Abstract
Aim
We investigated serum DNA methylation patterns in genomic repetitive elements, LINE-1 and Alu, for post-traumatic stress disorder (PTSD) cases and controls who were US military service members recently deployed to Afghanistan or Iraq.
Methods
Cases (n = 75) had a postdeployment diagnosis of PTSD. Controls (n = 75) were randomly selected service members with no postdeployment PTSD diagnosis. Pre- and post-deployment sera were accessed, DNA was extracted and DNA methylation (percentage 5-methyl cytosine) was quantifed via pyrosequencing. Conditional and unconditional logistic regressions were used to compare: cases post- to pre-deployment; controls post- to pre-deployment; cases to controls predeployment; cases to controls postdeployment.
Results
LINE-1 was hypermethylated in controls post- versus pre-deployment (odds ratio [OR]: 1.33; 95% CI: 1.06–1.65) and hypomethylated in cases versus controls postdeployment (OR: 0.82; 95% CI: 0.67–1.01). Alu was hypermethylated for cases versus controls predeployment (OR: 1.46; 95% CI: 1.08–1.97).
Conclusion
Patterns of hypermethylation of LINE-1 in controls postdeployment and of Alu in cases postdeployment are intriguing and may suggest resilience or vulnerability factors.
Keywords: Alu, combat, DNA methylation, epigenetics, LINE-1, Operation Enduring Freedom, Operation Iraqi Freedom, post-traumatic stress disorder, repetitive element
A significant cost of the wars in Iraq and Afghanistan has been the elevated incidence of post-traumatic stress disorder (PTSD) among returning soldiers. A recent study of combat troops following return from deployment found PTSD rates to range from 12.2 to 12.9% [1]. The underlying molecular mechanisms of PTSD are largely unknown. Epigenetic factors – inherited and acquired modifications of DNA and histones that regulate various genomic functions occurring without a change in nuclear DNA sequence – could offer new insights into PTSD.
Profiling using cDNA microarrays of peripheral blood during the triggering and development of PTSD in trauma survivors present at the emergency room and 4 months later has found differential gene-expression signatures in promoter regions of genes which distinguished PTSD patients [2,3]. An epigenetic mechanism, DNA methylation, may play a significant role in the pathophysiology of PTSD, since the process is intrinsically linked to regulation of gene expression – both transcriptional silencing and activation [4]. A recent study applying methylation microarrays to assay CpG sites in peripheral blood from PTSD cases and controls found differential methylation in genes related to immune system functions. Aberrant DNA methylation patterns have also been found in other psychiatric [5,6] and neurodegenerative [7–11] disorders. Since DNA is inherently stable compared with RNA, development of biomarkers based on DNA provide an attractive potential. In addition, understanding the role of DNA methylation in PTSD has the potential to fuel novel therapeutic approaches to PTSD therapy, particularly since modifications in DNA methylation can potentially be reversed.
The typical methylation pattern of mammalian genomes consists of short (<4 kb) unmethylated domains embedded in a matrix of long methylated domains [12,13]. These long methylated domains reside primarily in interspersed and tandem repetitive elements [12,13]. Repetitive elements comprise almost 50% of the human genome [14]. In this study, we focused on two repetitive elements, the long interspersed nucleotide element 1 (LINE-1) and the interspersed Alu. Since there are approximately 0.5 million copies of LINE-1 and 1.4 million copies of Alu repetitive elements in the human genome [15,16], the methylation status of these sequences is a major contributor of global DNA methylation patterns [16]. In young, healthy mammals non-CpG island cytosine predominantly located in repetitive genomic regions, such as LINE-1 and Alu, is almost universally methylated [17,18]. Lower global DNA methylation content (i.e., hypomethylation) has been associated with widespread alterations in gene expression and chromatin packaging control, as well as with higher genomic instability [19]. Previous studies have reported differential LINE-1 and Alu repetitive element expression in response to stress in horses and in various human cell lines, respectively [20,21].
Since human studies of brain tissue are highly invasive and in many cases impractical, it would be of great value to identify a low-invasive biomarker of epigenetic patterns of PTSD. Serum and cerebrospinal fluid have been found to have good correlation with respect to cytokine expression, indicating that serum may be a good biomarker. However, most of the studies which have measured expression signatures or methylation patterns in PTSD have been carried out using whole blood [2–6], and to date no human serum biomarkers for PTSD have been reported. In addition, the role of methylation of repetitive elements in PTSD has not been investigated in humans. We, therefore, carried out a case–control study to investigate DNA methylation patterns in LINE-1 and Alu repetitive elements and their potential association with PTSD in serum DNA from US military soldiers who deployed to Afghanistan (Operation Enduring Freedom [OEF]) or Iraq (Operation Iraqi Freedom [OIF]) between 2004 and 2006. The serum was housed at the Department of Defense Serum Repository (DoDSR; MD, USA), which stores serum remaining after mandatory HIV testing of all active and reserve service members of the US military. We measured LINE-1 and Alu DNA methylation as percentage of 5-methyl cytosine (%5-mC) prior to each participant’s first deployment to OEF or OIF and after their deployment. For cases, postdeployment measurement was synonymous with post-PTSD diagnosis measurement. Given the significance of PTSD burden in the US military population and availability of serial sera for every service member (1990 to the present), the DoDSR provides a unique opportunity to evaluate epigenetic patterns of this illness.
Methods & materials
Study population
All cases and controls were selected from among active duty Army and Marine service members with at least 2 years of continuous active duty prior to their first OEF/OIF deployment. All subjects carried out their first OEF/OIF deployment between 1 January 2004 and 31 December 2006, and were deployed for 6–18 months. Dating back to at least 2 years prior to first OEF/OIF deployment, there was an absence of any mental health diagnosis, ascertained via query of the International Classification of Diseases, 9th Revision (ICD-9) codes 290–320. To attempt to control for confounding by other psychiatric illnesses, postdeployment exclusion criteria for both cases and controls was ever having an ICD-9 diagnosis (either inpatient or outpatient) for any of the following mental health diagnoses: schizophrenia (ICD-9 code 295), bipolar disorders (ICD-9 code 296) and manic-phase bipolar disorder (also ICD-9 code 296).
PTSD cases (n = 75) with existing serum samples housed at the DoDSR were identified by the Armed Forces Health Surveillance Center (AFHSC; MD, USA). The cases met all the criteria above and had at least two outpatient records with a primary diagnosis of chronic PTSD, based on ICD-9 code 309.81 in the first diagnostic position. The first outpatient diagnosis was required to be between 4 and 6 months after the service member’s return from the first deployment. The second outpatient diagnosis was required to be any time after that, but within 2 years of return from the first deployment. Additional criteria for inclusion as a case to this study was having one serum sample drawn within 12 months prior to the first OEF/OIF deployment and one sample drawn within 6 months after return from the first OEF/OIF deployment. We randomly selected from among all cases meeting these criteria, within the ages of 20 and 35 years on their first day of first OEF/OIF deployment and of black or white race.
