Abstract
Following V(D)J cleavage, the newly liberated DNA signal ends can be either fused together into a signal joint or used as donor DNA in RAG-mediated transposition. We find that both V(D)J cleavage and release of flanking coding DNA occur before the target capture step of transposition can proceed; no coding DNA is ever detected in the target capture complex. Separately from its role in V(D)J cleavage, the DDE motif of the RAG1/2 active site is specifically required for target DNA capture. The requirement for cleavage and release of coding DNA prior to either physical target binding or functional target commitment suggests that the RAG1/2 transposase contains a single binding site for non-RSS DNA that can accommodate either target DNA or coding DNA, but not both together. Perhaps the presence of coding DNA may aid in preventing transpositional resolution of V(D)J recombination intermediates.
Keywords: pathway, RAG, V(D)J recombination, target capture, transposition
Introduction
During lymphoid development, a diverse array of functional immunoglobulin and T-cell receptor genes are combinatorially assembled from multiple, nonconsecutive gene segments in a process known as V(D)J recombination (Bassing et al, 2002; Gellert, 2002). Each of these gene segments is flanked by a recombination signal sequence (RSS) that consists of conserved heptamer and nonamer sequences separated by a spacer region of either 12 or 23 bp. Efficient recombination between gene segments only occurs when one segment is flanked by a 12-RSS and the other is flanked by a 23-RSS, a restriction termed the 12/23 rule (Tonegawa, 1983).
Biochemically, V(D)J recombination is a process of regulated DNA double-strand cleavage, followed by double-strand break repair. The generation of these double-strand breaks is catalyzed by the lymphoid-specific proteins RAG1 and RAG2, which together comprise the V(D)J recombinase. During the cleavage phase of the reaction, the RAG proteins first bind to both a 12-RSS and a 23-RSS, bringing these two RSSs together to form a synaptic paired complex (PC) (Hiom and Gellert, 1998). DNA double-strand breaks are generated within this PC via two phosphoryl transfer reactions. The RAG proteins first nick the top strand of each RSS, just 5′ of the heptamer sequence. These newly liberated 3′ hydroxyl groups on the top strand of the coding flank then attack the bottom strand via a direct transesterification, thereby generating the cleavage reaction products: two hairpinned coding ends and two blunt 5′-phosphoryl, 3′-hydroxyl signal ends (McBlane et al, 1995). PC formation requires a divalent metal ion (Ca2+, Mg2+, or Mn2+) (Hiom and Gellert, 1998). While all three of these divalent metal ions will support the initial step of RSS binding, only Mg2+ and Mn2+ will support the subsequent phosphoryl transfer steps of RSS cleavage. In the repair phase of the reaction, ubiquitous nonhomologous end-joining (NHEJ) factors such as Ku70/80, DNA-PKcs, XRCC4, DNA ligase IV, and Artemis collaborate to rejoin the cleavage reaction products to generate imprecise coding joints and precise signal joints (Bassing et al, 2002; Gellert, 2002; Schlissel, 2002).
Although the NHEJ factors repair the vast majority of RAG-generated double-strand breaks, the RAG proteins themselves can alter the postcleavage fate of signal ends by transposing these blunt, double-stranded DNA molecules into an unrelated piece of DNA (Agrawal et al, 1998; Hiom et al, 1998; Melek and Gellert, 2000). In this reaction, the RAG proteins catalyze another phosphoryl transfer reaction, enabling the exposed, nucleophilic 3′ hydroxyl group on the bottom strand of the signal end to attack a target DNA molecule. Both single-and double-ended insertion products can be generated depending on whether one or both signal ends attack the target. Ca2+, Mg2+, and Mn2+ can all support this postcleavage phosphoryl transfer reaction. RAG-mediated transposition has been observed in vitro with either core (Agrawal et al, 1998; Hiom et al, 1998) or full-length RAG proteins (Elkin et al, 2003), demonstrating that the proteins themselves are capable of catalyzing the chemical steps of transposition. Furthermore, RAG-mediated transposition can occur in yeast (Clatworthy et al, 2003), and rare events of RAG-mediated DNA transposition in mammalian cells have recently been reported (Messier et al, 2003).
RAG-catalyzed transposition represents an alternative postcleavage fate for the double-strand breaks generated during V(D)J recombination. By competing with the NHEJ pathway of double-strand break repair in vivo, transposition could not only cause insertional mutagenesis (Messier et al, 2003), but could also lead to genomic instability and the generation of potentially oncogenic chromosomal translocations (Hiom et al, 1998; Melek and Gellert, 2000). Furthermore, RAG-catalyzed transposition may have generated the Ig and TCR loci, thereby driving the evolution of the vertebrate adaptive immune system (Sakano et al, 1979; Oettinger et al, 1990; Thompson, 1995). For these reasons, it is important to understand the mechanism and regulation of RAG-catalyzed transposition.
As with all transposition reactions, certain events must take place during the course of RAG-mediated transposition. The donor DNA must be recognized and bound by the transposase (synaptic complex formation), the donor DNA must be cleaved and excised from the host genome (donor cleavage), a target DNA molecule must be bound by the transposase (target capture), the donor DNA must insert itself into the target DNA (strand transfer), and the resulting branched DNA molecules must be resolved. While the synaptic complex formation and cleavage steps of RAG-catalyzed transposition are rather well understood, the mechanism of target capture remains unclear.
Before strand transfer can occur, the RAG proteins must stably bind a target DNA, thereby generating a target capture complex (TCC), which consists of the RAG proteins, the donor strand RSSs, and the unrelated target DNA. However, it is not known whether the RAG1/2 transposase contains two distinct DNA-binding pockets—one for donor DNA and one for target DNA—or one DNA-binding pocket that accommodates both donor DNA and target DNA. Similarly, it is not clear at what stage of the transposition reaction the RAG proteins capture a target DNA molecule.
In theory, target capture could occur either prior to donor cleavage, in the presence of uncleaved coding DNA (‘uncleaved TCC'; Figure 1A, left panel), subsequent to cleavage but prior to the release of the hairpinned coding ends (‘hairpin TCC'; Figure 1A, center panel), or subsequent to both cleavage and release of the hairpinned coding ends (‘signal end TCC'; Figure 1A, right panel). If the RAG1/2 transposase contains a single DNA-binding pocket that accommodates both donor DNA and target DNA—as has been suggested for Tn10 (Sakai and Kleckner, 1997)—then one might expect this binding pocket to consist of two regions, one that accommodates the RSS and the other that accommodates non-RSS DNA. According to this model, the region that accommodates non-RSS DNA could either bind coding DNA or target DNA. If this were the case, one would not expect target capture to occur until the donor DNA has been cleaved and the resulting hairpinned coding ends have been released. If, however, the RAG1/2 transposase contains a separate DNA-binding pocket for target DNA, then one might expect target capture to occur either before or after donor cleavage.
