Abstract
The AAA ATPase p97/VCP is involved in many cellular events including ubiquitin-dependent processes and membrane fusion. In the latter, the p97 adaptor protein p47 is of central importance. In order to provide insight into the molecular basis of p97 adaptor binding, we have determined the crystal structure of p97 ND1 domains complexed with p47 C-terminal domain at 2.9 Å resolution. The structure reveals that the p47 ubiquitin regulatory X domain (UBX) domain interacts with the p97 N domain via a loop (S3/S4) that is highly conserved in UBX domains, but is absent in ubiquitin, which inserts into a hydrophobic pocket between the two p97 N subdomains. Deletion of this loop and point mutations in the loop significantly reduce p97 binding. This hydrophobic binding site is distinct from the predicted adaptor-binding site for the p97/VCP homologue N-ethylmaleimide sensitive factor (NSF). Together, our data suggest that UBX domains may act as general p97/VCP/CDC48 binding modules and that adaptor binding for NSF and p97 might involve different binding sites. We also propose a classification for ubiquitin-like domains containing or lacking a longer S3/S4 loop.
Keywords: AAA+ ATPase, p47, p97, ubiquitin, UBX
Introduction
p97/VCP is a member of the AAA+ family (ATPases associated with various cellular activities), comprising enzymes that are involved in a wide range of different cellular processes including proteolysis, DNA repair and membrane fusion (for reviews, see Ogura and Wilkinson, 2001; Lupas and Martin, 2002). One possible common feature of AAA+ function is a folding or unfolding step usually catalysed in an ATP-dependent manner. The ATPase domain is characterised by a conserved sequence of 200–250 residues that includes the Walker A and B motifs. Classical AAA proteins contain an additional ‘second region of homology' or SRH. AAA+ proteins can be further subdivided into type II containing two ATPase domains, referred to as D1 and D2 (for example, p97, N-ethylmaleimide sensitive factor (NSF), ClpA, ClpB, ClpC) or type I (e.g. ClpX, HslU) containing only one ATPase domain, termed D2. Additional less conserved domains are often found at the N-terminus (N domain) or the C-terminus and are often implicated in adaptor binding (for a review, see Dougan et al, 2002). Over the last few years, significant insight into the structures of AAA+ proteins has been obtained both by cryo-electron microscopy (cryo-EM) and crystallography (for a review, see Ogura and Wilkinson, 2001). Recently determined crystal structures include FtsH (Krzywda et al, 2002; Niwa et al, 2002) and ClpA (Guo et al, 2002b). However, much less is known about the nature of the interaction between AAA+ proteins and their adaptor molecules. To date, the only structure of such a complex is that of the ClpA N domain and its adaptor protein ClpS (Guo et al, 2002a; Zeth et al, 2002), although both structures are unrelated to p97 N domain and p47 C, respectively.
The hexameric mammalian p97/VCP interacts with various adaptor proteins, which are proposed to function in different cellular pathways (Kondo et al, 1997; Meyer et al, 2000; Hetzer et al, 2001). The adaptor complex of UFD1 and NPL4 is believed to be crucial for p97-mediated ubiquitin-dependent processes (Meyer et al, 2000; and for a review, see Bays and Hampton, 2002). For p97-mediated membrane fusion required for organelle biogenesis, the p47 and VCIP135 (valosin-containing protein [VCP][p97]/p47 complex-interacting protein, p135) proteins have been shown to be essential (Uchiyama et al, 2002). In addition, p97 also interacts with a large number of seemingly unrelated proteins, including SVIP (small VCP/p97-interacting protein) (Nagahama et al, 2003), clathrin (Pleasure et al, 1993), the breast/ovarian cancer susceptibility gene product, BRCA1 (Zhang et al, 2000a), a DNA unwinding factor (Yamada et al, 2000) and the ubiquitination factor UFD2 (Kaneko et al, 2003). The yeast homologue of p97, CDC48, also binds ubiquitin (Dai and Li, 2001; Rape et al, 2001) and can distinguish between native and non-native proteins in the absence of cofactors (Thoms, 2002). No adaptor proteins have been described for the archaeal homologue VAT, which has been suggested to act as a chaperone utilising its N domains for substrate binding (Golbik et al, 1999).
The p97 membrane fusion adaptor p47 is a conserved 370 residue eukaryotic protein that forms a tight complex with p97 (Kondo et al, 1997) and mediates the binding of p97 to its t-SNARE (soluble NSF attachment protein receptor) syntaxin5 (Rabouille et al, 1998). VCIP135 binds to the p97/p47/syntaxin5 complex dissociating it via p97-catalysed ATP hydrolysis, possibly preparing SNAREs for another round of membrane fusion (Uchiyama et al, 2002). The AAA protein NSF is essential for heterotypic membrane fusion by interacting with α-SNAP (soluble NSF attachment protein) during the process. Interestingly, α-SNAP competes with p47 for syntaxin5 binding (Rabouille et al, 1998); however, while p47 forms a stable complex with p97 inhibiting its ATPase activity (Meyer et al, 1998), α-SNAP has to recruit syntaxin5 first before it can bind to NSF, thereby stimulating its ATPase activity (for a review, see Whiteheart et al, 2001).
p47 contains two binding sites for p97 with the C-terminal region sufficient for tight p97 binding, while p97 N domain is essential for p47 binding (Uchiyama et al, 2002). The C-terminal domain of p47 (282–370) adopts a ubiquitin-like β-grasp fold (Yuan et al, 2001), generally referred to as a ubiquitin regulatory X domain (UBX) domain (Hofmann and Bucher, 1996; Buchberger et al, 2001). UBX domains are suggested to be involved in ubiquitin-related processes (Buchberger et al, 2001; Buchberger, 2002), but as yet no common function has been described.
In order to characterise the structural basis of one p97–adaptor complex, we have carried out a crystallographic analysis of hexameric p97 ND1 domains and the C-terminal region of p47 comprising the UBX domain (residues 244–370; p47 C). Our 2.9 Å structure represents the first structure of a UBX–protein complex, as well as AAA protein in its natural oligomeric state bound to an adaptor protein. Additional mutagenesis studies based on the structure reveal that a highly conserved loop specific to UBX domains mediates the p97 interaction. This interaction could represent a general mode of UBX domain recognition by p97/VCP/CDC48 or p97-like N domains.
Results
Overall structure
The structure of p97 ND1 in complex with p47 C was determined from crystals of space group P65, grown in low salt conditions. The structure was solved by the molecular replacement method employing ND1 as a search model. Within the asymmetric unit, the crystals contain an ND1 hexamer and two p47 C molecules bound to adjacent ND1 protomers. In our density maps, we also observe ill-defined density for a third p47 C molecule bound to another p97 protomer. The first 17–22 residues of the p97 N domain are not visible in our density maps. A ribbon diagram of the complex structure is shown in Figure 1A and a representative sample of electron density in Figure 1D. In the crystal packing, a plane of hexamers interlock in a ‘cog-like' manner, with the p47 C molecules serving as ‘bridges' between the different hexamers (Figure 1B). The p47 C-bound hexamers also spiral around the molecular axis, to obey the P65 symmetry. The p47 C fragment interacts with the N domain of one ND1 protomer and is positioned slightly below the plane of the hexameric ring (Figure 1A), pointing towards the D2 domain of full-length p97 (Huyton et al, 2003). The same p47 C molecule also makes contacts with an adjacent hexamer in the crystal lattice (Figure 1C). In the next packing layer below the plane, there are no major contacts.
Figure 1.
