Abstract
Prostate cancer has a range of clinical outcomes, from complete remission in response to treatment to death as a result of aggressive metastasis. Prognosis for individuals with prostate cancer is not readily predictable, and new diagnostics will be useful for treatment strategy determination. In this issue of the JCI, Haffner and colleagues use comprehensive tumor genome sequencing to investigate the origin of genetic mutations underlying a case of lethal prostate cancer. Surprisingly, the lethal clone in this individual arose from a tumor focus that is typically considered very low risk based on histology. Their report highlights the need to collect and curate “N of 1” cases to develop a database that can be used for clinical decision making.
The case of the lethal clone
Scientists are detectives at heart. When Haffner and colleagues learned about a case of lethal metastatic prostate cancer with evidence dating back 17 years, they had to take the case (1). Using an approach similar to one previously employed to follow pancreatic cancer progression (2), the authors began with comprehensive genome sequencing of metastatic tumor deposits recovered at autopsy. This analysis yielded evidence of mutations in several well-documented prostate cancer genes, such as tumor suppressor PTEN, tumor protein p53 (TP53), speckle-type POZ protein (SPOP), ATP-dependent helicase (ATRX), and androgen receptor (AR) (1), all of which are known to be recurrently altered in end-stage, castration-resistant prostate cancer. Because this patient’s primary tumor had been surgically excised (and saved) 17 years earlier, the authors had a unique opportunity to ask the “whodunit” question. Which, if any, of these mutations were present in the primary tumor? How did this constellation of mutations evolve over time as the patient suffered multiple relapses (with accompanying tissue biopsies) during 17 years of treatment with various interventions? Answering these questions offered the potential for new insights into prostate cancer progression, drug resistance mechanisms, and perhaps opportunities to develop molecular diagnostics.
Histological examination of the primary tumor revealed multiple regions of high-grade (Gleason 4) tumor, a small focus of lower-grade (Gleason 3) disease, and a single lymph node metastasis (1). It is well established that patients with high-volume, high-grade primary disease have an increased risk of recurrence; therefore, this patient’s subsequent clinical course of metastatic prostate cancer is not surprising. It was surprising that the lethal clone, defined by the presence of the same PTEN, TP53, and SPOP mutations recovered at autopsy, originated from the small, low-grade Gleason 3 focus, and not from the much more substantial, high-volume Gleason 4 tumors, which did not harbor PTEN, TP53, and SPOP mutations. Furthermore, the lymph node metastasis removed at the initial surgery was also negative for these mutations. Thus, this patient died from disease that, based on widely used (and validated) histologic criteria, would be considered low risk for metastasis.
Case closed?
We can certainly credit the authors for answering the “whodunit” question, but several clinical details of this case remain unexplained. For example, do the Gleason 3 (lethal) and Gleason 4 (nonlethal) foci represent independent primary tumors, or are they clonally related? This could have been addressed by comprehensive sequencing of multiple tumor foci, but the authors were unable to do so because of limitations in DNA quality (1). It is also unclear what molecular events led to the initial lymph node metastasis. Did this patient have a founder or “trunk” mutation present in all the primary tumor foci and the lymph node metastasis, with two distinct branches of tumor evolution, as recently described in kidney cancer (3)?
More importantly, are there broader lessons that we can take away from this case? For example, should we no longer trust the highly favorable prognosis associated with Gleason 3 cancer until we have evidence of a favorable molecular profile? If so, can we make a clinical decision based on the molecular profile obtained from the biopsy of a single tumor focus, knowing that an entirely different profile might be seen from the biopsy of an adjacent lesion? Of course, it is impossible to make any such conclusions from a single case. Indeed, the academic community typically frowns upon clinical anecdotes, and for good reason: attempts to generalize lack scientific validity. Therefore, it is notoriously difficult to publish anecdotes, and most go unreported.