Figure 1.
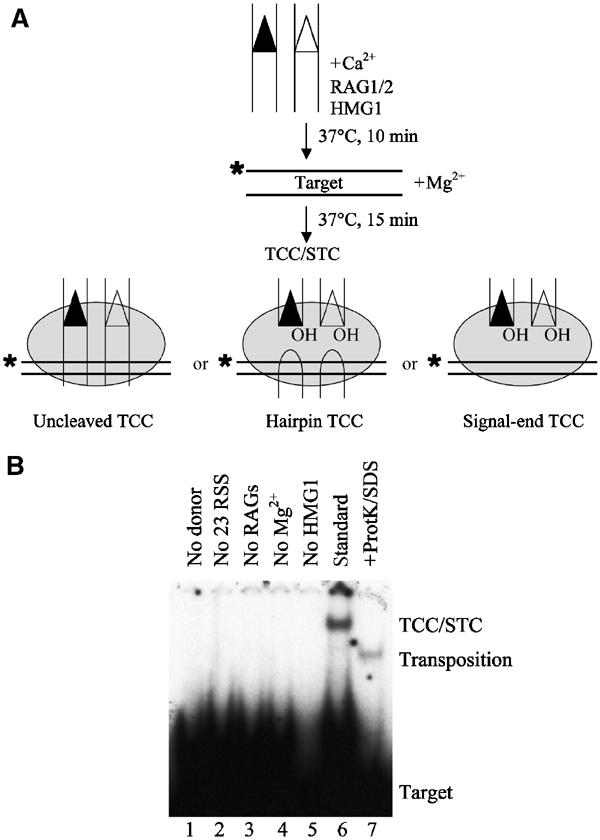
Physical detection of TCCs. (A) Schematic representation of the target capture assay using a labeled oligonucleotide target (position of the label is indicated by an asterisk) and intact oligonucleotide donors. In principle, TCCs detected in this assay could consist of uncleaved TCCs (left panel), hairpin TCCs (center panel), signal-end TCCs (right panel), or any combination of these three complexes. All three complexes would contain the RAG proteins, HMG, and target DNA but would differ in their coding DNA content (uncleaved TCC—intact coding flank; hairpin TCC—cleaved coding ends; signal end TCC—no coding DNA). (B) Requirements for target capture by the RAG1/2 transposase using uncleaved donor DNA. Factors omitted from each reaction are as indicated in lanes 1–5. Target capture occurring under standard conditions (12-RSS, 23-RSS, RAG1, RAG2, HMG1, target DNA, and Mg2+; see Materials and methods) is shown in lane 6. Transposition products can be liberated from the TCC/STC mixed band by treating the standard reaction products with Proteinase K and SDS (lane 7). The positions of the TCC/STC mixed band and the transposition products are shown.
While it has been suggested that the RAG1/2 transposase might contain a shared binding site for both coding DNA and target DNA (Tsai et al, 2002), another report implied that donor cleavage is not required for target capture (Neiditch et al, 2001). However, in the presence of uncleaved donor DNA, the interactions between the RAG proteins and target DNA appear unstable, suggesting that stable target capture might only occur after donor cleavage (Neiditch et al, 2001). Based on these reports, it is not clear when target DNA normally enters the RAG transposition reaction. Nonetheless, it is clear that the timing of target capture will have important implications for target site selection, RAG-mediated chromosomal translocations, and the evolution of the immune system.
In this paper, we directly analyze the DNA content of TCCs to determine whether the RAG1/2 transposase captures target DNA before or after donor cleavage. We demonstrate that the RAG1/2 transposase must cleave the RSS donors and release the hairpinned coding ends before physically binding to target DNA. We also demonstrate that functional commitment to target DNA only occurs after RSS cleavage. Finally, we show that all three of the DDE active site residues are required for target capture by the RAG proteins. Implications of these findings for the in vivo regulation of RAG transposition are discussed.
Results
Physical detection of target capture complexes
In order to define the components of the RAG1/2 TCC, we used a modified version of the target capture assay employed by others (Neiditch et al, 2001; Tsai et al, 2002) and incubated the RAG proteins with unlabeled, intact donor DNA, HMG1, and Ca2+ to allow formation of a synaptic PC. 32P-labeled target DNA and Mg2+ were added to allow donor cleavage and target capture (Figure 1A), and then the reaction mixture was separated on a native polyacrylamide gel. Consistent with previous studies (Neiditch et al, 2001; Tsai et al, 2002), target capture by the RAG1/2 transposase was 12/23 restricted (Figure 1B, see lane 2) and dependent on the presence of both donor DNA (Figure 1B, see lane 1) and the RAG proteins (Figure 1B, see lane 3). We also observed a requirement for HMG1 (Figure 1B, see lane 5), in keeping with the HMG dependence of PC formation (Hiom and Gellert, 1998) and transposition (Agrawal et al, 1998; Hiom et al, 1998).
The TCC is always found together with the strand transfer complex (STC), in which the covalent links between the donor and target have been made, but the RAG proteins remain bound to the DNA. Therefore, we wanted to quantitate the ratio of unreacted (TCC) to reacted (STC) product present in the mixed TCC/STC band. By dividing the target capture reaction mixtures in half, treating one half with proteinase K and SDS to remove any bound proteins, subjecting the other half to a mock treatment, and resolving these products on a native gel, we were able to detect TCC/STC in the mock-treated sample and the liberated transposition product (STC) in the treated sample. Consistent with previous observations (Neiditch et al, 2001; Tsai et al, 2002), approximately 70% of the material present in the TCC/STC band is unreacted TCC, while 30% is the covalent transposition product (STC) (Figure 1B, compare lanes 6 and 7).