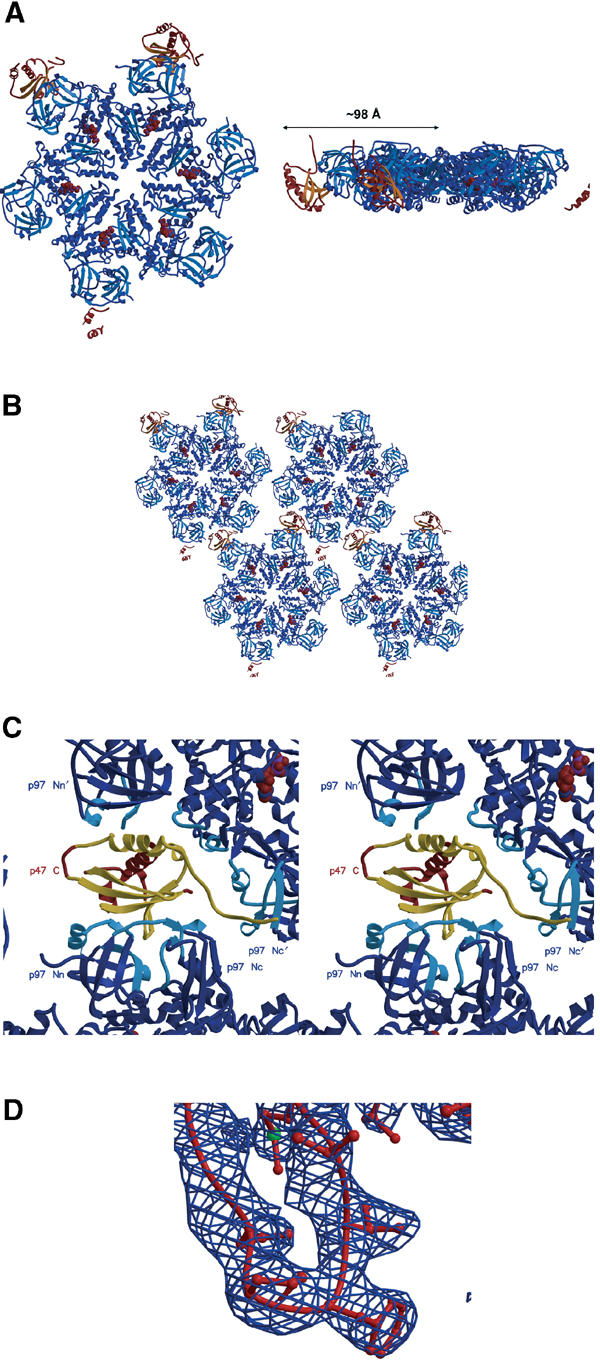
Overview of the p97 ND1–p47 C structure. (A) Ribbon representation of the p97 ND1 (blue)–p47 C (red/orange) crystal structure (top and side views). The radius of the complex structure is indicated. The disconnected secondary structure at the bottom (and right) of the p97 hexamer corresponds to parts of a third p47 C molecule we observe in the density maps. (B) Crystal packing arrangement of the p97 ND1–p47 C complex in one plane as viewed down the crystallographic 65 screw axis. (C) Stereo close-up view of one p47 C molecule (in red/gold) in the crystal lattice. The N domains from two ND1 hexamers (blue) are labelled. The ND1 secondary structure elements from the two hexamers that form the p47 C interface are depicted in light blue (‘binary' complex of p47 UBX with p97 Nn and p97 Nc; additional lattice contact between the N-terminal extended chain of p47 C with the p97' hexamer). p47 C secondary structure elements that interact with ND1 are depicted in gold. (D) Electron density (2Fo-Fc map contoured at 1.2σ) of the p47 C S3/S4 loop region.
In contrast to the crystal structure of ND1 (Zhang et al, 2000b), the six protomers of the ND1 hexamer in the complex structure are crystallographically independent copies. However, a comparison between both ND1 crystal structures shows no major domain movements, although both structures display totally different packing arrangements. The rmsd for different protomers superimposed on the P622 ND1 crystal structure ranges from 0.5 to 0.7 Å. Interestingly, the hydrogen-bonding interactions between the N domain and D1 are maintained, indicating that there is a preferred binding mode between the two domains. Differences between protomers in the complex are mainly observed in surface side-chain conformations and in the loop region 427–437, which is flexible and has poor-quality electron density. Overall, the Cα atoms can be superimposed with an rmsd ranging from 0.45 to 0.62 Å. In the active site of D1, bound ADP is observed, further confirming the preference of ND1 for ADP (Zhang et al, 2000b).
The two copies of p47 C observed in the complex superimpose with an rmsd of ∼0.9 Å, although there are slight differences at the end of β-strand 2 (S2). Both make nearly identical contacts with p97 ND1 covering a buried surface area of ∼1700 Å2. A comparison of p47 C bound to p97 ND1 with the solution structure of p47 UBX (Figure 2A, Table II) shows differences in the N-terminal part, which may be due to the shorter fragment used for the solution structure determination (282–370). The loop between β-strands 1 and 2 (S1 and S2) is shorter and in consequence results in a different hydrogen interaction pairing between the two strands. The loop region between strands 4 and 5 (S4 and S5) is flexible in solution and residues 349–352 are also not well defined in the crystal structure. Interestingly, residues N-terminal of S1 are ordered in the crystal complex, forming an additional helix (H1) that folds back onto the UBX domain between S2 and the main helix H2 of the UBX domain (Figure 2A). Pro294 facilitates this loop formation and there are hydrophobic contacts (Ala283 and Phe324) and a salt bridge (Glu280-Arg307) that stabilise the orientation of H1 with respect to the UBX core. At the N-terminal end of H1, Pro273 allows the extended chain to adopt a sharp bend, although no side-chain density can be observed for residues in this region, which in the final model are poly-Ala. The first ∼10 residues of the p47 C sequence as well as the His-tag are disordered and could not be observed. In summary, the secondary structure organisation for p47 C in complex with ND1 is αββαββαβ with an additional 310-type helix (labelled H3 in Figure 2A).
Figure 2.

Comparison of p47 UBX with other UBX domains/ubiquitin and p97 N domain with NSF. (A) Superpositions of p47 C (red) with p47 UBX solution structure (grey, pdb-code 1JRU), FAF1 UBX (blue, pdb-code 1H8C) and ubiquitin (yellow, pdb-code 1UBI). Note the shorter turn between S3 and S4 for ubiquitin and the additional helix H1 in p47 C. (B) Top view of a superposition of p97 ND1 and NSF N domain. The p97 N domain has 9% sequence identity with NSF (1.9 Å rmsd over 117 residues). The p97 N domain is coloured blue, p97 D1 grey and NSF N domain gold. Residues at the p97 N–p47 C complex interface are shown in ball-and-stick representation (red). Residues implicated in α-SNAP binding by NSF N are coloured magenta and are located on the opposite side of the p47-binding site.
Table 2.
