulin resistance in mice and humans (3–6), and increasing evidence suggests that this expansion is driven by replication of preexisting β cells (7–10). However, prior studies suggest that the capacity of β cells to replicate is age dependent, suggesting that restoring functional β mass in diabetic patients may require the manipulation of mechanisms that naturally limit the regenerative capacity of aging β cells (3, 11–13). Several studies have shown that increased transcription from the cyclin-dependent kinase inhibitor 2a (Ink4a) locus, which encodes the protein p16Ink4a, an inhibitor of CDK4 activity (hereafter referred to as Ink4a), curtails β cell replication during aging (14, 15). Further evidence for a role for Ink4a-mediated control of β cell expansion was provided by genome-wide association studies linking the Ink4a/Arf locus to insulin insufficiency and risk of type 2 diabetes in humans (16, 17). Thus, identifying and controlling the mechanisms that regulate transcription of Ink4a locus during aging are the focus of intensive efforts and could be useful for promoting β cell regeneration.
Prior studies from our laboratories have demonstrated that members of the polycomb group (PcG) of proteins are involved in regulating transcription from the Ink4a locus during aging (18). PcG proteins exist in distinct polycomb repressive complexes that function sequentially to repress expression of target genes. Polycomb repressive complex 1 (PRC1) contains Bmi/ubiquitin ligase-Ring1B proteins, and polycomb repressive complex 2 (PRC2) contains the histone methyltransferase called enhancer of zeste homolog 2 (EZH2; refs. 18–20). PRC2 catalyzes the trimethylation of lysine 27 in the tail of histone H3 (H3K27me3), signaling the recruitment of PRC1, which then mediates the ubiquitination of lysine K119 in histone H2 (H2AK119), resulting in the repression of the Ink4a locus (21, 22). ChIP analysis quantified association of PcG proteins with the Ink4a locus in pancreatic islet β cells and showed high levels of PcG protein enrichment in islets from young mice that were significantly higher than those in islets from aged mice. The age-dependent loss of PRC1 and PRC2 at the islet Ink4a locus was accompanied by declining levels of polycomb proteins in β cells (18, 19). Moreover, the Ink4a locus showed increased enrichment of H3K4me3 and increased binding of the trithorax group (TrxG) protein complex that contains Mll1, a histone methyltransferase that catalyzes trimethylation of H3K4 (23–26). Thus, chromatin changes at the Ink4a locus result in age-dependent increases of Ink4a expression to attenuate adult β cell replication.
Inactivation of a conditional Ezh2 allele in β cells leads to premature expression of Ink4a and severe reduction of β cell replication, demonstrating a crucial in vivo role for Ezh2 in repressing Ink4a in islets (18). Natural reductions of Ezh2 mRNA and protein levels in islets with aging could therefore account for derepression of the Ink4a locus in aged islets (19). Here, we tested whether replenishing Ezh2 in islets during aging could prevent Ink4a derepression and reverse age-dependent declines in β cell replication. If so, expression of Ezh2 in aged islets could rejuvenate the replicative capacity of β cells to promote regenerative expansion following β cell injury in adult mice.
Here, we report the generation and analysis of transgenic mice permitting conditional expression of Ezh2 in adult pancreatic β cells. Induction of Ezh2 in young adult mice was sufficient to repress Ink4a and stimulate β cell replication and regeneration. However, Ezh2 induction in β cells of aged mice failed to repress Ink4a and rejuvenate the capacity for replication. We show that this resistance to EZH2 results from enrichment of the Mll1-containing TrxG complex at the Ink4a locus. Combined knockdown of Mll1 and activation of Ezh2 was sufficient to repress Ink4a and increase β cell replication in aged mice. The principal elements of the PcG/TrxG/Ink4a pathway are conserved in human β cells, indicating that reprogramming of the Ink4a locus by modulating PcG-TrxG could be developed for use in diabetic patients.
Results
Inducible β cell–specific Ezh2 expression in vivo promotes β cell replication.
Prior studies have shown that Ezh2 mRNA and protein levels in islet β cells decline with advancing age in mice and humans (19, 27). In aging mouse islets, ChIP revealed reduced association of EZH2 and histone H3K27me3 levels at the Ink4a locus, accompanied by increased levels of H3K4me3 and Ink4a mRNA expression (19, 28). In islets from humans with advancing age, we observed similar changes (Supplemental Figure 1, A–C; supplemental material available online with this article; doi: 10.1172/JCI69468DS1). Thus, evolutionarily conserved mechanisms linked to EZH2 reduction may constrain β cell replication with age. To test the possibility that conditional Ezh2 activation might be sufficient to rejuvenate islet β cell proliferation, we constructed a transgenic mouse line permitting conditional Ezh2 reexpression in aged mouse β cells. First, we created mice harboring a bidirectional tetracycline-responsive element (TRE) that controlled expression of a transgene encoding myc-tagged Ezh2 (Myc-Ezh2TG, referred to herein as EzTG) and β-gal (Figure 1, A and B; see Methods). Intercross of EzTG mice with a mouse line expressing reverse tetracycline transactivator (rtTA) in β cells directed from rat insulin promoter elements (RIP-rtTA; refs. 18, 19) generated bitransgenic progeny (RIP-rtTA; EzTG, referred to herein as bEzTG). Exposure of bEzTG mice to doxycycline (Dox) for 1 week induced expression of EzTG mRNA and protein (Figure 1, C and J) and β-gal (Figure 1D) in pancreatic β cells. By contrast, bEzTG mice without Dox exposure (Figure 1, D and F, and Figure 2A) or single RIP-rtTA or EzTG transgenic mice exposed to Dox did not show increased EzTG expression levels or β-gal (data not shown). Thus, construction of the bEzTG mice permitted studies of conditional Ezh2 induction in islet β cells.
Figure 1. Conditional expression of EZH2 in 2-month-old bEzTG mice promotes β cell replication through repression of Ink4a.