RSS cleavage is required for target capture
In studying the requirements for target capture by the RAG1/2 transposase, we observed that TCC formation was also dependent on Mg2+ (Figure 1B, compare lanes 4 and 6). As Ca2+ supports binding and synaptic complex formation (Hiom and Gellert, 1998), the Mg2+-dependent step must occur after synapsis. Thus, either donor cleavage (which is Mg2+ dependent) is required for target capture or target capture itself requires Mg2+. In order to discriminate between these two possibilities, we asked whether Mg2+ is required for the signal end complex (SEC) (Agrawal and Schatz, 1997; Hiom and Gellert, 1998)—which is assembled from precleaved RSS donor DNA—to perform target capture. Using precleaved donor DNA that is 32P-labeled on the donor strand and an unlabeled plasmid target, TCC was readily detected in either Ca2+ or Mg2+ (Figure 2B, see lanes 13–14 and 15–16). Furthermore, in keeping with previous studies (Hiom et al, 1998), we observed that Ca2+ supports transposition of precleaved signal ends (Figure 2B, lane 14). Thus, when the requirement for donor cleavage is circumvented, target capture is Mg2+ independent. However, in the presence of intact donor DNA (Figure 2A), TCC formation required Mg2+ (Figure 2B, compare lanes 5 and 7; Figure 2C, compare lanes 1 and 3; Figure 1B, compare lanes 4 and 6). This requirement for Mg2+ was observed even though the substitution of plasmid target DNA for oligonucleotide target DNA consistently led to more efficient TCC formation (compare Figure 1B, lane 6 and Figure 2B, lane 7). The requirement for Mg2+ was also observed when we used altered coding flank substrates (Figure 2C, compare lanes 5 and 7). As the Mg2+ dependence of TCC formation was only observed in the presence of intact donor DNA, our results argue that RSS donor cleavage is required for target capture by the RAG1/2 transposase. Therefore, TCCs that contain no coding DNA (‘signal end TCC'; Figure 1A) are capable of forming, while those containing uncleaved coding DNA (‘uncleaved TCC'; Figure 1A) are not.
Figure 2.
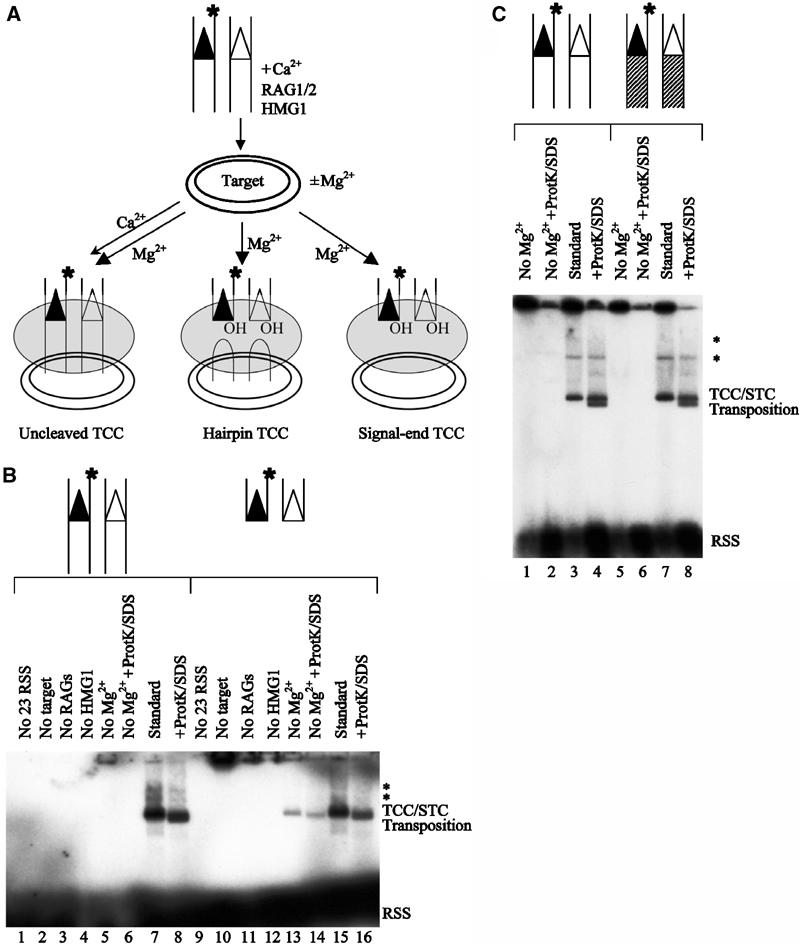
RSS cleavage is required for target capture. (A) Schematic representation of the target capture assay, using a plasmid target and oligonucleotide donors that are labeled on the donor strand (as indicated by the position of the asterisk). Reaction products that can, in principle, form under different reaction conditions are also shown. Using an intact RSS donor (as depicted here), TCCs detected under conditions that do not support donor cleavage (Ca2+ alone, thin arrowhead) can only consist of uncleaved TCCs. Using an intact RSS donor, TCCs detected under conditions that support donor cleavage (Mg2+, thick arrowhead) could theoretically consist of uncleaved TCCs, hairpin TCCs, signal-end TCCs, or any combination of these three complexes. Using a precleaved RSS donor (see (B), lanes 9–16), TCCs could only consist of signal-end TCCs. (B) Requirements for target capture by the RAG1/2 transposase using either uncleaved or precleaved donor DNA. Reactions with uncleaved (lanes 1–8) and precleaved (lanes 9–16) donors are shown. Factors omitted from the reactions are as indicated (lanes 1–6 and 9–14). Target capture under standard conditions is shown in lanes 7 and 15. Lanes 8, 14, and 16 show the transposition products that are liberated from the TCC/STC mixed band upon treatment with Proteinase K and SDS. The positions of the TCC/STC mixed band and the transposition products are shown. The symbol * marks higher molecular weight bands that represent TCCs and STCs that have used concatamerized plasmids as target DNA. (C) Metal ion requirements for target capture by the RAG1/2 transposase using two different RSS substrates. Reactions with our standard intact RSS substrate (VDJ100/101, VDJ132/133) (lanes 1–4) and altered coding flank substrates that differ in the length and sequence of the coding DNA (VDJ100.1/101.1, VDJ132.1/133.1) (lanes 5–8) are shown. Mg2+ was omitted from the reactions in lanes 1–2 and 5–6. Target capture under standard conditions is shown in lanes 3 and 7. Lanes 4 and 8 show the transposition products. The positions of the TCC/STC mixed band and the transposition products are shown. The symbol * marks higher molecular weight bands that represent TCCs and STCs that have used concatamerized plasmids as target DNA.
Release of hairpinned coding ends is required for target capture
After RSS cleavage, the reaction products—two blunt signal ends and two hairpinned coding ends—are held together by the RAG proteins in the cleaved signal complex (CSC) (Hiom and Gellert, 1998). Seeing that donor cleavage is required for target capture, we wanted to know whether coding ends are released from the CSC prior to target capture. To this end, we placed the 32P-label on the top strand coding end of an uncleaved 12-RSS and asked whether coding ends could be detected within TCCs. With the label in this position, TCC will only be detectable if coding DNA—either intact or cleaved—is present in the complex. Using either our standard RSS substrates (Figure 3A) or altered coding flank substrates (Figure 3B), TCC was not detected with the label on the coding strand (Figure 3A, lanes 3 and 4; Figure 3B, lanes 3 and 4), demonstrating that neither uncleaved coding DNA nor hairpinned coding ends are present in the TCC. Under the same conditions, TCC formation was readily observed when the 32P-label was placed on the donor strand of either an uncleaved or precleaved RSS (Figure 3A, lanes 1–2 and 5–6; Figure 3B, lanes 1–2 and 5–6). These results indicate that both RSS donor cleavage and release of the hairpinned coding ends must occur prior to target capture by the RAG1/2 transposase. Formation of TCCs containing hairpinned coding DNA (‘hairpin TCC'; Figure 1A) seems to be blocked.