Superpositions of p47 C with ubiquitin and ubiquitin-like domains
| Structure aligned with p47 C (PDB code) | No. of aligned residues at 3.5 Å distance cutoff (identical residues) | rmsd (Å) | p47 residue range aligned (identical residues) | Residue range of structure aligned |
|---|---|---|---|---|
| p47 (282–370) sol (1JRU) | 52 (39)a | 2.1 | 297–369 | 295–369 |
| (319–369)a | ||||
| FAF1 UBX (1H8C) | 68 (12) | 1.8 | 295–370 | 5–81 |
| UBI (1UBI) | 65 (12) | 1.5 | 296–370 | 1–72 |
| GATE-16 (1EO6) |
62 (4) |
1.8 |
294–369 |
27–111 |
| aDifferences in the N-terminal part due to a shorter loop between β-strands 1 and 2 in the crystal structure. Alignment of all residues gives rise to an rmsd of 2.7 Å. | ||||
Interactions between p47 C and p97 ND1
In our crystal structure, we observe a complex between the UBX domain of p47 C and a p97 N domain within the p97 hexamer. Additional interactions are mainly observed between the N-terminal extended chain of p47 C (modelled as poly-Ala) and an adjacent p97 hexamer (Figure 1C), which forms a lattice contact. The main β-sheet of the p47 UBX domain contacts the N domain of p97, resulting in a buried surface area of ∼1700 Å2, which is typical for a protein–protein interaction surface (Lo Conte et al, 1999). The most striking feature of the interaction is the insertion of a loop from p47 C (S3/S4 loop; residues 342–345) into a hydrophobic pocket formed between the two p97 N subdomains (p97 Nn and p97 Nc; see Figures 1C and 3). The aromatic ring of Phe343 inserts into the N-domain cleft and makes contacts with a hydrophobic patch comprising residues from the first ψ-loop (Asp35CG, Ser37CB, Val38) and Ile70 (Figure 3B). Pro344 allows for the sharp turn observed in the S3/S4 loop. In addition, the side chain of Asn345 forms a hydrogen bond with the main-chain carbonyl of residue Glu141 in the N domain. A number of other hydrophobic interactions between p47 C and ND1 are observed, which may contribute to the overall stability of the complex, including p47 C residues Leu308, Met340, Ala363, Val364 and Val366, and p97 N residues Phe52, Arg53, Gly54, Pro106, Tyr110 and Tyr143. Hydrogen-bonding interactions are also clearly seen between p47 C Arg301NH2 and p97 N Val108O, as well as p47 C Asn345ND2 and p97 N Glu141O (Figure 3B). p47 homologues from yeast to humans typically share 14–30% sequence identity. The majority of residues at the interface are conserved, although for Drosophila and yeast there are a number of exceptions (see Figure 4A). However, Phe343 and Pro344 within the S3/S4 loop are absolutely conserved, as is Arg301. The extended chain N-terminal of H1 is modelled as poly-Ala and forms a lattice contact. Part of this interaction consists of a short intermolecular β-sheet with a p97 Nc domain from a different hexamer in the plane of the crystal packing (p97 Nc′ in Figure 1C).
Figure 3.
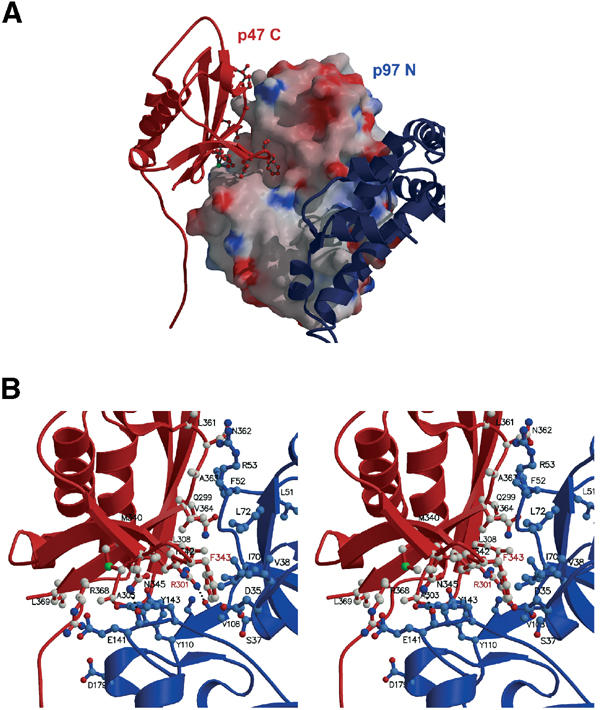
Binding of p47 C to p97 N domain. (A) Electrostatic surface representation of p97 N domain interacting with p47 C (ribbon coloured red). Key residues of p47 C at the N-domain interface are shown in ball-and-stick representation. D1 is depicted in blue. (B) Detailed view of specific interactions at the p47 C–p97 N interface. p47 C is shown as a ribbon representation (red) with key residues in ball-and-stick, p97 N is depicted in blue. Hydrogen-bonding interactions (p47 C Arg301NH2 and p97 N Val108O as well as p47 C Asn345ND2 and p97 N Glu141O) are indicated by a dotted line. Key residues conserved within UBX domains are labelled in red.
Figure 4.
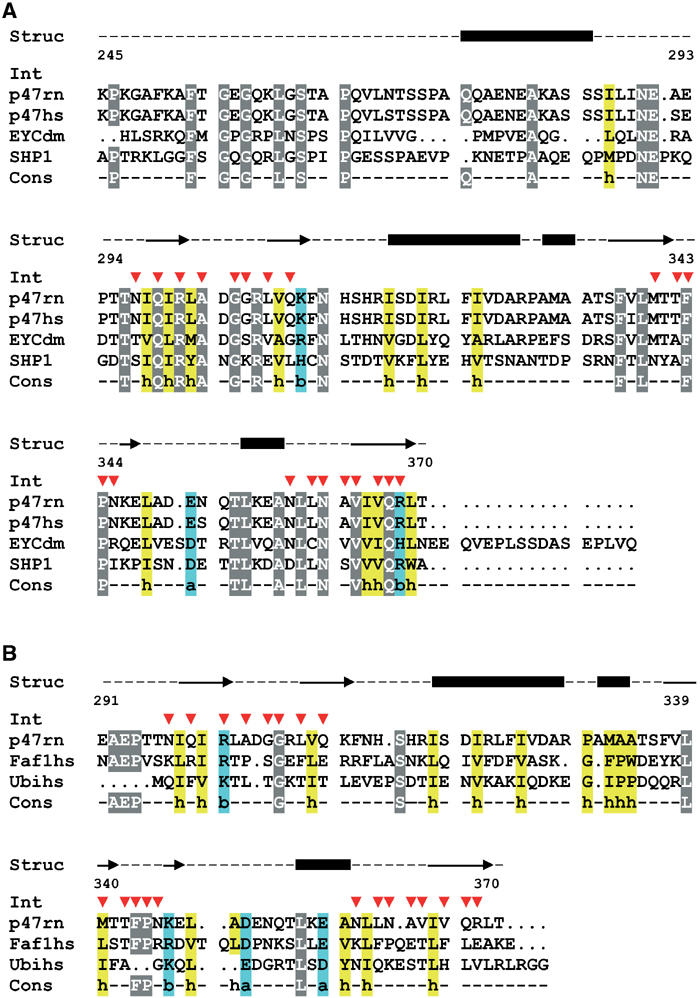
Sequence alignments showing p97–p47 interacting residues. (A) Sequence alignment of p47 homologues (rat p47: accession code tr∣O35987∣, human p47: accession code tr∣Q9UNZ2∣, Drosophila EYC: accession code tr∣Q9U9C9∣ and yeast SHP1: accession code sp∣P34223∣) starting at residue 245 of rat p47. (B) Structure-based sequence alignment of p47 and FAF1 UBX domains and ubiquitin. Residues located at the interface with p97 N in the ‘binary' complex are marked with a red arrow. Secondary structure elements of p47 C are shown above the alignment. Conserved residues are highlighted in grey, ‘h' stands for hydrophobic (yellow), ‘b' for basic and ‘a' for acidic residues (blue).
