Abstract
Malignant pleural mesothelioma (MPM) is an aggressive, asbestos-related malignancy of the thoracic pleura. Although, platinum-based agents are the first line of therapy, there is an urgent need for second-line therapies to treat the drug-resistant MPM. Cell cycle as well as apoptosis pathways are frequently altered in MPM and thus remain attractive targets for intervention strategies. Curcumin, the major component in the spice turmeric, alone or in combination with other chemotherapeutics has been under investigation for a number of cancers. In this study, we investigated the biological and molecular responses of MPM cells to curcumin treatments and the mechanisms involved. Flow-cytometric analyses coupled with western immunoblotting and gene-array analyses were conducted to determine mechanisms of curcumin-dependent growth suppression of human (H2373, H2452, H2461, and H226) and murine (AB12) MPM cells. Curcumin inhibited MPM cell growth in a dose- and time-dependent manner while pretreatment of MPM cells with curcumin enhanced cisplatin efficacy. Curcumin activated the stress-activated p38 kinase, caspases 9 and 3, caused elevated levels of proapoptotic proteins Bax, stimulated PARP cleavage, and apoptosis. In addition, curcumin treatments stimulated expression of novel transducers of cell growth suppression such as CARP-1, XAF1, and SULF1 proteins. Oral administration of curcumin inhibited growth of murine MPM cell-derived tumors in vivo in part by stimulating apoptosis. Thus, curcumin targets cell cycle and promotes apoptosis to suppress MPM growth in vitro and in vivo. Our studies provide a proof-of-principle rationale for further in-depth analysis of MPM growth suppression mechanisms and their future exploitation in effective management of resistant MPM.
Keywords: Malignant pleural mesothelioma, Curcumin, Apoptosis, Gene expression
Introduction
Malignant Pleural Mesothelioma (MPM) are the tumors of mesothelial or mesenchymal cells that line the thoracic cavity, peritoneum, or pericardium. MPM is an aggressive, locally recurrent cancer with no effective curative options [1, 2]. The median survival for patients diagnosed with MPM is between 8 and 18 months, despite aggressive multimodality strategies involving surgery, chemotherapy, and radiation [3–5]. The platinum-based agents are currently used either as monotherapy or in combination with other agents as first line therapeutics for MPM. However, the response rates remain unacceptably low.
MPM is characterized by a number of molecular events including chromosome 3p and neurofibromatosis 2 gene abnormalities [6–8], Wilms tumor [9], VEGF [10], thrombospondin [11], IGF receptor amplification [12], and inactivation of p53 and Rb by SV40 T antigen [13]. Simian virus 40 (SV40)-dependent Akt signaling has been shown to drive mesothelial cell transformation after asbestos exposure and knockdown of receptor tyrosine kinase EphB4 was found to reduce phosphorylation of Akt in murine model of mesothelioma [14]. In the absence of an effective standard second-line of treatment options, strategies to exploit aberrant growth and apoptosis signaling have been proposed for identification of new and more efficacious means to combat this disease [15].
A number of studies suggest that agents derived from dietary fruits and vegetables are helpful in either inhibiting or reversing the development of cancer [16, 17]. Curcumin, a phenolic compound isolated from the dietary plant Cur-cumina longa, has been demonstrated to possess antiinflammatory, anti-oxidant, and anti-cancer properties. Curcumin has been shown to suppress growth of cancer cells in vitro, while in animals, it interferes with tumor initiation as well as tumor promotion [18–22]. Although, the precise mechanisms of action of curcumin are yet to be elucidated, available evidence thus far indicates that it suppresses cancer cell growth in large part by down- regulating pathways of cell proliferation and survival. In this context, curcumin inhibition of the NF-κB as well as the P13K-Akt pathways of cell survival has been well documented. Since inhibition of the NF-κB and the PI3K–Akt signaling is often associated with increased apoptotic index in many cell types, the anti-cancer effects of curcumin also involve activation of the extrinsic and/or intrinsic pathways of apoptosis [21, 22].