Figure 3.
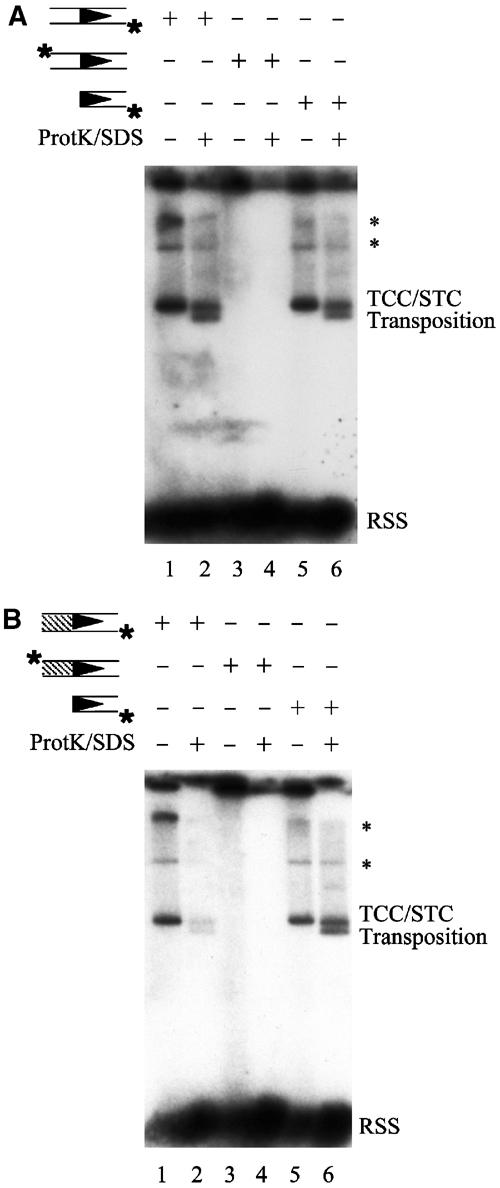
Release of hairpinned coding ends is required for target capture. (A) As indicated above the gel, target capture was carried out as in Figure 2B using either intact RSS substrates (VDJ100/101, VDJ132/133) labeled on the donor strand (lanes 1 and 2), intact RSS substrates (VDJ100/101, VDJ132/133) labeled on the coding strand (lanes 3 and 4), or precleaved RSS substrates (VDJ104/106, YD24/VDJ134) labeled on the donor strand (lanes 5 and 6). With intact RSS donors labeled on the donor strand, complexes detected in this assay could, in principle, consist of uncleaved TCCs, hairpin TCCs, signal-end TCCs, or any combination of these three complexes. With intact RSS donors labeled on the coding strand, complexes detected in this assay could, in principle, consist of uncleaved TCCs, hairpin TCCs, or a mixture of these two complexes. Using precleaved RSS donors labeled on the donor strand, complexes detected in this assay could only consist of signal-end TCCs. The positions of the TCC/STC mixed band and the transposition products are shown. The symbol * marks higher molecular weight bands that represent TCCs and STCs that have used concatamerized plasmids as target DNA. (B) Target capture was carried out as in (A), using either altered coding flank substrates (VDJ100.1/101.1, VDJ132.1/133.1) labeled on the donor strand (lanes 1 and 2), altered coding flank substrates (VDJ100.1/101.1, VDJ132.1/133.1) labeled on the coding strand (lanes 3 and 4), or precleaved RSS substrates (VDJ104/106, YD24/VDJ134) labeled on the donor strand (lanes 5 and 6). The positions of the TCC/STC mixed band and the transposition products are shown. The symbol * marks higher molecular weight bands that represent TCCs and STCs that have used concatamerized plasmids as target DNA.
A functional active site is required for target capture
We next used catalytically deficient RAG1 mutants (D600A, D708A, and E962A) to find out whether the active site of the RAG1/2 transposase plays a role in target capture. Using precleaved donors to bypass the step of RSS cleavage and specifically assess the role of the active site in target capture, we observed that target capture requires all three active site residues: D600 (Figure 4, lanes 3 and 4), D708 (Figure 4, lanes 5 and 6), and E962 (Figure 4, lanes 7 and 8). Under the same conditions, target capture was readily observed using catalytically active RAG1/2 transposases (Figure 4, lanes 1 and 2). Therefore, a functional DDE active site motif is required for target capture by the RAG1/2 transposase.
Figure 4.
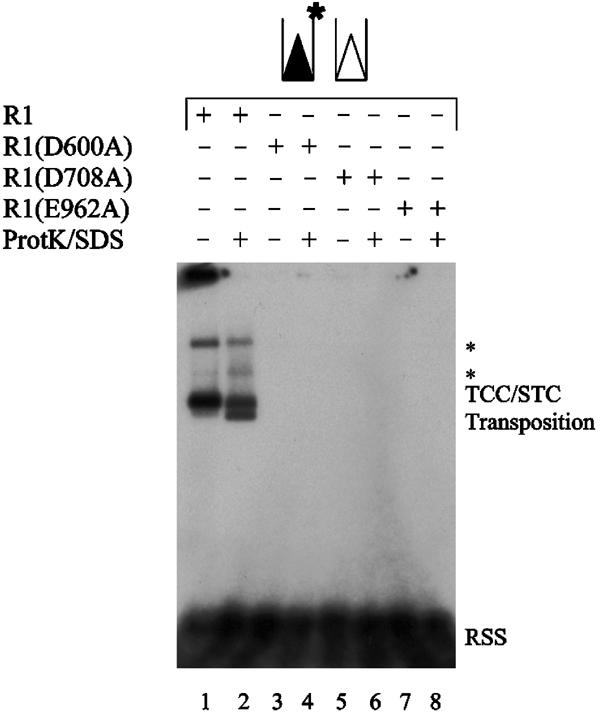
A functional DDE active site motif is required for target capture. Target capture assay was carried out as in Figure 2B, using precleaved RSS substrates labeled on the donor strand. Core RAG2 was used in all lanes with wild-type (WT) core RAG1 or the indicated mutant derivatives of core RAG1. The positions of the TCC/STC mixed band and the transposition products are shown. The symbol * marks higher molecular weight bands that represent TCCs and STCs that have used concatamerized plasmids as target DNA.
