The virally encoded Rev protein of human immunodeficiency virus type-1 (HIV-1) plays a critical role in viral replication by regulating the transport of unspliced and partially spliced viral RNA from the nucleus to the cytoplasm of infected cells.[1, 2] Rev binds first to a specific region of the HIV-1 mRNA, called the Rev responsive element (RRE) of stem loop IIb (Figure 1); subsequent to this initial binding event, approximately 10 additional Rev molecules oligomerize through protein–protein and protein–RNA interactions and coat the entire RRE.[3] Because of its essential role in viral replication, the interaction between Rev protein and the high-affinity RRE binding site represents an attractive yet unexploited target for antiviral therapy.
Figure 1.
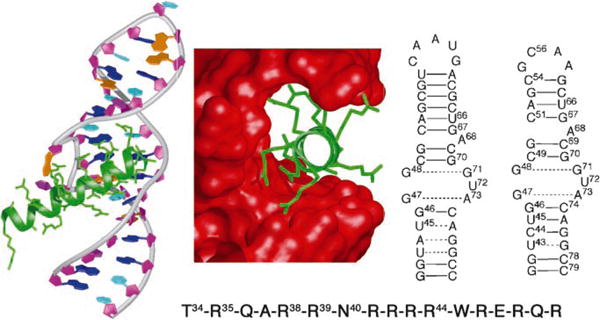
Left: X-ray structure of Rev (green) bound to HIV-1 RRE IIb (prepared from protein data base (PDB) number: 1ETF); middle: key residues in Rev (green) bound in the major groove of the RRE (red surface); right: secondary structure of RRE IIb Rev binding site, used in EMSA studies and the secondary structure of a modified RRE IIb used for NMR spectroscopic studies; bottom: sequence of the arginine-rich RNA binding domain (Rev34–50) of HIV-1 Rev protein.
Rev recognition of the high-affinity RRE binding site is mediated by a small N-terminal 17-residue Arg-rich RNA-binding motif (Figure 1, bottom).[4] NMR spectroscopic and biochemical studies have shown that the distorted A-form RRE RNA defines a deep binding pocket that is recognized by the peptide.[5–9] This protein segment is unfolded in isolation, but adopts a α-helical conformation upon binding the RRE.[6,9] To date, the search for drugs that target RRE has largely focused on derivatives of known aminoglycoside antibiotics, diphenylfurans, as well as related compounds that bind to the DNA minor groove,[10] such as proflavines.[11,12] However, this approach has failed to yield inhibitors of sufficient potency and selectivity to warrant further development.
Encouraged by recent success in using a β-hairpin structure to mimic an α-helical epitope in p53 and inhibit the p53-HDM2 interaction,[13] we explore herein a new approach to inhibitors of the Rev–RRE interaction based on conformationally constrained β-hairpin peptidomimetics. We reasoned that a β-hairpin might provide a robust scaffold upon which the groups critical for RRE recognition could be displayed, just as the a-helical scaffold of the basic domain of Rev presents the energetically important residues Thr34, Arg35, Arg38, Arg39, Asn40, and Arg44 to the RNA (Figure 1).[14] To test this hypothesis, we first assayed a small family of cyclic β-hairpin peptidomimetics (BIV-1 to BIV-8), prepared in earlier work, for their ability to mimic the α-helical Rev protein.[15]
The RNA-binding sequence of Rev is related to and belongs to the same class of protein (arginine-rich domain) as the Tat proteins of the bovine immunodeficiency virus (BIV) and HIV.Recently, we successfully mimicked BIV Tat using macrocyclic peptidomimetics containing a hairpin-inducing D-Pro-L-Pro template (Figure 2, left).[15,16] Owing to the high similarity of the arginine-rich regions of Tat and Rev proteins, the same BIV Tat-derived β-hairpin peptidomimetics were tested for in vitro binding to RRE, using polyacrylamide electrophoretic mobility shift assays (EMSA).[15,16] The Rev-derived peptide (Rev34–50) was used as a positive control in all binding assays.[4] The dissociation constant (Kd) of this Rev-derived peptide was determined by EMSA to be approximately 100 nM (Figure 3) in the presence of a large excess (280 μgmL–1) of E. coli tRNA that was used as a control for nonspecific binding.In the absence of excess tRNA, the Kd of Rev–RRE binding was about 20 nm, which is comparable to previous reports.[4]
Figure 2.