Comparison with other UBX domain-containing proteins
FAF1, a protein implicated in apoptosis, also contains a C-terminal UBX domain and represents the only other known UBX domain structure (Buchberger et al, 2001). Superposition of the p47 and FAF1 UBX domains reveals a close structural similarity (Figure 2A; Table II). Very few residues are conserved between both domains, as illustrated by a structure-based sequence alignment (Figure 4B), with the majority of these residues likely to contribute to the stability of the fold (Ile300, Gly306, Phe324, Leu339, Pro344, Leu355 and Leu360). Strikingly, residues that mediate the interaction between p97 N and p47 C are conserved in FAF1, namely Arg301 and Phe343 and two other hydrophobic residues (Leu308, Val366). The S3/S4 loop, although similar in sequence (Figure 4B), shows small differences in conformation compared to p47 (Figure 2A), which may be due to FAF1 being unbound. Nearly all UBX domains contain an arginine or less frequently a lysine at equivalent positions to Arg301 in p47 and a hydrophobic residue followed by a proline in the putative S3/S4 loop, indicating the functional importance of these residues (Buchberger et al, 2001). It is notable that VCIP135, which binds to p97 in a mutually exclusive manner with p47, also contains a predicted UBX domain (Uchiyama et al, 2002), and the main p97-interacting residues (Arg301 and Phe343) are conserved.
The S3/S4 loop is essential for binding to full-length p97
Despite a lack of sequence similarity, the UBX domain of FAF1 and ubiquitin were suggested to be evolutionary related (Buchberger et al, 2001; Buchberger, 2002). From our crystal structure, the p47 UBX domain is in fact more similar to ubiquitin (Ramage et al, 1994) than FAF1 (Buchberger et al, 2001), despite both having low respective sequence identities (see Figure 2A and Table II). This supports the notion that ubiquitin is the closest structural relative to the UBX domain known to date. In a structure-based sequence alignment (Figure 4B), the equivalent S3/S4 loop in ubiquitin is shortened by two residues, which interestingly correspond to Phe343 and Pro344. In ubiquitin, a glycine at the same position as Asn345 allows a sharp turn, which results in a shorter loop. To address the question as to whether the S3/S4 loop is important in mediating a full-length p97–p47 C interaction, we designed several mutants, including one that mimicked the shorter loop observed in ubiquitin (Thr342, Phe343, Pro344, Asn345 to Ala, Gly) and three single-point mutants of p97-interacting residues (Phe343Ser, Asn345Ala, Arg301Ala). In vitro binding studies clearly show that p47 C binding to full-length p97 is almost abolished for the loop mutant (Figure 5A) and significantly reduced for the single-point mutations (Figure 5B). Together, these data provide direct evidence for the importance of the S3/S4 loop in mediating p47 C–p97 binding.
Figure 5.
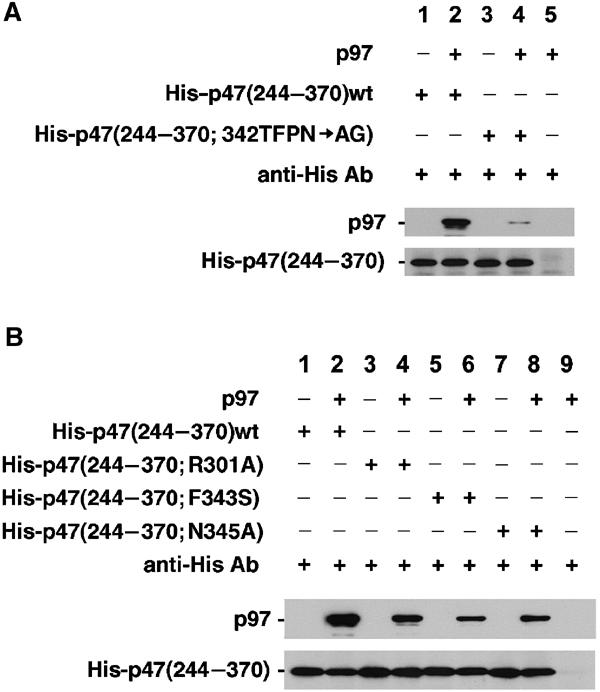
The S3/S4 loop is essential for binding to p97. (A) Binding studies of p47 C and mutant p47 C harbouring a shortened S3/S4 loop (Thr342, Phe343, Pro344, Asn345 to Ala, Gly) with full-length p97. The binding of p97 to the p47 C mutant (lane 4) is very much reduced compared to p47 C wild type (lane 2). (B) Effect of single-point mutations in p47 C upon p97 binding. S3/S4 loop mutants Phe343Ser (lane 6) and Asn345Ala (lane 8) as well as the hydrogen bonding interrupting mutant Arg301Ala (lane 4) show reduced binding to full-length p97 compared to wild-type p47 C (lane 2).
Comparison of N domains between p97 and its homologues
p97 homologues are found in species as diverse as archaebacteria and humans, whereas p47 homologues are only present in eukaryotes. The residues at the p97 N–p47 C binding interface are located around and within the cleft formed between the two p97 N subdomains Nn and Nc (Figure 2B). Residues that form part of the buried interaction surface in p97 Nn are located between strands S1 and S2 (referred to as one of the two ψ-loops), within H1, between H1 and S3, and on S4. The linker region between p97 Nn and p97 Nc is also involved as well as residues on S7, between H4 and S9, on S11 and on H5. Interestingly, all these residues are not conserved between p97, VAT and NSF. Moreover, a comparison of the p47 interaction regions in p97, VAT and NSF shows significant differences in secondary structure/and or chain length. Differences include an inserted long helix between strands S1 and S2, a lack of H1, a longer linker between the two subdomains and slightly different orientations of H4, S11 and H5 in NSF (Figure 2B). The loop between S10 and S11 in NSF is longer (16 residues) and is partly disordered in the crystal structure (May et al, 1999; Yu et al, 1999). In contrast, VAT N (27% sequence identity with p97 N, rmsd of 1.9 Å over 127 residues) has a shorter linker region between the two subdomains and the region between H4 and S9 is also shortened (Coles et al, 1999). However, the p47 contact residues are mostly conserved within eukaryotic p97 homologues, such as humans and rats (99% sequence identity), both of which are known to interact with p47, as well as the yeast homologue CDC48 (66% sequence identity).
Discussion
The p47 UBX domain binds to p97 N
The crystal structure of the p97 ND1–p47 C complex provides the first detailed description of the structural organisation of an AAA protein hexamer in complex with its adaptor molecule, as well as the mode of interaction between a UBX domain and a target protein. The p47 UBX domain predominantly binds to a single p97 N domain (‘binary complex') forming a large buried surface area, which is consistent with our previous NMR chemical shift perturbation studies on p47–p97 (Yuan et al, 2001) that implicated the p47 UBX β-sheet as forming part of the p97 N-domain interaction region. Peptide-mapping studies also show that residues from both p97 N subdomains are required for p47 (271–370) binding, which agrees with our crystal structure where residues from both subdomains contact p47 (Uchiyama et al, 2002). Furthermore, cryo-EM studies of the full-length p97–p47 complex show p47 at the periphery of the p97 hexamer (Rouiller et al, 2000; see Figure 6 and later discussion). The mainly hydrophobic nature of the p97 N–p47 UBX interface is also consistent with the observation that full-length p97–p47 complex is stable in the presence of high salt concentrations (up to 0.5 M KCl). Direct evidence, however, for the observed complex is provided by our mutagenesis studies of the p47 S3/S4 loop, which protrudes into the cleft formed between the p97 N subdomains. Mutant p47 C with a shortened loop shows significantly reduced full-length p97 binding in vitro, as do p47 C single-point mutants within the loop region. A further point mutant on β-strand S1 that disrupts a hydrogen-bonding interaction at the interface also shows reduced p97 binding in vitro. In conclusion, the binding of p47 to p97 is mediated primarily via the S3/S4 loop of the p47 UBX domain. This observed interaction is supported by mutagenesis data and in vitro full-length p97-binding studies.