Only the signal end complex can commit to a target DNA
Prior to the catalytic step of strand transfer, a transposase must not only physically bind an unrelated piece of DNA (target capture) but must also functionally commit to using that piece of DNA as its target for strand transfer (target commitment). By definition, once a transposase has functionally committed to using a particular target, its interactions with that target are competitor resistant (Sakai and Kleckner, 1997). Consequently, it will subsequently transpose the donor DNA into that target, even if it is challenged with a second target DNA.
In order to determine whether the stable, physical transposase–target interactions detected in the target capture assay were also functionally significant, we used a modified version of the target commitment assay employed by others (Sakai and Kleckner, 1997). As illustrated schematically in Figure 5A, staged in vitro reactions were carried out using either intact or precleaved RSS donors. In the first stage (stage I), either the PC or the SEC was assembled under conditions that do not support donor cleavage (Ca2+ alone). In the second stage (stage II), a plasmid target (target 1) was added and transposase–target interactions were allowed to equilibrate. In the third stage (stage III), a second competitor plasmid target of distinguishable size (target 2) was added along with Mg2+ to allow for donor cleavage and transposition. After incubating the reaction mixture further, the relative frequency of transposition into the two target DNAs was quantitated. If the RAG1/2 transposase commits to target 1 prior to donor cleavage and remains committed to this target during the subsequent challenge with target 2, then one would observe preferential transposition into target 1 despite the fact that both targets are present in equimolar amounts. If, on the other hand, the RAG1/2 transposase is incapable of functionally interacting with target DNA prior to donor cleavage, then one would observe no preference for transposition into either target.
Figure 5.
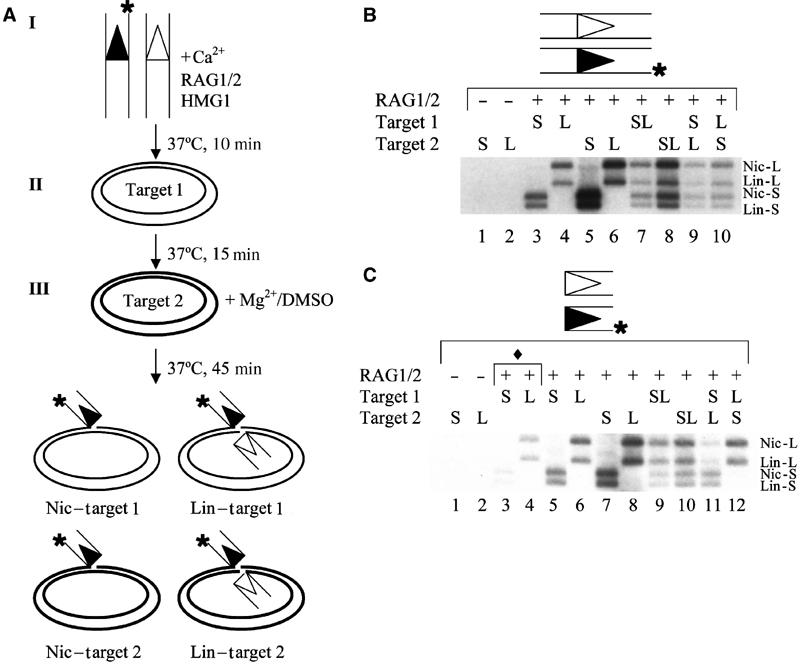
RSS cleavage is required for target commitment. (A) Schematic representation of the target commitment assay, using oligonucleotide donors that are labeled on the donor strand (as indicated by the position of the asterisk). Target 1 and Target 2 represent two distinct plasmids that differ in size. The nicked (Nic) and linearized (Lin) products resulting from single- and double-ended insertions, respectively, are diagrammed. (B) PCs do not exhibit target commitment. The stage at which a smaller plasmid target (S) or a larger plasmid target (L) was added to the reaction is indicated above the gel. The positions of the nicked smaller plasmid (Nic-S), linearized smaller plasmid (Lin-S), nicked larger plasmid (Nic-L), linearized larger plasmid (Lin-L) transposition products are shown. (C) SECs do exhibit target commitment. Target DNAs are identical to those in (B). The positions of the transposition products are also labeled as in (B). The symbol ♦ denotes reactions that were incubated with a single target for 15 min in the absence of Mg2+ and DMSO.
Incubation of the RAG proteins, intact RSS donor DNA, and a single target results in both single- and double-ended transposition events, generating nicked and linearized plasmids, respectively (Agrawal et al, 1998; Hiom et al, 1998) (Figure 5B, lanes 3 and 4). These products are observed regardless of whether the plasmid target is added during the second or third phase of the staged reaction (Figure 5B, compare lanes 3 and 4 to lanes 5 and 6). As expected, when both plasmid targets are added simultaneously, no target preference is observed (Figure 5B, lanes 7 and 8). However, when we used intact RSS donor DNA and added the plasmids sequentially, we still failed to observe a preference for the first target, regardless of whether the smaller or larger plasmid served as target 1 (Figure 5B, lanes 9 and 10). In contrast, we observed clear target commitment using precleaved RSS donors (Figure 5C, lanes 11 and 12). Although precleaved RSS donors can be transposed during both the Ca2+ (stage II) and Mg2+ (stage III) incubations, we observed that the majority (∼75%) of transposition events occur during stage III of the reaction (Figure 5C, compare lanes 3 and 4 to lanes 5 and 6). Consequently, the strong preference for target 1 (∼6–7 × bias) cannot be explained by the fact that target 1 was present during both stages II and III while target 2 was only present during stage III of the reaction. Therefore, the SEC can commit to a target while the PC cannot, suggesting that target capture and target commitment both occur after donor cleavage and coding end release.
Discussion
RAG synaptic complexes can capture target DNA only after donor cleavage and coding end release
In this report, we have shown that the RAG SEC, which contains cleaved RSS donor ends, forms stable, physically detectable, and functionally significant interactions with target DNA prior to strand transfer. However, no such interactions were observed for the RAG PC, which contains uncleaved RSS donor DNA with its flanking coding DNA. RAG complexes containing both cleaved signal ends and hairpinned coding ends also could not capture a target DNA. Taken together, these findings strongly suggest that target DNA normally enters the RAG transposition reaction only after cleavage of RSS donors and release of the resulting hairpinned coding ends. Based on our findings, we propose that RAG-mediated transposition proceeds via the pathway depicted in Figure 6.
Figure 6.