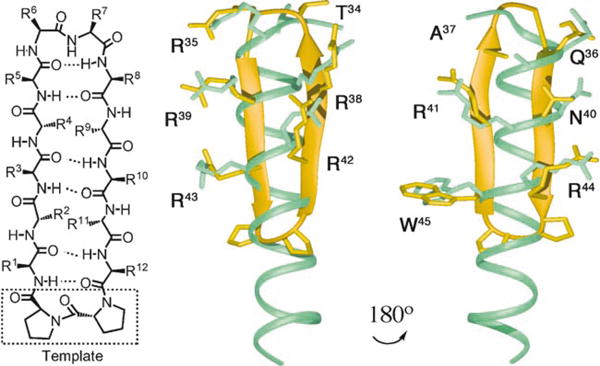
Left: a prototypical 2:2 β-hairpin mimetic (residues 1–12 with a D-Pro-l-Pro template); middle and right: a model β-hairpin (yel-low) superimposed (the C(β) atoms of the side chains shown were used as the anchor points for the superimposition) on the Rev helical peptide (cyan). The β-hairpin backbone can be used as a scaffold to pre-organize side chains in a geometry similar to that seen in the helical peptide.
Figure 3.
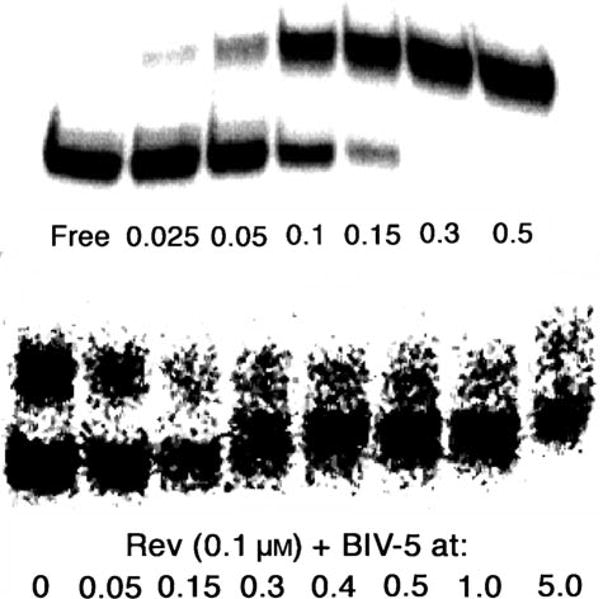
Top: binding assays (EMSA) for the wild-type Rev peptide to RRE (1 nm); bottom: inhibition of Rev peptide (0.1 μm) from a preformed complex with RRE (1 nm) by peptidomimetic BIV-5 (all concentrations in μm). The lower band corresponds to free RNA, the upper to bound RNA.
Among the peptidomimetics tested in the same way, BIV-5 exhibited the highest affinity for RRE with a Kd of 300 nm in the presence of tRNA (data not shown), only two-times higher than that measured for the wild-type Rev peptide under the same conditions (Table 1).Two additional mole-cules, BIV-2 and BIV-7, were also found to bind to the RRE with Kd values in the low micromolar range (data not shown). The other mimetics failed to bind even at high micromolar concentrations.It was shown earlier that BIV-2 was the tightest-binding ligand for BIV TAR (Kd = 0.15 μm), followed by BIV-5 (Kd = 1–2 μm), whereas for BIV-7 no binding to TAR was detected.[15]BIV-5, was also tested in an inhibition assay to measure its ability to displace Rev peptide in a preformed Rev–RRE complex. BIV-5 was found to displace the Rev peptide from the Rev–RRE complex with an IC50 of about 300 nm (Figure 3), which is similar to the estimated Kd for direct binding to the RRE.
Table 1.