(A) Schematics of the targeting cassette containing LacZ-TRE-EzTG. (B) Southern blotting to detect EzTG integration within the genome (lane 2–7), as compared with that in the WT control (lane 1). EzTG vectors were used as a positive control (lane 8–10). Expression level of (C) EzTG mRNA and (D) β-gal, as shown by LacZ staining in bEzTG islets treated with Dox for 1 week but not in littermate controls without Dox exposure (original magnification, ×20). (E) Dox feeding schemes: bEzTG mice were treated for 1 week with Dox food and drinking water, before the pancreata or the islets were harvested for experiments. For measuring β cell mass, 2- and/or 8-month-old (2M/8M) bEzTG mice were treated with an extended Dox procedure, with Dox treatment on week 1 and 3. Pancreatic tissue were harvest after 1 month. (F and I) Pancreatic sections from the control and Dox-treated 2-month-old bEzTG mice were immunostained with antibodies to insulin (green), (F) Ki67 (red), (I) Ink4a (red), and DAPI (blue) (original magnification, ×20). (G) Quantification of the percentages of Ki67-positive β cells with or without Dox exposure (5 pancreatic sections per animal). (H) β Cell mass (mg) measurement. (J) Western blotting and quantification of EzTG (Myc Ab) and Ink4a levels in isolated islets from indicated samples. n = 5–6 animals per group. *P < 0.05, ***P < 0.005.
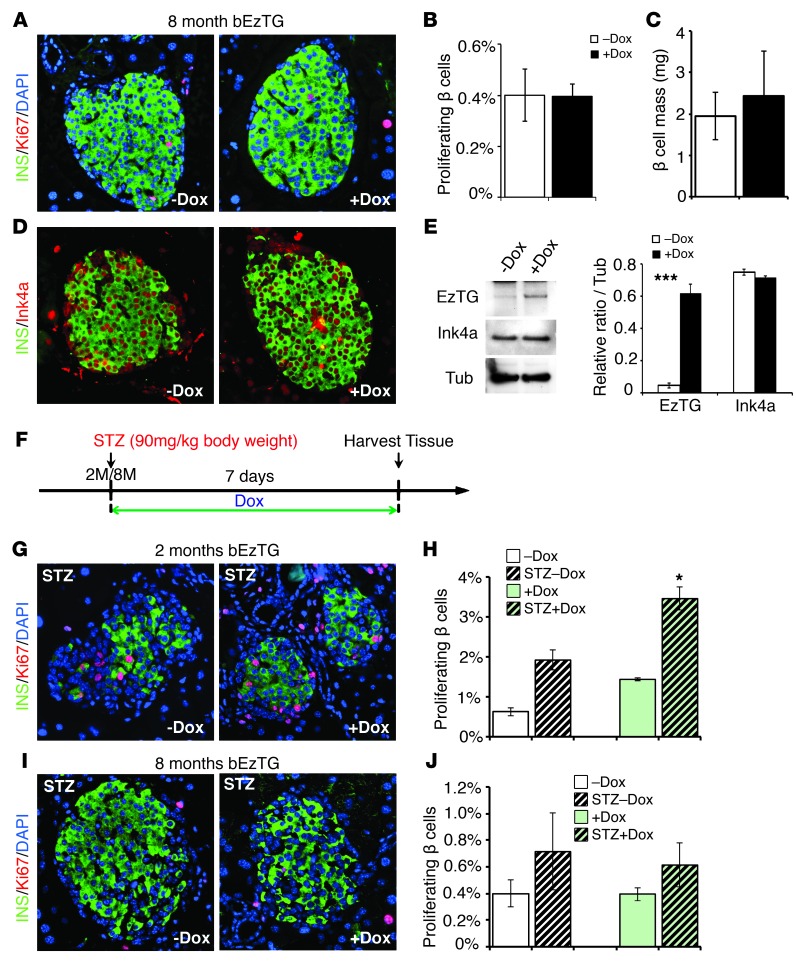
Figure 2. Activation of EZH2 expression using 8-month-old bEzTG mice did not show any changes in β cell replication or regeneration.
(A and D) Pancreatic sections from the control and Dox-treated 8-month-old bEzTG mice were immunostained with antibodies to insulin (green), (A) Ki67 (red), (D) Ink4a (red), and DAPI (blue) (original magnification, ×20). (B) Quantification of the percentages of Ki67-positive β cells with or without Dox exposure (5 pancreatic sections per animal). (C) β cell mass (mg) measurement. (E) Western blotting and quantification of EzTG (Myc Ab) and Ink4a levels in isolated islets. (F) STZ and Dox treatment scheme: Dox was administrated after single dose STZ (90 mg/kg) injection; tissues were harvested 7 days after injection. Immunostaining of insulin (green), Ki67 (red), and DAPI (blue) in pancreatic sections from bEzTG animals 1 week after STZ injection at (G) 2 months and (I) 8 month (original magnification, ×20). Percentages of Ki67-positive β cells with or without Dox exposure 1 week after mock or STZ treatment are quantified in (H) 2-month-old and (J) 8-month-old groups. n = 3–6 animals per group. *P < 0.05, ***P < 0.005.
To investigate whether Ezh2 reexpression was sufficient to stimulate adult β cell replication, we exposed 2-month-old bEzTG mice to Dox, when endogenous β cell EZH2 levels are nearly undetectable (29). In bEzTG mice exposed to Dox for 1 week (Figure 1E), we observed a 2-fold increase of the proliferation antigen Ki67 in islet insulin+ cells by immunostaining compared with that in littermate controls without Dox exposure (Figure 1, F and G). Exposure to Dox for 2 weeks (see Methods) led to a 50% increase in β cell mass, as assessed by morphometry (Figure 1H). Immunohistochemistry revealed a striking reduction of Ink4a in islet β cells from Dox-exposed bEzTG mice compared with controls (Figure 1I), a change confirmed by Western blotting (Figure 1J). Collectively, these data suggest that reexpression of Ezh2 in young adult β cells is sufficient to reduce Ink4a and promote β cell replication.
Inducible β cell–specific Ezh2 expression in vivo is insufficient to induce replication of β cells in aged mice.