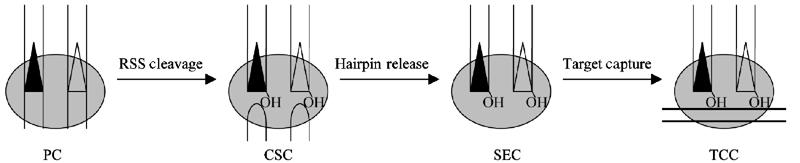
Proposed reaction pathway for RAG-mediated transposition. The RAG proteins first bind to both a 12-RSS and a 23-RSS, forming a PC. RAG-mediated coupled cleavage produces the CSC, containing two hairpinned coding ends and two blunt signal ends. Release of the hairpinned coding ends then generates the SEC. In the absence of coding DNA, the SEC can bind target DNA, generating a TCC. The subsequent steps of strand transfer and resolution are not shown here.
A recent report suggested that the RAG proteins can physically associate with target DNA prior to RSS cleavage, thereby implying that RSS cleavage is not required for target capture. Using target commitment assays similar to those employed here, the same report also argued that the RAG1/2 transposase could functionally commit to target DNA either before or after donor cleavage (Neiditch et al, 2001). However, competitor-resistant target commitment was only observed under reaction conditions that support donor cleavage, possibly confounding the results. Although it is hard to account for different experimental conditions, our novel observation that labeled coding DNA and target DNA cannot coexist in the same complex rules out the possibility of target capture before cleavage. Our finding that SECs support target commitment, while uncleaved PCs do not, is consistent with our observation that the RAG1/2 transposase performs target capture only after it has cleaved the RSS donors and released the hairpinned coding ends.
The RAG1/2 transposase contains a single binding pocket for coding ends and target DNA
It has recently been suggested that the RAG1/2 transposase contains a single binding site for non-RSS DNA that can accommodate either coding ends or target DNA (Tsai et al, 2002). Our finding that both RSS donor cleavage and coding end release precede target capture by the RAG1/2 transposase provides the first evidence for this model. As UV crosslinking studies suggest that RAG1 might directly bind to uncleaved coding DNA (Eastman et al, 1999; Swanson and Desiderio, 1999; Mo et al, 2000, 2001), the binding site for coding ends and target DNA might exist prior to RSS cleavage, where it functions to bind uncleaved coding DNA.
It is interesting to note that the RAG1/2 transposase requires a functional active site for target capture (Figure 4) but not for RSS donor recognition or synapsis (Kim et al, 1999; Landree et al, 1999; Fugmann et al, 2000). However, this finding is not unprecedented, as the DDE active site motif is required for divalent metal ion-dependent target capture by Tn10 (Junop and Haniford, 1997) even though it is dispensable for synaptic complex formation (Bolland and Kleckner, 1996; Kennedy and Haniford, 1996). As a functional active site is required for target capture by the RAG1/2 transposase, the shared binding site for coding DNA and target DNA might lie within the RAG active site.
We propose a model in which the RAG active site contains a single DNA-binding pocket consisting of two DNA-binding sites: one that accommodates RSS DNA and the other that accommodates non-RSS DNA. Active site residues might be positioned at the border between the RSS and non-RSS binding sites where they perform catalysis and either directly bind non-RSS DNA or coordinate divalent metal ions that bind non-RSS DNA. During the course of the transposition reaction, the non-RSS DNA-binding site first binds to uncleaved coding DNA, then binds the cleaved coding ends, and upon release of these hairpinned coding ends, then binds to target DNA. This model is attractive because non-RSS DNA must be in very close proximity to the 3′ OH groups of the RSS donors for either transposition or hybrid joining to occur. Furthermore, this model is consistent with the observation that the DDE motif within the RAG active site is required for all of the phosphoryl transfer reactions of transposition: nicking, hairpinning, and strand transfer.
Kleckner and colleagues have proposed a similar model for the Tn10 transposase, based on the finding that target capture and commitment only occur within the Tn10 double-end break complex (Sakai and Kleckner, 1997). However, to the best of our knowledge, this is the first study to provide evidence suggesting that release of flanking DNA is required for target capture by a transposase. As there are a number of similarities between RAG and Tn10, it will be interesting to see if Tn10 also releases flanking donor DNA prior to target capture and strand transfer.
Implications of postcleavage target capture for RAG-mediated transposition in vivo
As proposed by Roth and colleagues, the stage at which target DNA enters the RAG transposition reaction might affect target site selection and influence whether the RAG1/2 transposase inserts RSS DNA into nearby, relatively safe regions of the genome (i.e. the TCR or Ig loci) or into distant, comparatively dangerous regions of the genome, thereby generating oncogenic chromosomal translocations. That is, after binding both a 12-RSS and a 23-RSS to form a synaptic PC, the RAG1/2 transposase is physically tethered to the chromosome containing the RSS sequences. As each chromosome occupies its own distinct territory within the nucleus (Cremer and Cremer, 2001), tethering the RAG1/2 transposase to the actively rearranging chromosome may sequester the transposase away from other chromosomes. Consistent with this, IgH and Igκ loci that are poised to rearrange have been found to exhibit distinct subnuclear compartmentalization (Kosak et al, 2002). However, because the RAG1/2 transposase must release the hairpinned coding ends prior to target capture, the SEC should be able to diffuse freely throughout the nucleus (Chubb and Bickmore, 2003), encountering target DNA from other chromosomes. Thus, the RAG1/2 transposase may be capable of interchromosomal transposition and might generate mutagenic insertions or oncogenic chromosomal translocations in vivo. Indeed, a recent paper reported RAG-mediated transposition of TCRα signal ends from chromosome 14 into the X-linked HPRT gene in human peripheral T cells (Messier et al, 2003).
As transposition events can be deleterious to the host organism, causing insertional mutagenesis of essential genes and generating double-strand breaks that can lead to potentially oncogenic chromosomal translocations, RAG-mediated transposition must be tightly regulated in vivo. Whereas many transposons regulate their activity at the step of donor cleavage, RAG-mediated RSS cleavage is crucial to the assembly of functional antigen receptor genes during lymphoid development. As the RAG proteins are fully functional and perform RSS cleavage in mammalian lymphoid progenitors, RAG-mediated transposition must be regulated at a step subsequent to RSS donor cleavage. Here we show that the choice to commit to a transpositional resolution of the signal ends occurs postcleavage and that transposition is blocked by the presence of coding ends, providing one possible layer of regulation. By tethering the RAG proteins to the actively rearranging chromosome, the coding ends may temporarily sequester the RAG1/2 transposase from DNA on other chromosomes, thereby preventing interchromosomal transposition. Moreover, by occupying the non-RSS DNA-binding site, coding ends can also block target capture, thereby preventing transposition generally. However, as coding ends are processed more rapidly than signal ends in vivo (Ramsden and Gellert, 1995), other layers of regulation must also exist. As coding end release precedes target capture, it is possible that the NHEJ factors recruited to the postcleavage complex might serve to suppress transposition in vivo. For example, while interacting with the RAG proteins and repairing the postcleavage coding ends—thereby freeing up the target DNA-binding site—the NHEJ factors might directly block access to target DNA or perhaps induce a conformational change in the RAG proteins that impairs subsequent target capture or strand transfer. Additional layers of regulation may include a bias toward transposon disintegration at physiological Mg2+ concentrations (Melek and Gellert, 2000), inhibition of transposition by the C-terminus of RAG-2 (Elkin et al, 2003; Tsai and Schatz 2003), preferential transposition into hairpinned target DNA molecules (Lee et al, 2002), or GTP-mediated inhibition of target capture (Tsai and Schatz, 2003).