Sequences and binding affinities of peptides BIV-5 and R-01 to R-28 to RRE, as determined in EMSA.[a]
| Mimetic | 1 | 2 | 3 | 4 | 5 | Position | 8 | 9 | 10 | 11 | 12 | Kd | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 7 | [μm] | |||||||||||
| BIV-5 | R | R | G | T | R | G | K | R | R | I | G | R | 0.3 |
| R-01 | W | R | R | R | A | T | DR | Q | R | N | R | R | 1 |
| R-02 | W | R | R | R | A | P | DR | Q | R | N | R | R | 1 |
| R-03 | W | R | R | R | G | P | DR | Q | R | N | R | R | 0.5 |
| R-04 | W | R | R | R | V | P | DR | Q | R | N | R | R | 1 |
| R-05 | W | R | R | R | A | G | DR | Q | R | N | R | R | 0.4 |
| R-06 | W | R | R | R | A | G | K | Q | R | N | R | R | 0.3 |
| R-07 | W | R | R | R | A | T | DR | G | R | N | R | R | 0.3 |
| R-08 | W | R | R | R | A | P | DR | G | R | N | R | R | 0.5 |
| R-09 | W | R | R | R | G | P | DR | G | R | N | R | R | 0.5 |
| R-10 | W | R | R | R | G | T | DR | Q | R | N | R | R | 0.7 |
| R-11 | W | R | R | R | V | G | DR | Q | R | N | R | R | 0.5 |
| R-12 | W | R | R | R | A | S | DR | Q | R | N | R | R | 0.4 |
| R-13 | W | R | R | R | A | K | G | Q | R | N | R | R | 0.2 |
| R-14 | W | R | L | R | A | K | G | Q | R | N | R | R | nd |
| R-15 | W | R | Q | R | A | K | G | Q | R | N | R | R | nd |
| R-16 | W | R | R | R | A | K | G | Q | R | N | R | V | 0.5 |
| R-17 | W | R | L | R | A | DR | G | Q | R | N | R | R | 0.4 |
| R-18 | W | R | L | R | A | K | K | Q | R | N | R | R | 0.3 |
| R-19 | W | R | K | R | A | G | K | Q | R | N | R | R | 0.4 |
| R-20 | W | R | K | R | A | K | G | Q | R | N | R | R | 0.2 |
| R-21 | W | R | R | R | A | DR | G | Q | R | N | R | R | 0.5 |
| R-22 | W | R | L | R | A | DR | G | K | R | N | R | R | 0.25 |
| R-23 | W | R | R | R | A | G | R | Q | R | N | R | R | 0.15 |
| R-24 | R | R | L | R | A | DR | G | Q | R | N | R | R | 0.1 |
| R-25 | R | R | L | R | A | K | G | Q | R | N | R | R | 0.1 |
| R-26 | R | V | R | R | A | K | G | Q | R | N | R | R | 0.3 |
| R-27 | R | C | R | R | A | K | G | Q | R | R | C | R | 0.1 |
| R-28 | K | R | Q | R | T | K | G | R | R | L | O | R | 0.1 |
Positions 1–12 refer to residues 1–12 as mounted on the D-Pro-L-Pro template (Figure 2, left). The dissociation constants (Kd) were determined in the presence of a large excess (280 μgmL–1) of E. coli tRNA that was used as a control for nonspecific binding. Binding reactions were conducted in Tris-HCl buffer (50 mM, pH 8.0), KCl (50 mM), 1,4-dithiothreitol (DTT; 200 mM) and Triton X-100 (0.05%). nd = no binding detected.
To establish that binding of BIV-5 occurred at the Rev binding site on the RRE RNA, the BIV-5–RRE interaction was studied by NMR spectroscopy. Resonances in 2D TOCSY spectra in the free RNA were derived from the known assignment of RRE RNA.[17] From the TOCSY spectrum (in which pyrimidine H5–H6 resonances could be selectively identified, Figure 4), it was found that mainly resonances at or near the purine-rich internal RNA loop are shifted in the presence of a 1:1 ratio of BIV-5.In particular, the U72 resonance was greatly shifted downfield in the presence of BIV-5, in agreement with the structural changes observed upon Rev binding when U72 becomes bulged-out. Moreover, the C69 resonance disappeared and the C51, C74, and U66 resonances were shifted, further indicating the specific binding of BIV-5 to the internal loop region. In contrast, resonances arising from the apical loop and the stem region close to the apical loop and from the 5′ and 3′ ends either remain unaffected or are shifted slightly (Figure 4). These data are consistent with BIV-5 binding to an RNA structure which is similar to the RNA structure recognized by Rev.[6]
Figure 4.
