Abstract
High-level expression of many recombinant proteins in Escherichia coli leads to the formation of highly aggregated protein commonly referred to as inclusion bodies. Inclusion bodies are normally formed in the cytoplasm; however, if a secretion vector is used, they can form in the periplasmic space. Inclusion bodies can be recovered from cell lysates by low speed centrifugation. Following preextaction (or washing) protein is extracted from washed pellets using guanidine·HCl. The solubilized and unfolded protein is either directly folded as described in UNIT 6.1 or further purified by gel filtration in the presence of guanidine·HCl as described here. A support protocol describes the removal of guanidine·HCl from column fractions so they can be monitored by SDS-PAGE.
High-level expression of many recombinant proteins in Escherichia coli leads to the formation of highly aggregated protein commonly referred to as inclusion bodies (UNITS 5.1 & 6.1). Inclusion bodies are normally formed in the cytoplasm; alternatively, if a secretion vector is used, they can form in the periplasmic space. Inclusion bodies are not restricted to E. coli; they can also form in yeast, mammalian, and insect cells. Inclusion bodies recovered from cell lysates by low-speed centrifugation are heavily contaminated with E. coli cell wall and outer membrane components. The latter are largely removed by selective extraction with detergents and low concentrations of either urea or guanidine·HCl to produce so-called washed pellets. These basic steps result in a significant purification of the recombinant protein, which usually makes up ~60% of the washed pellet protein. The challenge, therefore, is not to purify the recombinant-derived protein, but to solubilize it and then fold it into native and biologically active protein.
Basic Protocol 1 describes preparation of washed pellets and solubilization of the protein using guanidine·HCl. The extracted protein, which is unfolded, is either directly folded as described in UNIT 6.5 or further purified by gel filtration in the presence of guanidine·HCl as in basic Protocol 2. A Support Protocol describes the removal of guanidine·HCl from column fractions so they can be monitored by SDS-PAGE (UNIT 10.1).
Other methods discussed in the Commentary section of this unit include the direct purification of polyhistidine-tagged proteins solubilized in guanidine·HCl, preparative removal of guanidine·HCl by reversed-phase chromatography as a prelude to protein folding, and the solubilization of inclusion bodies with anionic detergents.
BASIC PROTOCOL 1
Preparation and Extraction of Insoluble (Inclusion-Body) Proteins from Escherichia Coli
Bacterial cells are lysed using a French press, and inclusion bodies in the cell lysate are pelleted by low-speed centrifugation. The pellet fraction is washed (preextracted) with urea and Triton X-100 to remove E. coli membrane and cell wall material. Guanidine·HCl (8 M) and dithiothreitol (DTT) are used to solubilize the washed pellet protein. Extraction with the denaturant simultaneously dissociates protein-protein interactions and unfolds the protein. As a result, the extracted protein consists (ideally) of unfolded monomers, with sulfhydryl groups (if present) in the reduced state.
Materials
E. coli cells from fermentation (UNIT 5.3) containing the protein of interest
Lysis buffer (see recipe)
Wash buffer (see recipe), with and without urea and Triton X-100
Extraction buffer (see recipe)
250- and 500-ml stainless steel beakers
Waring blender
Polytron tissue-grinder homogenizer (Brinkmann: http://www.kinematica-inc.com)
French pressure cell (e.g., Thermo Electron Corp: http://www.thermo.com)
Probe sonicator
Beckman J2-21M centrifuge with JA-14 rotor (or equivalent)
Beckman Optima XL-90 ultracentrifuge with 45 Ti rotor (or equivalent)
0.22- μm syringe filters (e.g., Millex from Millipore)
20-ml disposable syringe
Additional equipment for breaking cells, homogenizing cells and pellets and centrifuging at low and high speeds (UNIT 6.2)
Break cells and prepare clarified lysate
-
1
Place thawed E. coli cells in a stainless steel beaker. Add 4 ml lysis buffer per gram wet weight of cells. Keep bacterial cells cool by placing the beaker on ice in an ice bucket.
The cells can be pretreated with lysozyme prior to lysis in the French press. Lysozyme treatment involves incubating cells –20 min at 20° to 25°C in lysis buffer supplemented with 200 μg/ml lysozyme, with intermittent homogenization using a tissue grinder. It should be emphasized that this optional step is carried out before French press breakage and is not simply an alternative method of cell breakage (compare the comments made in the annotation to step 4 of UNIT 6.2). Its purpose is to aid removal of the peptidoglycan and outer membrane protein contaminants during the washing steps (steps 6 to 9; for further details see unit 6.1 and Fig. 6.1.5). An example of this approach is given in Basic Protocol 1 of UNIT 6.5.
For sensitive proteins, replace benzamidine in the lysis buffer by a protease inhibitor cocktail that includes five protease inhibitors with broad specificity for the inhibition of aspartic proteases, cysteine proteases, serine proteases, and metalloproteases, as well as aminopeptidases. These are supplied by various companies including Calbiochem EMD Chemicals and Sigma.
-
2
Suspend cells using a Waring blender and homogenize using the Polytron tissue-grinder homogenizer until all clumps are disrupted, as described in UNIT 6.2, step 3.
-
3
Lyse cells with two passes through the French pressure cell operated at 16,000 to 18,000 psi (with the high-ratio setting, pressure gauge readings between 1011 and 1135), chilling the cell suspension to 4°C after each pass, as described in UNIT 6.2, steps 2 and 4.
-
4
Reduce the viscosity of the suspension by sonicating 5 min at full power with 50% duty cycle (on for 5 sec, off for 5 sec) using an ultrasonic homogenizer, as described in UNIT 6.2, step 5.
-
5
Clarify the lysed cell suspension by centrifuging 1 hr at 22,000 × g (12,000 rpm in a JA-14 rotor in a Beckman J2-21M centrifuge), 4°C.
Unbroken cells, large cellular debris, and the inclusion body protein will be pelleted.
The JA-14 rotor uses 250-ml centrifuge bottles. For processing smaller volumes the Beckman JA-20 rotor (or equivalent) with 50-ml tubes can be used, at 13,500 rpm (22,000 × g).
The procedure for dealing with insoluble inclusion-body proteins now diverges from that for purifying soluble proteins (UNIT 6.2).
Prepare washed pellets
-
6
Carefully decant the supernatant from the pellet. Using a tissue homogenizer, suspend the pellet with 4 to 6 ml wash buffer per gram wet weight cells.
Complete homogenization of the pellet is important to wash out soluble proteins and cellular components. Removal of cell wall and outer membrane material can be improved by increasing the amount of wash solution to 10 ml per gram cells.
The concentration of urea and Triton X-100 in the wash buffer can be varied. The urea concentration is usually between 1 and 4 M; higher concentrations may result in partial solubilization of the recombinant proteins. The usual detergent concentration is between 0.5% to 5%. Triton X-100 will not solubilize inclusion body proteins; it is included to help extract lipid and membrane-associated proteins.
-
7
Centrifuge the suspension 30 min at 22,000 × g (12,000 rpm in JA-14), 4°C. Discard supernatant and, using the tissue homogenizer, suspend the pellet in 4 to 6 ml wash buffer per gram wet weight of cells.
-
8
Repeat step 7 two more times.
If the supernatant is still cloudy or colored, continue washing the pellet until the supernatant is clear.
-
9
Suspend the pellet in wash buffer minus the Triton X-100 and urea, using 4 to 6 ml buffer per gram wet cells. Centrifuge 30 min at 22,000 × g (12,000 rpm in JA-14), 4°C.
The final wash removes excess Triton X-100 from the pellet.
If necessary the washed pellets can be stored at –80°C. It is better to store material at this stage rather than after the extraction stage (see annotation to step 13).
Extract recombinant protein from washed pellets with guanidine·HCl
-
10
Using the tissue homogenizer, suspend the pellet in guanidine·HCl-containing extraction buffer. Use 0.5 to 1.0 ml buffer per gram wet weight of original cells if the extract will be subjected to gel filtration, and 2 to 4 ml buffer if the extract will be used in protein folding procedures. Perform this step at room temperature.
To estimate the amount of recombinant protein in the washed pellets, use the following guidelines. (1) An expression level of 1% corresponds to ~1 mg recombinant protein per 1 g wet cells. (2) The recovery of highly aggregated recombinant protein in the washed pellets is ~75% that originally present in the cells. (3) About 60% of the total washed pellet protein is recombinant-derived. Thus, if 50 g cells is processed and the expression level is 5%, the washed pellets contain ~200 mg recombinant protein.
The total amount of recombinant-derived protein in washed pellets can be directly determined by measuring the total protein concentration or by analyzing the washed pellets via SDS-PAGE (see Support Protocol and UNIT10.1) to determine the proportions of the protein constituents.
For gel-filtration purposes, the pellets from 50 g wet weight E. coli cells are solubilized with 40 to 50 ml extraction buffer (see Basic Protocol 2); the concentration of recombinant protein in the extract will be 4 to 5 mg/ml. For direct protein folding (UNIT 6.5), the pellets are extracted with 100 to 200 ml buffer, and the concentration of recombinant protein 1 to 2 mg/ml. If the washed pellet is heavily contaminated with outer cell wall and peptidoglycan material, the extract must be diluted further with extraction buffer (usually 1:1 to 1:3) to reduce the viscosity before it can be used for chromatography.