We identified an appropriate control group (n = 75), who were randomly selected from among those active duty Army and Marine service members who met the same deployment, age, race and serum sample criteria as cases, but for whom there was never a diagnosis of PTSD (ICD-9 code: 309.81) or traumatic brain injury (TBI; ICD-9 codes: 800.0–801.9, 803.0–804.9 or 850.0–854.1). Controls were frequency matched to cases based on age group (20–26, 27–35 years), gender and race.
Sample preparation & laboratory methods
DNA extraction
For each PTSD case and control, The AFHSC identified a predeployment and a postdeployment serum sample (total samples = 300). The AFHSC permits the utilization of up to 0.5 ml of serum per sample, so genomic DNA (gDNA) was extracted from 0.5 ml serum. DNA was extracted using charge switch gDNA 0.2–1 ml serum kit from (Invitrogen Carlsbad, CA, USA) and quantified via quant-iT ds HS assay kit using a Qubit fluorometer (Invitrogen).
Quantification of DNA methylation
DNA methylation was quantified via bisulfite treatment, PCR and pyrosequencing. DNA was bisulfate treated using the Zymo DNA Methylation Kit (Zymo Research, CA, USA). Bisulfate treated DNA was eluted in 20 µl volume and 1 µl of it was used for each PCR. The PCR was performed with one of the PCR primers biotinylated to convert the PCR product to ssDNA templates. The PCR products (each 10 µl) were sequenced by Pyrosequencing PSQ96 HS System (Qiagen Pyrosequencing, Venlo, The Netherlands) following the manufacturer’s instructions (Qiagen Pyrosequencing). Pyrosequencing is a real-time sequencing method based on mutation analysis or methylation analysis technology. The methylation status of each locus was analyzed individually as a T/C SNP using QCpG software (Qiagen Pyrosequencing). The loci measured were four positions (−605, −593, −590 and −583 from translational start site of ORF1) in LINE-1 (GenBank Accession number: M80343) and four positions (nucleotide position at 114, 118, 123 and 126 based on GenBank Accession number X55933) in Alu. The percentage of methylation was expressed as 5-mC divided by the sum of methylated and unmethylated cytosine, thus yielding %5-mC. All measurements were run in duplicate and the mean of both runs was calculated for each position in LINE-1 and Alu. For both the LINE-1 and Alu assays, %5-mC was measured at four adjacent CpG sites. Four controls (low, medium, high methylated DNA [EpigenDx Inc., MA, USA] and a no DNA template) starting from bisulfite modification were included in every pyrosequencing run to ensure completion of bisulfite modification, specificity of PCR amplification, and success of pyrosequencing reactions.
For 10% of the samples, we included duplicates to which laboratory personnel were blinded for quality control. We calculated coefficients of variation (CV) for the duplicates (sample standard deviation [SD] divided by sample mean).
Statistical methods
Our statistical analyses considered the mean value of CpG methylation in the four LINE-1 and the four Alu CpG sites. This is standard practice and has been used in previous studies [22–27]. We carried out four major comparisons:
-
▪
Cases post- versus cases pre-deployment
-
▪
Controls post- versus controls pre-deployment
-
▪
Cases predeployment versus controls pre-deployment
-
▪
Cases postdeployment versus controls post-deployment
Paired t-tests were used to compare %5-mC levels for LINE-1 and Alu for case–case and control–control comparisons, while simple t-tests were used for the case–control comparisons. Likewise, conditional logistic regressions were employed to investigate the potential changes in LINE-1 and Alu methylation level (%5-mC) for cases post- versus pre-deployment and for controls post- versus pre-deployment. Unconditional logistic regressions were employed to investigate the potential differences in LINE-1 and Alu methylation level (%5-mC) between cases and controls predeployment and postdeployment. Odds ratios (ORs) and 95% CIs were calculated with %5-mC included in models as a continuous variable; for these models, the OR represents the per unit (1% methylation) change in estimate. We also carried out logistic regressions with %5-mC treated as a categorical (tertiles) variable, based on the controls’ predeployment levels. All models were adjusted for age, gender and race (black or white).
This study was approved by the Institutional Review Board at the Uniformed Services University of the Health Sciences (MD, USA).
Results
Baseline characteristics of the population studied are in Table 1. The age range of the population studied was narrow (20–35 years). This study included 100 males (66.7%) and 50 females (33.3%). Racial distribution was 80% white and 20% black. The distribution of these factors did not differ by case–control status, because of the selection and frequency matching criteria. Approximately 72% of cases and controls had deployments of 6 months to less than 12 months, while 25% had deployments of 12–18 months. The length of time between the deployment and serum draw was not standard for each study member. The number of days between end of deployment and postdeployment serum draw ranged from 1–170 (mean: 22.45; SD: 38.84; median: 7 days), and the number of days between predeployment serum draw and start of deployment ranged from 58–358 (mean: 87.46; SD: 84.49; median: 58.5 days). We did not find any difference between cases and controls, however, with respect to these time intervals. There were four cases who also had a postdeployment diagnosis of TBI (defined as an ICD-9 code of 800.0–801.9, 803.0–804.9 or 850.0–854.1; data not shown). We carried out all analyses including and excluding those four cases to ensure that none of our findings were confounded by TBI and found that results were very similar. We, therefore, present all results for analyses including those four TBI cases.
Table 1.
Baseline characteristics of population.
| Characteristic | Cases |
Controls |
p-value | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| Age | |||||
| Younger (20–23 years) | 40 | 53.3 | 39 | 52.0 | 0.87 |
| Older (24–35 years) | 35 | 46.7 | 36 | 48.0 | |
| Gender | |||||
| Male | 50 | 66.7 | 50 | 66.7 | 1.00 |
| Female | 25 | 33.3 | 25 | 33.3 | |
| Race | |||||
| White | 60 | 80.0 | 60 | 80.0 | 1.00 |
| Black | 15 | 20.0 | 15 | 20.0 | |
| Deployment length | |||||
| Short (6 to <12 months) | 55 | 73.0 | 57 | 76.0 | 0.71 |
| Long (12–18 months) | 20 | 27.0 | 18 | 24.0 | |
| Time between deployment end and postdeployment serum sample draw | |||||
| ≤7 days | 42 | 56.0 | 38 | 50.7 | 0.51 |
| >7 days | 33 | 44.0 | 7 | 49.3 | |
| Time between predeployment serum draw and start of deployment | |||||
| ≤90 days | 52 | 69.3 | 46 | 61.3 | 0.30 |
| >90 days | 23 | 30.7 | 29 | 38.7 | |
For the 10% of the samples for which we had duplicates, we calculated CVs for %5-mC measurements; the overall CV for LINE-1 was 5.46% and for Alu was 7.03%. Four samples failed the LINE-1 analysis and one sample failed the Alu analysis due to low yield of serum DNA resulting in no or low pyrosequencing signals.
