Abstract
Background
Numerous studies have examined gene × environment interactions (G×E) in cognitive and behavioral domains. However, these studies have been limited in that they have not been able to directly assess differential patterns of gene expression in the human brain. Here we assessed G×E interactions using two publically-available datasets to assess if DNA variation is associated with post-mortem brain gene expression changes based on smoking behavior, a biobehavioral construct that is part of a complex system of genetic and environmental influences.
Methods
We conducted an expression quantitative trait locus (eQTL) study on two independent human brain gene expression datasets assessing G×E for selected psychiatric genes and smoking status. We employed linear regression to model the significance of the Gene×Smoking interaction term, followed by meta-analysis across datasets.
Results
Overall, we observed that the effect of DNA variation on gene expression is moderated by smoking status. Expression of 16 genes were significantly associated with single nucleotide polymorphisms that demonstrated G×E effects. The strongest finding (p = 1.9×10−11) was neurexin 3-alpha (NRXN3), a synaptic cell-cell adhesion molecule involved in maintenance of neural connections (such as the maintenance of smoking behavior). Other significant G×E associations include four glutamate genes.
Conclusions
This is one of the first studies to demonstrate G×E effects within the human brain. In particular, this study implicated NRXN3 in the maintenance of smoking. The effect of smoking on NRXN3 expression and downstream behavior is different based upon SNP genotype, indicating that DNA profiles based on SNPs could be useful in understanding the effects of smoking behaviors. These results suggest that better measurement of psychiatric conditions, and the environment in post-mortem brain studies may yield an important avenue for understanding the biological mechanisms of G×E interactions in psychiatry.
Keywords: Genetics, environment, brain development, developmental psychopathology
Introduction
The effects of gene by environment (G×E) interaction on behavior are presumably mediated by differences in gene expression in the brain. There is a large and growing literature assessing measured G×E interactions to behaviors (Duncan and Keller, 2011), and less frequently with methylation or gene expression in peripheral tissues such as blood (Mehta et al., 2013, Boulle et al., 2012, Bagot and Meaney, 2010). That said, there is an emerging literature examining gene expression the prefrontal cortex to several important psychiatric and health related outcomes. Early life adversity is associated with widespread variation in gene expression in prefrontal cortex in rhesus macaques (Provencal et al., 2012). As well, evidence for G × E variation in gene expression in the brain from these monkeys indicates that serotonin transporter genotype interacts with early life stress to predict an epigenetic mark, H3K4me3 binding at the SLC6A4 promoter in hippocampus (Lindell et al., 2012). Generalization of brain research findings to human has been accomplished in with smaller hypothesis-driven replication study designs (McGowan et al., 2009). However, there has been no previous large sample study to directly examine G×E in brain-based gene expression for complex human behaviors. This is, in part, due to the impossibility of acquiring gene expression data in vivo and difficulty in acquiring appropriate behavioral and environmental data retrospectively from a post-mortem cohort. Thus, although child and adolescent psychiatry has made progress defining DNA variants and their relationship to behavior, the gene expression mechanisms that mediate these relationships have only recently begun being characterized (Naumova et al., 2013, Hebebrand et al., 2010). Understanding the neural underpinnings of behavior would improve if direct links from DNA variation to brain gene expression were to be studied in the context of behavior (Richards et al., 2012b, Kleinman et al., 2011).
This study is the first to examine gene expression in a gene × environment interaction study of smoking. Smoking is a leading cause of preventable death in the United States at about 1 in 5 deaths each year (US Surgeon General, 2012). And despite this fact, 88% of smokers begin by age 18 (Center for Behavioral Health Statistics and Quality, 2011). However, the neurobiological and epigenetic basis of smoking addiction is not well known. In this introduction, we first discuss the intersection of behavior and molecular genetics and discuss the advantage of gene expression studies on power to detect gene × environment interactions. Next we define the eQTL study paradigm and design while explaining relevant concepts. We then establish the basis of this present study, examining brain gene expression and smoking behaviors. Next, we establish relevance of the molecular underpinning of smoking maintenance effects on the brain as it relates to preventing smoking initiation in youth and possible avenues for intervention. In the discussion section, we further argue how the methods applied here may be generalized to other behavioral and psychiatric traits in children and adolescents.
Molecular genetic studies of gene × environment interactions require an assay of DNA variants, a defined and measured behavior of interest, and an environment that can be quantified or used for stratification/grouping (Caspi and Moffitt, 2006). Assaying DNA variation has become robust and routine. The choice of environment and selected metrics pose another barrier as these choices come from an unbounded pool of candidates, many of which may be related to behavior but not via interactions with genes; power is difficult to establish when designing a study and generally assumed to be low (Duncan and Keller, 2011). Finally, in psychiatric studies, the behavioral or clinical outcome is separated from processes involving DNA by many intervening biological steps.
By focusing in gene expression changes in human brain as the outcome, it is possible to reduce the number of biological steps between G × E and the outcome. Moreover, power is also enhanced by employing molecular variables that have strong a priori relationships (Johnson et al., 2005). By doing so, it is possible to model the principles of a G × E expression study in a setting that has greater signal to noise. However, we have chosen our variable set to provide more than a demonstration of a G×E study, but to also understand smoking behaviors, in a way that is relevant to prevention and intervention of smoking that could ideally be applied to adolescents before the cumulative and gradual psychological and medical consequences manifest (Audrain-McGovern et al., 2012, Baler and Volkow, 2011). This is the first molecular genetic study of smoking behavior, using gene × environment interactions directly associated with brain biology.
Mapping traits to DNA variation and gene expression
In animals and lower order model systems, it is possible to reverse engineer the role of genes by modifying or even removing a gene and observing the consequences called the reverse genetics paradigm (Lehner, 2013). However, the forward genetics paradigm applied to humans maps observed characteristics to specific chromosomal regions, with the ultimate goal of identifying specific genes and specific changes within those genes that influence the observed characteristics. In psychiatry, forward genetics is an observational design, where observed traits are measured in and DNA variation is assessed for an association with the observed trait through one of several different statistical genetic paradigms (Bailey-Wilson and Wilson, 2011, Dube and Hegele, 2013, Do et al., 2012). When the observed trait is quantitative in nature, mapping the molecular genetic basis can be called quantitative trait locus (QTL) mapping (Falconer and MacKay, 1996). The chromosomal location, or locus, is the quantity of interest in the calculations. One quantitative trait that perhaps the most fundamental in genetics is measurement of gene expression. This special type of QTL is called an expression quantitative trait locus or eQTL (Damerval et al., 1994, de Vienne et al., 1988). Therefore eQTL mapping is the paradigm for finding DNA variations that underlie variation in gene expression. Since DNA (i.e., the chromosomal location) is the direct template for the synthesis of RNA (i.e., gene expression), they represent a close biological relationship and therefore eQTL studies have much greater effect sizes than observed in behavioral studies.