Alternative Extraction of protein using a reduced concentration of chaotrope
Some recombinant protein may be extracted from an inclusion body with a lower concentration of chaotrope resulting in fewer contaminants being extracted as well. The effectiveness of using a low chaotrope extraction buffer needs to be evaluated on a case by case basis. If it is determined to be effective, substitute the extraction buffer with the low chaotrope extraction buffer.
-
11
Centrifuge the suspension 1 hr at 100,000 × g (30,000 rpm in 45 Ti rotor in a Beckman Optima XL-90 ultracentrifuge), 4°C.
For volumes <250 ml the Beckman 70 Ti rotor (capacity 6 × 39 ml) can be used at 32,000 rpm (–100,000 × g).
-
12
Carefully pour off the supernatant from the pellet. Filter the supernatant through a 0.22-μm syringe filter attached to a 20-ml disposable syringe.
The filter removes unpelleted large cell wall debris that will clog most chromatography columns.
-
13
Use the clarified inclusion body extract for preparing folded protein (UNIT 6.5) or purify further by gel filtration (see Basic Protocol 2).
Nickel-chelate affinity chromatography can also be directly used for His-tagged proteins (see Commentary in this unit and Basic Protocol 3 in UNIT 6.5).
The extract can be stored at –80°C until required. Freeze in plastic (or polyethylene) containers rather than glass. Divide sample into 10- to 20-ml aliquots instead of freezing in one large lot and fill containers to only 50% to 75% capacity.
BASIC PROTOCOL 2
MEDIUM-PRESSURE GEL-FILTRATION CHROMATOGRAPHY IN THE PRESENCE OF GUANIDINE HYDROCHLORIDE
Washed, extracted pellets (see Basic Protocol 1) contain >50% recombinant protein and are used as the starting material for purification of the protein of interest by gel-filtration chromatography. Superdex 200 gel-filtration medium, which allows high flow rates, is washed and packed into a column. The column is equilibrated at 4°C and the sample is applied.
Assay of column fractions by gel electrophoresis in the presence of SDS is complicated by the fact that guanidine·HCl forms a precipitate with SDS. Therefore, preparing samples for gel analysis involves selective precipitation of protein from guanidine·HCl prior to SDS-PAGE (see Support Protocol). The purified (or partially) purified protein is used as the starting material for procedures (e.g., UNIT 6.5) in which the denatured protein is folded into a native and biologically active structure.
Materials
Gel-filtration medium: Superdex 200 PG (preparative grade; GE Healthcare Life Sciences)
5% (v/v) ethanol
Gel-filtration buffer (see recipe)
Guanidine·HCl extract of E. coli cells containing the protein of interest (see Basic Protocol 1)
4- to 6-liter plastic beaker
Chromatography column: GE Healthcare Life Sciences XK 16/100, 26/100, or 50/100
Packing reservoir: GE Healthcare Life Sciences RK 16/26 (for 16- and 26-mm-i.d. columns) and RK 50 (for 50-mm-i.d. column)
Chromatography pump: GE Healthcare Life Sciences P-50 or P-900
Injection valve (to select between sample loop and pump)
UV monitor and fraction collector
Sample loop (volume determined by size of column; also see annotation to step 15)
NOTE: The various components of the chromatography system (pumps, valves, monitors, and sample loops) listed separately above are supplied as components of the ÄKTAexplorer chromatography system (GE Healthcare Life Sciences), which is used to run the XK 50/100 column. The smaller XK columns (2.6 and 2.5 cm i.d.) are run using the ÄKTA-FPLC chromatography system (also from GE Healthcare Life Sciences), which is designed for small- to medium-scale work. For further details on this equipment see the manufacturer's literature (e.g., Process Products, GE Healthcare Life Sciences).
NOTE: Perform steps 1 to 11 at room temperature. After the column is packed, equilibrate and elute at 4°C.
Pack the column
-
1
Wash the gel-filtration medium in a large plastic beaker with 5% ethanol. Let the medium settle and adjust the volume of liquid to yield a gel slurry concentration of 65% to 75%.
The XK 16/100, 26/100, and 50/100 columns are 100 cm long and have inner diameters of 16, 26, and 50 mm, respectively. Hence, for an XK 50/100 column, column volume = radius (2.5 cm) 2 × 3.1416 × bed height (97 cm) ≅ 1900 ml, and ~2 liters preparative-grade Superdex 200 is required. To pack this column, the gel medium is suspended in 5% ethanol to give a total volume of 3 liters which corresponds to ~70% gel slurry (it should be noted that the RK 50 reservoir has a capacity of 1 liter, so the 3 liters of gel slurry can be poured in a single operation).
-
2
Fix the chromatography column in an upright position, using a level to adjust the position. Attach the packing reservoir.
-
3
Add sufficient 5% ethanol to displace the air from a few centimeters of the bottom of the column. Clamp off the bottom of the column.
-
4
Gently mix the gel-filtration medium in the plastic beaker using a glass rod or plastic paddle to an even slurry of 70% medium suspended in 5% ethanol.
Do not use a magnetic stirrer, as it could damage the medium.
-
5
Degas the suspension 5 to 10 min using a vacuum flask and laboratory vacuum.
The ethanol is included to reduce the surface tension and density of the solvent, thus allowing air bubbles that form to rise to the surface more quickly.
-
6
Carefully pour the slurry of medium into the column, introducing material along the side of the column to avoid creating air bubbles.
-
7
Let the column stand 5 min and then unclamp the bottom of the column.
-
8
Attach the chromatography pump to the packing reservoir and pump 5% ethanol (degassed) into the column at an appropriate flow rate (based on manufacturer's instructions). Pack the column at a pressure greater than the pressure at which the column will be run (up to twice as high), but not greater than the maximum pressure rating of the column.
For example, the XK 50/100 column (rated to 0.5 MPa) is packed at ~20 to 30 ml/hr and ~0.4 MPa.
-
9
After the medium has settled, turn off the pump and close the bottom of the column. Pipet fluid from the reservoir and remove the reservoir.
Once the column has been packed, be careful to prevent air from entering the column bed. Air will disturb the bed and reduce the column separation resolution.
-
10
Attach the column top adapter to the column. Place the top of the adapter onto the top of the packed medium and gently compress the medium.
-
11
Reattach the pump to the column and wash the column with water at a flow rate that will generate the maximum pressure to be used. If the medium continues to settle, readjust the top adapter to maintain a firm fit against the gel.
From this point onward, perform all steps at 4°C.
Equilibrate the column
-
12
Equilibrate the column with at least 1 column volume of 4°C gel-filtration buffer.
Although the proteins were extracted with buffer containing 8 M guanidine·HCl (see Basic Protocol 1), the gel-filtration buffer contains only 4 M guanidine·HCl. The concentration is reduced to allow faster flow rates and for reasons of economy. Most proteins remain unfolded at the lower guanidine·HCl concentration. If, however, the protein elutes in an anomalous manner (e.g., in more than one peak or at an elution position not consistent with its size), and assuming there is adequate reducing agent present, then try increasing the guanidine·HCl concentration in the gel-filtration buffer.
-
13
Measure the actual flow rate while running the column at a flow rate that generates a back pressure about one-half of that generated when packing the column (step 8).
For an XK 50/100 column packed using Superdex 200 at 0.4 MPa, a running pressure of ~0.2 MPa is used, which generates flow rates of 5 to 10 ml/min that are equivalent to linear flow rates of 15.3 to 30.6 cm/hr. The linear flow rate equals the flow rate (ml/hr)/cross-sectional area (cm2). At these flow rates it takes between 3 and 6 hr to complete the chromatography.
-
14
Connect tubing from the end of the column to the UV monitor and the fraction collector.
Apply the sample
-
15
Load the sample loop with the guanidine·HCl extract to be separated.
Avoid loading a sample volume >5% of the total column volume; the optimum sample size is 2% (~40 ml for the XK 50/100 column). The sample consists of washed pellets extracted with guanidine·HCl (see Basic Protocol 1). A sample size of 40 to 50 ml is usually derived from ~50 g wet weight cells. With smaller sample sizes, use columns with proportionally smaller diameters (e.g., XK 16/100 or 26/100 columns). If purchase of only one column is possible, a 2.5 × 100–cm size is a good compromise for variable sample loading.
-
16
Monitor column effluent with the UV monitor and collect fractions with the fraction collector.
For an XK 50/100 column, collect 15- to 20-ml fractions in 16 × 20–mm culture tubes.
The eluent from the column is usually monitored at 280 nm or, if the protein has a particularly low extinction coefficient, at 230 nm (guanidine·HCl strongly absorbs below 225 nm). For an XK 50/100 column, fractions need only be collected after ~500 ml of elution. The excluded volume (void volume) is ~570 ml. Run a total of one column volume (1900 ml) to ensure all of the load material is eluted from the column.
-
17
Prepare the fractions to be assayed for SDS-PAGE (see Support Protocol and UNIT 10.1).
SUPPORT PROTOCOL
PREPARATION OF SAMPLES CONTAINING GUANIDINE HYDROCHLORIDE FOR SDS-PAGE
Because guanidine·HCl forms a precipitate with SDS, it is necessary to remove the former before carrying out SDS-PAGE. Protein in column fractions is separated from guanidine·HCl by precipitation using 90% ethanol (Pepinsky, 1991).