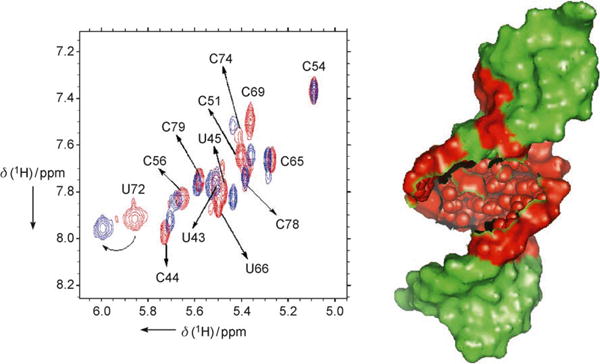
Left: superimposition of TOCSY spectra of free RRE-RNA in red (sequence shown in Figure 1 top, right) and 1:1 BIV-5:RRE complex in blue. Resonances at or near the purine-rich internal loop are affected by peptide binding, while the rest of the RNA is not. Homonuclear 1H TOCSY and NOESY experiments of the free RNA and BIV-5-RRE complex were recorded at 750 MHz (Bruker DMX-750) at RNA concentrations of 0.5–1.5 mM in D2O at pH 6.6 in phosphate buffer; right: schematic representation of the region of RRE affected by peptide binding (red) and the region that remained unaffected (green).
These promising results prompted the synthesis of a second library of β-hairpin peptidomimetics (R-01 to R-28, Table 1), which were made to test a new design concept.This concept is based on the premise that, in the Rev–RRE complex, the key RNA-interacting side chains in Rev are displayed around almost the entire circumference of the Rev a-helix (Figure 1, middle). Superimposing a model β-hairpin (yellow, in Figure 2, right) on this Rev helical peptide, revealed that the side chains of residues 1, 12, 3, 10, 5, and 8 on one face of the 12-mer hairpin have approximately the same orientations as the side chains of residues W45, R44, N40, R41, A37, and Q36 in Rev. In a similar way, residues R35, T34, R38, R39, R42, and R43 in the Rev helix might be spatially mimicked by corresponding residues at positions 6, 7, 9, 4, 2, and 11, respectively, on the other face of the hairpin. Moreover, the mimicry appears optimal when Arg7 in the hairpin (which should mimic Arg35 in Rev) has the D-configuration (consistent with this hypothesis, switching from D- to L-Arg reduced activity). Thus, the first mimetic (R-01, Table 1) has the sequence W1R2R3R4A5T6R7Q8R9N10R11R12 mounted upon the D-Pro-L-Pro template (Figure 2, left). A series of related mimetics (R-02 to R-23) were also studied, which differ in sequence mainly at the tip of the β-hairpin, in an effort to identify a motif that promotes a stable β-turn in this region.
The peptidomimetics R-01 to R-23 were synthesized, purified (see Supporting Information) and assayed for bind-ing to RRE by EMSA. Their Kd values measured in the presence of excess tRNA are shown in Table 1. With the exception of peptides R-14 and R-15 that did not retain binding, all the other peptides exhibited a strong affinity to RRE under the conditions tested. NMR spectroscopic studies on several of these mimetics in unbound form in aqueous solution, however, suggested that few regular β-hairpin structures were present, that is, that they are disordered.For example, 3J(NH,C(α)H) values were all in the range 6–8 Hz, and few characteristic long-range NOEs could be detected. Although the binding assays show high-affinity binding to the RNA, the lack of stable hairpin structure in these free ligands in solution prompted fur-ther studies aimed at stabilizing the hairpin fold.
The final group of mimetics tested (R-24 to R-28) combine features of BIV-5 and R-01 to R-23, and include a K6G7 motif at the hairpin tip which is expected to promote a stable β-turn, as seen in a closely related TAR-binding mimet-ics.[18] In addition, they have a disulfide bridge at a non-hydrogen-bonding posi-tion in R-27, to further stabilize a β-hairpin structure. Peptides R-24, R-25, R-27, and R-28 were found to bind to RRE with Kd≈ 0.1 μm, similar to the Rev-derived peptide (see Figure 5 for representative data for R- 24). When these molecules were assayed in the absence of excess tRNA, a Kd of 10 nm was observed for R-24 (Figure 5) and R-25, whereas R-27 showed a Kd of 1–2 nm. The affinity of this latter peptide for the RNA is thus significantly stronger than that of the Rev-derived a-helical peptide.
Figure 5.