Figure 6.
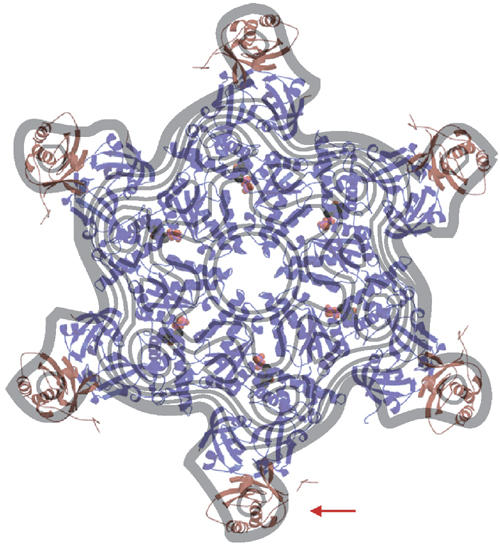
Model of p47–p97 as a physiological complex. Fitting of a model based on our crystal structure with six p47 C molecules bound to the periphery of the p97 ND1 hexamer onto a cryo-EM projection map of the full-length p97–p47 complex (figure adapted from Rouiller et al, 2000). Note the remarkable agreement in both dimensions and overall fit of the ND1–p47 C structure. An arrow indicates the position of the p47 C domain bound to the ND1 hexamer, which fits well in density attributed to p47 by Rouiller et al (2000).
There is at present no evidence to support the binding of the N-terminal extended chain to p97, modelled as poly-Ala in our crystal structure. This interaction, which forms a lattice contact with a different p97 ND1 hexamer, could mimic an additional p47–p97 interaction, positioning the remaining p47 chain at the top of the hexamer. Interestingly, in the related ubiquitin-like structure, GABARAP (γ-aminobutyric acid receptor type A receptor-associated protein), a proline (Pro10) located at the N-terminal end of the equivalent H1 helix, mediates two conformations, which are likely to be biologically relevant (Coyle et al, 2002). In p47, this proline (Pro273) and/or Pro265 and three glycines (Gly255, Gly257, Gly261) could be responsible for different conformations in this region. Parts of this sequence motif are also found in UFD1 (Meyer, personal communication), a known p97-binding protein, although the exact mapping of the interaction is not known. p97 also interacts with UFD1, SVIP and p47 in a mutually exclusive manner (Meyer et al, 2000; Nagahama et al, 2003), which implies that they share part of the same interface.
A common function for UBX domains?
Although UBX domains were suggested to play a role in ubiquitin-related processes such as protein degradation, endocytosis and DNA repair, their role as a ubiquitin-like modifier seems unlikely, given that they lack a C-terminal extension required for conjugation to target proteins (Buchberger et al, 2001). In the nonredundant SMART database (Letunic et al, 2002), 166 proteins contain a UBX domain. UBX domains share little overall sequence homology and can be found either at the C-terminus or in the middle of a protein. However, the few highly conserved residues (Arg301, Phe343, Pro344 in p47) within the UBX family are proposed to be functionally important (Buchberger et al, 2001). We now demonstrate that all these residues are involved in p97 N-domain binding and that mutating either Phe343 or Arg301 results in reduced full-length p97 binding in vitro. It is noteworthy that insertions and deletions in UBX domains are mainly restricted to loops after H1 (H2 in p47 C) and S4 (Buchberger et al, 2001), which should not interfere with p97 binding. This leads us to suggest that UBX domains may act as general binding modules for p97 and/or p97 homologues, possibly representing a first common role for UBX domains.
UBX domains and ubiquitin—possible implications for ubiquitin-like domains
The UBX domain and ubiquitin adopt a β-grasp fold, which is characterised by the presence of a β–β–α–β–β unit (Lo Conte et al, 2000). Proteins of that fold have been shown to form protein–protein complexes via intermolecular β-sheet interactions utilising strand S2. Examples are the ElonginB–ElonginC complex (Stebbins et al, 1999) and the RalGDS–Ras complex (Huang et al, 1998). However, for the p47 UBX domain, the S3/S4 loop is important for p97 interaction. Another protein containing a β-grasp fold domain, namely jak1, also uses this loop for interaction with a cytokine receptor, as demonstrated by homology modelling and mutagenesis (Haan et al, 2001). Compared to ubiquitin, ElonginB and GATE-16, p47 C/jak1 contains a longer S3/S4 loop. It is tempting to speculate that during the evolution of the β-grasp fold a longer loop has developed at this position for interaction with other proteins. A survey of all β-grasp fold structures known to date reveals about half that contain a longer S3/S4 loop than ubiquitin. Interestingly, β-grasp fold proteins that act as modifiers (RUB1/NEDD8, HUB, GATE-16, SUMO, for a review, see Jentsch and Pyrowolakis, 2000) tend to have shortened S3/S4 loops similar to ubiquitin. Therefore, a classification of ubiquitin-like domains could take the length of the S3/S4 loop into account.
For ubiquitin, two hydrophobic clusters constitute the main functional/binding sites and not directly the S3/S4 loop (Sloper-Mould et al, 2001). Interestingly, CDC48, in the absence of nucleotide or the presence of ATP, binds efficiently to tetra-ubiquitin and requires the N domain for binding (Dai and Li, 2001). The binding of mono-ubiquitin, however, is still a matter of debate (Dai and Li, 2001; Rape et al, 2001; Meyer et al, 2002). p97 has been suggested to function as a proteasome-independent chaperone facilitating proteolysis (Dai et al, 1998; Thrower et al, 2000), and ubiquitin binding to p97 may provide insights into how poly-ubiquitin chains bind to the proteasome (Lam et al, 2002). Apart from a longer S3/S4 loop, ubiquitin also lacks some of the UBX hydrophobic residues at its C-terminus. Hence, our proposal suggests that, despite a very similar fold, ubiquitin is unlikely to bind to p97 N in the same manner as p47 UBX. However, for poly-ubiquitin, it cannot be ruled out that additional interactions between poly-ubiquitin chains and CDC48 compensate for the shortened S3/S4 loop.
Other proteins that adopt a β-grasp fold and bind to AAA proteins
There is at least one other protein that adopts a β-grasp fold and binds to a type II AAA protein, namely GATE-16. GATE-16 aids in intra-Golgi transport and binds to NSF (but not p97) to enhance its ATPase activity, and can associate with the Golgi v-SNARE GOS-28 (Sagiv et al, 2000). In contrast, p47 is known to inhibit p97 ATPase activity (Meyer et al, 1998). Additionally, GATE-16 only consists of 117 residues and has been shown to act as a covalent modifier protein (Tanida et al, 2003). From a comparison of the structures of GATE-16 and p47 C (Table II), as well as p97 N and NSF N, it is likely that GATE-16 binds to NSF in a manner different from that of p47 to p97, since GATE-16 has a short S3/S4 loop and the corresponding binding surface of NSF differs from that seen in p97 N.