Gene expression may vary by cell and tissue type, environmental conditions covering everything from moment to moment changes in metabolic activities to fully extrinsic variables like temperature and past experience (Maranville et al., 2012). Some genes are stably expressed regardless of other factors while others may be expressed so quickly or in such a complicated pattern that they cannot be studied using current technology. In between these two boundary conditions lies genes that show variation in expression to some degree. eQTL studies, when conducted on gene expression from the human brain will clearly not be sensitive to genes with rapid responses to the environment as the logistics of tissue dotation preclude the necessary speed in sample processing. However, many of the genes expressed in brain do seem to have variability from person to person that is stable enough and can be reliably measured for eQTL studies to successfully map eQTL’s (Colantuoni et al., 2011, Gibbs et al., 2010, Heinzen et al., 2008, Kang et al., 2011, Liu et al., 2010, Myers et al., 2007, Webster et al., 2009).
To accommodate eQTL studies, datasets must include genotyping and expression profiling in the same subjects, though only two such datasets have meaningful measures of behavior (e.g., smoking status) at the time of death (Colantuoni et al., 2011, Liu et al., 2010). Here, we interrogated those two studies within a meta-analysis framework, to increase power, since the use of different microarray platforms prevented a single unified reanalysis. Our study is able to find novel eQTLs using SNP, gene expression, and environmental measures, in this case smoking status, and it quantifies the interaction between SNPs and the environment that may affect gene expression in the brain.
Gene expression as an outcome of smoking
Aside from demonstrating the veracity of human brain eQTL studies in understanding the brain basis of G×E, smoking behavior represents an important outcome in its own right from genetic, environmental, and biological perspectives. Smoking behavior and nicotine dependence have strong genetic and environmental components, with genetics likely playing a more significant role in dependence (Vink et al., 2005). A number of genetic loci have been previously associated with smoking behaviors. For example, a recent meta-analysis (Tobacco and Genetics Consortium, 2010) identified SNPs associated with the quantitative phenotype number of cigarettes per day within the nicotinic receptor gene CHRNA3 (chromosome 15q25.1), in LOC100188947 (10q15) and in EGLN2 (19q13). In addition, eight SNPs in the gene encoding brain-derived neurotrophic factor (BDNF) were associated with smoking initiation. There is evidence for the involvement of a variety of other genes in smoking-related behaviors, including those encoding the class of cell adhesion proteins known as neurexins, specifically neurexin 3 (Bierut et al., 2007, Docampo et al., 2012, Nussbaum et al., 2008). Neurexins are important developmentally and are associated with dopamine neurons in experimental data in cell culture (Noelker et al., 2012) and indirectly from their association protein-protein interactions with CASK (calcium/calmodulin-dependent serine protein kinase), a protein where mutations in a binding partner causes Parkinson’s disease (Houlden and Singleton, 2012) – a disorder to dopamine depletion.
Twin studies in humans also provide evidence that the genetics of smoking behavior in adulthood is informative about the genetics in adolescence. Several meta-analyses of the heritability of tobacco use report 37-60% of variation in smoking initiation behavior and 46-59% in persistence of smoking to be accounted by additive genetics (Hall et al., 2002, Li et al., 2003, Sullivan and Kendler, 1999). When considering smoking initiation longitudinally, the heritability was not found to change significantly from ages 13 to 46 in a meta-analysis consisting of only longitudinal datasets (Bergen et al., 2007. More recent data on the heritability of nicotine dependence (defined using DSM-III-R criteria), use here as an indicator of smoking persistence, increased modestly from age 15 to 18, and thereafter stabilized from age 18 to 21. However, the additive genetic correlation between ages 15 and 18, and between ages 18 and 21 was quite stable at 0.22 and 0.27, respectively (Tully et al., 2010). Using cigarettes per day as a quantitative metric of smoking behavior assessed at ages 14, 17, 20 and 24 showed that the majority of smoking persistence across those ages was due to additive genetics (Vrieze et al., 2012).
The genetics of smoking from adolescence into adulthood also shows a longitudinally-stable common risk factor with illicit drug use and alcohol use (Baker et al., 2011). The heritability of this common genetic factor was consistent, estimated between 52% and 54% at each age range 13-14, 16-17, and 19-20. Importantly, the heritability of the common variance across ages was >40% for all time points, indicating that the majority of additive genetic effects common to smoking, alcohol and illicit drug use is longitudinally stable.
Smoking is an environmental factor that impacts the brain, resulting in, for example, a greater risk of dementia and cognitive impairments (Anstey et al., 2007). Animal studies have demonstrated that exposure to nicotine can drive changes in gene expression within the cerebral cortex and other brain structures (e.g., (Kenny et al., 2001, Konu et al., 2001, Trauth et al., 1999, Trauth et al., 2000)), and particularly genes associated with the mesocorticolimbic dopaminergic pathway known to play a role in addiction (Flatscher-Bader and Wilce, 2009). Thus, a natural question that we address in the present analysis is whether the expression levels of certain genes in the human frontal cortex, treated as quantitative phenotypes in the eQTL approach, are driven by interactions between genotype and smoking history.
Gene by smoking interactions in child and adolescent psychology
In this study, we examined whether smoking behavior changes the expression of genes in the frontal lobe and whether these changes can be explained by interactions between smoking status and SNPs. It is important to note, that since the environmental variable measured here is smoking at the time of death, this study design may not be sensitive to genetic factors related to the initiation of smoking (Thorgeirsson et al., 2010, Tobacco and Genetics Consortium, 2010, Yoon et al., 2012), but will be important for understanding the effect of smoking on the genetic regulation of the maintenance of smoking behavior. For this reason, we are interested in the extent to which the reward pathways typically studied in the addiction literature are similar to or distinct from neural pathways related to learning and stabilization of synaptic connections to maintain a behavior.
To accomplish the goal of the study, to understand which genes are involved in smoking maintenance in a gene × environment analysis, smoking status at time of death was used here as a measure of the environment. Regular doses of nicotine to the brain alter the environment for neurons and drive changes in gene expression to adapt to that environment (Flatscher-Bader and Wilce, 2009). The outcome of interest was the statistical significance of the interaction term from a linear regression of gene expression values on SNP×Smoking. A significant interaction term indicates a non-additive effect of smoking and SNP, whereby the effects of smoking on gene expression are not the same across all levels of SNP genotype. We focused our study to genes most commonly studied in psychiatric genetics since these genes have the greatest a priori likelihood for association.
Methods
In this section we include a general outline of the methods, leaving more specific details, mostly from genomic and bioinformatics aspects of the study, for the online appendix (Online supporting information: Methodology). The greater detail included in the online supplementary information provides additional data and citations to allow for replication of our work. Note that we used publicly available deidentified datasets where informed consent was obtained by the primary dataset investigators.
We began with a text mining of the psychiatric genetics literature to determine the most commonly studied genes (see Online supplementary information: Methodology). The top 100 genes from this text mining approach were initially selected for study inclusion. We also expanded the list to include additional neurotransmitter family genes including receptors and metabolism genes for the major neurotransmitter systems: acetylcholine, dopamine, GABA, glutamate and serotonin. Table 1 shows the 158 genes selected based on these criteria. However, after filtering expression data (see below) for data quality, suitable variability for analysis, and number of subjects with expression values above background detection levels, as well as requiring that gene expression data were available in both datasets, only 29 genes remained (bolded in Table 1).
Table 1.