Materials and methods
DNA substrates
Oligonucleotide substrates VDJ100/101 (12-RSS with flanking coding sequence), VDJ132/133 (23-RSS with flanking coding DNA), VDJ104/106 (12-RSS, precleaved), YD24/VDJ134 (23-RSS, precleaved), and mm30t/mm30b (a target for TCC assay) (Neiditch et al, 2001) were gel purified, annealed, and 5′-end labeled with 32P-ATP as described (Cuomo et al, 1996). Oligonucleotide sequences for VDJ100, VDJ101, VDJ132, VDJ133, VDJ104, VDJ106, YD24, and VDJ134 are described in Cuomo et al (1996) and Kim et al (1999). Altered coding flank substrates VDJ100.1 (5′-GAACGTCTTGCAGACCTGCAGCACAGTGCTACAGACTGGAA CAAAAACCCAGGTCTC-3′), VDJ101.1 (5′-TGAGACCTGGGTTTTTGTTCCAGTCTGT A GCACTGTGCTGCAGGTCTGCAAGACGTT-3′), VDJ132.1 (5′-GAACGTCTTGCAGACCT GCAGCACAGTGGTAGTACTCCACTGTCTGGCTGTACAAAA ACCCAGGT-3′), and VDJ133.1 (5′-TACCTGGGTTTTTGTACAGCCAGACAGTGGAGTACTACCA CTGTGCTG CAGGTCTGCAAGACGTT-3′) were designed by taking the heptamer, spacer, and nonamer sequences from our standard RSS substrates (VDJ100, VDJ101, VDJ132, and VDJ133) and changing the length and sequence of the flanking coding DNA. The altered coding flank substrates were also gel purified, annealed, and 5′-end labeled with 32P-ATP as described (Cuomo et al, 1996). Plasmid substrates pUC19 and pBR322 were obtained from NEN.
Proteins
Recombinant core RAG2 (aa 1–383) was expressed by vaccinia infection of HeLa cells and purified as described previously (McBlane et al, 1995). Recombinant wild-type core RAG1 (aa 384–1040) and catalytically deficient core RAG1 (D600A, D708A, and E962A) were purified from Escherichia coli as described previously (Kim et al, 1999). The activity of each protein preparation was validated by EMSA and single-site cleavage assays (Supplementary Figure 2). The ability of RAG1(D600A), RAG1(D708A), and RAG1(E962A) to form the SEC was validated by EMSA (Supplementary Figure 1).
Target capture assay
For detection of the TCC and STC transposition intermediates using labeled target DNA, initial binding reactions contained 25 mM K-morpholinepropanesulfonic acid (MOPS) (pH 7.0), 4 mM dithiothreitol, 75 mM potassium glutamate, 5 mM CaCl2, 100 μg/ml of bovine serum albumin, 80 ng of core RAG1, 20 ng of core RAG2, 50 ng of HMG1, and 0.02 pmol each of uncleaved 12- and 23-RSS oligonucleotide donor (VDJ100/101 and VDJ132/133, respectively) in a final volume of 6.5 μl. Binding was allowed to proceed at 37°C for 10 min after which MgCl2 (5 mM, final concentration), dimethyl sulfoxide (DMSO) (10%, final wt/vol ratio), and 0.1 pmol of labeled oligonucleotide target (mm30t/mm30b) were added (10 μl final reaction volume), after which reaction mixtures were incubated for an additional 15 min. The reactions were divided in half, with SDS (1%, final wt/vol ratio) and Proteinase K (200 ng/ml final concentration) added to one half but not the other. After both samples were incubated for 15 min at 37°C, 2 μl of 50% glycerol was added to each and the samples were then loaded directly onto a 6% nondenaturing, DNA-retardation polyacrylamide gel (Novex) and run at 90 V for 90 min in 0.5 × Tris–borate–EDTA at 4°C. Dried gels were visualized by PhosphorImager analysis and autoradiography.
For STC/TCC capture with labeled donor DNA, reaction conditions were as described above except that a labeled 12-RSS oligonucleotide was substituted for the unlabeled 12-RSS, and 0.1 pmol of unlabeled pUC19 plasmid target was substituted as the target. Entire reaction mixtures were separated on a 1% agarose-ME gel, run at 80 V for 120 min in 1 × Tris–acetate–EDTA at 4°C.
Target commitment assay
Unless otherwise indicated, initial binding conditions were as described above and contained 25 mM MOPS (pH 7.0), 4 mM dithiothreitol, 75 mM potassium glutamate, 5 mM CaCl2, 100 μg of bovine serum albumin/ml, 80 ng of core RAG1, 20 ng of core RAG2, 50 ng of HMG1, 0.02 pmol of 32P-labeled 12-RSS oligonucleotide donor (VDJ100/101 or VDJ104/106), and 0.02 pmol of unlabeled 23-RSS oligonucleotide donor (VDJ132/133 or YD24/VDJ134) in a final volume of 6.5 μl. Binding was allowed to proceed at 37°C for 10 min. A measure of 0.1 pmol of unlabeled plasmid target or Tris–EDTA (TE) was then added and the reaction mixture was incubated for an additional 15 min at 37°C. Following this incubation, 0.1 pmol of a second unlabeled plasmid target of distinguishable size, or an equal volume of TE, was added along with MgCl2 (5 mM, final concentration) and DMSO (10%, final wt/vol ratio) for a final reaction volume of 11 μl. In these reactions, pUC19 was used as the smaller plasmid and pBR322 was used as the larger plasmid. The reaction mixtures were incubated for an additional 45 min. The reaction mixtures were then supplemented with SDS (1%, final wt/vol ratio) and Proteinase K (200 ng/ml final concentration) and incubated for 15 min at 37°C. A volume of 2 μl of 50% glycerol was added to each of the samples and entire reaction mixtures were separated on a 1% agarose–ME gel, run at 80 V for 120 min in 1 × Tris–acetate–EDTA at 4°C. Dried gels were exposed to X-ray film and bands were quantitated by Phosphorimager analysis.
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We thank Meni Melek for assistance in developing the target capture assay, Nadja Patenge for helpful discussions, and Katrina Morshead for critical reading of the manuscript. AGW Matthews is a Howard Hughes Medical Institute Predoctoral Fellow. This work was supported by the National Institutes of Health grant R01 GM48026.