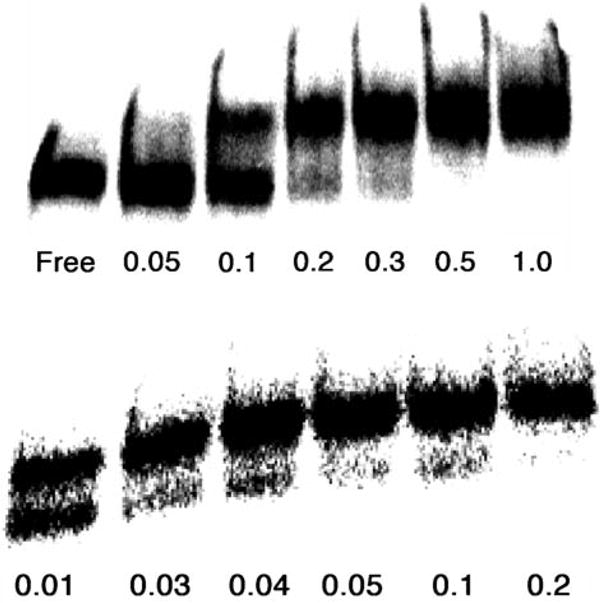
Top: binding assay of R-24 to RRE (1 nm) in the presence of excess tRNA; bottom: binding assay of R-24 to RRE (1 nm) in the absence of tRNA (concentrations of R-24 in μm).
NMR spectroscopic studies on R-27 free in solution revealed a relatively stable β-hairpin structure for this peptide (see Supporting Information), in which the two strands of antiparallel β-sheet are connected by a type I′ turn at K6G7. In particular, cross-strand NOEs characteristic of a β-hairpin are now observed, and the 3J(NH,C(α)H) values are predom-inantly greater then 8 Hz for residues within the β-strands. Thus the disulfide cross-link is clearly important in stabilizing the regular β-hairpin structure.
A further key issue in RNA ligand design is the specificity of RNA binding, which could clearly be influenced by the flexibility of the ligand as well as by the overall positive charge of this class of peptides. As a very stringent test of binding specificity to RRE, we also evaluated binding of the mimetics to the structurally very similar HIV TAR RNA. As a control, we also tested binding of a peptide containing 7 consecutive Arg residues (Ac-Arg7-NH2), which is expected to interact strongly but nonspecifically with both RNAs, mainly through electrostatic interactions. Consistent with this hypothesis, and demonstrated by EMSA, the Arg7 peptide binds nonspecifically with a Kd of approximately 2 nm to both HIV TAR and RRE (Table 2). Among the ligands tested R- 26 and R-28 discriminate only modestly (less than tenfold) between RRE and TAR.However, R-27 discriminates by approximately 50-fold between the two closely related RNA structures. Given how closely related these RNA targets are, and the relatively poor specificity typically seen in the interactions of other RNA-binding molecules with more diverse RNA targets, this result represents a notable level of RNA-binding specificity.
Table 2.
Binding affinities (in μm) of selected peptides to HIV RRE and TAR RNAs.[a]
| Mimetic | Kd (RRE) | Kd (TAR) |
|---|---|---|
| R-24 | 0.010 | 0.005 |
| R-25 | 0.010 | 0.010 |
| R-26 | 0.005 | 0.010 |
| R-27 | 0.002 | 0.100 |
| R-28 | 0.002 | 0.010 |
| Arg7 | 0.002 | 0.002 |
The affinities were determined by EMSA in the absence of tRNA.
The structural and binding data together provide strong evidence supporting our hypothesis that this family of β-hairpin peptidomimetics can mimic the α-helical Rev peptide in its binding to RRE RNA. Although we cannot yet rationalize the quantitative effects of individual side-chain substitutions on the binding energy, we were able to trans-plant the molecular interactions observed on the α-helical Rev-derived peptide onto a β-hairpin scaffold and very rapidly discover ligands that are more potent than the Rev protein. Furthermore, the results suggest that not just the number of positively charged groups, but also their relative orientations, as determined by the flexibility of the β-hairpin mimetic, have a decisive influence on the affinity and specificity of RNA binding. These important conclusions merit further investigation, not least since this family of mimetics represents a relatively new class of RNA-binding molecules, with potential for development into novel drugs against HIV.
Supplementary Material
Footnotes
This work was supported by grants from the Swiss National Science Foundation (to JAR) and the NIH (to GV).
Supporting information for this article is available on the WWW under http://www.angewandte.org or from the author.
Contributor Information
Kerstin Moehle, Department of Chemistry, University of Zürich, Winterthurerstrasse 190, 8057 Zürich (Switzerland), Fax: (+41) 44-1635-6833.
Zafiria Athanassiou, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA).
Krystyna Patora, Department of Chemistry, University of Zürich, Winterthurerstrasse 190, 8057 Zürich; (Switzerland), Fax: (+41) 44-1635-6833.