Materials
Sample containing the protein of interest
100% and 90% ethanol, 0° to 4°C
1× SDS sample buffer (UNIT 10.1)
Gilson Pipetman (http://www.gilson.com)
Additional reagents and equipment for gel electrophoresis (UNIT 10.1)
-
1
Pipet 25 μl sample containing the protein of interest into a 1.5-ml microcentrifuge tube.
-
2
Add 225 μl cold (0° to 4°C) 100% ethanol to the sample in the tube.
The final ethanol concentration is 90% by volume.
-
3
Mix the sample and ethanol by vortexing. Chill 5 to 10 min at –20°C or colder (e.g., –80°C).
Do not use a magnetic stirrer, as it could damage the medium.
-
4
Microcentrifuge the sample 5 min at maximum speed (~15,000 × g), 4°C. Carefully withdraw the supernatant and retain the pellet.
The pellet may be difficult to see. Be careful not to draw the pellet out of the microcentrifuge tube with the supernatant.
-
5
Suspend the pellet in 250 μl cold 90% (v/v) ethanol. Mix thoroughly using a vortex mixer.
The 90% ethanol is prepared by mixing 225 μl ethanol and 25 μl H2O.
-
6
Microcentrifuge the sample 5 min at maximum speed, 4°C. Carefully pipet off the supernatant and suspend the pellet in 25 μl of 1× SDS sample buffer.
Some proteins are more difficult than others to suspend from an ethanol precipitate. Electrophoresis sample buffer containing 8 M urea is helpful for such proteins (UNIT 10.1). Sonication with a microtip probe can also be used to disperse the sample. A volume of sample buffer >25 μl may be required in this case (e.g., 50 μl), and great care must be taken to prevent foaming of the sample caused by excessive sonication power.
-
7
Heat the sample 3 to 5 min at 90° to 100°C. Load on an SDS-polyacrylamide gel (UNIT 10.1).
REAGENTS AND SOLUTIONS
Use Milli-Q-purified water or equivalent in all recipes and protocol steps. For common stock solutions, see APPENDIX 2E; for suppliers, see SUPPLIERS APPENDIX.
Extraction buffer
50 mM Tris·Cl, pH 7.0
5 mM EDTA
8 M guanidine·HCl (764 g/liter)
5 mM DTT (770 mg/liter)
If the buffer is cloudy, filter through a 0.45- to 0.5-μm filter (the solution should be clear if high-quality guanidine·HCl—e.g., ultrapure grade, Invitrogen Life Technologies—is used; see APPENDIX 3A). Buffer can be stored without DTT for at least 1 month at 4°C.
Low Chaotrope Extraction buffer
50 mM Tris·Cl, pH 7.0
5 mM EDTA
3 M guanidine·HCl (287 g/liter)
5 mM DTT (770 mg/liter)
If the buffer is cloudy, filter through a 0.45- to 0.5-μm filter (the solution should be clear if high-quality guanidine·HCl—e.g., ultrapure grade, Invitrogen Life Technologies—is used; see APPENDIX 3A). Buffer can be stored without DTT for at least 1 month at 4°C.
Gel-filtration buffer
50 mM Tris·Cl, pH 7.5
4 M guanidine·HCl (382 g/liter; ultrapure)
5 mM DTT (770 mg/liter)
Buffer can be stored without DTT at least 1 month at 4°C. Filter (as for extraction buffer; see recipe) and degas before use.
Higher concentrations of guanidine·HCl (up to 8 M) may be required for some proteins (see annotation at step 12 of basic Protocol 2).
Lysis buffer
100 mM Tris·Cl, pH 7.0
5 mM EDTA
5 mM DTT (770 mg/liter)
5 mM benzamidine·HCl (780 mg/liter)
Prepare immediately before use
The Tris·Cl and EDTA are diluted from concentrated stock solutions. The other components are added to the diluted buffer before use.
Wash buffer
100 mM Tris·Cl, pH 7.0
5 mM EDTA
5 mM DTT (770 mg/liter)
2 M urea (120 g/liter; ultrapure (Invitrogen Life Technologies)
2% (w/v) Triton X-100 (20 g/liter; Calbiochem EMD Chemicals)
Add DTT, urea, and Triton X-100 to the other components immediately before use. Prepare this buffer in two forms: one with and one without the urea and Triton X-100 (the latter for use in Basic Protocol 1, step 9).
COMMENTARY
Background Information
The decision of whether to work with insoluble recombinant protein or to put more effort into generating soluble protein (e.g., by modifying the expression vector, changing the host strain and fermentation conditions, or coexpressing chaperones etc.) is dictated by the nature of the protein. Enhancement of soluble protein expression has also been achieved by the use of fusion tags (Esposito and Chatterjee, 2006). If protein folding is attempted, a small protein (10 to 17 kDa) with, for example, one or two cysteine residues might be expected to fold in reasonable yield (>25%) from extracted inclusion bodies. Larger proteins (>25 kDa) with many cysteine residues may be more problematic, and lower folding yields (< 20%) can normally be expected. In the latter case, if only small amounts of material are needed then lower folding yields will still provide adequate for study.
It should be emphasized that, unless proven otherwise, a protein folded from insoluble inclusion bodies will have the same structural and conformational integrity as the same protein directly purified from soluble extracts (also see UNIT 6.1). It is similarly true that a purified soluble protein can be denatured and renatured (reversible denaturation) without structural or conformational modifications (reviewed by Anfinson, 1973; Fersht, 1998; Pain, 2001, Tsumoto et al., 2003). The main danger of working with an unfolded protein is that it vulnerable to chemical modification, especially oxidations of methionine and cysteine and also is very sensitive to any contaminating protease activity.
Basic Protocol 1 describes the preparation of inclusion bodies suitable for extraction purposes. The insoluble protein is washed in buffers containing various additives by suspension and low-speed centrifugation. The resultant washed pellets (inclusion bodies), although highly enriched in the recombinant protein (Figure 6.3.1), are still contaminated with bacterial proteins, lipids, and nucleic acids. This is not an issue if the protein is going to be partially purified, e.g., by gel filtration (Basic Protocol 2) prior to folding. However, if the solubilized protein is going to be directly used in folding scenarios, there may be an advantage in further purifying the inclusion bodies, as the bacterial contaminants, especially the nonproteinacous ones, may influence protein folding (Darby and Creighton, 1990; Maachupalli-Reddy et al., 1997). Inclusion bodies have also been purified from bacterial lysates by sucrose step-gradient centrifugation (Georgiou and Valax, 1999).
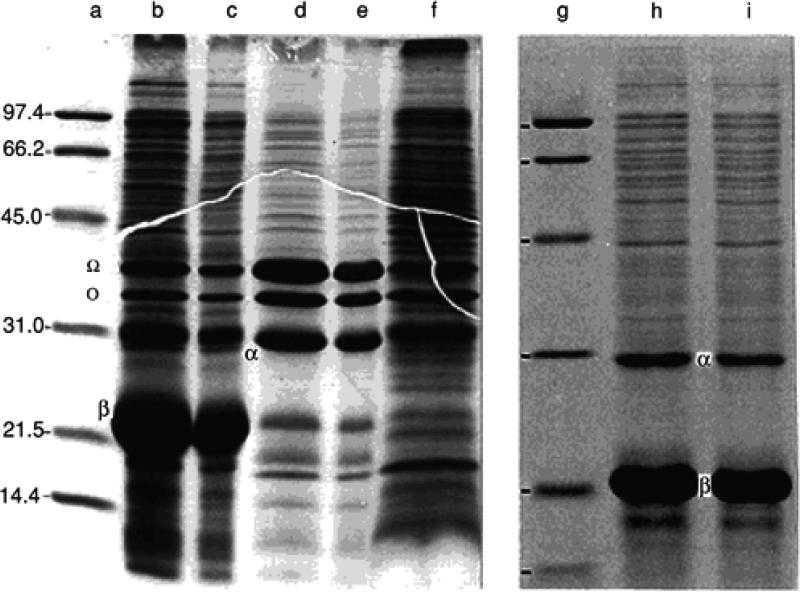
Figure 6.3.1.
Analysis by SDS-PAGE of fractions from low-speed centrifugation of E. coli cell lysates containing aggregated bovine growth hormone. A 12.5% acrylamide gel of dimensions 12 cm × 16 cm × 1.5 mm was used with the Laemmli buffer system (UNIT 10.1). Lanes a and g contain molecular weight standards (low-range standards, Bio-Rad) in order of increasing migration distance: phosphorylase b (97.4 kDa), bovine serum albumin (66.2 kDa), hen egg white ovalbumin (45 kDa), bovine carbonic anhydrase (31 kDa), soybean trypsin inhibitor (21.5 kDa), and hen egg white lysozyme (14.4 kDa). After low-speed centrifugation of the clarified lysate and of the washed pellet homogenate (see Basic Protocol 1, steps 5 and 7), the supernatants will be cloudy (lane f) and the pellets usually consist of two layers (see Fig. 6.1.5). The bottom layer is inclusion body protein plus unbroken cells (lanes b and c) and the top layer consists of outer membrane and peptidoglycan fragments (lanes d and e). The outer membrane proteins OmpA (35 kDa) and OmpF/C (38 kDa) are indicated by Ω; and o, respectively. After the washing steps, the growth hormone (marked β, 21 kDa) is the major constituent (lanes h and i) together, in this example, with another plasmid-encoded protein, namely kanamycin phosphotransferase (marked α, 30.8 kDa), the product of the gene conferring resistance to the antibiotic kanamycin.