Genes of Interest from Text Mining Psychiatric Literature*
| AANAT | ABAT | ACHE | ADRA1D | ADRA2A |
| ADRA2B | ADRB1 | ADRB2 | ADRB3 | AKT1 |
| APOE | APP | BDNF | CHAT | CHRFAM7A |
| CHRM1 | CHRM2 | CHRM3 | CHRM4 | CHRM5 |
| CHRNA1 | CHRNA10 | CHRNA2 | CHRNA3 | CHRNA4 |
| CHRNA5 | CHRNA6 | CHRNA7 | CHRNA9 | CHRNB1 |
| CHRNB2 | CHRNB3 | CHRNB4 | CHRND | CHRNE |
| CHRNG | CLOCK | COMT | CRELD2 | CRHR1 |
| DAG1 | DAOA | DBH | DBI | DDC |
| DISC1 | DISC2 | DRD1 | DRD2 | DRD3 |
| DRD4 | DRD5 | DTNBP1 | DUSP27 | DUT |
| EFNB3 | FOXP2 | GABBR1 | GABBR2 | GABRA1 |
| GABRA2 | GABRA3 | GABRA4 | GABRA5 | GABRA6 |
| GABRB1 | GABRB2 | GABRB3 | GABRD | GABRE |
| GABRG1 | GABRG2 | GABRG3 | GABRP | GABRQ |
| GABRR1 | GABRR2 | GABRR3 | GAD1 | GALR3 |
| GRIA1 | GRIA2 | GRIA3 | GRIA4 | GRID1 |
| GRID2 | GRID2IP | GRIK1 | GRIK2 | GRIK3 |
| GRIK4 | GRIK5 | GRIN1 | GRIN2A | GRIN2B |
| GRIN2C | GRIN2D | GRIN3A | GRIN3B | GRINA |
| GRM1 | GRM2 | GRM3 | GRM4 | GRM5 |
| GRM6 | GRM7 | GRM8 | HTR1A | HTR1B |
| HTR1D | HTR1E | HTR1F | HTR2A | HTR2B |
| HTR2C | HTR3A | HTR3B | HTR3C | HTR3D |
| HTR3E | HTR4 | HTR5A | HTR6 | HTR7 |
| HTR7P1 | ISOC2 | KIAA0125 | MAOA | NOTCH4 |
| NRXN1 | NRXN3 | NTRK2 | OPRM1 | PPP1R1B |
| PRODH | RGS4 | RIC3 | SLC18A2 | SLC18A3 |
| SLC22A2 | SLC29A4 | SLC32A1 | SLC36A1 | SLC5A7 |
| SLC6A1 | SLC6A11 | SLC6A12 | SLC6A2 | SLC6A3 |
| SLC6A4 | SLC6A7 | SNAI3 | SNAP25 | SOD2 |
| TPH1 | TPH2 | YIF1B |
Bolded genes passed probe filtration and were included in the analysis
The data consisted of two publicly available human brain eQTL datasets, (Colantuoni et al., 2011, Liu et al., 2010), COLANTUONI and LIU respectively, that include smoking status at time of death, genome-wide SNP microarray genotypes that assay genetic variants distributed across the entire human genome, and microarray-based gene expression profiles from frontal lobe samples. Each dataset, COLANTUONI and LIU, was pre-processed independently. For gene expression data, we performed covariate correction, including correction for multiple batches run at different times, tissue source, sex, ancestry, age, postmortem interval (PMI), RNA integrity number (RIN), brain pH and psychiatric status for LIU subjects (see online supplementary information: Methodology for statistical details on covariate correction). A sudden cause of death was reported in 86% of the subjects and no agonal states were reported to be neurologically based. These datasets then underwent quality control checks to ensure the processed data were appropriate for downstream eQTL analysis (see sections COLANTUONI gene expression and LIU gene expression in the online supporting information: Methodology). Genotypes were filtered for quality before use in (1) estimating ancestry principal component covariates, which are critical for controlling false positives related to ethnic stratification, and (2) input for genotype imputation, a method that makes meta-analysis possible by ensuring that both datasets are genotyped using the same set of SNPs.
After the above pre-processing workflow was carried out, G×E analysis included N=144 subjects from COLANTUONI (38 smokers and 106 non-smokers; ages 18-77, avg=43.1; 68% male) and N=84 subjects from LIU (56 smokers and 28 non-smokers; ages 19-65, avg=44.6; 70% male). These datasets were used for the primary analysis in the study. Secondary analysis of the effects of SNPs without regard to smoking was carried out using larger sample sizes, since information on smoking status was not a requirement in the filtering process (N=186 for COLANTUNI and N=127 for LIU). Within or nearby the genes selected for analysis there were 405,875 SNPs in common across both datasets.
A meta-analysis of the two eQTL analyses (see Statistical Analysis in online supporting information: Methodology) to test for G×E effects was conducted by assessing the significance of the linear regression interaction term (SNP genotype × smoking status) as a predictor of gene expression. After correcting for multiple testing (see online supporting information: Methodology), the threshold for significance is 8.1×10−8. Additional follow-up analyses to assess if smoking alone or SNP genotype alone (i.e., main effects) accounted for additional variance in gene expression. Meta-analysis R2 values were obtained by pooling sums of squares for residuals and sums of squares for error across studies and applying the standard adjusted R2 formula with pooled df to account for both studies. Lastly, properties of the SNPs and the loci were examined to interpret the main findings (see SNP annotations in the online supporting information: Methodology).
Results
Analysis of SNPs (G) × Smoking Status (E) on gene expression
G×E analysis was conducted on all SNP-gene pairs (405,875 SNPs and 29 genes; with multiple gene measurements present in some COLANTUONI genes). The metaanalysis yielded 312 SNPs (328 SNP-gene pairs) meeting the typical cut-off for significance in a genome-wide association study, p = 5×10−8, with the most significant result found for expression of the neurexin 3 (NRXN3) gene at SNP rs12411798 (p = 1.9×10−11). There were 16 genes associated with significant G×E interactions (see Table 2), including four glutamate related genes (GRIK1, GRIK2, GRIK5, are kainite-sensitive ionotropic receptor subunits and GRM3, a metabotropic glutamate receptor) and three gamma-aminobutyric acid (GABA) related genes (ABAT, GAD1, involved in synthesis and GABBR2, a GABA receptor subunit), in addition to another member of the neurexin family, NRXN1, implicated in stabilizing synaptic proteins and possibly a special role in dopamine reward pathways. Most SNPs were more than 1 Mb distant from the associated gene, indicating an indirect effect on transcription is most likely (i.e., the effect is mediated by at least one other gene). Many associated SNPs are close together and represent the same underlying eQTL. We examined this by assuming correlation patterns among nearby SNPs (called linkage disequilibrium) represent the same locus (i.e., the same cause in gene expression change). Table 2 lists the number of independent G×E loci for the top 16 genes. The proportion of variance accounted for by SNPs and G×E in this study was determined by forward selection of regression terms with R2 values presented in Table 2. While the average R2 was 0.16, comparison of the two studies indicated a average shrinkage of 60% indicating that additional studies will be needed to more accurately estimate the true effect sizes. A regression of eQTL number on gene length indicates a relationship (R2 = 0.50, p = 0.002), whereby longer genes have more eQTLs that remains significant when only trans SNPs are included. This indicates that when a gene is important for G×E, length offers more opportunities for trans regulation of gene expression.
Table 2.