Diabetes incidence increases with aging (19, 28), but the basis for this observation is unclear. Thus, we studied whether EzTG induction could lead to a similar β cell phenotype of increased replications in mice at more advanced age. Prior studies suggest that, by 8 months of age, islets lose their capacity to replicate β cells to compensate for increased metabolic demands (30). In contrast to our findings with 2-month-old bEzTG mice, though exposure of 8-month-old bEzTG mice to Dox clearly induced EzTG mRNA and increased protein levels were evident, no detectable increase in β cell Ki67 staining or β cell mass was observed (Figure 2, A–C). Furthermore, Ink4a levels were unaltered (Figure 2, D and E). Together, these data reveal that age-dependent mechanisms restrict the capacity of EZH2 to repress β cell Ink4a expression and induce replication. Increased EZH2 levels have been associated with β cell regeneration (13), suggesting that Ezh2 might play a key regulatory role in the β cell regenerative response. We considered whether EzTG induction would promote β cell regeneration in aged mice under conditions that would reflect a metabolic need to expand β cell mass. We therefore studied bEzTG mice following β cell injury from streptozotocin (STZ) injection, in which a significant mass of β cells would be destroyed. To test whether Ezh2 induction stimulated this regeneration, we injected 2-month-old bEzTG mice with STZ and then exposed them to Dox to induce EZH2 (Figure 2F). Quantification of the Ki67+ β cells 7 days after STZ challenge accompanied by Dox revealed a 2-fold increase in regeneration compared with that in STZ-challenged bEzTG controls not exposed to Dox (Figure 2, G and H). Thus, reexpression of Ezh2 in young adult mice can stimulate regeneration after β cell destruction. By contrast, a similar experimental strategy using 8-month-old bEzTG mice injected with STZ did not result in increased Ki67 labeling in β cells (Figure 2, I and J). Together, these findings reveal a distinctive age-dependent β cell response to Ezh2 reexpression and suggest that Ezh2-independent regulatory mechanisms accrue in aging to restrict the regenerative capacity of β cells.
EzTG recruitment to the Ink4a locus in islets is age dependent.
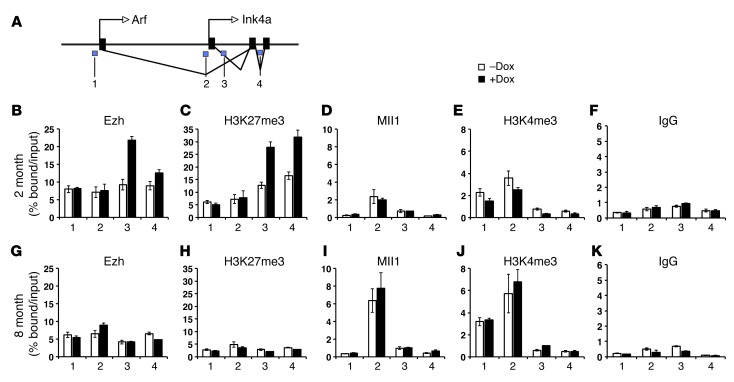
Based on resistance of islet β cell replication to Ezh2 induction in aged bEzTG mice, we postulated that EZH2 association at loci such as Ink4a might be restricted by age. To test this possibility, we performed ChIP to assess the Ink4a locus in 2-month-old and 8-month-old bEzTG mice, respectively. As expected, ChIP revealed increased EZH2 association at the Ink4a locus in islets from Dox-exposed 2-month-old bEzTG mice compared with islets from control bEzTG mice not exposed to Dox (Figure 3A; appropriate ChIP antibody and primer controls and validation are presented in Supplemental Figure 2A). Increased histone H3K27me3 levels at the Ink4a locus accompanied this EZH2 increase (Figure 3, B and C). Consistent with prior studies, we also measured increased association of the PRC2 complex protein EED, the PRC1 protein Bmi-1, and ubiquitinated H2A at the Ink4a locus in islets from Dox-exposed 2-month-old bEzTG mice (Supplemental Figure 2, B–D). By contrast, we failed to detect increased EZH2, EED, Bmi-1, or H3K27me3 at the Ink4a locus in islets from Dox-exposed 8-month-old bEzTG mice (Figure 3, G–K, and Supplemental Figure 2, E–G), despite increased Ezh2 transgene expression (Figure 2E). Thus, the Ink4a locus is resistant to EZH2 association and Ezh2-dependent chromatin changes in aged islets.
Figure 3. EZH2 transgene is recruited to Ink4a locus in 2-month-old, but not 8-month-old, bEzTG islets.
(A) Schematic representation of the Ink4a locus, with blue regions marked 1–4 indicating the amplified regions in the ChIP studies. Representative ChIP analysis for the indicated antibodies at the Ink4a locus in islets isolated from (B–F) 2-month-old and (G–K) 8-month-old groups.
We postulated that increased association of H3K4 methyltransferase Mll1 with the Ink4a locus could antagonize association of EZH2. ChIP analysis revealed relatively low levels of Mll1 and H3K4me3 association at the Ink4a locus in islets from 2-month-old mice, and treatment of bEZTG mice with Dox did not alter this association (Figure 3, D and E). Eight-month-old control and bEzTG mice had increased association of Mll1 and H3K4me3 at the Ink4a locus compared with 2-month-old mice (Figure 3, I and J). These observations support the hypothesis that increased Mll1 and/or H3K4me3 levels at the Ink4a locus locus in aged islets could impair association of EZH2 with that locus and prevent repression of Ink4a by transgenic EZH2.
Mll1 occupancy at the Ink4a locus restricts Ezh2 recruitment in aged islets.
We postulated that disrupting Mll1 association with the Ink4a locus might permit EZH2 recruitment to that locus in aged β cells. To test this possibility, we used siRNA to knockdown Mll1 levels in cultured islets from 8-month-old bEzTG mice (see Methods). To enhance assessment of β cells in cultured islets, we used nuclear Pdx1 immunostaining to quantify proliferating β cells that incorporated BrdU. Following exposure of islets to Dox, quantification of BrdU+Pdx1+ cells revealed that EZH2 induction was insufficient to increase β cell replication, similar to the observations in vivo (Figure 4, A and B). Knockdown of Mll1 alone without Ezh2 induction led to a modest reduction of Ink4a and increase in replication, but the combination of Ezh2 induction and Mll1 siRNA led to a 2.5-fold increase in BrdU incorporation (Figure 4, A and B). Consistent with these data, qRT-PCR and Western blotting studies demonstrated that Ink4a mRNA and protein levels were clearly reduced by the combination of Ezh2 induction and Mll1 siRNA in 8-month-old bEzTG islets (Figure 4, C and D). In these Dox-exposed bEzTG islets, ChIP analysis revealed that simultaneous Ezh2 induction and Mll1 knockdown led to increased EZH2 association and H3K27m3 levels at the Ink4a locus locus, accompanied by reduced Mll1 and H3K4me3 levels (Figure 4, E–H). Together, these data support our view that a complex containing Mll1 protein restricts association of EZH2 with Ink4a in islets from aged mice.