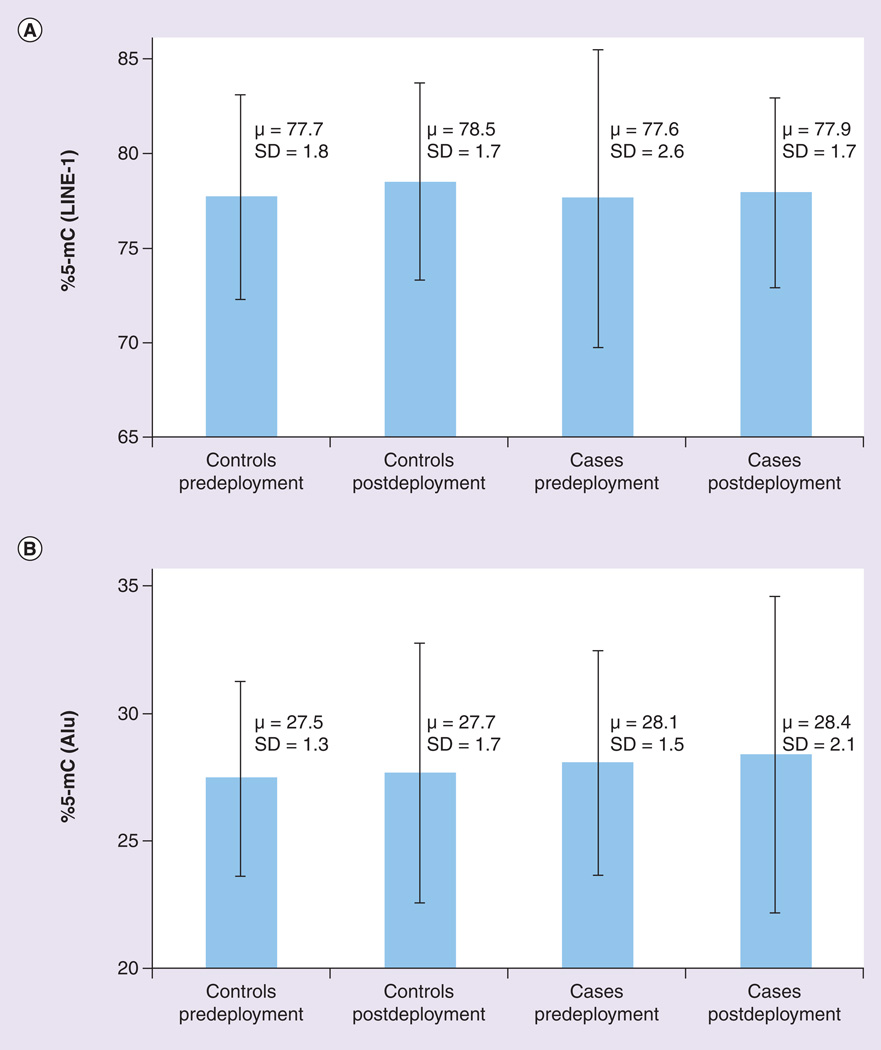
Mean levels of %5-mC for cases and controls, both pre- and post-deployment, for LINE-1 and Alu are presented in Figure 1A & B, respectively. Results from paired t-tests for the control–control and case–case comparisons as well as the results from simple t-test for the case–control comparisons are presented in the footnotes of each figure. Figure 1a shows that mean levels of LINE-1 %5-mC were higher for controls post-deployment (µ: 78.5; SD: 1.7) than for controls predeployment (µ: 77.7; SD: 1.8; p-value paired t-test = 0.01). There were no statistically significant differences between cases post- and pre-deployment, and there were no differences in LINE-1 between cases and controls, either pre-or post-deployment. Figure 1B shows that mean levels of Alu %5-mC were higher for cases (µ: 28.1; SD: 1.5) versus controls (µ: 27.5; SD: 1.3), predeployment (p-value t-test = 0.01). Cases and controls did not differ postdeployment, and there were also no significant differences between controls pre- and post-deployment or between cases pre- and post-deployment.
Figure 1. Comparison of mean levels of methylation (percentage 5-methyl-cytosine) in repetitive elements for controls predeploment, controls postdeployment, cases predeployment and cases postdeployment.
(A) LINE-1. Comparison of controls pre- and post-deployment via paired t-test: p = 0.01. Comparison of cases pre- and post-deployment via paired t-test: p = 0.15. Comparison of cases and controls predeployment via simple t-test: p = 0.74. Comparison of cases and controls postdeployment via simple t-test: p = 0.07. (B) Alu. Comparison of controls pre- and post-deployment via paired t-test: p = 0.64. Comparison of cases pre- and post-deployment via paired t-test: p = 0.20. Comparison of cases and controls predeployment via simple t-test: p = 0.01. Comparison of cases and controls postdeployment via simple t-test: p = 0.09. %5-mC: Percentage 5-methyl-cytosine; LINE-1: Long interspersed nucleotide element 1; SD: Standard deviation.
ORs and 95% CIs from conditional logistic regressions comparing cases post- versus pre-deployment and controls post- versus pre-deployment are presented in Table 2. These ORs represent the change in estimate per unit (1%) change in DNA methylation. For the case–case comparisons (i.e., cases postdeployment vs cases predeployment), we found no statistically significant associations for LINE-1, and although the ORs for Alu were slightly elevated for the total population (OR: 1.26; 95% CI: 0.90–1.78) and for various subgroups, none of the estimates was statistically significant. For the control–control comparisons (i.e., controls postdeployment vs controls predeployment), we found statistically significant elevated ORs for LINE-1 for the total population (OR: 1.33; 95% CI: 1.06–1.65), and for younger age (OR: 2.45; 95% CI: 1.31– 4.57), male (OR: 1.38; 95% CI: 1.04–1.83) and white race (OR: 1.42; 95% CI: 1.10–1.84) subgroups. When we further limited this analysis to young, white, males, we found a more pronounced statistically significant elevated OR for LINE-1 (OR: 2.05; 95% CI: 1.11–3.82; data not shown). There were also elevated ORs if time between deployment end and postdeployment serum draw was ≤7 days (OR: 1.47; 95% CI: 1.01–2.14) and if time between pre-deployment serum draw and deployment start was ≤90 days (OR: 1.71; 95% CI: 1.13–2.58). Control–control comparisons of Alu were relatively null. There were no striking differences for any of the comparisons with respect to length of deployment. Including %5-mC as a categorical variable in the models yielded similar results (data not shown).
Table 2.
Conditional logistic regressions comparing cases post- versus predeployment and controls post- versus predeployment: 5-methyl-ytosine long interspersed nucleotide element 1 and Alu levels.