How inclusion bodies are formed
Inclusion bodies are noncrystalline, amorphous structures; however, there is some evidence that the constituent densely packed proteins may have native-like secondary structures (Oberg et al., 1994). This suggests that the aggregates are formed by the association of partially folded protein or misfolded protein. Folded subdomains could presumably associate with complementary binding surfaces in an intermolecular manner to form oligomers and eventually aggregates (Fink, 1998). During in vitro folding studies with a mixture of proteins, folding intermediates do not coaggregate with each other, but only with themselves. This indicates that aggregation occurs by specific interaction of certain conformations of folding intermediates rather than by nonspecific coaggregation, providing a rationale for recovering relatively pure protein from the inclusion body state (Speed et al., 1996).
The propensity of a protein to form inclusion bodies does not appear to be directly related to the presence of sulfhydryl residues, as proteins without sulfhydryls still form such aggregates. It has been found that aggregates derived from proteins that in their native state contain disulfides consisting mainly of reduced protein (Langley et al., 1987). The reduction of disulfides in native proteins can dramatically reduce their solubility and even result in unfolding. Hence the reducing environment of E. coli may contribute to the instability and low solubility of these types of protein. Once the protein has been solubilized, reducing agents must be included to prevent the formation of nonnative disulfide bonds.
Some of the other factors that may contribute to aggregate formation include: lack of post-translational modification (especially glycosylation), lack of access to chaperones, and enzymes catalyzing folding (e.g., cis-trans isomerase) and the very high concentration of protein coupled with limited solubility of folding intermediates.
Although inclusion body formation and its prevention are of great academic and commercial interest, the pragmatic situation is clearly summarized by Seckler and Jaenicke (1992), who state: “That inclusion bodies are aggregates of otherwise intact polypeptides in nonnativelike conformations has been proven repeatedly by the successful refolding of active proteins after dissociation of the aggregates by chemical dissociation.” It must be noted that the aforementioned findings of Oberg et al. (1994) do not invalidate this statement. It is simply a matter of terminology: the protein in aggregates may have nativelike secondary structures, but compared with soluble folded protein it must still be considered nonnative. For an updated discussion of inclusion bodies formation and their potential application in nanotechnology see García-Fruitós et al., (2012).
Extracting Inclusion Body Protein
Proteins are extracted from inclusion bodies using strong protein denaturants. Protein denaturation can be induced by the following solvent conditions or reagents.
1. pH
Protein denaturation occurs because of the ionization of side chains. Generally, proteins retain less residual structure (are more denatured) when exposed to high pH (e.g., >10.5) compared to low pH (<4.5). Acidic or basic pH may be used in conjunction with urea or inorganic salts. An example of acidic pH extraction is given in UNIT 6.5.
2. Organic solvents
Examples of organic solvents include ethanol and propanol. Generally, proteins are not completely unfolded in these solvents. Organic solvents are infrequently used for the primary extraction of inclusion bodies but can be useful cosolvents to enhance protein folding (see Table 1 in De Bernardez Clark, et al., 1999 and references cited therein).
3.Organic solutes
Examples of organic solutes include guanidine·HCl (used at 6 to 8 M) and urea (used at 6 to 9 M). The effectiveness of urea is modulated by pH and ionic strength, whereas that of guanidine is not. Both solutes are more effective at elevated temperatures (see additional discussion of temperature under point 6, below). Organic solutes are the most versatile and most commonly used denaturants for solubilizing inclusion bodies. The denaturing power of guanidinium salts increases in the order Cl– < Br– < I– < SCN-. Arginine (0.1- 1M) has long been used to facilitate refolding of recombinant proteins and at higher concentrations (0.5-2 M) can be effective in extracting proteins from inclusion bodies (Tsumoto et al., 2004).
Some Recombinant proteins may be extracted with lower concentrations of guanidinium salts as shown by Daumy et al., (1991). The primary benefit of this method is fewer cellar containments are extracted from the pellet. It is impossible to predict which proteins may be extracted this way. We have also observed some proteins which appear to form inclusion bodies by virtue of recovery in the low speed pellet fraction following cell breakage. These are in fact folded proteins “caught up” with cellular debris through weak associations with both protein and nonprotinacious components. This can be tested by small-scale exactions with, for example, urea (1-3M), gaunidinium-HCl (1-3M) and arginine (0.5-2M).
4.Detergents
The most commonly used protein-denaturing detergents are sodium dodecyl sulfate (SDS, an anionic detergent) and cetyltrimethylammonium bromide (CTAB, a cationic detergent). SDS is a very effective protein denaturant, as it binds directly to the protein; however, it should be used with caution to solubilize inclusion bodies as it can interfere with the recovery of folded protein in reasonable yields. Recently, membrane proteins extracted with SDS from inclusion bodies have been successfully folded (Schroder-Tittmann et al., 2010) and methods have been described for refolding SDS-denatured proteins using amphiphatic cosolvents (Michaux et al., 2007) and α-cyclodextrin (Otzen and Oliveberg, 2001). Solubilization with the related anionic detergent N-lauroylsarcosine (Sarkosyl) has also been used (Burgess, 1996). Sarkosyl, with a critical micelle concentration (CMC) of ~14.5 mM and micellar weight of ~600 has the advantage over SDS (CMC, ~8.5 mM; micellar weight, ~18,000) that it can be more easily removed, especially by dialysis. Proteins solubilized in either Sarkosyl or SDS can be purified by gel filtration in a manner analogous to Basic Protocol 2; they can also be chromatographed on hydroxylapatite (Rosenblum and Moss, 1972). A suitable matrix for this purpose is CHT ceramic hydroxylapatite supplied by Bio-Rad. It should be noted that His-tagged proteins solubilized with SDS or Sarkosyl do not bind very efficiently, if at all, to metal chelate affinity resins. There have been some noted successes using <10mM SDS (Schröder-Tittmann et al., 2010). Dilution of anionic detergent extract with an excess of nonionic detergent such as Triton X-100 (to form mixed micelles) may improve the binding characteristics. On the other hand, His-tagged protein solubilized with guanidine·HCl (6M) and urea (8M) bind very efficiently to metal chelate matrices.
5. Inorganic salts
Salts at high concentration (>1 M) often denature proteins. The denaturing power increases in the following order for anions: SO42– < CH3COO– < Cl– < Br– < ClO4– < SCN–; for cations, the order is (CH3)4N+, NH4+, K+, Na+ < Li+ < Ca2+ < Gdn+ (reviewed by von Hippel and Schleich, 1969). The organic guanidinium ion (Gdn) is included for comparative purposes. Salts have not been widely used for solubilizing inclusion bodies. On the other hand, they are frequently used for selective extraction of extrinsic membrane proteins and in principle should be useful for preextracting inclusion bodies.
6. Temperature
Thermally induced extraction is rarely used as it often results in irreversible protein denaturation (Zale and Klibanov, 1986). However, denaturants such as guanidine·HCl and urea are more effective at elevated temperatures (e.g., 37° to 60°C). Care must be taken when heating urea solutions because of the increased rate of cyanate formation, which will covalently modify amino groups on the protein, especially at pH >6.
A comparison of some protein denaturants and their relative effectiveness is described by Pace and Marshall (1980, and references cited therein). Monera et al. (1994) provide an explanation of why guanidine·HCl is usually two to three times per mole (2- to 3-fold) more effective than urea at unfolding proteins. The mechanism of protein denaturation is reagent-specific; this topic has been reviewed by Tanford (1968), Ghelis and Yon (1982), and Creighton (1993). It is noteworthy that despite over 40 years of use in protein folding experiments the mechanistic basis of urea and guanidine-HCl denaturation is still controversial (Lim, 2009).
Extraction conditions
The extraction conditions can be empirically determined on a small scale (<1 ml) by taking aliquots of washed pellet suspension and microcentrifuging. If preweighed microcentrifuge tubes are used, the extraction conditions can be related to a given wet weight of pellet. A fixed volume of extraction buffer is added plus a reducing agent (5 to 20 mM DTT). Initially, guanidine·HCl (4 to 8 M) or urea (6 to 9 M) should be tried. The pellets are resuspended by vortexing, then incubated for various times at different temperatures. (Suspension of the pellet is aided by brief sonication or by using a tissue homogenizer.) After equilibration, solutions are clarified either by filtration or by centrifugation. For the latter, a benchtop ultracentrifuges such as the Beckman Optima TLX and Airfuge CLS are ideal. The comparative effectiveness of extractants can determined by measuring the amount of protein solubilized from the washed pellets using standard protein estimation method (UNIT 3.4) and/or SDS-PAGE (UNIT 10.1).