Amy Davidson, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA).
Gabriele Varani, Email: varani@chem.washington.ed, Department of Chemistry, University of Washington, Seattle, WA 98195 (USA); Department of Biochemistry, University of Washington, Seattle, WA 98195 (USA).
John A. Robinson, Email: robinson@oci.unizh.ch, Department of Chemistry, University of Zürich, Winterthurerstrasse 190, 8057 Zürich (Switzerland), Fax: (+41) 44-1635-6833.
References
- 1.Emerman M, Vazeux R, Peden K. Cell. 1989;57:1155. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 2.Malim MH, Hauber J, Le SY, Maizel JV, Cullen BR. Nature. 1989;338:254. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 3.Mann DA, Mikaelian I, Zemmel RW, Green SM, Lowe AD, Kimura T, Singh M, Butler G, Gait M, Karn J. J Mol Biol. 1994;241:193. doi: 10.1006/jmbi.1994.1488. [DOI] [PubMed] [Google Scholar]
- 4.Kjems J, Calnan BJ, Frankel AD, Sharp PA. EMBOJ. 1992;11:1119. doi: 10.1002/j.1460-2075.1992.tb05152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Battiste JL. Biochemistry. 1994;33:2741. doi: 10.1021/bi00176a001. [DOI] [PubMed] [Google Scholar]
- 6.Battiste JL, Mao H, Rao NS, Tan R, Muhandiram DR, Kay LE, Frankel AD, Williamson JR. Science. 1996;273:1547. doi: 10.1126/science.273.5281.1547. [DOI] [PubMed] [Google Scholar]
- 7.Gosser Y, Hermann T, Majumdar A, Hu W, Frederick R, Jiang F, Xu W, Patel D. Nat Struct Biol. 2001;409:146. doi: 10.1038/84138. [DOI] [PubMed] [Google Scholar]
- 8.Ippolito JA, Steitz TA. J Mol Biol. 2000;295:711. doi: 10.1006/jmbi.1999.3405. [DOI] [PubMed] [Google Scholar]
- 9.Ye X, Gorin A, Ellington AD, Patel DJ. Nat Struct Biol. 1996;3:1026. doi: 10.1038/nsb1296-1026. [DOI] [PubMed] [Google Scholar]
- 10.Boykin DW, Kumar J, Spychala J, Zhou M, Lombardy RJ, Wilson JD, Dykstra CC, Jones SK, Hall JE, Tidwell RR, Laughton C, Nunn CM, Neidle S. J Med Chem. 1995;38:912. doi: 10.1021/jm00006a009. [DOI] [PubMed] [Google Scholar]
- 11.Chapman RL, Stanley TB, Hazen R, Garvey EP. Antiviral Res. 2002;54:149. doi: 10.1016/s0166-3542(01)00222-4. [DOI] [PubMed] [Google Scholar]
- 12.DeJong ES, Chang CE, Gilson MK, Marino JP. Biochemistry. 2003;42:8035. doi: 10.1021/bi034252z. [DOI] [PubMed] [Google Scholar]
- 13.Fasan R, Dias RLA, Moehle K, Zerbe O, Vrijbloed JW, Obrecht D, Robinson JA. Angew Chem. Angew Chem Int Ed. 2004;2004;11643:2161. 2109. doi: 10.1002/anie.200353242. [DOI] [PubMed] [Google Scholar]
- 14.Tan R, Frankel AD. Biochemistry. 1994;33:14579. doi: 10.1021/bi00252a025. [DOI] [PubMed] [Google Scholar]
- 15.Athanassiou Z, Dias RLA, Moehle K, Dobson N, Varani G, Robinson JA. J Am Chem Soc. 2004;126:6906. doi: 10.1021/ja0497680. [DOI] [PubMed] [Google Scholar]
- 16.Leeper TC, Athanassiou Z, Dias RLA, Robinson JA, Varani G. Biochemistry. 2005;44:12362. doi: 10.1021/bi0510532. [DOI] [PubMed] [Google Scholar]
- 17.Battiste JL, Tan R, Frankel AD, Williamson JR. J Biomol NMR. 1995;6:375. doi: 10.1007/BF00197637. [DOI] [PubMed] [Google Scholar]
- 18.Athanassiou Z, Patora K, Dias RLA, Moehle K, Robinson JA, Varani G. Biochemistry. 2007;46:741. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