| Population | Cases (post- vs pre-deployment) |
Controls (post- vs pre-deployment) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
LINE-1 |
Alu |
LINE-1 |
Alu |
|||||||||
| Npost/pre | OR† | 95% CI | Npost/pre | OR† | 95% CI | Npost/pre | OR† | 95% CI | Npost/pre | OR† | 95% CI | |
| Total population | 66/71 | 1.11 | 0.94–1.31 | 50/52 | 1.26 | 0.90–1.78 | 70/74 | 1.33 | 1.06–1.65* | 51/60 | 1.07 | 0.82–1.39 |
| Age groups | ||||||||||||
| Younger (20–23 years) | 37/38 | 1.04 | 0.79–1.37 | 28/28 | 1.35 | 0.85–2.16 | 35/39 | 2.45 | 1.31–4.57* | 25/33 | 0.93 | 0.61–1.42 |
| Older (24–35 years) | 29/33 | 1.16 | 0.92–1.46 | 22/24 | 1.12 | 0.62–2.02 | 35/35 | 1.06 | 0.82–1.36 | 26/27 | 1.17 | 0.82–1.67 |
| Gender groups | ||||||||||||
| Males | 45/48 | 1.09 | 0.83–1.43 | 36/35 | 1.31 | 0.87–1.98 | 45/49 | 1.38 | 1.04–1.83* | 32/41 | 0.96 | 0.65–1.42 |
| Females | 21/23 | 1.12 | 0.90–1.40 | 14/17 | 1.14 | 0.57–2.27 | 25/25 | 1.23 | 0.85–1.78 | 19/19 | 1.17 | 0.80–1.71 |
| Race groups | ||||||||||||
| Black race | 10/15 | 1.94 | 0.82–4.58 | 7/11 | 1.29 | 0.64–2.63 | 13/15 | 0.93 | 0.54–1.60 | 10/10 | 1.68 | 0.62–4.54 |
| White race | 56/56 | 1.06 | 0.90–1.25 | 43/41 | 1.26 | 0.85–1.85 | 57/59 | 1.42 | 1.10–1.84* | 41/50 | 1.01 | 0.77–1.34 |
| Deployment length | ||||||||||||
| Short (6 to <12 months) | 51/51 | 1.12 | 0.94–1.34 | 39/39 | 1.16 | 0.84–1.61 | 54/57 | 1.24 | 0.97–1.57 | 39/45 | 1.07 | 0.76–1.50 |
| Long (12–18 months) | 15/20 | 1.04 | 0.64–1.70 | 11/13 | 3.20 | 0.44–22.97 | 16/17 | 1.93 | 0.94–3.93 | 12/15 | 1.06 | 0.69–1.63 |
| Time between deployment end and postdeployment serum draw | ||||||||||||
| ≤7 days | 39/38 | 1.18 | 0.91–1.54 | 31/29 | 1.66 | 0.85–3.22 | 36/37 | 1.47 | 1.01–2.14 | 30/32 | 1.12 | 0.81–1.57 |
| >7 days | 27/33 | 1.06 | 0.87–1.30 | 19/23 | 1.13 | 0.80–1.60 | 34/37 | 1.25 | 0.95–1.63 | 21/28 | 0.96 | 0.61–1.52 |
| Time between predeployment serum draw and deployment start | ||||||||||||
| ≤90 days | 46/50 | 1.12 | 0.94–1.34 | 36/38 | 1.26 | 0.81–1.96 | 42/45 | 1.71 | 1.13–2.58 | 32/38 | 1.23 | 0.83–1.80 |
| >90 days | 20/21 | 1.03 | 0.65–1.65 | 14/14 | 1.27 | 0.74–2.17 | 28/29 | 1.09 | 0.82–1.46 | 19/22 | 0.92 | 0.62–1.36 |
ORs for the continuous DNA methylation variable represent the OR per unit (1%) change in methylation.
Statistically signifiant at p ≤ 0.05.
LINE-1: Long interspersed nucleotide element 1; Npost/pre: Number of post- or pre-deplyoment cases or controls; OR: Odds ratio.
ORs and 95% CIs from unconditional logistic regressions comparing cases versus controls, predeployment and cases versus controls, post-deployment are presented in Table 3. For the pre-deployment comparisons of cases and controls, LINE-1 estimates were relatively null, while Alu, predeployment comparisons of cases and controls showed a positive association for the total population (OR: 1.46; 95% CI: 1.08– 1.97) and for older age (OR: 1.62; 95% CI: 1.02–2.57), females (OR: 1.97; 95% CI: 1.02–3.79) and black race (OR: 2.93; 95% CI: 1.01–8.53). For postdeployment comparisons of cases and controls a negative association was found for LINE-1 for the total population (OR: 0.82; 95% CI: 0.67–1.01) and for younger age (OR: 0.71; 95% CI: 0.51–0.98), male (OR: 0.71; 95% CI: 0.54–0.93) and white race (OR: 0.75; 95% CI: 0.59–0.95) subgroups. When we further limited the postdeployment analysis to young, white, males, we found a more pronounced statistically significant negative association for LINE-1 (OR: 0.63; 95% CI: 0.43–0.94; data not shown). Postdeployment, the elevated ORs found between the predeployment case and control groups for Alu %5-mC persisted among the total population (OR: 1.24; 95% CI: 0.96–1.60), however this estimate and the estimates of various subgroups were not statistically significant. Including %5-mC into the models as a categorical variable yielded more pronounced associations (negative for LINE-1 and positive for Alu) in the third versus first tertile comparisons than in the second versus first tertile c omparisons (data not shown).
Table 3.
Unconditional logistic regressions† comparing cases versus controls predeployment and cases versus controls postdeployment: 5-methyl-cytosine long interspersed nucleotide element 1 and Alu.
| Population | Predeployment (cases vs controls) |
Postdeployment (cases vs controls) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
LINE-1 |
Alu |
LINE-1 |
Alu |
|||||||||
| Nca/co | OR | (95%CI) | Nca/co | OR | (95%CI) | Nca/co | OR | (95%CI) | Nca/co | OR | (95%CI) | |
| Total population | 71/74 | 0.97 | 0.84–1.13 | 52/60 | 1.46 | 1.08–1.97* | 66/70 | 0.82 | 0.67–1.01 | 50/51 | 1.24 | 0.96–1.60 |
| Age groups | ||||||||||||
| Younger (20–23 years) | 38/39 | 1.14 | 0.88–1.46 | 28/33 | 1.36 | 0.89–2.08 | 37/35 | 0.71 | 0.51–0.98* | 28/25 | 1.42 | 0.94–2.15 |
| Older (24–35 years) | 33/35 | 0.88 | 0.71–1.10 | 24/27 | 1.62 | 1.02–2.57* | 29/35 | 0.92 | 0.70–1.20 | 22/26 | 1.11 | 0.76–1.62 |
| Gender groups | ||||||||||||
| Males | 48/49 | 0.98 | 0.78–1.24 | 35/41 | 1.31 | 0.91–1.86 | 45/45 | 0.71 | 0.54–0.93* | 36/32 | 1.39 | 0.95–2.03 |
| Females | 23/25 | 0.96 | 0.79–1.18 | 17/19 | 1.97 | 1.02–3.79* | 21/25 | 1.06 | 0.74–1.50 | 14/19 | 1.09 | 0.74–1.62 |
| Race groups | ||||||||||||
| Black race | 15/15 | 0.82 | 0.51–1.31 | 11/10 | 2.93 | 1.01–8.53* | 10/13 | 1.26 | 0.75–2.14 | 7/10 | 1.41 | 0.69–2.86 |
| White race | 56/59 | 1.00 | 0.85–1.17 | 41/50 | 1.30 | 0.93–1.81 | 56/57 | 0.75 | 0.59–0.95* | 43/41 | 1.22 | 0.93–1.60 |
| Deployment length | ||||||||||||
| Short (6 to <12 months) | 51/57 | 0.92 | 0.78–1.09 | 39/45 | 1.45 | 1.02–2.07* | 51/54 | 0.85 | 0.67–1.08 | 39/39 | 1.20 | 0.90–1.60 |
| Long (12–18 months) | 20/17 | 1.83 | 0.98–3.43 | 13/15 | 1.55 | 0.81–2.94 | 15/16 | 0.75 | 0.46–1.23 | 11/12 | 1.11 | 0.62–1.98 |
| Time between deployment end and postdeployment serum sample draw | ||||||||||||
| ≤7 days | 38/37 | 0.92 | 0.66–1.29 | 29/32 | 1.51 | 0.98–2.32 | 39/36 | 0.87 | 0.67–1.13 | 31/31 | 1.30 | 0.92–1.86 |
| >7 days | 33/37 | 0.99 | 0.84–1.17 | 23/28 | 1.55 | 0.99–2.43 | 27/34 | 0.71 | 0.50–1.02 | 19/21 | 1.17 | 0.82–1.67 |
| Time between predeployment serum draw and start of deployment | ||||||||||||
| ≤90 days | 50/45 | 1.00 | 0.85–1.19 | 38/38 | 1.45 | 0.99–2.12 | 46/42 | 0.85 | 0.67–1.09 | 36/32 | 1.11 | 0.77–1.58 |
| >90 days | 21/29 | 0.94 | 0.67–1.33 | 14/22 | 1.45 | 0.85–2.46 | 20/28 | 0.76 | 0.52–1.11 | 14/19 | 1.46 | 0.91–2.36 |
ORs for total population adjusted for age, gender and race (black, white); ORs for age groups adjusted for gender and race; ORs for gender groups adjusted for age and race; ORs for race groups adjusted for age and gender; ORs for deployment length, time between deployment and postdeployment serum sample draw, and time between predeployment serum draw and start of deployment were all adjusted for age, gender and race.