List of top 16 genes and their characteristics
| Gene |
Significant
eQTLs |
Significant
SNPs |
Peak SNP |
Peak p-
value |
Peak
adjusted R 2 |
| NRXN3 | 17 | 150 | rs12411798 | 1.87×10−11 | 0.25 |
| ABAT | 4 | 23 | rs6558338 | 9.30×10−10 | 0.14 |
| GRIK1 | 3 | 5 | rs2268132 | 9.50×10−10 | 0.12 |
| GRIK5 | 2 | 22 | rs118031442 | 1.09×10−9 | 0.12 |
| NRXN1 | 4 | 20 | rs1646239 | 1.94×10−9 | 0.17 |
| GRIK2 | 4 | 21 | rs2585459 | 1.98×10−9 | 0.13 |
| NTRK2 | 1 | 5 | rs11135168 | 3.95×10−9 | 0.08 |
| RGS4 | 1 | 1 | rs35472486 | 8.42×10−9 | 0.17 |
| APP | 3 | 22 | rs4608331 | 8.74×10−9 | 0.29 |
| ISOC2 | 7 | 18 | rs1046695 | 9.77×10−9 | 0.15 |
| GAD1 | 1 | 4 | rs149541349 | 1.08×10−8 | 0.16 |
| PRODH | 6 | 14 | rs73735512 | 1.27×10−8 | 0.15 |
| GABBR2 | 3 | 7 | rs216137 | 1.30×10−8 | 0.13 |
| SNAP25 | 4 | 14 | rs8073177 | 1.54×10−8 | 0.22 |
| HTR2A | 1 | 1 | rs35472486 | 1.95×10−8 | 0.13 |
| GRM3 | 1 | 1 | rs74005241 | 4.08×10−8 | 0.11 |
We examined if the SNPs in our study had previously described biological functions (i.e., database annotations) to determine if the eQTLs had a common underlying functional characteristic. No such trends were observed. Only 0.3% of SNPs were located in a transcription factor binding site, where the SNP could possibly disrupt binding as a mechanism to alter transcription. Only one SNP was predicted to have functional consequences at the protein level using standard prediction tools SIFT and PolyPhen 2 (Kumar et al., 2009, Ramensky et al., 2002). We examined CHiP-seq data from all neutrally derived cell lines from the whole genome survey known as the Encyclopedia of DNA Elements (ENCODE) database to see if any of our SNPs fell within a experimentally determined DNA binding proteins (presumed regulatory regions) or open markers of chromatin (presumed transcriptionally active regions). A total of 69% of the genome fell within ENCODE regions while 75% of the significant SNPs fell within those regions. This enrichment was significant, (binomial test, P=.009) though the mechanism for how our eQTL SNPs affect transcription requires additional experimental work.
The effect of smoking alone
We examined if smoking is associated with changes in gene expression independent of SNP genotype. Analysis of each dataset yielded no significant genes with differential expression by smoking status. Top genes in COLANTUONI included two genes with prior evidence for a relationship with smoking, DUSP27 (Nielsen et al., 2010) and YIF1B (Carrel et al., 2008), though the best false discovery rate (FDR) was 0.57, far from the necessary cut-off of 0.05. The fact that two genes previously identified as having expression changes associated with smoking were highlighted by the analysis but not at an appropriate significance level may indicate low power. Top hits within the LIU dataset had an FDR of 0.37, with no overlap of the top 100 genes from COLANTUONI.
NRXN3
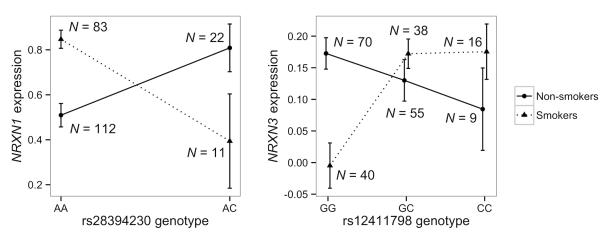
SNPs associated with NRXN3 expression comprised 6 of the top 10 SNPs based on p-value from the eQTL analysis, including the top SNP, and thus we sought to further characterize results related to this gene. Upon visual inspection of the data, the G×E loci display the classical “X” shape (Figure 1), which can cause assessment of the main effect of genotype and the main effect of smoking to be absent, since averaging over either factor minimizes the remaining effect. In agreement with this observation, regression of gene expression on these SNPs does not yield significant SNP effects. To assess the possibility that the additive linear model was only approximating a more genetically complex model, we performed Bayesian genetic modeling to estimate parameters that can differentiate between the additive model and models that have dominance (i.e., where one copy of an allele has the same effect as two copies of that allele, thus introducing a non-linearity). Parameter estimates were fully consistent with the additive linear model for the top 100 G×E SNPs, indicating that the linear regression model was adequate for these loci.
Figure 1.
Examples of the observed SNP×Smoking interactions. Mean gene expression from the SCAN normalization method (y-axis) with standard error bars are plotted by SNP genotype (x-axis) and smoking status (symbols). A gene expression value of 0 indicates background levels (no expression). The pattern of results indicates an interaction effect that implies little to no main effects for either SNP or Smoking since averaging over either predictor variable will largely eliminate the effect of the other.
Discussion
This study represents the first attempt to examine G × E effects on gene expression in the human brain. In this study we have demonstrated a significant interaction between genotype and smoking status on the expression of a set of 16 genes that were of previous interest in psychiatric genetics, though none had been shown to be involved in smoking persistence through G × E effects. These novel findings for psychiatrically important genes suggest that those genes may predispose a variety of behaviors specifically through gene expression changes as demonstrated by our results. The most significant results were for six SNPs in the neurexin 3 gene (NRXN3), one of three genes in the neurexin family. Neurexins are pre-synaptic cell adhesion proteins that play an important role in the formation and function of synapses. The three neurexin genes are each transcribed from two promoter regions, resulting in alpha and beta neurexin proteins, which are alternatively spliced, yielding more than 1000 distinct isoforms that are differentially expressed in the brain (Ullrich et al., 1995). As a whole, neurexins are widely expressed in the mammalian brain, including in the prefrontal cortical areas studied here. Neurexin genes have been previously associated with smoking (Bierut et al., 2007, Docampo et al., 2012) and also with alcohol (Hishimoto et al., 2007) and opioid (Lachman et al., 2007) dependence. SNPs in NRXN3 specifically have been previously associated with susceptibility to smoking (Docampo et al., 2012), and with degree of smoking in schizophrenic patients (Novak et al., 2009). The present results add new evidence indicating that NRXN3 expression in the human prefrontal cortex is differentially affected by smoking behavior depending on an individual SNP profile. And like previous studies, the association is exclusively with the membrane-bound NRXN3-alpha transcript and not in the soluble NRXN3-beta isoform (Docampo et al., 2012, Novak et al., 2009). The association of both glutamatergic and GABAergic genes may imply that the balance between excitation and inhibition neural circuitry of prefrontal corticolimbic circuitry associated with addiction, specifically prefrontal cortex-striatal circuitry. In addition to neurotransmitter findings, neurexins, involved in stabilizing synaptic proteins to perhaps modulate transmission efficiency, might effect the balance of excitation and inhibition to alter smoking persistence.