References
- Agrawal A, Eastman QM, Schatz DG (1998) Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature 394: 744–751 [DOI] [PubMed] [Google Scholar]
- Agrawal A, Schatz DG (1997) RAG1 and RAG2 form a stable postcleavage synaptic complex with DNA containing signal ends in V(D)J recombination. Cell 89: 43–53 [DOI] [PubMed] [Google Scholar]
- Bassing CH, Swat W, Alt FW (2002) The mechanism and regulation of chromosomal V(D)J recombination. Cell 109 (Suppl): S45–S55 [DOI] [PubMed] [Google Scholar]
- Bolland S, Kleckner N (1996) The three chemical steps of Tn10/IS10 transposition involve repeated utilization of a single active site. Cell 84: 223–233 [DOI] [PubMed] [Google Scholar]
- Chubb JR, Bickmore WA (2003) Considering nuclear compartmentalization in the light of nuclear dynamics. Cell 112: 403–406 [DOI] [PubMed] [Google Scholar]
- Clatworthy AE, Valencia MA, Haber JE, Oettinger MA (2003) V(D)J recombination and RAG-mediated transposition in yeast. Mol Cell 12: 489–499 [DOI] [PubMed] [Google Scholar]
- Cremer T, Cremer C (2001) Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat Rev Genet 2: 292–301 [DOI] [PubMed] [Google Scholar]
- Cuomo CA, Mundy CL, Oettinger MA (1996) DNA sequence and structure requirements for cleavage of V(D)J recombination signal sequences. Mol Cell Biol 16: 5683–5690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastman QM, Villey IJ, Schatz DG (1999) Detection of RAG protein–V(D)J recombination signal interactions near the site of DNA cleavage by UV cross-linking. Mol Cell Biol 19: 3788–3797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkin SK, Matthews AG, Oettinger MA (2003) The C-terminal portion of RAG2 protects against transposition in vitro. EMBO J 22: 1931–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fugmann SD, Villey IJ, Ptaszek LM, Schatz DG (2000) Identification of two catalytic residues in RAG1 that define a single active site within the RAG1/RAG2 protein complex. Mol Cell 5: 97–107 [DOI] [PubMed] [Google Scholar]
- Gellert M (2002) V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem 71: 101–132 [DOI] [PubMed] [Google Scholar]
- Hiom K, Gellert M (1998) Assembly of a 12/23 paired signal complex: a critical control point in V(D)J recombination. Mol Cell 1: 1011–1019 [DOI] [PubMed] [Google Scholar]
- Hiom K, Melek M, Gellert M (1998) DNA transposition by the RAG1 and RAG2 proteins: a possible source of oncogenic translocations. Cell 94: 463–470 [DOI] [PubMed] [Google Scholar]
- Junop MS, Haniford DB (1997) Factors responsible for target site selection in Tn10 transposition: a role for the DDE motif in target DNA capture. EMBO J 16: 2646–2655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy AK, Haniford DB (1996) Isolation and characterization of IS10 transposase separation of function mutants: identification of amino acid residues in transposase that are important for active site function and the stability of transposition intermediates. J Mol Biol 256: 533–547 [DOI] [PubMed] [Google Scholar]
- Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA (1999) Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev 13: 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H (2002) Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162 [DOI] [PubMed] [Google Scholar]
- Landree MA, Wibbenmeyer JA, Roth DB (1999) Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev 13: 3059–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GS, Neiditch MB, Sinden RR, Roth DB (2002) Targeted transposition by the V(D)J recombinase. Mol Cell Biol 22: 2068–2077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBlane JF, van Gent DC, Ramsden DA, Romeo C, Cuomo CA, Gellert M, Oettinger MA (1995) Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell 83: 387–395 [DOI] [PubMed] [Google Scholar]
- Melek M, Gellert M (2000) RAG1/2-mediated resolution of transposition intermediates: two pathways and possible consequences. Cell 101: 625–633 [DOI] [PubMed] [Google Scholar]
- Messier TL, O'Neill JP, Hou SM, Nicklas JA, Finette BA (2003) In vivo transposition mediated by V(D)J recombinase in human T lymphocytes. EMBO J 22: 1381–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Bailin T, Noggle S, Sadofsky MJ (2000) A highly ordered structure in V(D)J recombination cleavage complexes is facilitated by HMG1. Nucleic Acids Res 28: 1228–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo X, Bailin T, Sadofsky MJ (2001) A C-terminal region of RAG1 contacts the coding DNA during V(D)J recombination. Mol Cell Biol 21: 2038–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiditch MB, Lee GS, Landree MA, Roth DB (2001) RAG transposase can capture and commit to target DNA before or after donor cleavage. Mol Cell Biol 21: 4302–4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettinger MA, Schatz DG, Gorka C, Baltimore D (1990) RAG-1 and RAG-2, adjacent genes that synergistically activate V(D)J recombination. Science 248: 1517–1523 [DOI] [PubMed] [Google Scholar]
- Ramsden DA, Gellert M (1995) Formation and resolution of double-strand break intermediates in V(D)J rearrangement. Genes Dev 9: 2409–2420 [DOI] [PubMed] [Google Scholar]
- Sakai J, Kleckner N (1997) The Tn10 synaptic complex can capture a target DNA only after transposon excision. Cell 89: 205–214 [DOI] [PubMed] [Google Scholar]
- Sakano H, Huppi K, Heinrich G, Tonegawa S (1979) Sequences at the somatic recombination sites of immunoglobulin light-chain genes. Nature 280: 288–294 [DOI] [PubMed] [Google Scholar]
- Schlissel MS (2002) Does artemis end the hunt for the hairpin-opening activity in V(D)J recombination? Cell 109: 1–4 [DOI] [PubMed] [Google Scholar]
- Swanson PC, Desiderio S (1999) RAG-2 promotes heptamer occupancy by RAG-1 in the assembly of a V(D)J initiation complex. Mol Cell Biol 19: 3674–3683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson CB (1995) New insights into V(D)J recombination and its role in the evolution of the immune system. Immunity 3: 531–539 [DOI] [PubMed] [Google Scholar]
- Tonegawa S (1983) Somatic generation of antibody diversity. Nature 302: 575–581 [DOI] [PubMed] [Google Scholar]
- Tsai CL, Drejer AH, Schatz DG (2002) Evidence of a critical architectural function for the RAG proteins in end processing, protection, and joining in V(D)J recombination. Genes Dev 16: 1934–1949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CL, Schatz DG (2003) Regulation of RAG1/RAG2-mediated transposition by GTP and the C-terminal region of RAG2. EMBO J 22: 1922–1930 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2