Statistically signifiant at p ≤ 0.05.
LINE-1: Long interspersed nucleotide element 1; Nca/co: Number of cases/controls; OR: Odds ratio.
Discussion
We carried out four major comparisons in this study:
-
▪
Cases post- versus cases pre-deployment;
-
▪
Controls post- versus controls pre-deployment;
-
▪
Cases versus controls, predeployment;
-
▪
Cases versus controls, postdeployment.
As predeployment cases had not had any previous diagnosis of PTSD and were equivalent to healthy controls in that respect, we predicted that comparisons one and two would give us insight into deployment related stresses potentially associated with DNA methylation, while comparison two would additionally provide insight into any potential associations of PTSD and DNA methylation in repetitive elements, as would comparison four. Comparison three would enable us to determine if there were any fundamental underlying differences with respect to DNA methylation between cases and controls prior to deployment, which may indicate vulnerability to stress.
We found that controls postdeployment had higher LINE-1 %5m–C than they did predeployment. Interestingly, LINE-1 levels did not signifiantly change after deployment/diagnosis for cases. Although we found that there was no difference for LINE-1 between cases and controls predeployment, postdeployment LINE-1 was hypomethylated in cases versus controls. However, these statistically signifiant reduced ORs for cases versus controls, postdeployment, cannot be evaluated in isolation of the fact that controls’ postdeployment LINE-1 methylation increased (from predeployment levels), and this may be largely what drove the negative association between LINE-1 and PTSD. It is unclear why controls’ levels increased postdeployment. A possible explanation is that psychological stress incurred during deployment may be associated with a response in LINE-1, and people with that response may be protected against PTSD. For the Alu repetitive element, the case–case comparisons revealed nonstatistically significant elevated ORs with increasing Alu %5-mC postdeployment/diagnosis. In addition, we found that the people who later became cases (i.e., predeployment cases), when compared with the controls, predeployment, had elevated ORs with increasing Alu %5-mC. Postdeployment ORs were also elevated for Alu, although not statistically signifiant. These results are suggestive of a potential protective effect from lower levels of Alu and a potential for vulnerability to stress with higher levels of Alu, even before exposure to a potentially traumatic event.
DNA hypomethylation in repetitive elements is generally associated with chromosomal instability and, for various cancers, the expression of genes which would normally be methylated and therefore silenced (i.e., oncogenes) [28,29]. The role of global methylation has also been implicated in memory formation [8] and plasticity [30]. We found hypomethylation of LINE-1 for cases versus controls, postdeployment, although this fiding may have been driven by increases in controls’ LINE-1 methylation after deployment. In this study DNA methylation in the Alu repetitive element was hypermethylated for cases postdeployment compared with predeployment and for cases compared with controls predeployment. A recent epidemiologic study found that night-shift workers had elevated Alu %5-mC in their blood compared with day workers, suggesting that Alu hypermethylation may represent a response to psychological stress [31]. Indeed Alu has been reported to have a physiological role during responses to stress, [32] and it is hypothesized that PTSD develops as a result of an inability to control a normal stress response [4,33]. Although both LINE-1 and Alu have been implicated in the regulation of cell stress responses of the immune system, the strongest data on regulation of these repetitive elements by cell stress are for Alu [21]. It is important to acknowledge that the study of DNA methylation in association with trauma exposure and PTSD is in its very early stages, and it is still not clear how to interpret the direction of measures of DNA methylation in repetitive elements. It could be the case that hypomethylation or hypermethylation of repetitive elements is pathogenomic.
Although a handful of recent studies have evaluated gene-specific DNA methylation and PTSD [4,34–36], to our knowledge, this is the first study to investigate the association between DNA methylation in repetitive elements (LINE-1 and Alu) and PTSD in humans. A cross-sectional study of PTSD-affected and -unaffected individuals enrolled in a longitudinal study applied methylation microarrays to investigate methylation and immune function profiles in DNA derived from whole blood [4]. The investigators reported that immune system functions were significantly over-represented among the genes uniquely unmethylated in those with PTSD [4]. Another recent study in humans found that individuals with more traumatic events were at increased risk for PTSD, but only at lower methylation levels of a serotonin transporter gene, SLC6A4. At higher methylation levels, individuals with more traumatic events were protected from PTSD [37].
Serum has not previously been evaluated as a biomarker for DNA methylation patterns associated with PTSD. The few human studies which have investigated DNA methylation in PTSD have utilized predominantly whole blood-derived DNA [4,34–37]. It is not clear how well serum or whole blood DNA methylation levels correlate with levels in brain or other CNS tissues. Although relevant to a different class of disease, many of the aberrations that have been detected in the DNA of primary tumor tissue can also be detected in DNA present in serum [38–49], and cell-free DNA in the circulation has increasingly been recognized as a valuable diagnostic tool in various diseases [50–53]. Compared with cultured cells, clinical specimens, such as whole blood, serum, and even brain tissue and other CNS tissues, contain a heterogeneous mixture of cell types, each contributing its own unique methylation profile to the final analysis. We are, therefore, not able to assess serum cell-specific differences in methlyation status.
A limitation of this study is that we do not have detailed data on deployment exposures for either the cases or controls. Deployment was used as a proxy for the potentially traumatic event, and the exact timing of the potentially traumatic event is not known. Likewise, the timing of sample collection was not standardized, so there was heterogeneity in the length of time between deployment and serum draw. Although we tried to minimize this time interval, the design of the study was such that we had to rely on when a service member had serum drawn for HIV testing. We tried to control for differences in deployment experience by ensuring that all cases and controls had not been previously deployed, that they were all active duty Army or Marines, and that they were deployed from between 6 and 18 months, but there is still potential for significant variation among all our subjects with respect to intensity of combat during deployment. We did, however, evaluate a few variables which could be indicators of increased probability for exposure to more intense combat, such as lower rank and occupation in a combat-related job (i.e., infantry, lineman, tracked vehicle operator and others), assessed via military occupational codes. We found that service member rank and occupation were similar between cases and controls, indicating that controls were potentially as likely as cases to have been exposed to combat trauma. This provides additional confidence that the control group was actually also exposed to combat but did not develop PTSD.