The lack of a main effect of smoking on expression of NRXN3 in this study is accounted for by the nature of the observed cross-over interactions (Dick and Kendler, 2012), between a large set of SNPs and the categorical smoking variable on NRXN3 expression. In particular, interactions such as those depicted in Figure 1 (X-shaped), result in the effect of either factor (genotype or smoking status) being largely eliminated when averaging across levels of the other. Such a result indicates, for instance, that even the direction of the effect of smoking on NRXN3 expression in these brain areas depends on specific genomic variations. The lack of any observable main effects of smoking (i.e., genes that are differentially expressed in smoking group vs. non-smoking group) may reflect heterogeneity within the two groups of subjects or lack of statistical power.
Our analyses used only a single categorical variable, which was coded in the existing eQTL datasets, to determine smoking behavior. Such a binary variable cannot account for the full complexity of smoking-related phenotypes, and quantitative measures (e.g., number of cigarettes per day) might provide additional power if they were available. Indeed, it has been suggested that smoking initiation, number of cigarettes per day, and nicotine dependence each reflect distinct stages of smoking behavior, and may involve distinct biological and genetic mechanisms (Mayhew et al., 2000, Breitling et al., 2011). However, the lack of more detailed smoking behavioral indicators would only reduce study power and would not induce false positive results. Therefore, our findings are likely to be an underestimate of the number of specific genetic loci that are involved in smoking and gene expression in the human brain.
The overall results of this study reflect the utility of analyzing eQTLs, in which associations are tested between individual SNP genotypes and gene expression levels in brain tissue samples, and of G×E analyses, in which the effects of genotype and environmental variables on the phenotypic measure (gene expression in this case) are simultaneously modeled. In particular, we have shown that the effects of smoking to alter expression of psychiatric genes in the brain can depend on particular genotypes, based on two independent studies. These data imply that G × E for smoking persistence is driven by differential expression of genes based upon the genotype at nearby SNP loci. Or, to reframe the conclusion, expression of psychiatric genes relevant to smoking depends upon the environment and genotypes of the individual.
An issue when interpreting the lack of main effects on gene expression for G×E SNPs is the possibility that smoking causes epigenetic changes, here referring to modifications of DNA that change the regulatory potential of the DNA without changing the sequence (The ENCODE Project Consortium, 2012). Therefore, a SNP that is normally silent can induce changes in gene expression when one of the two alleles is more likely to be epigenetically modified than the other. Without assessing the known markers for DNA modification, such as methylation or histone binding, it is not possible here to assert positive evidence for this hypothesis. Regardless of the specific mechanism, however, understanding how the environment changes gene expression will shed light on which genes to study in more detail in conjunction with other behavioral designs. Because no main effect of smoking was found on NRXN3 expression, nor was there a significant main effect of genotype, it is clear that many standard statistical models, which do not explicitly account for G×E effects, will miss such results. Thus, we suggest that the combination of the eQTL approach with modeling of G×E interactions is a promising direction for establishing the relationships between genes, brain, and behavior in complex psychiatric disorders. These results also suggest the importance of – where feasible – including environmental and behavioral measures from individual subjects as future eQTL datasets are gathered.
Brain gene expression profiling has only recently been used in the domain of psychiatric disease etiology (Geschwind and Konopka, 2009, Luykx et al., 2013, Mexal et al., 2005, Richards et al., 2012a), but this study extends that approach to G×E and indicates that future studies using this approach may be powerful for addressing other types of questions. This study suggests that human brain gene expression datasets, when coupled with behavioral and genetic data, provide useful phenotypes that can be incorporated in psychiatric genetics studies, and provides the first demonstration of G×E effects on gene expression in the human brain. As brain banks for child and adolescent subjects continue to grow, it would be invaluable to the field to collect environmental data wherever possible to facilitate additional studies on issues more widely appreciated in younger developmental stages.
Supplementary Material
Key Points.
Gene by environment interaction effects on human behavior are assumed to be mediated by changes in gene expression patterns in the brain, though no empirical demonstrations exist.
An expression quantitative trait locus study assesses the relationship between DNA loci and gene expression values and can be extended to include a linear regression interaction term for DNA locus by environment (e.g., smoking).
We observed that the effect of DNA variation on gene expression changes depends on smoking status, demonstrating that behavioral gene by environment interactions can affect brain gene expression.
Several psychiatrically important genes involving neural connectivity and glutamate transmission associate with DNA variants that interact with smoking, suggesting an important role for these genes in the maintenance of smoking.
Acknowledgements
We gratefully acknowledge NIH funding from the National Institute of Deafness and Other Communication Disorders R01 DC009453 (to C.W.B.) and the National Institute of General Medical Sciences U01 GM092655 (to K.H.). This work was also supported in part by an allocation of computing time from the Ohio Supercomputer Center Grant PCCR0001 (C.W.B.).
Abbreviations
- eQTL
expression quantitative trait locus
- G×E
gene by environment interaction
- PMI
post-mortem interval
- RIN
RNA integrity number
- SNP
single nucleotide polymorphism
Footnotes
Conflict of interest statement: No conflicts of interest declared.
References
- Anstey KJ, Von Sanden C, Salim A, O’kearney R. Smoking as a risk factor for dementia and cognitive decline: a meta-analysis of prospective studies. Am J Epidemiol. 2007;166:367–378. doi: 10.1093/aje/kwm116. [DOI] [PubMed] [Google Scholar]