It should be noted that solubilized protein must at some stage be folded into the native conformation; hence, the most effective solubilizing conditions might not necessarily be the best, especially if they cause irreversible denaturation. Irreversible denaturation appears to result from chemical modification of the protein and is induced by such factors as high temperature, extremes of pH, and tight binding of denaturants such as SDS.
The extraction process (Basic Protocol 1) should result in protein that is both monomeric (assumed to be unfolded) and contains cysteine residues in the reduced state. This provides a defined starting point from which to develop a reproducible folding protocol or from which to further purify the protein by, for example, gel filtration under denaturing conditions (Basic Protocol 2). Extraction of inclusion bodies with some denaturants, such as urea, may not completely convert the protein to monomers, resulting in physically heterogeneous mixtures. As pointed out in UNIT 6.1, extraction can be accomplished with strong denaturants (e.g., guanidine·HCl) that can then be exchanged by dialysis or gel filtration for weaker ones (e.g., urea). This particular solvent exchange often results in much better yields of folded protein compared to that obtained by the direct removal of guanidine·HCl (e.g., UNIT 6.5).
Gel-filtration chromatography (Basic Protocol 2) is not generally considered a high-resolution separation technique. However, as the recombinant-derived protein content of well-prepared washed pellets will be >50% of the total (see Fig. 6.3.1, lanes h and i), only a 2-fold purification is required to obtain pure protein.
Protein extracted with guanidine·HCl in the presence of reductant will ideally be in a random coil conformation with all sulfhydryl residues in the reduced state. Under such conditions, the order in which proteins elute from a gel-filtration matrix in guanidine·HCl can be directly correlated with molecular size (Mann and Fish, 1972).
Selection of the proper chromatography resin is critical for success (for detailed discussion, see Critical Parameters and Troubleshooting). The main disadvantage of gel filtration in guanidine·HCl, especially when using some of the (soft) agarose-based resins (e.g., Sepharoses and Bio-Gels), is that flow rates can be very slow due to the high viscosity of the gel-filtration buffer. Basic Protocol 2 uses a Superdex matrix that permits fast flow rates in the presence of high guanidine·HCl (or urea). A column size of 5 × 100 cm allows ~40 to 50 ml guanidine·HCl-containing extract to be processed. For smaller sample sizes, columns with proportionally smaller diameters are used.
Because the proteins separated by gel filtration in the presence of guanidine·HCl are unfolded, they have no biological activity, so the column fractions are usually assayed by SDS-PAGE. Direct addition of SDS to samples containing guanidine·HCl results in the formation of the insoluble guanidinium salt of dodecyl sulfate. The guanidine·HCl must therefore be removed before addition of the sample buffer used for SDS-PAGE analysis (UNIT 10.1). The approach used in Support Protocol takes advantage of the fact that guanidine·HCl is soluble in 90% ethanol whereas the protein is not. An alternative is to simply remove the guanidine·HCl by dialysis. Small samples (10 to 100 μl) can be dialyzed using one of various commercially available microdialyzing systems (e.g., the Microdialyzer System 100 from Pierce (Thermo Scientific) which allows simultaneous dialysis of up to 12 samples) or a simple homemade device that uses modified microcentrifuge tubes (Falson, 1992).
Critical Parameters and Troubleshooting
Breaking Cells and Preextracting Inclusion Bodies
We have used the French Press for cell breakage as it is still the best system and any laboratory performing routine cell breakage at the small-medium scale would be advised to invest in this equipment. (The French Press is also effective for breaking yeast cells etc., although two passes are usually required). Other methods for breaking bacterial cells are summarized in Unit 6.1
Once it has been established that the recombinant protein is insoluble (UNIT 6.1), extra care should be taken to ensure complete cell lysis. If unbroken cells are present in the low-speed pellets, they will leach contaminants when the pellets are extracted with strong protein denaturants. Proper operation of the French press is described in UNIT 6.2. Other major sources of contamination are outer membrane and cell wall material, most of which can be preextracted with detergents such as Triton X-100 (0.5% to 5%) and urea (1 to 4 M).
The proper concentration of urea in the wash buffer is determined empirically in small-scale extractions. It is probably safe to use 2 M urea for most aggregates. The effectiveness of the wash process at removing the cell wall and outer membrane material can be improved by increasing the volume of wash buffer (see Basic Protocol 1, step 6: use 10 ml instead of 6 ml buffer per gram cells). Treatment of the cells with lysozyme prior to homogenization with the French press will also help remove this material (UNIT 6.5, Basic Protocols 1 and 2).
Although insoluble proteins are less susceptible than native proteins to proteolysis, inclusion of EDTA and a serine protease inhibitor such as benzamidine·HCl or AEBSF is recommended (many manufactures conveniently sell protease inhibitor cocktails) The choice of cell lysis buffer is not critical; however, the buffer pH should be >6.5, as many soluble E. coli proteins precipitate at slightly acidic pH values.
After preextraction, wash the aggregates with buffer alone to remove excess detergent. When attempting to develop a reproducible folding protocol, note that varying amounts of detergent carryover may influence the outcome. In fact, nonionic detergents can be used as cosolvents to aid folding (UNIT 6.1); even so, it is important to know how much detergent is present.
Extracting Inclusion Bodies with Guanidine·HCl
The washed pellets containing the insoluble inclusion body proteins are extracted with guanidine·HCl (see Basic Protocol, step 1). To obtain complete dissolution of the pellets, some form of mechanical dispersion is often required. Homogenization at room temperature with a tissue grinder (as described) is often adequate; however, sonication can be used if the pellet is especially recalcitrant to dissolution. Heating the solution will also aid protein solubilization; 10 to 15 min at 50° to 60°C is usually a good starting point. Excessive heating, whether direct or caused by sonication without adequate cooling, should be avoided. It is worth remembering that if extraction involves extremes of heat and or pH, such conditions will favor deamidation of Asn and may also promote cleavage at the labile Asp-Pro bond and other chemical modifications of the protein (Zale and Klibanov, 1986; Daniel et al., 1986).
It is best to use the extract directly after preparation rather than storing it frozen. In general, an unfolded protein in solution is much more susceptible to proteolytic degradation and chemical modification than its native (folded) counterpart.
Polyhistidine-tagged proteins
Polyhistidine-tagged fusion proteins that form inclusion bodies can be extracted with guanidine·HCl and purified directly by nickel-chelate affinity chromatography. Extraction of the inclusion pellet is the same procedure described in Basic Protocol 1, with the only change being that DTT is omitted. DTT reacts with nickel to form a black precipitate and will ruin the affinity resin. If a reductant is required, then 2 mM β-mercaptoethanol (2-ME) can be used or 0.1 – 2 mM TCEP. A buffer of 100 mM sodium phosphate/10 mM Tris·Cl, pH 8.0, or 50 mM HEPES, pH 7.5, should be used for the extraction with guanidine·HCl. After extraction of the protein, the solution is diluted with buffer to final guanidine·HCl concentration of 4 M to 6 M. The extract is loaded onto the nickel chelation column equilibrated with buffer containing guanidine·HCl and washed with column buffer until no UV absorbance is observed in the elute. The protein may be eluted from the column by either lowering the pH (gradient: pH 8.0 to 3.0) or by imidazole (gradient: 0 to 300 mM). Further details can be found elsewhere especially from manufacturers, for example, Qiagen (which publishes QIAexpressionist, a handbook for expression and purification of 6xHis-tagged proteins: http://www.qiagen.com). Many of the manufacturers list protocols with a batch elution of the his-tagged proteins. A Gradient will yield cleaner results as any protein containing an exposed histidine pair, such as for example, the E.coli OmpL and OmpW outer membrane proteins, or proteins containing a Zinc-finger, as reported by Voráčková et al., (2011) will bind to the Ni-resin. These contaminating proteins will not bind as tightly as a 6x his-tagged protein, but may not be always be removed by wash steps and thus co-elute in a batch elution. The affinity step can be used for purification instead of gel filtration or in combination with it. Gel filtration is often used after the affinity column in order to exchange solvents and perform further purification.
Selecting Medium and Column for Gel Filtration in Guanidine·HCl
Selecting the proper gel-filtration resin is one of the most critical steps. First of all, the chromatography medium must have high chemical stability that permits prolonged exposure to high concentrations of denaturing solvents such as guanidine·HCl and urea. It must also have high physical strength that allows high flow rates with viscous solvents. Suitable media are listed in Table 6.3.1. The Sepharoses and Bio-Gels are cross-linked agaroses, Sephacryl is dextran cross-linked with bisacrylamide, and Superdex is cross-linked agarose with bonded dextran. All the matrices listed are stable in high concentrations of urea and guanidine·HCl. Details on the properties of various media can be obtained from the manufacturers.
Table 6.3.1.