Figure 4. Knockdown of Mll1 facilitates EZH2 recruitment to the Ink4a locus and increased β cell replication.
(A) 8-month-old bEzTG islets were treated with scrambled siRNA (100 pM), Dox (1 μg/ml), Mll1 siRNA (100 pM), and both Dox and Mll1 siRNA for 48 hours. Islets sections were immunostained for Ki67 (red) and Pdx1 (green) (original magnification, ×20). (B) BrdU was added in culture 48 hours before harvest to measure proliferation. Percentage of BrdU-positive β cells in each experiment group (5 pancreatic sections per animal). (C) Real-time qRT-PCR of Ink4a, Ezh2, and Mll1 mRNA in each group. (D) Western blotting and quantification of EzTG (detected by Myc Ab), Ink4a, and β-tublin protein levels. (E–I) Representative ChIP result for the indicated antibodies at the Ink4a locus in islets treated as in A. Data represents 3 experiments. *P < 0.05.
Targeting TrxG complex by JmjD3 knockdown enhances Ezh2-induced β cell replication in aged islets.
MLL proteins interact with histone H3K27 demethylases (19), including UTX and JmjD3 (31, 32). JmjD3 associates with established PcG targets, including Ink4a, and regulates H3K27me3 levels (33). However, it remains unclear whether Mll1 associates with JmjD3 in β cells. We observed a modest increase in H3K27me3 at the Ink4a locus and reduced Ink4a mRNA levels in 8-month-old bEzTG islets with Mll1 knockdown (Figure 4, C and F), consistent with the possibility that reduction of TrxG proteins might affect H3K27 demethylases and Ink4a expression in aged islets. To identify proteins associated with Mll1 in the TrxG complex, we performed Mll1 immunoprecipitation in mouse insulinoma Min6 cells. Mll1 immunoprecipitation revealed its association with JmjD3, along with another core component of the TrxG complex, RbBP5 (Supplemental Figure 3, A and B). Since the recruitment of Mll1 at the Ink4a locus increases with age, we postulated that, as an interaction partner of Mll1, the association of JmjD3 with the Ink4a locus might follow a similar pattern. We found that the levels of JMJD3 mRNA increased with age in human and mouse islets (Figure 5A and data not shown). Consistent with this finding, ChIP analysis with JmjD3 antibody revealed increased association of JmjD3 with the Ink4a locus of human and mouse islets, as a function of age (Figure 5B and Supplemental Figure 3, C and D). Thus, increased JmjD3 demethylase association accompanies reduced H3K27me3 levels at the Ink4a locus in aging islets. To test the possibility that JMJD3 might regulate induced EZH2 activity in islets, we used siRNA targeting to knockdown JMJD3 in aged bEzTG islets. Following exposure of 8-month-old bEzTG islets to Dox (resulting in Ezh2 induction) and to an siRNA targeting JMJD3, immunostaining of Ki67 and Pdx1 demonstrated a 5-fold increase in β cell replication (Figure 5, C and D, and Supplemental Figure 3D). This effect on β cell replication is similar to that observed following simultaneous knockdown of Mll1 and Ezh2 induction in these islets. Similar to Mll1 knockdown experiments, ChIP analysis revealed that simultaneous Ezh2 induction and JMJD3 knockdown led to increased EZH2 association and H3K27m3 levels at the Ink4a locus, accompanied by reduced Mll1 and H3K4me3 levels (Figure 5, E–J). However, a combined knockdown of Mll1 and JmjD3 in islets from 2-month-old mice did not show any difference in proliferation, indicating that this mechanism is age dependent (Supplemental Figure 3, E and F). Together, our findings suggest a 2-step mechanism of Ink4a-mediated aging that includes attenuation of PcG protein levels and accumulation of TrxG complexes, including JmjD3, at the Ink4a chromatin in aged β cells.
Figure 5. Knockdown of JmjD3 facilitates EZH2 recruitment to the Ink4a locus and increased β cell replication.
(A) JMJD3 mRNA levels in juvenile (<10 years) and adult (≥10 years) human islets, as show by quantitative RT-PCR. (B) ChIP analysis showing JmjD3 binding to the Ink4a locus in human islets. (C) 8-month-old bEzTG islets were treated with scrambled siRNA (100 pM), Dox (1 μg/ml), JMJD3 siRNA (100 pM), and both Dox and JMJD3 siRNA for 48 hours. Islet sections were immunostained for Ki67 (red) and Pdx1 (green) (original magnification, ×20). (D) Percentage of Ki67-positive endocrine cells in each experiment group (5 pancreatic sections per animal). *P < 0.05. (E–J) Representative ChIP results for the indicated antibodies at the Ink4a locus in islets treated with the same experiment conditions as in E. n = 3 experiments.
Dominant function of TrxG at the Ink4a locus is a general mechanism in aging cells.