In our complex structure, we observe an N-terminal helix (H1) as an extension to the ubiquitin fold of p47 UBX, which is not observed in the solution structure (Yuan et al, 2001). To our knowledge, only GATE-16 and its close orthologue GABARAP contain N-terminal helical extensions (Paz et al, 2000; Knight et al, 2002), which for GABARAP has been implicated in tubulin binding (Knight et al, 2002). However, the sequence that forms H1 in p47 is only conserved in mammalian p47 homologues (Figure 4A) and the orientation of the corresponding helix in GATE-16 is different.
p97–p47 as a functional complex
It has been suggested that either three (Kondo et al, 1997) or six (Rouiller et al, 2000) p47 molecules can bind to a p97 hexamer. Although the p47 fragment used in our studies is sufficient for full-length p47–p97 binding (Uchiyama et al, 2002), our p97 ND1–p47 C crystal structure cannot confirm the stoichiometry of the complex, since we only observe two p47 C domains bound to the ND1 hexamer and a third partially ordered domain. Preliminary isothermal titration calorimetry experiments, however, using p97 ND1 and p47 C, indicate that ND1 can bind six p47 C molecules when saturated (unpublished data), conditions that are similar to those used for complex formation. The binding arrangement that we observe in our crystal structure is therefore probably influenced by the crystallisation process, since crystal packing favours p47 C binding to certain protomers over others. By using the observed p47 C N-domain binding arrangement in the crystal structure, we can construct a ‘six to six model' of the p47 C–ND1 complex. Strikingly, such a model is in near perfect agreement with a contour plot of a cryo-EM projection map of full-length p97–p47 complex (Rouiller et al, 2000; Figure 6). This even extends to the location of the p47 C domain within the EM map, which is attributed to p47 by difference maps of unbound and bound p97 (Rouiller et al 2000). This fit strongly argues that p47 in the full-length complex binds to p97 as we observe in our crystal structure, and represents at least one functional state.
Interestingly, when compared to the ND1 structure alone, the N domains in the p47 C complex are located in similar positions. This suggests that the observed ND1 conformation in both structures is biologically relevant, possibly representing a distinct nucleotide-dependent state. For NSF, the adaptor α-SNAP also binds to N domains, although EM shows that it binds on top of the NSF hexamer (Hanson et al, 1997; Hohl et al, 1998). The equivalent residues in p97 required for α-SNAP binding (Matveeva et al, 2002) are located in close proximity to D1 on the opposite side of the p97 N–p47 C interface (Figure 2B). This suggests a different binding mode for α-SNAP to NSF, which is plausible given that the structures of α-SNAP (Rice and Brunger, 1999) and p47 C are unrelated. Assuming a similar arrangement of ND1 domains in NSF, these residues would be inaccessible. For NSF, it has therefore been proposed that these α-SNAP-binding residues become exposed in the presence of ATP, since ATP is required for α-SNAP–SNARE binding (May et al, 2001). This would suggest that N domains have distinct nucleotide-dependent conformations, which is consistent with previous EM studies (Rouiller et al, 2002; Beuron et al, 2003). The proposed flexibility of N domains argues that N-domain motions when bound to adaptors could mediate p97 function by transmitting the energy required for disassembly and/or unfolding of p97/VCP target proteins.
Materials and methods
Expression and purification of the p97 ND1–p47 C complex
The cDNA corresponding to residues 2–458 of murine p97 (ND1 domains) was cloned into pET22b (NdeI/BamHI) (Novagen) and the cDNA corresponding to the C-terminal p47 fragment (residues 244–370) was cloned into pPRO-EX Hta (SfoI/BamHI) (Life Tech). Proteins were expressed in Escherichia coli Rosetta (DE3) cells (Novagen) at 37°C by inducing a mid-log phase culture with 1 mM IPTG. Cell pellets were lysed by sonication and the His-tagged p47 C was purified on a chelating HiTrap column (Pharmacia) charged with NiSO4. The obtained protein fractions were pooled, dialysed and reloaded onto the column before adding the ND1 lysate. Elution of the complex was performed with an imidazole gradient and resulted in a fraction of ND1–p47 C complex, which was further purified on a Superdex-200 column (Pharmacia) equilibrated with 50 mM Tris–Cl, 5 mM MgCl2, 5% glycerol, 150 mM KCl, pH 7.4. The pure peak fractions were pooled and concentrated to 10 mg/ml.
Protein crystallisation and data collection
Single crystals suitable for crystallographic analysis were obtained at 20°C by the hanging drop vapour diffusion method using 5 μl of the protein mixed with 5 μl of a mother liquor containing 10% PEGmme 5 K, 100 mM citrate, 50 mM KSCN, pH 5.5. The crystals typically grew to a size of 0.5 × 0.05 × 0.05 mm3. For data collection, crystals were transferred to different drops containing mother liquor with increasing amounts of PEGmme 5K and PEG400 and flash frozen in liquid nitrogen. Data were collected at the European Synchrotron Radiation Facility in Grenoble on beamline ID-14-4 equipped with a MAR CCD detector. The crystals belong to space group P65, as confirmed by the molecular replacement procedure with cell parameters of a=b=157.7 Å, c=243.2 Å, α=β=90°, γ=120° and diffracted to a resolution of 2.9 Å. Data were processed with Denzo and Scalepack (Otwinowski and Minor, 1997). The summary of the data collection statistics is shown in Table I.
Table 1.
Data collection and refinement statistics
| Data collection statistics | |
|---|---|
| Cell parameters | 157.7 Å; 157.7 Å; 243.2 Å; 90°; 90°; 120° |
| Space group | P65 |
| Wavelength | 0.9394 Å |
| Resolution max. | 2.9 Å |
| Completeness (last shell) | 96.3% (99.3%) |
| Rmerge (last shell) | 9.6% (58.0%) |
| Redundancy | 6.9 |
| 〈I/σI〉 | 10.5 (3.1) |
| Refinement | |
| Resolution range | 40–2.9 Å |
| No. of reflections | 72860 (6732 in test set) |
| R-factor | 24.8% |
| Rfree | 29.5% |
| No. of residues | 2871 |
| No. of ADP | 6 |
| Solvent | 63 |
| Ramachandran plot | |
| Most favoured regions | 81.7% |
| Add. allowed regions | 16.8% |
| Disallowed regions |
0.2% |
| Rmerge=∑hkl∑i∣I(hkl)i−〈I(hkl)〉∣/∑hkl∑iI(hkl)i. | |
| R-factor=∑hkl∣Fo(hkl)−Fc(hkl)∣/∑hkl∣Fo(hkl)∣, where Fo and Fc are observed and calculated structure factors, respectively. | |
Structure solution and refinement
The ND1–p47 C structure was solved by the molecular replacement method employing the full ND1 hexamer as a search model using the program CNS (Brunger et al, 1998). The correct solution was the highest peak in the rotation and translation function in space group P65. Initial rigid body refinement of this solution resulted in an Rfree of 39%. Additional density near two protomers of the p97 ND1 hexamer was clearly visible in the initial electron density map. Several rounds of manual building and rebuilding were carried out using the program O (Jones et al, 1991) with careful inspection of 2Fo-Fc, Fo-Fc and ‘omit' electron density maps. Due to the unfavourable observation to parameter ratio (Table I), refinement was restricted to rigid body refinement and a few rounds of NCS-restrained refinement followed by further NCS-unrestrained rigid body refinement (defining either N domains and D1 domains as well as p47 C as rigid bodies or the whole protomers and p47 C) using CNS (Brunger et al, 1998). A final round of grouped B-factor refinement was carried out and a few ordered water molecules were added, resulting in a final R factor of 24.8% and Rfree of 29.5%. Some residual uninterpretable density was still visible at the end of refinement, which we attribute to a third p47 C molecule, as well as the residues at the N-termini of ND1 and p47 C. Due to a lack of side chain density, a few surface residues and residues N-terminal of Pro273 (p47 C) were modelled as alanines. For one of the p47 C copies, only parts of the N-terminal extended chain were built. The final refinement statistics are given in Table I. Figures were prepared using GRASP (Nicholls, 1993), MOLSCRIPT (Kraulis, 1991) and RASTER3D (Merritt and Bacon, 1997).