Gel-Filtration Matrices Suitable for Use with Solutions Containing Guanidine Hydrochloride
| Matrixa | Mass range (kDa) |
Reference | |
|---|---|---|---|
| Native proteinsb | Unfolded proteins | ||
| Sepharose CL-6B | 10-4,000 | 1-80 | Mann and Fish (1972) |
| Bio-Gel A-5m | 10-5,000 | 1-80 | Mann and Fish (1972) |
| Sepharose CL-4B | 60-20,000 | 10-300 | Mann and Fish (1972) |
| Sephacryl S-100 HR | 1-100 | <1-30c | — |
| Sephacryl S-200 HR | 5-250 | 1-50 | Belew et al. (1978) |
| Sephacryl S-300 HR | 10-1,500 | 1-100c | — |
| Sephacryl S-400 HR | 20-8,000 | 1->100c | — |
| Superdex 75 | 3-70 | <1-25 | I.P. and P.T.W. (unpub. observ.) |
| Superdex 200 | 10-600 | 1-80 | I.P. and P.T.W. (unpub. observ.) |
All resins are from GE Healthcare Life Sciences except Bio-Gel A-5m, which is from Bio-Rad. The Sepharose and Bio-Gel matrices are normally run under low pressure; all other resins can be run under low or medium pressure. Medium pressure is achieved using one of the chromatography pumps indicated in Basic Protocol 2; the pumps are normally included in the GE Healthcare Life Sciences ÄKTA-FPLC or ÄKTAexplorer systems.
Data on the fractionation range in the unfolded state refer to proteins unfolded with guanidineHCl; however, the guidelines also apply to proteins unfolded and eluted with urea (assuming they are random coils).
Estimates based on fractionation range for native proteins.
Second, selecting a medium with the proper pore size is important for good separation. Manufacturers provide information about the separation ranges of their products. These ranges are determined for proteins in an aqueous buffer. In guanidine·HCl, denatured proteins (assuming they are random coils) will have a radius 3- to 4-fold greater than an average folded globular protein of the same mass. Empirical relationships between the measured hydrodynamic radius (Rh) and the number of residues in the polypeptide chain (N) have been established: for native folded proteins Rh = 4.75N0.29 Å and for highly denatured states Rh = 2.21N0.57 Å (Wilkins et al, 1999). Thus, a protein of 200 residues will have a folded radius of 7.3 Å and an unfolded one of 32.2 Å: a 4- fold increase in radius. Thus, the apparent mass range values for unfolded proteins in Table 6.3.1 should be used rather than the native protein values.
Third, the pressure rating of the column must be adequate. Because 4 M guanidine HCl has a higher viscosity than most aqueous buffers, higher pressures are needed to achieve the same flow rates. The column should be capable of safely withstanding moderate pressures of around 0.5 MPa (5 bar) or ~75 psi (the XK columns are rated to 0.5 MPa). The proteins separate as they pass through the column, so column length is important. A column 60 to 100 cm in length will generally be suitable. Some manufacturers offer prepacked columns with a choice of media. If column packing is to be done in the laboratory, then selecting a column with a packing reservoir is helpful.
Prepacked columns (diameters 1.6, 2.6, 3.5, and 6.0 cm and length 60 cm) prepacked with Sephacryl resin (Sephacryl S-100, S-200, or S-300) or Superdex resin (Superdex 200 and 75) are commercially available from GE Healthcare Life Sciences. Superdex 200 and 75 PG (preparative grade) 60/600 columns (6.0 cm i.d. × 60 cm length) are used with the ÄKTAexplorer system (GE Healthcare Life Sciences) either individually or sequentially to obtain, in many cases, pure protein (for example, see Fig. 6.3.1).
Performing Gel Filtration in Guanidine·HCl
The general guidelines and critical parameters that apply to gel-filtration chromatography in general and are detailed elsewhere (UNIT 8.3) and also apply here. The main distinction from typical gel filtration is the use of a highly viscous solvent, which requires robust matrices to obtain reasonable flow rates. On the other hand, it should be noted that the presence of high concentrations of guanidine·HCl yields higher-resolution separations than those obtained with normal solvents because the high viscosity of the solvent reduces diffusion, which otherwise tends to broaden elution peaks.
It is important to ensure that all connections and tubing are leak-free. Concentrated guanidine·HCl is highly corrosive to metal and electronic components.
The sample derived from extraction of highly aggregated protein must be clarified before being applied to the column. Furthermore, to prevent the sample from precipitating near or at the top of the column, it should be equilibrated to the same temperature as the column matrix and buffer. This is especially important if chromatography is carried out at 4°C and the sample was heated during extraction.
The column must be equilibrated with freshly made buffer before use. The column buffer contains the sulfhydryl reagent DTT, which will oxidize over time, thus lessening its effectiveness and increasing the background UV absorbance of the buffer (Iyer and Klee, 1973).
If the recombinant protein has been effectively solubilized, it will be physically homogeneous and its elution position will be proportional to its mass. If the protein elutes anomalously (i.e., is located in more than one peak), there are several possible reasons.
1. Incomplete protein solubilization
Try more drastic extraction conditions—in particular, increase the denaturant concentration and temperature during extraction. Use a stronger chemical denaturant, especially if urea was originally used.
2. Aggregation/precipitation of protein(s) in the column
Ensure that the concentration of guanidine·HCl in the column buffer will maintain the solubility of all proteins during chromatography. The column can be run at room temperature (as opposed to 4°C), and reasonable flow rates can still be obtained with buffers containing 6 to 7 M guanidine·HCl (4 M is used in Basic Protocol 2).
3. Conformational heterogeneity
Related to the above two situations is the case where the protein, although monomeric, contains both unfolded and partially folded species. The folded protein will elute later (appear smaller) than the unfolded protein. Conformational heterogeneity is usually not a major problem in high (>4 M) guanidine·HCl concentrations.
4. Oxidation of sulfhydryl residues
The formation of intermolecular disulfide bonds is indicated by the appearance of protein eluting earlier than expected and corresponds to the formation of multimers (dimers, trimers, etc.). Disulfide interchange occurs more readily at high pH; thus, use of a slightly acidic buffer (pH 6.0 to 5.0) and inclusion of fresh reductant in both the sample and column buffer are recommended. For analytical separations, cysteine residues are normally capped by alkylation with iodoacetamide; cysteines can also be reversibility modified by S-sulfonation (Cowley and MacKin, 1997).
5. Proteolysis
(a) The sample contains partially proteolyzed protein, with proteolysis by E. coli proteases occurring before or after cell breakage: In fully denatured protein, it should be possible to separate clipped protein from unmodified protein if there is sufficient mass difference. Once the protein has been separated by gel filtration, it should be checked by mass spectrometry to confirm its integrity. If the protein is not fully denatured, clipped protein may have similar elution properties as unmodified protein; however, the processing will be revealed when the protein is boiled in SDS and analyzed by gel electrophoresis.
(b) Where the recombinant protein is itself a protease: As pointed out by Fish et al. (1969), “Since denaturation is a kinetic process, the dissolution of proteases in guanidine·HCl might result in a condition where the fraction of the protein not denatured at a given instance may digest the unfolded protein leading to low molecular weight estimate.” The converse of this situation is also true: if proteases are extracted under conditions where they are fully denatured and, thus, inactive (e.g., 8 M guanidine·HCl), changes in the solvent conditions (e.g., reducing the guanidine·HCl concentration or exchanging for urea) might lead to a fraction of the protein folding into an active conformation. If autolytic processing is suspected, include an appropriate inhibitor or choose a solvent pH where the enzyme has minimal activity.
Anticipated Results
Cell Lysis and Preparation of Washed Pellets
Pelleted aggregates after washing contain ~30% dry weight, of which 90% is protein. SDS-PAGE of a typical washed pellet preparation (Fig. 6.3.1, lanes h and i) indicates that recombinant bovine growth hormone (21 kDa) makes up >60% of the total protein. The washed pellets analyzed in Figure 6.3.1 are typical starting materials for the protein folding and purification described in UNIT 6.5 (Basic Protocol 1). The multiple bands in the background are either derived from unbroken cells (most likely) or are E. coli cytoplasmic proteins coprecipitated or trapped during aggregate formation. The gel analysis indicates the presence of another plasmid-encoded protein, namely, kanamycin phosphotransferase (30.8 kDa), a product of the gene conferring resistance to the antibiotic kanamycin (Kane and Hartley, 1991). The washing procedure described only partially extracts the phosphotransferase but removes most of the outer membrane proteins. For further details, see the legend to Figure 6.3.1.