We postulated that the regulation of Ink4a by PcG and TrxG proteins might be a general mechanism mediating Ink4a-based replicative senescence. To test this, we studied mouse embryonic fibroblasts (MEFs); like islet β cells, MEFs accumulate Ink4a with passaging (34, 35). In serially passaged C57BL/6 MEFs, levels of EZH2 declined, whereas Ink4a mRNA and protein levels increased, changes accompanied by reduced proliferation, as quantified by Ki67 immunodetection (Supplemental Figure 4, A and B). Like those in pancreatic β cells, EZH2 and H3K27me3 levels at the Ink4a locus decreased in late-passage MEFs, while Mll1 and H3K4me3 levels increased, confirming prior studies (Supplemental Figure 4, C–G, and ref. 27). To test whether TrxG restricts EZH2-dependent Ink4a repression in late-passage MEFs, we transfected these MEFs with DNA vectors permitting myc-EZH2 misexpression, Mll1 siRNA-mediated knockdown, or both. Consistent with the results in pancreatic β cells, we observed that simultaneous Mll1 knockdown and Ezh2 induction increased fibroblast proliferation, accompanied by a 40% reduction of Ink4a mRNA levels. (Figure 6). Mll1 knockdown alone slightly enhanced BrdU incorporation, while Ezh2 induction alone did not detectably alter BrdU labeling (Figure 6B). Therefore, rejuvenation of the replicative capacity of β cells to promote regenerative expansion may require a 2-step approach, including dissociation of TrxG proteins and replenishment of the PcG proteins at the Ink4a chromatin. Taken together, these data support our findings and conclusions from studies on pancreatic β cells and suggest that combined activity of TrxG and PcG to regulate Ink4a transcription could be a general approach to promote cell regeneration.
Figure 6. Knockdown of Mll1 combined with EZH2 expression results in increased proliferation of later passage MEFs.
(A) Passage 13 MEFs were transfected with scrambled siRNA (100 pM), pcDNA Myc-EZH2 (2 μg), Mll1 siRNA (100 pM), and both pcDNA Myc-EZH2 and Mll1 siRNA for 48 hours. BrdU was added in culture 48 hours before immunostaining for BrdU (red) and DAPI (blue) (original magnification, ×20). (B) Percentage of BrdU-positive MEFs in each experiment group. (C) Real-time qRT-PCR of Ink4a, Ezh2, and Mll1 mRNA in each group. n = 3 experiments. *P < 0.05, **P < 0.01.
Discussion
To achieve functional β cell restoration in diabetic patients, the mechanisms regulating β cell replication have been intensively investigated. Using transgenic mice to offset the loss of endogenous EZH2, we have advanced the concept of regenerative plasticity in adult β cells. In young adulthood, activation of EZH2 expression in β cells was sufficient to repress Ink4a and increase the replication of β cells. This is consistent with previous observations that the β cell mass in young adults can expand to adapt to metabolic demands accompanying pregnancy and obesity (27). Thus, results here and in prior work suggest that, even in the absence of ongoing replication, β cells in young adults retain the capacity for replication and expansion. By contrast, aged mice are resistant to adaptive changes in β cell mass, and replenishing EZH2 alone in aged mice was not sufficient to repress the Ink4a locus. Our finding that chromatin changes at the Ink4a locus accurately reflect β cell replicative capacity validates investigations of Ink4a expression to promote β cell regeneration and, more generally, justifies studies of age-dependent chromatin changes in islet β cells.
Our findings also suggest that efforts to regenerate β cells will be advanced by identifying the signaling pathways that regulate expression and activity of PcG and TrxG factors in aging β cells. Previously, we showed that PDGF signaling regulates β cell EZH2 levels and is an important conserved mechanism in sustaining β cell replication during neonatal expansion of mouse and human islets (13). Here, we observed increased recruitment of the Mll1-JmjD3 protein complex to the Ink4a locus, coinciding with reduced levels of PDGF signaling and EZH2 decline, accompanied by increased levels of JmjD3 in the β cells. Moreover, findings with bEzTG mice here suggest that activation of Ink4a expression by the Mll1-JmjD3 complex may prevent EZH2-dependent repression of the Ink4a locus in aged β cells. Further studies are needed to determine whether PDGF signaling or other pathways regulates the age-dependent changes in Mll1 and JmjD3 that limit β cell replication.
The loss of H3K27me3-repressive marks at the Ink4a locus could not be solely attributed to the decline in the binding of EZH2, and our study highlights an active mechanism of removal of H3K27me3 marks involving JmjD3. We show that JmjD3 and Mll1 act together to modify 2 distinct lysines: Jmjd3 demethylated H3K27, while Mll1 methylated H3K4, resulting in transcriptional activation of the Ink4a locus. This mechanism of H3K27me3 removal appears to be a very tightly regulated process, highlighted by the fact that, while the levels of H3K27 methyltransferase EZH2 decline with aging, there is a concomitant increase in the levels of H3K27 demethylase, JmjD3, in β cells. The importance of JmjD3 is underscored by studies in MEFs, which show that ectopic expression of JmjD3 leads to increased expression of Ink4a and induction of replicative senescence (28). Our experiments indicate that loss of JmjD3 in β cells from aged animals leads to repression of Ink4a and induction of β cell replication, suggesting a critical role for active removal of H3K27me3 marks in the regulation of Ink4a locus.
Previous studies have demonstrated that Ink4a repression by PcG proteins is a well-conserved mechanism across cell types (35) as well as during iPS cell generation (36). Our experiments with late-passage MEFs show that reexpression of EZH2 was insufficient to repress Ink4a unless coupled with loss of the Mll1 complex, in a manner similar to pancreatic β cells. Thus, TrxG-PcG mechanisms governing replicative senescence elucidated here may be generally relevant to creating regenerative strategies for multiple types of tissues and cells.
Methods
pBi-G-Myc-Ezh2 construct.
pBI-G tet vector is used to express EZH2, which contains a bidirectional tet-responsive promoter containing TRE between 2 minimal CMV promoters that control the expression of β-gal and EZH2. The plasmid was cloned as follows. A mouse EZH2 full-length cDNA fragment was obtained by BssHII digestion and blunt-ended with T4 DNA polymerase, followed by a NotI digestion from the parental plasmid from an ATCC clone (Image clone ID: 6817366). This sequence was inserted into pcDNA3-Myc vector in frame with Myc-tag at N-terminal, with the vector treated with EcoRI digestion, blunt-ended with T4 DNA polymerase, and then digested with NotI. pcDNA3-Myc vector was generated by insertion of the Myc tag into into KpnI/BamHI sites. The myc-EZH2 sequence was then isolated from pcDNA3-MycEZH2 by KpnI digestion, blunt-ended with T4 DNA polymerase, digested with XhoI and then cloned into pBi-g Tet vector with the vector treated with PstI digestion, blunt-ended with T4 DNA polymerase, and followed with a SalI digestion.