- Audrain-Mcgovern J, Rodriguez D, Rodgers K, Cuevas J, Sass J. Longitudinal variation in adolescent physical activity patterns and the emergence of tobacco use. J Pediatr Psychol. 2012;37:622–633. doi: 10.1093/jpepsy/jss043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagot RC, Meaney MJ. Epigenetics and the biological basis of gene × environment interactions. J Am Acad Child Adolesc Psychiatry. 2010;49:752–771. doi: 10.1016/j.jaac.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Bailey-Wilson JE, Wilson AF. Linkage analysis in the next-generation sequencing era. Hum Hered. 2011;72:228–236. doi: 10.1159/000334381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker JH, Maes HH, Larsson H, Lichtenstein P, Kendler KS. Sex differences and developmental stability in genetic and environmental influences on psychoactive substance consumption from early adolescence to young adulthood. Psychol Med. 2011;41:1907–1916. doi: 10.1017/S003329171000259X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baler RD, Volkow ND. Addiction as a systems failure: focus on adolescence and smoking. J Am Acad Child Adolesc Psychiatry. 2011;50:329–339. doi: 10.1016/j.jaac.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett CW, Vieland VJ. Two novel quantitative trait linkage analysis statistics based on the posterior probability of linkage: application to the COGA families. BMC Genet. 2005;6(Suppl 1):S121. doi: 10.1186/1471-2156-6-S1-S121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett CW, Vieland VJ, Bartlett J, Bell JT, Bhattacharjee S, Clerget-Darpoux F, Bush WS, Edwards TL, Gao G, Halder I, Huang Y, Kotti S, Larkin EK, Li H, Motsinger AA, Mukhopadhyay N, Namkung J, Park T, Ritchie MD, Stein CM, Zhou JY. Discussing gene-gene interaction: warning--translating equations to English may result in jabberwocky. Genetic epidemiology. 2007;31(Suppl 1):S61–67. doi: 10.1002/gepi.20281. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society: Series B (Statistical Methodology) 1995;57:289–300. [Google Scholar]
- Bergen SE, Gardner CO, Kendler KS. Age-related changes in heritability of behavioral phenotypes over adolescence and young adulthood: a meta-analysis. Twin Res Hum Genet. 2007;10:423–433. doi: 10.1375/twin.10.3.423. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, Fugman D, Goate AM, Hinrichs AL, Konvicka K, Martin NG, Montgomery GW, Saccone NL, Saccone SF, Wang JC, Chase GA, Rice JP, Ballinger DG. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulle F, Van Den Hove DL, Jakob SB, Rutten BP, Hamon M, Van Os J, Lesch KP, Lanfumey L, Steinbusch HW, Kenis G. Epigenetic regulation of the BDNF gene: implications for psychiatric disorders. Mol Psychiatry. 2012;17:584–596. doi: 10.1038/mp.2011.107. [DOI] [PubMed] [Google Scholar]
- Breitling LP, Yang R, Korn B, Burwinkel B, Brenner H. Tobacco-smoking-related differential DNA methylation: 27K discovery and replication. Am J Hum Genet. 2011;88:450–457. doi: 10.1016/j.ajhg.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel D, Masson J, Al Awabdh S, Capra CB, Lenkei Z, Hamon M, Emerit MB, Darmon M. Targeting of the 5-HT1A serotonin receptor to neuronal dendrites is mediated by Yif1B. The Journal of Neuroscience. 2008;28:8063–8073. doi: 10.1523/JNEUROSCI.4487-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi A, Moffitt TE. Gene-environment interactions in psychiatry: joining forces with neuroscience. Nat Rev Neurosci. 2006;7:583–590. doi: 10.1038/nrn1925. [DOI] [PubMed] [Google Scholar]
- Center for Behavioral Health Statistics and Quality . Results from the 2010 National Survey on Drug Use and Health: Summary of National Findings. US Department of Health and Human Services; Rockville, MD: 2011. [Google Scholar]
- Chanda P, Yuhki N, Li M, Bader JS, Hartz A, Boerwinkle E, Kao WH, Arking DE. Comprehensive evaluation of imputation performance in African Americans. Journal of Human Genetics. 2012;57:411–421. doi: 10.1038/jhg.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelala C, Khan A, Lemoine NR. SNPnexus: a web database for functional annotation of newly discovered and public domain single nucleotide polymorphisms. Bioinformatics. 2009;25:655–661. doi: 10.1093/bioinformatics/btn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, Colantuoni EA, Elkahloun AG, Herman MM, Weinberger DR, Kleinman JE. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, Watson SJ, Meng F. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damerval C, Maurice A, Josse JM, De Vienne D. Quantitative trait loci underlying gene product variation: a novel perspective for analyzing regulation of genome expression. Genetics. 1994;137:289–301. doi: 10.1093/genetics/137.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayem Ullah AZ, Lemoine NR, Chelala C. SNPnexus: a web server for functional annotation of novel and publicly known genetic variants (2012 update) Nucleic Acids Res. 2012;40:W65–70. doi: 10.1093/nar/gks364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayem Ullah AZ, Lemoine NR, Chelala C. A practical guide for the functional annotation of genetic variations using SNPnexus. Briefings in Bioinformatics. 2013 doi: 10.1093/bib/bbt004. [DOI] [PubMed] [Google Scholar]
- De Vienne D, Leonardi A, Damerval C. Genetic aspects of variation of protein amounts in maize and pea. Electrophoresis. 1988;9:742–750. doi: 10.1002/elps.1150091110. [DOI] [PubMed] [Google Scholar]
- Dick DM, Kendler KS. The impact of gene-environment interaction on alcohol use disorders. Alcohol Res. 2012;34:318–324. [PMC free article] [PubMed] [Google Scholar]
- Do R, Kathiresan S, Abecasis GR. Exome sequencing and complex disease: practical aspects of rare variant association studies. Hum Mol Genet. 2012;21:R1–9. doi: 10.1093/hmg/dds387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Docampo E, Ribases M, Gratacos M, Bruguera E, Cabezas C, Sanchez-Mora C, Nieva G, Puente D, Argimon-Pallas JM, Casas M, Rabionet R, Estivill X. Association of Neurexin 3 polymorphisms with smoking behavior. Genes, Brain and Behavior. 2012;11:704–711. doi: 10.1111/j.1601-183X.2012.00815.x. [DOI] [PubMed] [Google Scholar]
- Dube JB, Hegele RA. Genetics 100 for cardiologists: basics of genome-wide association studies. Can J Cardiol. 2013;29:10–17. doi: 10.1016/j.cjca.2012.10.011. [DOI] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. Am J Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. Benjamin Cummings; London, England: 1996. [Google Scholar]
- Flatscher-Bader T, Wilce PA. The effect of alcohol and nicotine abuse on gene expression in the brain. Nutrition Research Reviews. 2009;22:148–162. doi: 10.1017/S0954422409990114. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Konopka G. Neuroscience in the era of functional genomics and systems biology. Nature. 2009;461:908–915. doi: 10.1038/nature08537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JR, Van Der Brug MP, Hernandez DG, Traynor BJ, Nalls MA, Lai SL, Arepalli S, Dillman A, Rafferty IP, Troncoso J, Johnson R, Zielke HR, Ferrucci L, Longo DL, Cookson MR, Singleton AB. Abundant quantitative trait loci exist for DNA methylation and gene expression in human brain. PLoS Genet. 2010;6:e1000952. doi: 10.1371/journal.pgen.1000952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haibe-Kains B, Desmedt C, Sotiriou C, Bontempi G. A comparative study of survival models for breast cancer prognostication based on microarray data: does a single gene beat them all? Bioinformatics. 2008;24:2200–2208. doi: 10.1093/bioinformatics/btn374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall W, Madden P, Lynskey M. The genetics of tobacco use: methods, findings and policy implications. Tob Control. 2002;11:119–124. doi: 10.1136/tc.11.2.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebebrand J, Scherag A, Schimmelmann BG, Hinney A. Child and adolescent psychiatric genetics. European child & adolescent psychiatry. 2010;19:259–279. doi: 10.1007/s00787-010-0091-y. [DOI] [PubMed] [Google Scholar]