Despite the limitations of a single case study, enthusiasm for reporting single cases appears to be on the rise, as evidenced by other “N of 1” anecdotes in which comprehensive genomic sequencing has led to surprising insights. One recent example comes from tumor genome sequencing of an extraordinary responder to an experimental cancer drug. This patient was the only person enrolled in a clinical trial to test the mTOR kinase inhibitor everolimus in metastatic bladder cancer who had a complete remission (4). Remarkably, the patient’s tumor had a mutation in TSC1, a tumor suppressor known to regulate mTOR kinase activity. Once this anecdotal evidence was obtained, the authors examined tumor tissue from other study patients. Notably, three patients whose tumors had TSC1 mutations had partial responses, whereas all those whose tumors were WT for TSC1 were unresponsive to everolimus treatment. Despite the small number of patients, this report provided enough evidence for the commercial sponsor to initiate a new clinical trial of everolimus in TSC1 mutant cancers, regardless of histology. Such biomarker-based clinical trials (called “basket studies”) are becoming increasingly common in oncology and reflect growing confidence that molecular diagnostics can predict treatment response.
Future detective work
To harness the potential effect of these anecdotes on a larger scale, the National Cancer Institute recently announced the Exceptional Cases Initiative to identify and sequence the tumors from 100 extraordinary responders to any form of cancer therapy (5). If successful, this initiative could legitimize a new field of “N of 1” science. Should we also consider an initiative of “diagnosis-to-death” autopsy cases with longitudinal genomic analysis of tumor progression, as in the example reported here (1)? Such a project would be the 21st-century version of a much older, highly successful “initiative” led by William Osler and colleagues in the late 19th century, when autopsies were routinely performed and yielded new insights into disease pathophysiology. Importantly, these autopsies were not just a series of independent case studies. Only through the collection and cross comparison of hundreds of cases did common patterns emerge, culminating in Osler’s landmark text, The Principles and Practice of Medicine.
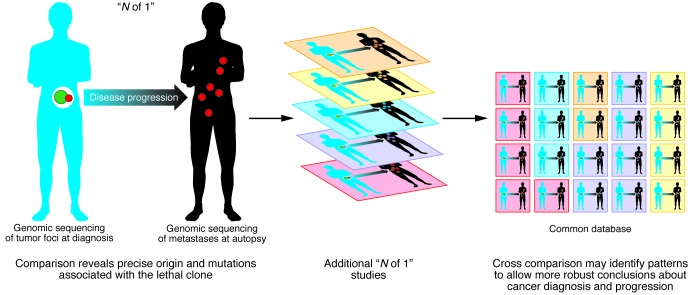
Haffner and colleagues should be commended for reporting such a fascinating anecdote (1), but the effect of their work will only be realized if we can collect and assemble this type of data across hundreds of cases (Figure 1). The growing penetration of genomic sequencing into clinical medicine has led to increased calls for the creation of various types of medical “data commons” that will allow comparisons between individual cases (6). Perhaps we are entering an era in which “N of 1” cases are here to stay.
Figure 1. The potential for “N of 1” studies to benefit clinical practice will require collection of these cases into databases.
Genomics is a powerful tool for mapping tumor progression/evolution with great precision in individuals. Collecting these “N of 1” cases into a common database will help identify common patterns and enable more robust conclusions about cancer diagnosis and progression.
Acknowledgments
Support was provided by the National Cancer Institute, NIH, under award numbers 5T32CA160001, R01-CA155169, P50-CA092629, and U01 CA141502. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of interest: Charles L. Sawyers serves on the Board of Directors of Novartis Pharmaceuticals and on the Scientific Advisory Boards of Agios, Beigene, Blueprint, Housey Pharmaceuticals, Nextech, and Tracon.
Citation for this article: J Clin Invest. 2013;123(11):4568–4570. doi:10.1172/JCI70935.
See the related Brief Report beginning on page 4918.
References
- 1.Haffner MC, et al. Tracking the clonal origin of lethal prostate cancer. J Clin Invest. 2013;123(11):4918–4922. doi: 10.1172/JCI70935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yachida S, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467(7319):1114–1117. doi: 10.1038/nature09515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iyer G, et al. Genome sequencing identifies a basis for everolimus sensitivity. Science. 2012;338(6104):221. doi: 10.1126/science.1226344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Eastman P. Online first: New FDA ‘breakthrough’ designation likely to speed cancer drug approvals.Oncology Times Web site. http://journals.lww.com/oncology-times/blog/onlinefirst/pages/post.aspx?PostID=578 . Updated November 17, 2012. Accessed September 25, 2013.
- 6. National Research Council (US) Committee on A Framework for Developing a New Taxonomy of Disease.Toward precision medicine: Building a knowledge network for biomedical research and a new taxonomy of disease. Washington DC, USA: The National Academies Press; 2011. [PubMed] [Google Scholar]