Generating inducible bEzTG mice.
A 9-Kb DNA fragment that contains LacZ-TRE-MycEZH2 sequences was then obtained by AseI digestion of pBiG-MycEZH2 and injected into F2 donor embryos of C57BL/6 × DBA/2 and FVB/N animals to generate transgenic animals. Founder mice with the correct insert were then crossed into Rip-rtTA mice to generate inducible bEzTG mice with bEzTG double transgene.
Mouse husbandry and genotyping.
Animals were fed ad libitum and kept under a 12-hour-light/dark cycle. DNA extracted from the tails was used for PCR-based genotyping using standard methods. Primers used for genotyping bEzTG were as follows: forward 5′-CACGCTGTTTTGACCTCCATAG-3′ and reverse 5′-TGGAAAATCCAAGTCACTGGTG-3′.
Dox administration.
Dox was administrated both in drinking water (2 g/l) supplemented with sucrose (0.5%) shielded from light in red bottles and food pellets (200 mg/kg, Bio-Serv). Dox food and water were changed every 2 to 3 days. Gender-matched littermates with bEzTG genotype on normal diet and drinking water were used as controls. For proliferation index analysis, pancreata were harvested after 2 weeks of Dox treatment. For β cell mass analysis, pancreata were harvested after 1 month of feeding, with Dox treatment throughout week 1 and 3 and normal chow and water throughout week 2 and 4 (Figure 2A).
Regeneration studies.
A single dose of 90 mg/kg body weight STZ (Sigma-Aldrich) was prepared fresh in 0.1 M citrate buffer (pH 4.5) and injected intraperitoneally into animals in different age groups. STZ-treated animals were exposed to Dox or control diet/water immediately after the injection for 1 week before the pancreata were harvested for proliferation index analysis.
Southern blotting.
Genomic DNA from control and transgenic mice was digested with PvuII, transferred to nylon membrane, and incubated with Myc-EZH2 full-length probe. pcDNA-Myc-EZH2 was digested with KpnI/Xhol, and 2.6-kb length Myc-EZH2 sequence was purified from the gel and used as a template to generate the probe. Probe preparation and detection were performed using the DIG High Primer DNA Labeling and Detection Starter Kit (Roche) according to manufacture’s instructions.
LacZ staining.
LacZ staining of isolated islets was performed in islets from bEzTG and littermate control mice 1 week after Dox exposure. In brief, freshly isolated islets were fixed at 4°C in 4% paraformaldehyde, prepared in 100 mM PBS for 10 minutes, followed by 3 washes of cold PBS with 0.1% Triton X-100 and 2-hour incubation in PBS solution containing 5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM MgCl2, and 1 mg/ml X-gal at 4°C. After staining, images were collected with a dissecting microscope equipped with a Zeiss camera.
RNA isolation, RT-PCR, and real-time qPCR.
RNA from islets was isolated with the RNeasy Mini/Micro Kit (Qiagen) according to the manufacturer’s instructions. 200 ng–1 μg RNA was used for preparation of cDNA using SuperScript III Reverse Transcriptase (Invitrogen) with the oligo dT primers. The cDNA was then analyzed with regular PCR or real-time quantitative PCR (qPCR). Real-time qPCR was performed using ABI7900HT (Applied Biosystems), with initial denaturation at 95°C for 20 seconds, followed by 50 cycles of 94°C for 1 second and 60°C for 20 seconds, which was then followed by a dissociation stage. The expression levels of each transcript were normalized to the housekeeping gene cyclophilin A (PPIA). Each real-time PCR experiment shown is a representative from at least 3 independent experiments; for each experiment, islets were pooled from 3 to 4 mice per specified group.
For lists of RT-PCR and real-time qRT-PCR primers used for mRNA expression analysis, see Supplemental Tables 1 and 2.
Immunoprecipitation and Western blotting.
2.5 μg anti-Mll1 antibody (05-765, Millipore) or control IgG was used for each immunoprecipitation. Antibodies were immobilized to protein G Sepharose (Millipore) for 2 to 3 hours and washed with cold nondenaturing buffer (20 mM Tris-HCl [pH 8], 137 mM NaCl, 10% glycerol, 1% NP-0, 2 mM EDTA). The antibody-coated beads were then incubated with Min6 cell extracts prepared in nondenaturing buffer overnight at 4°C on a rotator. Beads were washed 3 times in ice-cold nondenaturing buffer and eluted in 1X SDS-PAGE gel-loading buffer. The immunoprecipitates were analyzed by Western blotting with antibodies against JmjD3 (07-1434, Millipore), RbBP5 (A300-109A, Bethyl Laboratories), and control Dnmt3a (IMG-268A, Imgenex).
For Western blotting, isolated islets were lysed in cell dissociation buffer (Invitrogen) supplemented with protease inhibitor cocktail (Calbiochem). Protein concentration was measured with Dc protein assay (Bio-Rad), and protein samples (or immunoprecipitates) (10 μg–30 μg) were loaded to each lane of the SDS-PAGE gel as previously described (18, 19, 27, 37). Target protein level was quantified against the housekeeping protein β-tublin using ImageJ. Each Western blot shown is a representative from 3 independent experiments using different islet preparations.
Immunohistochemistry.
Immunohistochemistry was performed as previously described (8). Briefly, pancreatic tissue was fixed in 4% formaldehyde and processed for paraffin embedding. 5-μm sections were deparaffinized and rehydrated and then subjected to antigen unmasking and permeabilization. The slides were then blocked with 3% BSA in TBST (0.1% Tween 20) and incubated with primary antibody overnight at 4°C. Primary antibodies used for study included guinea pig anti-insulin (1:400; Dako), mouse anti–Ki-67 (1:40; BD Biosciences), mouse anti-BrdU (1:2; RPN-202, GE Healthcare/Amersham), mouse anti-p16 (diluted 1:250; sc-1661, Santa Cruz Biotechnology Inc.). The slides were then washed and incubated with donkey- and goat-derived secondary antibodies conjugated to FITC or Cy3 (1:200; Jackson ImmunoResearch Laboratories). Slides were mounted with Vectashield with DAPI (Vector Laboratories) prior to imaging using a Leica DM6000 microscope and Openlab software (Improvision). The immunofluorescence data presented are representatives of at least 5 animals per group in each group.