Gel Filtration in Guanidine·HCl
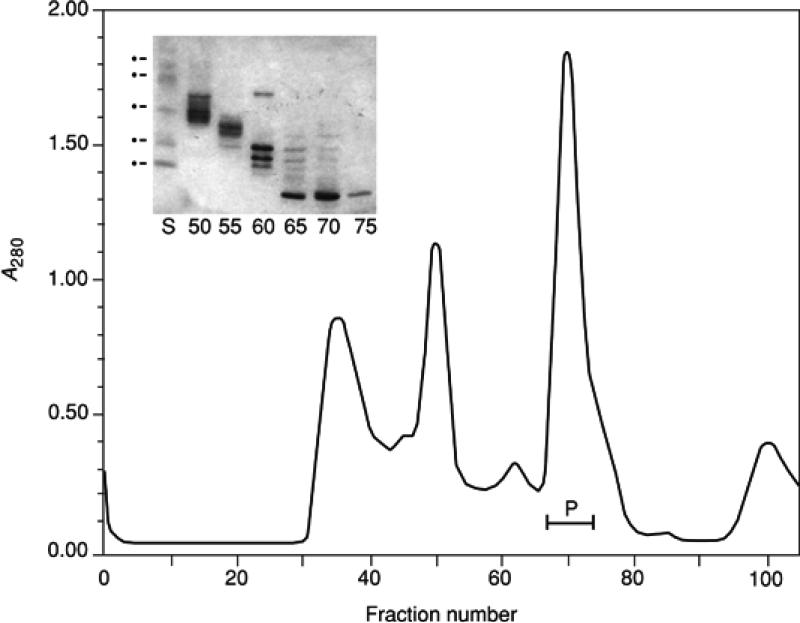
Fractionation of human immunodeficiency virus type 1 (HIV-1) protease illustrates expected results from gel-filtration chromatography. The protein is expressed in E. coli as a 17-kDa precursor that at some stage undergoes autolytic processing to form the mature-sized protease (10.7 kDa). The protease and undigested precursor accumulate as insoluble inclusions in the E. coli cytoplasm. Washed pellets prepared and extracted with 6 M guanidine·HCl as described (see Basic Protocol 1) were applied to a Superdex 200 column. The protease eluted in a single peak (Fig. 6.3.2, fractions 66 to 72), well separated from unprocessed protein (e.g., fraction 60) and larger molecular mass proteins (e.g., fractions 50 to 55). The fractions indicated in Figure 6.3.2 by P were pooled and the protein further purified by repeat gel-filtration chromatography on a Superdex 75 column under the same conditions. The protein at this stage is >95% pure and after solvent exchange is folded into active protein. (As it happens, the native protease is a 20-kDA homodimer that is very susceptible to autolytic digestion; hence, purification in the denatured, inactive state is highly advantageous).
Figure 6.3.2.
Gel filtration of an extract containing HIV-1 protease, using Superdex 200 in 4 M guanidine·HCl. Column dimensions, 6 × 60 cm; buffer, 50 mM Tris·Cl (pH 7.5)/4 mM guanidine·Cl/2 mM DTT; flow rate, 5 ml/min (300 ml/hr). The sample has a mass of 10 kDa. Protein fractions 66 to 72 (pool P) was further purified under the same conditions using a Superdex 75 matrix. The inset shows SDS-PAGE analysis of selected fractions. The protein standard markers (lane S) correspond to mass values of 66.2, 45, 30, 21.5, and 14.4 kDa, respectively (migration order top to bottom).
In the inset of Figure 6.3.2, analysis by SDS-PAGE of fraction 60 illustrates incomplete dissociation of the sample, a problem commonly observed when electrophoresing samples precipitated with ethanol and trichloroacetic acid (TCA). If necessary, repeat the analysis using SDS sample buffer (UNIT 10.1; see Support Protocol).
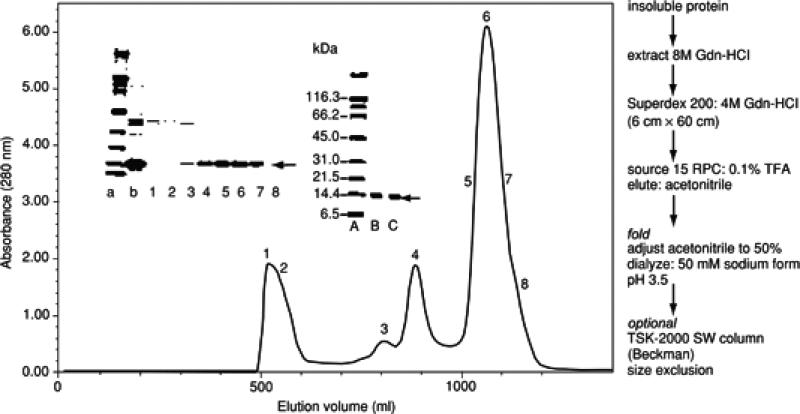
In another example, the extracellular domain of the SIV gp41 (residues 27 to 149; molecular weight ~14,000) was extracted from inclusion bodies and chromatographed on Superdex 200 (Figure 6.3.3). Analysis of the highly purified protein in the main peak (apex labeled 6) by analytical ultracentrifugation and circular dichroism indicated that it was monomeric and unfolded. Guanidine·HCl was removed from the protein in this peak by reversed-phase chromatography as described below, and the protein folded by dialysis (Wingfield et al., 1997).
Figure 6.3.3.
Superdex 200 chromatography in guanidine·HCl of SIV gp4127-149. SDS-PAGE of the numbered fractions is shown in the first inset (upper left); lane “a” contains molecular weight standards (bottom to top: 6.5, 14.4, 21.5, 31, 45, 66.2 kDa), and the purified protein migrates close to the 14.4 kDa standard. Lane “b” represents starting material loaded to column corresponding to guanidine·HCl-extracted inclusion bodies. Protein in the main peak (fractions 5 to 7) marked with arrow was used for protein folding after removal of guanidine·HCl by reversed-phase chromatography. The second inset (upper middle) refers to protein expression in minimal medium; lane A contains the same molecular weight standards as lane “a” in the first inset; lanes B and C correspond to insoluble protein and purified protein, respectively. Protein is labeled with 15N and 13C for NMR analysis. A summary of the protein purification is indicated in the right-hand part of the figure. Adapted from Wingfield et al. (1997).
The fractionation of HIV-1 protease is somewhat of a best-case scenario due to the small size of the protein. Usually it is not possible to purify the recombinant protein completely, especially for proteins close in size to the main E. coli contaminants (30 to 45 kDa). In general, small proteins (<25 kDa) are best suited for the method; however, any protein will be purified to some extent, and further purification can be attempted using some of the methods discussed in UNIT 6.1. Moreover, partial purification may be all that is required to enhance protein folding significantly.
Recovery of recombinant protein from gel-filtration chromatography is close to quantitative, and the purified (or partially purified) protein can be folded or can be stored in aliquots at –80°C. On thawing the protein, add fresh DTT (an additional 1 or 2 mM) and warm the solution at 37° to 40°C for several minutes before use. The protein concentration can be conveniently estimated by UV spectroscopy (UNIT 3.1).
Removal of the Guanidine·HCl by Reversed-Phase Chromatography
A useful method for removing the guanidine·HCl prior to folding proteins is reversed-phase chromatography (also see UNIT 11.6). An HPLC system such as the Beckman Coulter system Gold can used for this procedure. A reversed-phase column using a polystyrene/divinylbenzene matrix (Source 15RPC from GE Healthcare Life Sciences) works well for most proteins. The binding capacity of the matrix is ~10 to 30 mg protein per ml of matrix. The solutions used for gradient elution are 0.1% TFA in water (solvent A) and 0.1% TFA in 100% acetonitrile (solvent B). High-quality chromatography-grade reagents are essential for this procedure. The protein, in guanidine·HCl, is loaded onto the column by means of an injection loop. The column should be washed with at least 2 to 5 column volumes of 0.1% TFA. The protein is then eluted with a 10-column-volume gradient of solvent A to B. The optimization of the gradient profile for a given protein is determined empirically. The eluted protein may be refolded directly from the reversed phase TFA/acetonitrile solution or can be lyophilized for storage and refolding at a future time.
Time Considerations
Cell Lysis and Preparation of Washed Pellets
French press lysis of ~150 to 200 ml cell suspension will take ~30 to 35 min. Preparation and extraction of the washed pellet take about half a day.
Gel Filtration in Guanidine·HCl
Using Superdex resins in conjunction with an ÄKTA-FPLC or ÄKTA explorer, gel-filtration chromatography takes 3 to 6 hr and the gel analysis 1.5 to 2 hr. With low-pressure resins, elution takes much longer. For example, a Sephacryl S-300 column (2.5 × 98 cm) eluted with 6 M guanidine·HCl runs at ~25 to 30 ml/hr. An average-sized protein (15 to 30 kDa) is eluted after 10 to 12 hr. A column can be loaded in late afternoon and conveniently run overnight.
Final Comment
In a joint publication of various structural genomics consortia (Nature Methods. 2008 5: 135-146: http://www.nature.com/naturemethods) strategies for the expression and purification of recombinant proteins are summarized and recommendations are made based on the consensus findings of the many groups. This paper is well worth reading and has plenty of useful guidelines, however; they badly overstate the problems of “rescuing” insoluble inclusion bodies as folded purified proteins. The aim of these consortia is the mass production of proteins for structural analysis and you win some and lose some. This is in contrast to individual groups were much effort can be focused on a particular protein including working out a simple extraction and folding scheme as many others have over the last 25 years or so.