Mutation of p47 C and binding studies with full-length p97
To obtain His-tagged p47 (244–370) fragment, the corresponding cDNA with stop codon was subcloned into pQE30 using BamHI and KpnI sites. Mutations (Thr342, Phe343, Pro344, Asn345 to Ala, Gly and single-point mutations Arg301Ala, Phe343Ser or Asn345Ala) were directly introduced into the cDNA in pQE30 by PCR reactions, using the Quick-change mutagenesis kit (Stratagene), as described previously (Uchiyama et al, 2003). The clones were verified by DNA sequencing. The fragments were expressed in E. coli and purified with Ni beads, followed by further rounds of purification with gel filtration. Recombinant p97 without a tag was expressed in E. coli and purified using standard biochemical procedures. His-tagged p47 C (1.5 μg) was incubated with p97 (9.0 μg) in 200 μl of buffer (20 mM HEPES, 0.2 M KCl, 1 mM MgCl2, 0.2 mM ATP, 1 mM DTT, 0.1% Triton X100, 10% glycerol, pH 7.4). After 1 h incubation on ice, the p47 fragment was isolated using anti-His antibodies and protein G-beads, and bound proteins were fractionated by SDS–PAGE. Blots were probed with anti-His and anti-p97 antibodies, respectively.
Accession codes
Coordinates have been deposited in the Protein Data Bank with accession code 1S3S.
Acknowledgments
We thank Melanie Thein for assisting in the early work of the project and Fabienne Beuron, Trevor Huyton, Steve Matthews, Ciaran McKeown, Valerie Pye and Xuemei Yuan for stimulating discussions and critically reading the manuscript, Catherine Keetch and Carol Robinson for fruitful discussions, John Ladbury and Matthew Cliff for help with the ITC experiment and Suhail Islam for help with the figures. We also thank the beamline scientists at ID-14-4 for help with data collection. We thank the Wellcome Trust for their generous funding of this work.
References
- Bays NW, Hampton RY (2002) Cdc48-ufd1-npl4: stuck in the middle with ub. Curr Biol 12: R366–R371 [DOI] [PubMed] [Google Scholar]
- Beuron F, Flynn TC, Ma J, Kondo H, Zhang X, Freemont PS (2003) Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantised elastic deformational model. J Mol Biol 327: 619–629 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, Delano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Warren GL (1998) Crystallography and NMR system (CNS): a software system for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Buchberger A (2002) From UBA to UBX: new words in the ubiquitin vocabulary. Trends Cell Biol 12: 216–221 [DOI] [PubMed] [Google Scholar]
- Buchberger A, Howard MJ, Proctor M, Bycroft M (2001) The UBX domain: a widespread ubiquitin-like module. J Mol Biol 307: 17–24 [DOI] [PubMed] [Google Scholar]
- Coles M, Diercks T, Liermann J, Groger A, Rockel B, Baumeister W, Koretke KK, Lupas A, Peters J, Kessler H (1999) The solution structure of VAT-N reveals a ‘missing link' in the evolution of complex enzymes from a simple betaalphabetabeta element. Curr Biol 9: 1158–1168 [DOI] [PubMed] [Google Scholar]
- Coyle JE, Qamar S, Rajashankar KR, Nikolov DB (2002) Structure of GABARAP in two conformations: implications for GABA(A) receptor localization and tubulin binding. Neuron 33: 63–74 [DOI] [PubMed] [Google Scholar]
- Dai RM, Chen E, Longo DL, Gorbea CM, Li CC (1998) Involvement of valosin-containing protein, an ATPase co-purified with IkappaBalpha and 26 S proteasome, in ubiquitin-proteasome-mediated degradation of IkappaBalpha. J Biol Chem 273: 3562–3573 [DOI] [PubMed] [Google Scholar]
- Dai RM, Li CC (2001) Valosin-containing protein is a multi-ubiquitin chain-targeting factor required in ubiquitin-proteasome degradation. Nat Cell Biol 3: 740–744 [DOI] [PubMed] [Google Scholar]
- Dougan DA, Mogk A, Zeth K, Turgay K, Bukau B (2002) AAA+ proteins and substrate recognition, it all depends on their partner in crime. FEBS Lett 529: 6–10 [DOI] [PubMed] [Google Scholar]
- Golbik R, Lupas AN, Koretke KK, Baumeister W, Peters J (1999) The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol Chem 380: 1049–1062 [DOI] [PubMed] [Google Scholar]
- Guo F, Esser L, Singh SK, Maurizi MR, Xia D (2002a) Crystal structure of the heterodimeric complex of the adaptor, ClpS, with the N-domain of the AAA+ chaperone, ClpA. J Biol Chem 277: 46753–46762 [DOI] [PubMed] [Google Scholar]
- Guo F, Maurizi MR, Esser L, Xia D (2002b) Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J Biol Chem 277: 46743–46752 [DOI] [PubMed] [Google Scholar]
- Haan C, Is'harc H, Hermanns HM, Schmitz-Van De Leur H, Kerr IM, Heinrich PC, Grotzinger J, Behrmann I (2001) Mapping of a region within the N terminus of Jak1 involved in cytokine receptor interaction. J Biol Chem 276: 37451–37458 [DOI] [PubMed] [Google Scholar]
- Hanson PI, Roth R, Morisaki H, Jahn R, Heuser JE (1997) Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy. Cell 90: 523–535 [DOI] [PubMed] [Google Scholar]
- Hetzer M, Meyer HH, Walther TC, Bilbao-Cortes D, Warren G, Mattaj IW (2001) Distinct AAA-ATPase p97 complexes function in discrete steps of nuclear assembly. Nat Cell Biol 3: 1086–1091 [DOI] [PubMed] [Google Scholar]
- Hofmann K, Bucher P (1996) The UBA domain: a sequence motif present in multiple enzyme classes of the ubiquitination pathway. Trends Biochem Sci 21: 172–173 [PubMed] [Google Scholar]
- Hohl TM, Parlati F, Wimmer C, Rothman JE, Sollner TH, Engelhardt H (1998) Arrangement of subunits in 20 S particles consisting of NSF, SNAPs, and SNARE complexes. Mol Cell 2: 539–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Hofer F, Martin GS, Kim SH (1998) Structural basis for the interaction of Ras with RalGDS. Nat Struct Biol 5: 422–426 [DOI] [PubMed] [Google Scholar]
- Huyton T, Pye V, Briggs L, Flynn TC, Beuron F, Kondo H, Ma J, Zhang X, Freemont PS (2003) The crystal structure of murine p97/VCP at 3.6 Å. J Struct Biol 144: 337–348 [DOI] [PubMed] [Google Scholar]
- Jentsch S, Pyrowolakis G (2000) Ubiquitin and its kin: how close are the family ties? Trends Cell Biol 10: 335–342 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan S, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kaneko C, Hatakeyama S, Matsumoto M, Yada M, Nakayama K, Nakayama KI (2003) Characterization of the mouse gene for the U-box-type ubiquitin ligase UFD2a. Biochem Biophys Res Commun 300: 297–304 [DOI] [PubMed] [Google Scholar]
- Knight D, Harris R, McAlister MS, Phelan JP, Geddes S, Moss SJ, Driscoll PC, Keep NH (2002) The X-ray crystal structure and putative ligand-derived peptide binding properties of gamma-aminobutyric acid receptor type A receptor-associated protein. J Biol Chem 277: 5556–5561 [DOI] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G (1997) p47 is a cofactor for p97-mediated membrane fusion. Nature 388: 75–78 [DOI] [PubMed] [Google Scholar]
- Kraulis PJ (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr 24: 946–950 [Google Scholar]
- Krzywda S, Brzozowski AM, Verma C, Karata K, Ogura T, Wilkinson AJ (2002) The crystal structure of the AAA domain of the ATP-dependent protease FtsH of Escherichia coli at 1.5 A resolution. Structure (Camb) 10: 1073–1083 [DOI] [PubMed] [Google Scholar]
- Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart CM (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416: 763–767 [DOI] [PubMed] [Google Scholar]
- Letunic I, Goodstadt L, Dickens NJ, Doerks T, Schultz J, Mott R, Ciccarelli F, Copley RR, Ponting CP, Bork P (2002) Recent improvements to the SMART domain-based sequence annotation resource. Nucleic Acids Res 30: 242–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res 28: 257–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Conte L, Chothia C, Janin J (1999) The atomic structure of protein–protein recognition sites. J Mol Biol 285: 2177–2198 [DOI] [PubMed] [Google Scholar]
- Lupas AN, Martin J (2002) AAA proteins. Curr Opin Struct Biol 12: 746–753 [DOI] [PubMed] [Google Scholar]
- Matveeva EA, May AP, He P, Whiteheart SW (2002) Uncoupling the ATPase activity of the N-ethylmaleimide sensitive factor (NSF) from 20S complex disassembly. Biochemistry 41: 530–536 [DOI] [PubMed] [Google Scholar]
- May AP, Misura KM, Whiteheart SW, Weis WI (1999) Crystal structure of the amino-terminal domain of N-ethylmaleimide-sensitive fusion protein. Nat Cell Biol 1: 175–182 [DOI] [PubMed] [Google Scholar]
- May AP, Whiteheart SW, Weis WI (2001) Unraveling the mechanism of the vesicle transport ATPase NSF, the N-ethylmaleimide-sensitive factor. J Biol Chem 276: 21991–21994 [DOI] [PubMed] [Google Scholar]
- Merritt EA, Bacon DJ (1997) Raster3D: photorealistic molecular graphics. Meth Enzymol 277: 505–524 [DOI] [PubMed] [Google Scholar]
- Meyer HH, Kondo H, Warren G (1998) The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS Lett 437: 255–257 [DOI] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G (2000) A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. EMBO J 19: 2181–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G (2002) Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. EMBO J 21: 5645–5652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahama M, Suzuki M, Hamada Y, Hatsuzawa K, Tani K, Yamamoto A, Tagaya M (2003) SVIP is a novel VCP/p97-interacting protein whose expression causes cell vacuolation. Mol Biol Cell 14: 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls AJ (1993) GRASP: Graphical Representation and Analysis of Surface Properties. New York, NY, USA: Columbia University [Google Scholar]
- Niwa H, Tsuchiya D, Makyio H, Yoshida M, Morikawa K (2002) Hexameric ring structure of the ATPase domain of the membrane-integrated metalloprotease FtsH from Thermus thermophilus HB8. Structure (Camb) 10: 1415–1423 [DOI] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ (2001) AAA+ superfamily ATPases: common structure—diverse function. Genes Cells 6: 575–597 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of x-ray diffraction data collected in oscillation mode. Meth Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Paz Y, Elazar Z, Fass D (2000) Structure of GATE-16, membrane transport modulator and mammalian ortholog of autophagocytosis factor Aut7p. J Biol Chem 275: 25445–25450 [DOI] [PubMed] [Google Scholar]
- Pleasure IT, Black MM, Keen JH (1993) Valosin-containing protein, VCP, is a ubiquitous clathrin-binding protein. Nature 365: 459–462 [DOI] [PubMed] [Google Scholar]
- Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G (1998) Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell 92: 603–610 [DOI] [PubMed] [Google Scholar]
- Ramage R, Green J, Muir TW, Ogunjobi OM, Love S, Shaw K (1994) Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin. Biochem J 299 (Part 1): 151–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S (2001) Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107: 667–677 [DOI] [PubMed] [Google Scholar]
- Rice LM, Brunger AT (1999) Crystal structure of the vesicular transport protein Sec17: implications for SNAP function in SNARE complex disassembly. Mol Cell 4: 85–95 [DOI] [PubMed] [Google Scholar]
- Rouiller I, Butel VM, Latterich M, Milligan RA, Wilson-Kubalek EM (2000) A major conformational change in p97 AAA ATPase upon ATP binding. Mol Cell 6: 1485–1490 [DOI] [PubMed] [Google Scholar]
- Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM (2002) Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol 9: 950–957 [DOI] [PubMed] [Google Scholar]
- Sagiv Y, Legesse-Miller A, Porat A, Elazar Z (2000) GATE-16, a membrane transport modulator, interacts with NSF and the Golgi v-SNARE GOS-28. EMBO J 19: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloper-Mould KE, Jemc JC, Pickart CM, Hicke L (2001) Distinct functional surface regions on ubiquitin. J Biol Chem 276: 30483–30489 [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG Jr, Pavletich NP (1999) Structure of the VHL–ElonginC–ElonginB complex: implications for VHL tumor suppressor function. Science 284: 455–461 [DOI] [PubMed] [Google Scholar]
- Tanida I, Komatsu M, Ueno T, Kominami E (2003) GATE-16 and GABARAP are authentic modifiers mediated by Apg7 and Apg3. Biochem Biophys Res Commun 300: 637–644 [DOI] [PubMed] [Google Scholar]
- Thoms S (2002) Cdc48 can distinguish between native and non-native proteins in the absence of cofactors. FEBS Lett 520: 107–110 [DOI] [PubMed] [Google Scholar]
- Thrower JS, Hoffman L, Rechsteiner M, Pickart CM (2000) Recognition of the polyubiquitin proteolytic signal. EMBO J 19: 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, Kondo H (2002) VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. J Cell Biol 159: 855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Lindman M, Jackman M, Kano F, Murata M, Zhang X, Kondo H (2003) The localization and phosphorylation of p47 are important for Golgi disassembly–assembly during the cell cycle. J Cell Biol 161: 1067–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteheart SW, Schraw T, Matveeva EA (2001) N-ethylmaleimide sensitive factor (NSF) structure and function. Int Rev Cytol 207: 71–112 [DOI] [PubMed] [Google Scholar]
- Yamada T, Okuhara K, Iwamatsu A, Seo H, Ohta K, Shibata T, Murofushi H (2000) p97 ATPase, an ATPase involved in membrane fusion, interacts with DNA unwinding factor (DUF) that functions in DNA replication. FEBS Lett 466: 287–291 [DOI] [PubMed] [Google Scholar]
- Yu RC, Jahn R, Brunger AT (1999) NSF N-terminal domain crystal structure: models of NSF function. Mol Cell 4: 97–107 [DOI] [PubMed] [Google Scholar]
- Yuan X, Shaw A, Zhang X, Kondo H, Lally J, Freemont PS, Matthews S (2001) Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly. J Mol Biol 311: 255–263 [DOI] [PubMed] [Google Scholar]
- Zeth K, Ravelli RB, Paal K, Cusack S, Bukau B, Dougan DA (2002) Structural analysis of the adaptor protein ClpS in complex with the N-terminal domain of ClpA. Nat Struct Biol 9: 906–911 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang Q, Kajino K, Greene MI (2000a) VCP, a weak ATPase involved in multiple cellular events, interacts physically with BRCA1 in the nucleus of living cells. DNA Cell Biol 19: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS (2000b) Structure of the AAA ATPase p97. Mol Cell 6: 1473–1484 [DOI] [PubMed] [Google Scholar]