For analysis of the β cell mass and proliferation index, 3–6 mice were analyzed in each group. For β cell mass, pancreatic sections were stained for insulin and scanned using a Leica DM6000 microscope. Montages of the pancreatic area by DAPI staining and β cell area by insulin staining were made with ImageJ. The cross-sectional areas of pancreata and β cells were determined using an automated program on Openlab software. Proliferation index was calculated using Ki67 (or BrdU) staining as a marker for proliferation and insulin (or Pdx1) staining for β cells.
ChIP protocol.
ChIP was performed as previously described (38). In brief, isolated or cultured islets (150–200 islets per ChIP) were treated with 1% paraformaldehyde at room temperature for 10 minutes for crosslinking. The islets were washed and suspended in ChIP lysis buffer with protease inhibitors and sonicated with Bioruptor (Diagenode). The chromatin was then precleared with protein A/G beads. Meanwhile, 10 μl protein A or G beads were incubated with 2 to 5 mg of the antibodies anti-H3K27me3 (Diagenode), anti–Ub-H2A (05-678, Millipore; the antibody is an IgM and therefore requires a bridging antibody [12-488, Millipore]), anti-H3K4me3 (Diagenode), anti–Bmi-1 (05-637, Millipore), anti-EZH2 (clone AC22, no. 3147, Cell Signaling), anti-Mll1 (05-765, Millipore), anti-JmjD3 (07-1534, Millipore), or normal mouse IgG as a control at 4°C for 2 hours on a rotator. The antibody-bound bead mix was then washed and incubated with precleared chromatin overnight at 4°C on a rotator. After immunoprecipitation, the chromatin was eluted from the beads and subjected to reverse crosslinking, and the DNA was purified. The ChIPed DNA was quantified as percentage bound/input with real-time PCR, performed using the ABI Master Mix Kit (Life Tech) and the ABI 7900 HT Fast Real-Time PCR Machine (Life Tech). Each ChIP experiment was performed 3–5 times, using independent pooled islet isolations from 4 to 8 mice. 21. The real-time PCR primers used to amplify the Ink4a locus in the ChIP DNA spanned throughout the locus and were labeled 1–4, as indicated in Figure 3A.
Mouse islet isolation and culture.
Islets were isolated using the Liberase/DNase Enzyme Mix (Roche Diagnostics) as described previously (39). In brief, the pancreas was inflated with the enzyme mix through the common bile duct and digested at 37°C. Islets were enriched by gradient and hand picked. 100–200 islets were cultured in Easy-Grip Tissue Culture Dishes (35 mm × 10 mm, Falcon) in DMEM (Cellgro) supplemented with 10% FBS. Islet cells were transfected after overnight resting in culture using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. The islets were harvested 48 hours after transfection and processed for immunostaining, protein extraction, RNA extraction, and ChIP analysis. For proliferation analyses in islets, cultured islets were incubated with BrdU (1 μg/ml) and/or Dox (2 μg/ml) after transfection 48 hours before they were harvested. The islets were then fixed in 4% paraformaldehyde, embedded in HistoGel (Thermo Scientific), and processed for paraffin-embedded sections.
Human islet studies.
Human islet samples were obtained from healthy, nondiabetic organ donors, deceased due to acute traumatic or anoxic death and obtained from the National Diseases Resource Interchange or Integrated Islet Distribution Program as previously described (8). Equilibrated human islets from different age groups, ranging from 8 months to 70 years, were processed for ChIP and real-time qRT-PCR analysis similar to the mouse islets. ChIP results in humans were normalized to GAPDH locus. For JMJD3 mRNA and ChIP results (Figure 5, C and D), samples were grouped into juvenile (<10 years) and adult (≥10 years). ChIP results for EZH2 and histones are shown for each age group. qRT-PCR results were normalized to Actb. For Ezh2 mRNA, samples were grouped into young (<20 years) and old (≥20 years).
MEF culture.
MEFs were isolated from E13.5 C57BL/6 embryos as previously described (28). In brief, embryos were harvested in cold sterile PBS, and embryo head and organs (everything inside body cavity) were carefully removed. The remainder tissue was minced and trypsinized to make single cell suspension. Trypsin was quenched by adding MEF medium (DMEM with 10% FBS). The cells at P0 were centrifuged and cultured in dishes coated with gelatin. Early-passage MEFs were passaged every 2 to 3 days using trypsin; however, the proliferation slowed down over a few passages and could be passed every 4 to 5 days or weekly toward the later passages.
Statistics.
All data are expressed as mean ± SEM. Mean and SEM values were calculated from triplicates (at least) of representative experiments. Statistical significance was determined by an unpaired, 2-tailed Student’s t test. A P value of less than 0.05 indicated statistical significance.
Study approval.
All animal experiments were approved by the Animal Research Committee of the Office for the Protection of Research Subjects at UCLA.
Supplementary Material
Acknowledgments
We are grateful to Senta Georgia, Shuen-ing Tschen, and Murtaza Kanji (UCLA) for helpful discussions; H. Chen, W. Goodyer, R. Banerjee, J. Wang, and Xueying Gu (Stanford University) for studies of young human islets and/or work on bEzTG mice; and the Integrated Islet Distribution Program for adult human islets. S. Dhawan is supported by a Juvenile Diabetes Research Foundation (JDRF) Advanced Postdoctoral Fellowship (10-2011-184) and a pilot grant from NIH/NIA (P30-AG028748). S. Kundu was supported by a JDRF postdoctoral fellowship. S.K. Kim is an Howard Hughes Medical Institute investigator. Work by the Kim group was supported by the NIDDK (UO1-089532), JDRF, the Snyder Foundation, and the Howard Hughes Medical Institute and work by the Bhushan group was supported by grants from the NIDDK (DK-080996, DK-068763, UO1-089532), JDRF, and the Helmsley Trust.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2013;123(11):4849–4858. doi:10.1172/JCI69468.