We also have no data on other relevant exposures which are known to affect DNA methylation, such as dietary factors (folate and vitamin B12 intake) [54,55], smoking [56] and alcohol consumption [27,56,57]. Ascertainment of PTSD via query of medical encounter coded via the ICD-9 is not ideal. Although we attempted to restrict the definition of PTSD to a scenario which would minimize misclas-sification of disease, by requiring the ICD-9 code 309.81 be present in the first diagnostic position for two outpatient records spaced at a reasonable calendar time distance, this type of case ascertainment is still prone to misclas-sification. However, this is the PTSD case definition developed in September 2008 by the Department of Defense Interagency PTSD and TBI Standardization Committee, and it has been accepted by Military Health Affairs for surveillance [101]. The DNA yields from the sera in this study were small, another potential limitation. Four samples failed the LINE-1 analysis and one sample failed the Alu analysis due to low yield of serum DNA resulting in no or low pyrosequencing signals.
A major strength of this study is that we had predeployment and prediagnosis samples. While most case–control studies would not be able to infer whether the observed methylation patterns were a consequence of PTSD or whether they indicated vulnerabilities that existed among the cases before the onset of PTSD, our study was able to address both possibilities.
PTSD is unique among psychiatric disorders since there is an explicit requirement for a well-defined environmental event, the potentially traumatic event. This suggests that adaptable molecular processes such as DNA methylation are highly relevant. Indeed, there is growing evidence that the molecular mechanisms that regulate DNA methylation are involved in synaptic plasticity, learning and memory [9]. Understanding the role of repetitive elements in PTSD is imperative, since they comprise approximately 50% of the human genome [58]. The results of the present study should be considered as preliminary and future studies are needed to confirm these findings. The Department of Defense Serum Repository, which contains longitudinally collected sera of US military service members will provide a vast resource for carrying out future investigations in larger populations.
Conclusion
Postdeployment, LINE-1 was hypomethylated in cases versus controls; however, this is mainly a function of controls’ levels increasing significantly during postdeployment. Patterns of hypermethylation of LINE-1 in controls postdeployment and of Alu in cases post deployment are intriguing and may be suggestive of resilience or vulnerability factors. These findings are preliminary and should be investigated in larger studies.
Future perspective
We anticipate an increased interest in investigation of the epigenetic patterns of mental illness, in particular PTSD. The research in this area has lagged behind that of cancer and other diseases, but a few emerging studies in human populations have underscored the importance of evaluating epigenetics, in particular DNA methylation patterns, in PTSD. There are already studies which have discovered differential methylation of genes involved in immune system responses, and methylation of repetitive elements, in particular, is of relevance because of the high composition of repetitive elements in the human genome.
Executive summary.
Background
-
▪
The underlying molecular mechanisms of post-traumatic stress disorder (PTSD) are largely unknown; profiling using cDNA microarrays of peripheral blood found differential gene-expression signatures in promoter regions of genes which distinguished PTSD patients.
-
▪
Understanding the role of DNA methylation in PTSD has the potential to fuel novel therapeutic approaches to PTSD therapy, particularly since modifications in DNA methylation can potentially be reversed.
-
▪
We focused on two repetitive elements, the long interspersed nucleotide element 1 (LINE-1) and the interspersed Alu; decreases in DNA methylation in repetitive elements (i.e., hypomethylation) have been associated with genomic instability.
-
▪
We carried out a case–control study to investigate DNA methylation patterns in LINE-1 and Alu and their association with PTSD in serum DNA from US military soldiers who deployed to Iraq or Afghanistan between 2004 and 2006; serum was housed at the Department of Defense Serum Repository.
Study population
-
▪
PTSD cases (n = 75) with existing serum samples housed at the Department of Defense Serum Repository were identified by the Armed Forces Health Surveillance Center; cases had at least two outpatient records with a primary diagnosis of chronic PTSD, based International Classification of Diseases, 9th Revision code 309.81. Controls (n = 75) met the same deployment, age, race and serum sample criteria as cases, but had no diagnosis of PTSD (International Classification of Diseases, 9th Revision code: 309.81).
Study population & sample preparation & laboratory methods
-
▪
For each PTSD case and control, The Armed Forces Health Surveillance Center identified a predeployment and a postdeployment serum sample (total samples = 300); DNA was extracted from 0.5 ml serum and DNA methylation was quantified via bisulfite treatment, PCR and pyrosequencing; methylation was measured as percentage 5-methyl-cytosine.
Statistical methods
-
▪Using t-tests and logistical regressions, we carried out four major comparisons:
-
–Cases post- versus cases pre-deployment
-
–Controls post- versus controls pre-deployment
-
–Cases predeployment versus controls predeployment
-
–Cases postdeployment versus controls postdeployment
-
–
-
▪
Odds ratios (ORs) and 95% CIs were calculated.
Results
-
▪
LINE-1 was hypermethylated in controls post- versus pre-deployment (OR: 1.33; 95% CI: 1.06–1.65); cases showed no change postdeployment; LINE-1 was hypomethylated in cases versus controls postdeployment (OR: 0.82; 95% CI: 0.67–1.01).
-
▪
Alu was hypermethylated for cases versus controls predeployment (OR: 1.46; 95% CI: 1.08–1.97).
Discussion
-
▪
Patterns of hypermethylation of LINE-1 in controls postdeployment and of Alu in cases postdeployment are intriguing and may be suggestive of resilience or vulnerability factors. These findings are preliminary and should be investigated in larger studies.
Acknowledgments
This study was funded by a Congressionally Directed Medical Research Program Grant, W81XWH-08-2-0053. A Baccarelli received support from the Search Results National Institute of Environmental Health Sciences – Harvard School of Public Health Center for Environmental Health New Investigator Fund (P30ES000002).
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Hoge C, Castro C, Messer SC, McGurk D, Cotting D, Koffman R. Combat duty in Iraq and Afghanistan, mental health problems, and barriers to care. N. Engl. J. Med. 2004;351:13–22. doi: 10.1056/NEJMoa040603. [DOI] [PubMed] [Google Scholar]
- 2.Segman R, Shefi N, Goltser-Dubner T, Friedman N, Kaminski N, Shalev A. Peripheral blood mononuclear cell gene expression profiles identify emergent post-traumatic stress disorder among trauma survivors. Mol. Psychiatry. 2005;10:500–513. doi: 10.1038/sj.mp.4001636. [DOI] [PubMed] [Google Scholar]
- 3. Zieker J, Zieker D, Jatzko A, et al. Differential gene epxression in peripheral blood of patients suffering from post-traumatic stress disorder, letter to the editor. Mol. Psychiatry. 2007;12:116–119. doi: 10.1038/sj.mp.4001905.▪ This study found that using cDNA microarrays, there were clear changes on the mRNA level in post-traumatic stress disorder (PTSD) patients even 16 years after the traumatic event. Based on studies like this one, we hypothesized that aberrant DNA methylation, a process intrinsically linked to regulation of gene expression - both transcriptional silencing and activation - may be present in PTSD.