- Heinzen EL, Ge D, Cronin KD, Maia JM, Shianna KV, Gabriel WN, Welsh-Bohmer KA, Hulette CM, Denny TN, Goldstein DB. Tissue-specific genetic control of splicing: implications for the study of complex traits. PLOS Biology. 2008;6:e1. doi: 10.1371/journal.pbio.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hishimoto A, Liu QR, Drgon T, Pletnikova O, Walther D, Zhu XG, Troncoso JC, Uhl GR. Neurexin 3 polymorphisms are associated with alcohol dependence and altered expression of specific isoforms. Hum Mol Genet. 2007;16:2880–2891. doi: 10.1093/hmg/ddm247. [DOI] [PubMed] [Google Scholar]
- Hou L, Phillips C, Azaro M, Brzustowicz LM, Bartlett CW. Validation of a cost-efficient multi-purpose SNP panel for disease based research. PLoS One. 2011;6:e19699. doi: 10.1371/journal.pone.0019699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou L, Wang K, Bartlett CW. Evaluation of a bayesian model integration-based method for censored data. Hum Hered. 2012;74:1–11. doi: 10.1159/000342707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlden H, Singleton AB. The genetics and neuropathology of Parkinson’s disease. Acta Neuropathol. 2012;124:325–338. doi: 10.1007/s00401-012-1013-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Bartlett CW, Segre AM, O’connell JR, Mangin L, Vieland VJ. Exploiting gene × gene interaction in linkage analysis. BMC Proceedings. 2007;1(Suppl 1):S64. doi: 10.1186/1753-6561-1-s1-s64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Vieland VJ. Association statistics under the PPL framework. Genetic epidemiology. 2010;34:835–845. doi: 10.1002/gepi.20537. [DOI] [PubMed] [Google Scholar]
- Johnson AD, Wang D, Sadee W. Polymorphisms affecting gene regulation and mRNA processing: broad implications for pharmacogenetics. Pharmacol Ther. 2005;106:19–38. doi: 10.1016/j.pharmthera.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, Sousa AM, Pletikos M, Meyer KA, Sedmak G, Guennel T, Shin Y, Johnson MB, Krsnik Z, Mayer S, Fertuzinhos S, Umlauf S, Lisgo SN, Vortmeyer A, Weinberger DR, Mane S, Hyde TM, Huttner A, Reimers M, Kleinman JE, Sestan N. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny PJ, File SE, Rattray M. Nicotine regulates 5-HT(1A) receptor gene expression in the cerebral cortex and dorsal hippocampus. European Journal of Neuroscience. 2001;13:1267–1271. doi: 10.1046/j.0953-816x.2001.01501.x. [DOI] [PubMed] [Google Scholar]
- Kleinman JE, Law AJ, Lipska BK, Hyde TM, Ellis JK, Harrison PJ, Weinberger DR. Genetic neuropathology of schizophrenia: new approaches to an old question and new uses for postmortem human brains. Biol Psychiatry. 2011;69:140–145. doi: 10.1016/j.biopsych.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konu O, Kane JK, Barrett T, Vawter MP, Chang R, Ma JZ, Donovan DM, Sharp B, Becker KG, Li MD. Region-specific transcriptional response to chronic nicotine in rat brain. Brain Res. 2001;909:194–203. doi: 10.1016/s0006-8993(01)02685-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lachman HM, Fann CS, Bartzis M, Evgrafov OV, Rosenthal RN, Nunes EV, Miner C, Santana M, Gaffney J, Riddick A, Hsu CL, Knowles JA. Genomewide suggestive linkage of opioid dependence to chromosome 14q. Hum Mol Genet. 2007;16:1327–1334. doi: 10.1093/hmg/ddm081. [DOI] [PubMed] [Google Scholar]
- Lehner B. Genotype to phenotype: lessons from model organisms for human genetics. Nat Rev Genet. 2013;14:168–178. doi: 10.1038/nrg3404. [DOI] [PubMed] [Google Scholar]
- Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- Lindell SG, Yuan Q, Zhou Z, Goldman D, Thompson RC, Lopez JF, Suomi SJ, Higley JD, Barr CS. The serotonin transporter gene is a substrate for age and stress dependent epigenetic regulation in rhesus macaque brain: potential roles in genetic selection and gene × environment interactions. Dev Psychopathol. 2012;24:1391–1400. doi: 10.1017/S0954579412000788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Cheng L, Badner JA, Zhang D, Craig DW, Redman M, Gershon ES. Whole-genome association mapping of gene expression in the human prefrontal cortex. Mol Psychiatry. 2010;15:779–784. doi: 10.1038/mp.2009.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luykx JJ, Bakker SC, Lentjes E, Neeleman M, Strengman E, Mentink L, Deyoung J, De Jong S, Sul JH, Eskin E, Van Eijk K, Van Setten J, Buizer-Voskamp JE, Cantor RM, Lu A, Van Amerongen M, Van Dongen EP, Keijzers P, Kappen T, Borgdorff P, Bruins P, Derks EM, Kahn RS, Ophoff RA. Genome-wide association study of monoamine metabolite levels in human cerebrospinal fluid. Mol Psychiatry. 2013 doi: 10.1038/mp.2012.183. [DOI] [PubMed] [Google Scholar]
- Maranville JC, Luca F, Stephens M, Di Rienzo A. Mapping gene-environment interactions at regulatory polymorphisms: insights into mechanisms of phenotypic variation. Transcription. 2012;3:56–62. doi: 10.4161/trns.19497. [DOI] [PubMed] [Google Scholar]
- Mayhew KP, Flay BR, Mott JA. Stages in the development of adolescent smoking. Drug Alcohol Depend. 2000;59(Suppl 1):S61–81. doi: 10.1016/s0376-8716(99)00165-9. [DOI] [PubMed] [Google Scholar]
- Mcgowan PO, Sasaki A, D’alessio AC, Dymov S, Labonte B, Szyf M, Turecki G, Meaney MJ. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–348. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Klengel T, Conneely KN, Smith AK, Altmann A, Pace TW, Rex-Haffner M, Loeschner A, Gonik M, Mercer KB, Bradley B, Muller-Myhsok B, Ressler KJ, Binder EB. Childhood maltreatment is associated with distinct genomic and epigenetic profiles in posttraumatic stress disorder. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1217750110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mexal S, Frank M, Berger R, Adams CE, Ross RG, Freedman R, Leonard S. Differential modulation of gene expression in the NMDA postsynaptic density of schizophrenic and control smokers. Molecular Brain Research. 2005;139:317–332. doi: 10.1016/j.molbrainres.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Holmans P, Heward CB, Reiman EM, Stephan D, Hardy J. A survey of genetic human cortical gene expression. Nat Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- Naumova OY, Lee M, Rychkov SY, Vlasova NV, Grigorenko EL. Gene expression in the human brain: the current state of the study of specificity and spatiotemporal dynamics. Child Dev. 2013;84:76–88. doi: 10.1111/cdev.12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen DA, Ji F, Yuferov V, Ho A, He C, Ott J, Kreek MJ. Genome-wide association study identifies genes that may contribute to risk for developing heroin addiction. Psychiatric Genetics. 2010;20:207–214. doi: 10.1097/YPG.0b013e32833a2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noelker C, Schwake M, Balzer-Geldsetzer M, Bacher M, Popp J, Schlegel J, Eggert K, Oertel WH, Klockgether T, Dodel RC. Differentially expressed gene profile in the 6-hydroxy-dopamine-induced cell culture model of Parkinson’s disease. Neurosci Lett. 2012;507:10–15. doi: 10.1016/j.neulet.2011.11.035. [DOI] [PubMed] [Google Scholar]