References
- 1.Butler AE, Janson J, Bonner-Weir S, Ritzel R, Rizza RA, Butler PC. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52(1):102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 2.Gepts W. Pathologic anatomy of the pancreas in juvenile diabetes mellitus. Diabetes. 1965;14(10):619–633. doi: 10.2337/diab.14.10.619. [DOI] [PubMed] [Google Scholar]
- 3.Wang RN, Bouwens L, Kloppel G. Beta-cell growth in adolescent and adult rats treated with streptozotocin during the neonatal period. Diabetologia. 1996;39(5):548–557. doi: 10.1007/BF00403301. [DOI] [PubMed] [Google Scholar]
- 4.Montanya E, Nacher V, Biarnes M, Soler J. Linear correlation between beta-cell mass and body weight throughout the lifespan in Lewis rats: role of beta-cell hyperplasia and hypertrophy. Diabetes. 2000;49(8):1341–1346. doi: 10.2337/diabetes.49.8.1341. [DOI] [PubMed] [Google Scholar]
- 5.Pick A, et al. Role of apoptosis in failure of beta-cell mass compensation for insulin resistance and beta-cell defects in the male Zucker diabetic fatty rat. Diabetes. 1998;47(3):358–364. doi: 10.2337/diabetes.47.3.358. [DOI] [PubMed] [Google Scholar]
- 6.Van Assche FA, Gepts W, Aerts L. Immunocytochemical study of the endocrine pancreas in the rat during normal pregnancy and during experimental diabetic pregnancy. Diabetologia. 1980;18(6):487–491. doi: 10.1007/BF00261705. [DOI] [PubMed] [Google Scholar]
- 7.Georgia S, Bhushan A. Beta cell replication is the primary mechanism for maintaining postnatal beta cell mass. J Clin Invest. 2004;114(7):963–968. doi: 10.1172/JCI22098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhong L, Georgia S, Tschen SI, Nakayama K, Bhushan A. Essential role of Skp2-mediated p27 degradation in growth and adaptive expansion of pancreatic beta cells. J Clin Invest. 2007;117(10):2869–2876. doi: 10.1172/JCI32198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429(6987):41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 10.Teta M, Rankin MM, Long SY, Stein GM, Kushner JA. Growth and regeneration of adult beta cells does not involve specialized progenitors. Dev Cell. 2007;12(5):817–826. doi: 10.1016/j.devcel.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 11.Teta M, Long SY, Wartschow LM, Rankin MM, Kushner JA. Very slow turnover of beta-cells in aged adult mice. Diabetes. 2005;54(9):2557–2567. doi: 10.2337/diabetes.54.9.2557. [DOI] [PubMed] [Google Scholar]
- 12.Meier JJ, et al. Beta-cell replication is the primary mechanism subserving the postnatal expansion of beta-cell mass in humans. Diabetes. 2008;57(6):1584–1594. doi: 10.2337/db07-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tschen SI, Dhawan S, Gurlo T, Bhushan A. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009;58(6):1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114(9):1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443(7110):453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 16.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127(2):265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Sharpless NE, DePinho RA. The INK4A/ARF locus and its two gene products. Curr Opin Genet Dev. 1999;9(1):22–30. doi: 10.1016/S0959-437X(99)80004-5. [DOI] [PubMed] [Google Scholar]
- 18.Dhawan S, Tschen SI, Bhushan A. Bmi-1 regulates the Ink4a/Arf locus to control pancreatic beta-cell proliferation. Genes Dev. 2009;23(8):906–911. doi: 10.1101/gad.1742609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen H, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23(8):975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao R, Tsukada Y, Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol Cell. 2005;20(6):845–854. doi: 10.1016/j.molcel.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Francis NJ, Kingston RE, Woodcock CL. Chromatin compaction by a polycomb group protein complex. Science. 2004;306(5701):1574–1577. doi: 10.1126/science.1100576. [DOI] [PubMed] [Google Scholar]
- 22.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 23.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17(15):1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee MG, et al. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318(5849):447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- 26.Mujtaba S, Manzur KL, Gurnon JR, Kang M, Van Etten JL, Zhou MM. Epigenetic transcriptional repression of cellular genes by a viral SET protein. Nat Cell Biol. 2008;10(9):1114–1122. doi: 10.1038/ncb1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21(5):525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, et al. PDGF signalling controls age-dependent proliferation in pancreatic beta-cells. Nature. 2011;478(7369):349–355. doi: 10.1038/nature10502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Heit JJ, et al. Calcineurin/NFAT signalling regulates pancreatic beta-cell growth and function. Nature. 2006;443(7109):345–349. doi: 10.1038/nature05097. [DOI] [PubMed] [Google Scholar]
- 30.Cowie CC, et al. Full accounting of diabetes and pre-diabetes in the U.S. population in 1988–1994 and 2005–2006. Diabetes Care. 2009;32(2):287–294. doi: 10.2337/dc08-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chi P, Allis CD, Wang GG. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat Rev Cancer. 2010;10(7):457–469. doi: 10.1038/nrc2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130(6):1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 33.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449(7163):731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 34.Barradas M, et al. Histone demethylase JMJD3 contributes to epigenetic control of INK4a/ARF by oncogenic RAS. Genes Dev. 2009;23(10):1177–1182. doi: 10.1101/gad.511109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Agger K, et al. The H3K27me3 demethylase JMJD3 contributes to the activation of the INK4A-ARF locus in response to oncogene- and stress-induced senescence. Genes Dev. 2009;23(10):1171–1176. doi: 10.1101/gad.510809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460(7259):1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kotake Y, Cao R, Viatour P, Sage J, Zhang Y, Xiong Y. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene. Genes Dev. 2007;21(1):49–54. doi: 10.1101/gad.1499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgia S, Soliz R, Li M, Zhang P, Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev Biol. 2006;298(1):22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 39.Dhawan S, Georgia S, Tschen SI, Fan G, Bhushan A. Pancreatic beta cell identity is maintained by DNA methylation-mediated repression of Arx. Dev Cell. 2011;20(4):419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.