- 4. Uddin M, Aiello A, Wildman D, et al. Epigenetic and immune function profiles associated with posttraumatic stress disorder. Proc. Natl Acad. Sci. 2010;107(20):9470–9475. doi: 10.1073/pnas.0910794107.▪▪ One of the first studies to measure DNA methylation related to PTSD, this study used blood samples to identify specific epigenetic profiles underlying immune system changes associated with PTSD. Although it did not focus on repetitive elements, it demonstrated that genes whose methylation levels are significantly and negatively correlated with traumatic burden show a similar strong signal of immune function among the PTSD affected.
- 5.Kuratomi G, Iwamoto K, Bundo M, et al. Aberrant DNA methyltaion associated with bipolar disorder identified from discordant monozygotic twins. Mol. Psychiatry. 2007;13(4):429–441. doi: 10.1038/sj.mp.4002001. [DOI] [PubMed] [Google Scholar]
- 6.Tamura Y, Kunugi H, Ohashi J, Hohjoh H. Epigenetic aberration of the human REELIN gene in psychiatric disorders. Mol. Psychiatry. 2007;12:593–600. doi: 10.1038/sj.mp.4002014. [DOI] [PubMed] [Google Scholar]
- 7. Bollati V, Galimberti D, Pergoli L, et al. DNA methylation in repetitive elements and Alzheimer disease. Brain Behav. Immun. 2011;25(6):1078–1083. doi: 10.1016/j.bbi.2011.01.017.▪▪ The results of this study found that long interspersed nucleotide element 1 (LINE-1) was increased in Alzheimer’s disease patients compared with healthy volunteers. This suggests that LINE-1 methylation may lead to a better understanding of Alzheimer’s disease pathogenesis and course, and may contribute to identify novel markers useful to assess risk stratification
- 8.Levenson J, Sweatt J. Epigenetic mechanisms: a common theme in vertebrate and invertebrate memory formation. Cell Mol. Life Sci. 2006;63(9):1009–1016. doi: 10.1007/s00018-006-6026-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sananbenesi F, Fischer A. The epigenetic bottleneck of neurodegenerative and psychiatric diseases. Biol. Chem. 2009;390(11):1145–1153. doi: 10.1515/BC.2009.131. [DOI] [PubMed] [Google Scholar]
- 10.Urdinguio RG, Sanchez-Mut JV, Esteller M. Epigenetic mechanisms in neurological diseases: genes, syndromes, and therapies. Lancet Neurol. 2009;8(11):1056–1072. doi: 10.1016/S1474-4422(09)70262-5. [DOI] [PubMed] [Google Scholar]
- 11.Zawia NH, Lahiri DK, Cardozo-Pelaez F. Epigenetics, oxidative stress, and Alzheimer disease. Free Radic. Biol. Med. 2009;46(9):1241–1249. doi: 10.1016/j.freeradbiomed.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rollins RA, Haghighi F, Edwards JR, et al. Large-scale structure of genomic methylation patterns. Genome Res. 2006;16(2):157–163. doi: 10.1101/gr.4362006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9(6):465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 14.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21(35):5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 15.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32(3):e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson KD. DNA methylation and human disease. Natre Rev. Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 18.Moore LE, Huang WY, Chung J, Hayes RB. Epidemiologic considerations to assess altered DNA methylation from environmental exposures in cancer. Ann. NY Acad. Sci. 2003;983:181–196. doi: 10.1111/j.1749-6632.2003.tb05973.x. [DOI] [PubMed] [Google Scholar]
- 19.Dean W, Lucifero D, Santos F. DNA methylation in mammalian development and disease. Birth Defects Res. C Embryo Today. 2005;75(2):98–111. doi: 10.1002/bdrc.20037. [DOI] [PubMed] [Google Scholar]
- 20.Capomaccio S, Verini-Supplizi A, Galla G, et al. Transcription of LINE-derived sequences in exercise-induced stress in horses. Anim. Genet. 2010;41(Suppl. 2):23–27. doi: 10.1111/j.1365-2052.2010.02094.x. [DOI] [PubMed] [Google Scholar]
- 21.Li TH, Schmid CW. Differential stress induction of individual Alu loci: implications for transcription and retrotransposition. Gene. 2001;276(1–2):135–141. doi: 10.1016/s0378-1119(01)00637-0. [DOI] [PubMed] [Google Scholar]
- 22.Baccarelli A, Wright R, Bollati V, et al. Ischemic heart disease and stroke in relation to blood DNA methylation. Epidemiology. 2010;21(6):819–828. doi: 10.1097/EDE.0b013e3181f20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cash HL, Tao L, Yuan JM, et al. LINE-1 hypomethylation is associated with bladder cancer risk among nonsmoking Chinese. Int. J. Cancer. 2011;28(10):26098. doi: 10.1002/ijc.26098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabris S, Bollati V, Agnelli L, et al. Biological and clinical relevance of quantitative global methylation of repetitive DNA sequences in chronic lymphocytic leukemia. Epigenetics. 2011;6(2):188–194. doi: 10.4161/epi.6.2.13528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kile ML, Baccarelli A, Tarantini L, Hoffman E, Wright RO, Christiani DC. Correlation of global and gene-specific DNA methylation in maternal-infant pairs. PLoS ONE. 2010;5(10):e13730. doi: 10.1371/journal.pone.0013730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Madrigano J, Baccarelli A, Mittleman MA, et al. Prolonged exposure to particulate pollution, genes associated with glutathione pathways, and DNA methylation in a cohort of older men. Environ. Health Perspect. 2011;8:8. doi: 10.1289/ehp.1002773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu ZZ, Hou L, Bollati V, et al. Predictors of global methylation levels in blood DNA of healthy subjects: a combined analysis. Int. J. Epidemiol. 2010 doi: 10.1093/ije/dyq154. Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dunn BK. Hypomethylation: one side of a larger picture. Ann. NY Acad. Sci. 2003;983:28–42. doi: 10.1111/j.1749-6632.2003.tb05960.x. [DOI] [PubMed] [Google Scholar]
- 29.Gaudet F, Hodgson G, Eden A, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 30.Bredy T. Behavioural epigenetics and psychiatric disorders. Med. Hypotheses. 2007;68(2):453. doi: 10.1016/j.mehy.2006.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Bollati V, Baccarelli A, Sartori S, et al. Epigenetic effects of shiftwork on blood DNA methylation. Chronobiol. Int. 2010;27(5):1093–1104. doi: 10.3109/07420528.2010.490065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz W, Steinhoff C, Florl A. Methylation of endogenous human retroelements in health and disease. Curr. Top. Microbiol. Immunol. 2006;310:211–250. doi: 10.1007/3-540-31181-5_11. [DOI] [PubMed] [Google Scholar]