- Novak G, Boukhadra J, Shaikh SA, Kennedy JL, Le Foll B. Association of a polymorphism in the NRXN3 gene with the degree of smoking in schizophrenia: a preliminary study. The World Journal of Biological Psychiatry. 2009;10:929–935. doi: 10.1080/15622970903079499. [DOI] [PubMed] [Google Scholar]
- Nussbaum J, Xu Q, Payne TJ, Ma JZ, Huang W, Gelernter J, Li MD. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum Mol Genet. 2008;17:1569–1577. doi: 10.1093/hmg/ddn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo SR, Sun Y, Campbell JD, Lenburg ME, Bild AH, Johnson WE. A single-sample microarray normalization method to facilitate personalized-medicine workflows. Genomics. 2012;100:337–344. doi: 10.1016/j.ygeno.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D, Bennett AJ, Pierre PJ, Friedman DP, Cote SM, Hallett M, Tremblay RE, Suomi SJ, Szyf M. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci. 2012;32:15626–15642. doi: 10.1523/JNEUROSCI.1470-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. The American Journal of Human Genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30:3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, Sanders AR, Purcell S, Visscher PM, Craddock N, Owen MJ, Holmans P, O’donovan MC. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012a;17:193–201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AL, Jones L, Moskvina V, Kirov G, Gejman PV, Levinson DF, Sanders AR, Purcell S, Visscher PM, Craddock N, Owen MJ, Holmans P, O’donovan MC. Schizophrenia susceptibility alleles are enriched for alleles that affect gene expression in adult human brain. Mol Psychiatry. 2012b;17:193–201. doi: 10.1038/mp.2011.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, Prokunina-Olsson L, Ding CJ, Swift AJ, Narisu N, Hu T, Pruim R, Xiao R, Li XY, Conneely KN, Riebow NL, Sprau AG, Tong M, White PP, Hetrick KN, Barnhart MW, Bark CW, Goldstein JL, Watkins L, Xiang F, Saramies J, Buchanan TA, Watanabe RM, Valle TT, Kinnunen L, Abecasis GR, Pugh EW, Doheny KF, Bergman RN, Tuomilehto J, Collins FS, Boehnke M. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics. 2012;28:1353–1358. doi: 10.1093/bioinformatics/bts163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons TR, Flax JF, Azaro MA, Hayter JE, Justice LM, Petrill SA, Bassett AS, Tallal P, Brzustowicz LM, Bartlett CW. Increasing genotype-phenotype model determinism: application to bivariate reading/language traits and epistatic interactions in language-impaired families. Hum Hered. 2010;70:232–244. doi: 10.1159/000320367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- Sullivan PF, Kendler KS. The genetic epidemiology of smoking. Nicotine Tob Res. 1999;1(Suppl 2):S51–57. doi: 10.1080/14622299050011811. discussion S69-70. [DOI] [PubMed] [Google Scholar]
- The Encode Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, Gieger C, Rawal R, Mangino M, Prokopenko I, Magi R, Keskitalo K, Gudjonsdottir IH, Gretarsdottir S, Stefansson H, Thompson JR, Aulchenko YS, Nelis M, Aben KK, Den Heijer M, Dirksen A, Ashraf H, Soranzo N, Valdes AM, Steves C, Uitterlinden AG, Hofman A, Tonjes A, Kovacs P, Hottenga JJ, Willemsen G, Vogelzangs N, Doring A, Dahmen N, Nitz B, Pergadia ML, Saez B, De Diego V, Lezcano V, Garcia-Prats MD, Ripatti S, Perola M, Kettunen J, Hartikainen AL, Pouta A, Laitinen J, Isohanni M, Huei-Yi S, Allen M, Krestyaninova M, Hall AS, Jones GT, Van Rij AM, Mueller T, Dieplinger B, Haltmayer M, Jonsson S, Matthiasson SE, Oskarsson H, Tyrfingsson T, Kiemeney LA, Mayordomo JI, Lindholt JS, Pedersen JH, Franklin WA, Wolf H, Montgomery GW, Heath AC, Martin NG, Madden PA, Giegling I, Rujescu D, Jarvelin MR, Salomaa V, Stumvoll M, Spector TD, Wichmann HE, Metspalu A, Samani NJ, Penninx BW, Oostra BA, Boomsma DI, Tiemeier H, Van Duijn CM, Kaprio J, Gulcher JR, Mccarthy MI, Peltonen L, Thorsteinsdottir U, Stefansson K. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobacco and Genetics Consortium Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauth JA, Seidler FJ, Mccook EC, Slotkin TA. Adolescent nicotine exposure causes persistent upregulation of nicotinic cholinergic receptors in rat brain regions. Brain Res. 1999;851:9–19. doi: 10.1016/s0006-8993(99)01994-0. [DOI] [PubMed] [Google Scholar]
- Trauth JA, Seidler FJ, Slotkin TA. An animal model of adolescent nicotine exposure: effects on gene expression and macromolecular constituents in rat brain regions. Brain Res. 2000;867:29–39. doi: 10.1016/s0006-8993(00)02208-3. [DOI] [PubMed] [Google Scholar]
- Tsuruoka Y, Tsujii J, Ananiadou S. FACTA: a text search engine for finding associated biomedical concepts. Bioinformatics. 2008;24:2559–2560. doi: 10.1093/bioinformatics/btn469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tully EC, Iacono WG, Mcgue M. Changes in genetic and environmental influences on the development of nicotine dependence and major depressive disorder from middle adolescence to early adulthood. Dev Psychopathol. 2010;22:831–848. doi: 10.1017/S0954579410000490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich B, Ushkaryov YA, Sudhof TC. Cartography of neurexins: more than 1000 isoforms generated by alternative splicing and expressed in distinct subsets of neurons. Neuron. 1995;14:497–507. doi: 10.1016/0896-6273(95)90306-2. [DOI] [PubMed] [Google Scholar]
- Us Surgeon General . Preventing Tobacco Use Among Youth and Young Adults. US Deparment of Health and Human Services; Rockville, MD: 2012. [Google Scholar]
- Vieland VJ, Huang Y, Seok SC, Burian J, Catalyurek U, O’connell J, Segre A, Valentine-Cooper W. KELVIN: a software package for rigorous measurement of statistical evidence in human genetics. Hum Hered. 2011;72:276–288. doi: 10.1159/000330634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink JM, Willemsen G, Boomsma DI. Heritability of smoking initiation and nicotine dependence. Behav Genet. 2005;35:397–406. doi: 10.1007/s10519-004-1327-8. [DOI] [PubMed] [Google Scholar]
- Vrieze SI, Mcgue M, Iacono WG. The interplay of genes and adolescent development in substance use disorders: leveraging findings from GWAS meta-analyses to test developmental hypotheses about nicotine consumption. Human genetics. 2012;131:791–801. doi: 10.1007/s00439-012-1167-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster JA, Gibbs JR, Clarke J, Ray M, Zhang W, Holmans P, Rohrer K, Zhao A, Marlowe L, Kaleem M, Mccorquodale DS, 3rd, Cuello C, Leung D, Bryden L, Nath P, Zismann VL, Joshipura K, Huentelman MJ, Hu-Lince D, Coon KD, Craig DW, Pearson JV, Heward CB, Reiman EM, Stephan D, Hardy J, Myers AJ. Genetic control of human brain transcript expression in Alzheimer disease. The American Journal of Human Genetics. 2009;84:445–458. doi: 10.1016/j.ajhg.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitlock MC. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. Journal of Evolutionary Biology. 2005;18:1368–1373. doi: 10.1111/j.1420-9101.2005.00917.x. [DOI] [PubMed] [Google Scholar]
- Yoon D, Kim YJ, Cui WY, Van Der Vaart A, Cho YS, Lee JY, Ma JZ, Payne TJ, Li MD, Park T. Large-scale genome-wide association study of Asian population reveals genetic factors in FRMD4A and other loci influencing smoking initiation and nicotine dependence. Human genetics. 2012;131:1009–1021. doi: 10.1007/s00439-011-1102-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.