Literature Cited
- Anfinson CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Belew M, Fohlman J, Janson J-C. Gel filtration on Sephacryl S-200 superfine in 6 M guanidine·HCl. FEBS Lett. 1978;91:302–304. doi: 10.1016/0014-5793(78)81197-1. [DOI] [PubMed] [Google Scholar]
- Burgess RR. Purification of overproduced E.coli RNA polymerase sigma factors by solubizing inclusion bodies and refolding from Sarkosyl. Meth. Enzymol. 1996;273:144–145. doi: 10.1016/s0076-6879(96)73014-8. [DOI] [PubMed] [Google Scholar]
- Cowley DJ, Mackin RB. Expression, purification and characterization of recombinant human proinsulin. FEBS Letters. 1997;402:124–130. doi: 10.1016/s0014-5793(96)01511-6. [DOI] [PubMed] [Google Scholar]
- Creighton TE. Proteins: Structures and Molecular Properties. (2nd ed.) 1993:293–296. [Google Scholar]
- Daniel RM, Dines M, Petach The denaturation and degradation of stable enzymes at high temperature. Biochem J. 1996;317:1–11. doi: 10.1042/bj3170001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby NJ, Creighton TE. Folding proteins. Nature. 1990;344:715–716. doi: 10.1038/344715b0. [DOI] [PubMed] [Google Scholar]
- Daumy GO, Merenda JM, McColl AS, Andrews GC, Franke AE, Geoghegan KF, Otterness IG. Isolation and characterization of biologically active murine interleukin-1α derived from expression of a synthetic gene in Escherichia coli. Biochimica et Biophysica Acta (BBA) - Protein Structure and Molecular Enzymology. 1989;998:32–42. doi: 10.1016/0167-4838(89)90115-5. [DOI] [PubMed] [Google Scholar]
- De Bernardez Clark E, Schwartz E, Rudolph R. Inhibition of aggregation side reactions during in vitro protein folding. Methods in Enzymology. 1999;309:217–236. doi: 10.1016/s0076-6879(99)09017-5. [DOI] [PubMed] [Google Scholar]
- Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr. Opin. Biotechnol. 2006;17:353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- Falson P. An efficient procedure to dialyze volumes in the range of 10-200 microliters. BioTechniques. 1992;13:20. [PubMed] [Google Scholar]
- Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. W. H. Freeman; New York: 1998. [Google Scholar]
- Fink AL. Protein aggregation: Folding aggregates, inclusion bodies and amyloid. Folding and Design. 1998;3:R9–R23. doi: 10.1016/S1359-0278(98)00002-9. [DOI] [PubMed] [Google Scholar]
- Fish WW, Mann KG, Tanford C. The estimation of polypeptide chain molecular weights by gel filtration in 6 M guanidine hydrochloride. J. Biol. Chem. 1969;244:4989–4994. [PubMed] [Google Scholar]
- García-Fruitós E, Vázquez E, Díez-Gil C, Corchero JL, Seras-Franzoso J, Ratera I, Veciana J, Villaverde A. Bacterial inclusion bodies: making gold from waste. Trends in Biotechnology. 2012;30:65–70. doi: 10.1016/j.tibtech.2011.09.003. [DOI] [PubMed] [Google Scholar]
- Georgiou G, Valax P. Isolating inclusion bodies from bacteria. Meth. Enzymol. 1999;309:48–58. doi: 10.1016/s0076-6879(99)09005-9. [DOI] [PubMed] [Google Scholar]
- Ghelis C, Yon Y. Simulation of protein folding: Studies of in-vitro denaturation-renaturation. In: Horecker B, editor. Protein Folding. Academic Press; San Diego: 1982. pp. 225–243. [Google Scholar]
- Iyer KS, Klee W. Direct spectroscopic measurement of the rate of reduction of disulfide bonds J. Biol. Chem. 1973;248:707–710. [PubMed] [Google Scholar]
- Kane JK, Hartley DL. Properties of recombinant protein-containing inclusion bodies in E. coli. In: Seetharam R, Sharma SK, editors. Purification and Analysis of Recombinant Proteins. Marcel Dekker; New York: 1991. pp. 121–145. [PubMed] [Google Scholar]
- Langley KE, Berg TF, Strickland TW, Fenton DM, Boone TC, Wypych J. Recombinant-DNA-derived growth hormone from Escherichia coli: Demonstration that the hormone is expressed in the reduced form, and isolation of the hormone in the oxidized, native form. Eur. J. Biochem. 1987;163:313–321. doi: 10.1111/j.1432-1033.1987.tb10802.x. [DOI] [PubMed] [Google Scholar]
- Lim WK, Rösgen J, Englander SW. Urea, but not guanidinium, destabilizes proteins by forming hydrogen bonds to the peptide group. PNAS. 2009;106:2595–2600. doi: 10.1073/pnas.0812588106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maachupalli-Reddy J, Kelley BD, De Bernardez Clark E. Effect of inclusion body contaminants on the oxidative renaturation of hen white lysozyme. Biotech. Prog. 1997;13:144–150. doi: 10.1021/bp970008l. [DOI] [PubMed] [Google Scholar]
- Mann KG, Fish WW. Protein polypeptide chain molecular weights by gel chromatography in guanidinium chloride. Methods Enzymol. 1972;26:28–42. doi: 10.1016/s0076-6879(72)26004-9. [DOI] [PubMed] [Google Scholar]
- Michaux C, Pomroy NC, Prive GG. Refolding SDS-denatured proteins by the addition of amphipathic cosolvents. J.Mol.Biol. 2007;375:1477–1488. doi: 10.1016/j.jmb.2007.11.026. [DOI] [PubMed] [Google Scholar]
- Monera OD, Kay CM, Hodges RS. Protein denaturation with guanidine hydrochloride provides a different estimate of stability depending on the contributions of electrostatic interactions. Protein Sci. 1994;3:1984–1991. doi: 10.1002/pro.5560031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberg K, Chrunyk BA, Wetzel R, Fink AL. Nativelike secondary structure in interleukin-1β inclusion bodies by attenuated total reflectance FTIR. Biochemistry. 1994;33:2628–2634. doi: 10.1021/bi00175a035. [DOI] [PubMed] [Google Scholar]
- Otzen DE, Oliveberg M. A simple way to measure protein refolding in water. J.Mol.Biol. 2001;313:479–483. doi: 10.1006/jmbi.2001.5039. [DOI] [PubMed] [Google Scholar]
- Pace CN, Marshall HF. A comparison of protein denaturants for β-lactoglobulin and ribonuclease. Arch. Biochem. Biophys. 1980;199:270–276. doi: 10.1016/0003-9861(80)90281-7. [DOI] [PubMed] [Google Scholar]
- Pain RH, editor. Mechanisms of Protein Folding. 2nd ed. Oxford University Press; 2001. [Google Scholar]
- Pepinsky BR. Selective precipitation of proteins from guanidine hydrochloride-containing solutions with ethanol. Anal. Biochem. 1991;195:177–181. doi: 10.1016/0003-2697(91)90315-k. [DOI] [PubMed] [Google Scholar]
- Rosenblum EN, Moss B. Hydroylapaptite chromatography of protein-sodium dodecyl sulfate complexes. J. Biol. Chem. 1972;247:5194–5198. [PubMed] [Google Scholar]
- Schröder-Tittmann K, Bosse-Doenecke E, Reedtz-Runge S, Ihling C, Sinz A, Tittmann K, Rudolph R. Recombinant expression, in vitro refolding, and biophysical characterization of the human glucagon-like peptide-1 receptor. Biochemistry. 2010;49:7956–7965. doi: 10.1021/bi101159s. [DOI] [PubMed] [Google Scholar]
- Seckler R, Jaenicke R. Protein folding and protein refolding. FASEB J. 1992;6:2545–2552. doi: 10.1096/fasebj.6.8.1592207. [DOI] [PubMed] [Google Scholar]
- Speed MA, Wang DIC, King J. Specific aggregation of partially folded polypeptide chains: The molecular basis of inclusion body composition. Nature Biotech. 1996;14:1283–1287. doi: 10.1038/nbt1096-1283. [DOI] [PubMed] [Google Scholar]
- Tanford C. Protein denaturation. Adv. Prot. Chem. 1968;23:122–275. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- Tsumoto K, Ejima D, Kumagai I, Arakawac T. Practical considerations in refolding proteins from inclusion bodies. Protein Expression and Purification. 2003;28:1–8. doi: 10.1016/s1046-5928(02)00641-1. [DOI] [PubMed] [Google Scholar]
- Tsumoto K, Umetsu M, Kumagai I, Ejima D, Philo JS, Arakawa T. Role of Arginine in Protein Refolding, Solubilization, and Purification. Biotechnol. Prog. 2004;20:1301–1308. doi: 10.1021/bp0498793. [DOI] [PubMed] [Google Scholar]
- von Hippel PH, Schleich T. Ion effects on the solution structure of biological macromolecules. Acc. Chem. Res. 1969;2:257–265. [Google Scholar]
- Voráčková I, Suchanová S, Ulbrich P, Diehl WE, Ruml T. Purification of proteins containing zinc finger domains using immobilized metal ion affinity chromatography. Protein Expression and Purification. 2011;79:88–95. doi: 10.1016/j.pep.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins DK, Grimshaw SB, Receveur V, Dobson CA, Jones JA, Smith LJ. Hydrodynamic Radii of Native and Denatured Proteins Measured by Pulse Field Gradient NMR Techniques. Biochemistry. 1999;38:16424–16431. doi: 10.1021/bi991765q. [DOI] [PubMed] [Google Scholar]
- Wingfield PT, Stahl SJ, Kaufman J, Zlotnick A, Hyde CC, Gronenborn AM, Clore GM. The extracellular domain of immunodeficiency virus gp41 protein: expression in Escherichia coli, purification and crystallization. Protein Sci. 1997;6:1653–1660. doi: 10.1002/pro.5560060806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zale SE, Klibanov M. Why does ribonuclease irreversibly inactivate at high temperature? J. Biol. Chem. 1986;25:5432–5444. doi: 10.1021/bi00367a014. [DOI] [PubMed] [Google Scholar]