- 33.Yehuda R, Ledoux J. Response variation following trauma: a translational neuroscience approach to understanding PTSD. Neuron. 2007;56(1):19–32. doi: 10.1016/j.neuron.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Ressler KJ, Mercer KB, Bradley B, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470(7335):492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trollope AF, Gutierrez-Mecinas M, Mifsud KR, Collins A, Saunderson EA, Reul JM. Stress, epigenetic control of gene expression and memory formation. Exp. Neurol. 2011 doi: 10.1016/j.expneurol.2011.03.022. Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 36. Uddin M, Galea S, Chang SC, et al. Gene expression and methylation signatures of MAN2C1 are associated with PTSD. Dis. Markers. 2011;30(2–3):111–121. doi: 10.3233/DMA-2011-0750. ▪The authors investigated PTSD-associated methylation differences in 33 genes previously shown to differ in whole blood-derived gene expression levels between those with versus without the disorder. The results indicate that MAN2C1 methylation levels modify cumulative traumatic burden on risk of PTSD, and suggest that both gene expression and epigenetic changes at specific loci are associated with this disorder
- 37.Koenen KC, Uddin M, Chang SC, et al. SLC6A4 methylation modifies the effect of the number of traumatic events on risk for post traumatic stress disorder. Depress. Anxiety. 2011;28(8):639–647. doi: 10.1002/da.20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson PJ. A framework for the molecular classification of circulating tumor markers. Ann. NY Acad. Sci. 2001;945:8–21. doi: 10.1111/j.1749-6632.2001.tb03859.x. [DOI] [PubMed] [Google Scholar]
- 39.Abbaszadegan MR, Moaven O, Sima HR, et al. p16 promoter hypermethylation: a useful serum marker for early detection of gastric cancer. World J. Gastroenterol. 2008;14(13):2055–2060. doi: 10.3748/wjg.14.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.An Q, Liu Y, Gao Y, et al. Detection of p16 hypermethylation in circulating plasma DNA of non-small cell lung cancer patients. Cancer Lett. 2002;188(1–2):109–114. doi: 10.1016/s0304-3835(02)00496-2. [DOI] [PubMed] [Google Scholar]
- 41.Bastian PJ, Palapattu GS, Lin X, et al. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin. Cancer Res. 2005;11(11):4037–4043. doi: 10.1158/1078-0432.CCR-04-2446. [DOI] [PubMed] [Google Scholar]
- 42.Hsu HS, Chen TP, Hung CH, et al. Characterization of a multiple epigenetic marker panel for lung cancer detection and risk assessment in plasma. Cancer. 2007;110(9):2019–2026. doi: 10.1002/cncr.23001. [DOI] [PubMed] [Google Scholar]
- 43.Lavon I, Refael M, Zelikovitch B, Shalom E, Siegal T. Serum DNA can define tumor-specific genetic and epigenetic markers in gliomas of various grades. Neuro. Oncol. 2010;12(2):173–180. doi: 10.1093/neuonc/nop041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Leung WK, To KF, Chu ES, et al. Potential diagnostic and prognostic values of detecting promoter hypermethylation in the serum of patients with gastric cancer. Br. J. Cancer. 2005;92(12):2190–2194. doi: 10.1038/sj.bjc.6602636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reibenwein J, Pils D, Horak P, et al. Promoter hypermethylation of GSTP1 AR, and 14-13-3sigma in serum of prostate cancer patients and its clinical relevance. Prostate. 2007;67(4):427–432. doi: 10.1002/pros.20533. [DOI] [PubMed] [Google Scholar]
- 46.Tan SH, Ida H, Lau QC, et al. Detection of promoter hypermethylation in serum samples of cancer patients by methylation-specific polymerase chain reaction for tumour suppressor genes including RUNX3. Oncol. Rep. 2007;18(5):1225–1230. [PubMed] [Google Scholar]
- 47.Ulivi P, Zoli W, Calistri D, et al. p16INK4A and CDH13 hypermethylation in tumor and serum of non-small cell lung cancer patients. J. Cell. Physiol. 2006;206(3):611–615. doi: 10.1002/jcp.20503. [DOI] [PubMed] [Google Scholar]
- 48.Wakabayashi T, Natsume A, Hatano H, et al. p16 promoter methylation in the serum as a basis for the molecular diagnosis of gliomas. Neurosurgery. 2009;64(3):455–461. doi: 10.1227/01.NEU.0000340683.19920.E3. discussion 461–452. [DOI] [PubMed] [Google Scholar]
- 49.Wang YC, Yu ZH, Liu C, et al. Detection of RASSF1A promoter hypermethylation in serum from gastric and colorectal adenocarcinoma patients. World J. Gastroenterol. 2008;14(19):3074–3080. doi: 10.3748/wjg.14.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Atamaniuk J, Hsiao YY, Mustak M, et al. Analysing cell-free plasma DNA and SLE disease activity. Eur. J. Clin. Invest. 2011;41(6):579–583. doi: 10.1111/j.1365-2362.2010.02435.x. [DOI] [PubMed] [Google Scholar]
- 51.Koffler D, Agnello V, Winchester R, Kunkel HG. The occurrence of single-stranded DNA in the serum of patients with systemic lupus erythematosus and other diseases. J. Clin. Invest. 1973;52(1):198–204. doi: 10.1172/JCI107165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Leon SA, Ehrlich GE, Shapiro B, Labbate VA. Free DNA in the serum of rheumatoid arthritis patients. J. Rheumatol. 1977;4(2):139–143. [PubMed] [Google Scholar]
- 53.Shapiro B, Chakrabarty M, Cohn EM, Leon SA. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer. 1983;51(11):2116–2120. doi: 10.1002/1097-0142(19830601)51:11<2116::aid-cncr2820511127>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 54.Fenech M. The role of folic acid and vitamin B12 in genomic stability of human cells. Mutat. Res. 2001;475:57–67. doi: 10.1016/s0027-5107(01)00079-3. [DOI] [PubMed] [Google Scholar]
- 55.Piyathilake CJ, Johanning GL. Cellular vitamins, DNA methylation and cancer risk. J. Nutr. 2002;132(Suppl. 8):2340S–2344S. doi: 10.1093/jn/132.8.2340S. [DOI] [PubMed] [Google Scholar]
- 56.Toh Y, Oki E, Ohgaki K, et al. Alcohol drinking, cigarette smoking, and the development of squamous cell carcinoma of the esophagus: molecular mechanisms of carcinogenesis. Int. J. Clin. Oncol. 2010;15(2):135–144. doi: 10.1007/s10147-010-0057-6. [DOI] [PubMed] [Google Scholar]
- 57.Seitz HK, Stickel F. Molecular mechanisms of alcohol-mediated carcinogenesis. Nat. Rev. Cancer. 2007;7(8):599–612. doi: 10.1038/nrc2191. [DOI] [PubMed] [Google Scholar]
- 58.Schulz W. L1 retrotransposons in human cancers. J. Biomed. Biotechnol. 2006;2006(83672):1–12. doi: 10.1155/JBB/2006/83672. [DOI] [PMC free article] [PubMed] [Google Scholar]
Website
- 101.Armed Forces Health Surveillance Center. 2011 www.afhsc.mil/viewDocument?file=CaseDefs/Web_12_MENTAL%20HEALTH_MAR11.pdf.