In this study, we investigated effects of curcumin on growth of MPM cells in vitro. Consistent with observations in other models, curcumin treatments caused cell growth inhibition that involved elevated apoptosis. Acombination of curcumin and cisplatin were superior in suppressing growth of the MPM cells in vitro. Although curcumin stimulated expression of pro-apoptotic Bax and activated p38 SAPK and caspase-9, our gene-array-based analysis revealed that curcumin induced expression of novel transducers of apoptosis signaling such as XIAP-associated factor1 (XAF1) and CARP-1/CCAR1 [23, 24]. Curcumin treatments also stimulated expression of a putative tumor suppressor SULF1 protein [25]. Further oral administration of curcumin suppressed growth of murine mesothelioma allografts in part by enhancing apoptosis. Our proof-of-concept studies reveal, for the first time, MPM inhibitory properties of curcumin and are expected to facilitate utilization of this agent or its potent derivatives as potential adjuvants for treatment and perhaps chemoprevention of MPM.
Materials and methods
Cells and reagents
Three MPM patient-derived cell lines [H2373, H2452, and H2461] established in our laboratory and characterized in detail [26] were cultured in RPMI 1640 (Mediatech Inc., Herndon, VA) supplemented with 100 units/ml of penicillin, 100 µg/ml streptomycin, 4 mM L-glutamine, and 10% fetal calf serum. H226 MPM cells were obtained from ATCC (Manassas, VA) and maintained following vendor’s guidelines. The AB12 murine malignant mesothelioma cell line was derived from BALB/c mice and was shown to form subcutaneous tumors when implanted in mice [27, 28]. This cell line was cultured and maintained in high-glucose DMEM supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 ug/ml streptomycin. Cells were incubated at 37°C in a humidified atmosphere of 5% CO2 in air and were passaged weekly. Curcumin was obtained from Sigma-Aldrich (St. Louis, MO). Mouse monoclonal antibody against human poly (ADP-ribose) polymerase (PARP) was from BIOMOL International LP (Plymouth Meeting, PA). Anti-XAF1 antibodies were obtained from Abcam Inc., (Cambridge, MA). Anti-Bax, anti-caspase-3, anti-caspase-9, anti-Akt, anti-p-p38 antibodies were purchased from Cell Signaling Technology (Beverly, MA). Anti-IkB-α and anti-Bcl2 antibodies were obtained from Santa Cruz Biotechnology Inc. (Santa Cruz, CA), while anti-β actin antibody was purchased from Sigma-Aldrich (St. Louis, MO). Anti-HSulf-1 rabbit polyclonal antibodies were purchased from Abcam. Generation and characterization of the anti-CARP-1/CCAR1 rabbit polyclonal antibodies have been described before [24]. 3-(4,5-dimethyltiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO).
Cell growth inhibition studies by MTT assay
MPM (H2373, H2452, H2461, H226 and AB12) cells (5 × 103) were seeded in a 96-well culture plate and subsequently treated with indicated agents at different concentrations for noted times. Control cells were treated with 0.1% dimethyl sulfoxide (DMSO) in culture medium. After treatment, the cells were incubated with 1 mg/ml of MTT reagent at 37°C for 4 h and then MTT was removed and 100 µl of DMSO was added, followed by colorimetric analysis using a multilabel plate reader at 560 nm (Victor3; PerkinElmer, Wellesley, MA, USA). Results were plotted as the mean from triplicate experiments.
Western blot analysis
Cells were harvested and lysed in RIPA buffer (50 mM Tris-HCI, pH 8.0, 150 mM sodium chloride, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, and 0.1% of protease inhibitor cocktail) for 20 min at 4°C. The lysates were centrifuged at 14,000 rpm at 4°C for 15 min to remove debris. Protein concentrations of whole cell lysates were determined using the Protein Assay Kit. Supernatant proteins, 50 µg from each sample, were separated by SDS-10% polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membrane (Bio-rad, Hercules, CA) by standard procedures. The membranes were hybridized with primary antibodies followed by incubation with appropriate secondary antibodies. The antibody-bound proteins were visualized by treatment with the chemiluminescence detection reagent (Pierce) according to manufacturer’s instructions, followed by exposure to film (Kodak X-Omat). The same membrane was reprobed with the anti-β actin antibody, which was used as an internal control for protein loading.
Flow cytometry and cell cycle analysis
The cell cycle was analyzed by flow cytometry. In brief, 1 × 106 cells were untreated or treated with cisplatin, curcumin, or a combination of both, and harvested and washed in PBS, then fixed in 70% alcohol for 30 min at 4°C. After washing in cold PBS thrice, cells were resus-pended in 1 ml of PBS solution with 50 µg of propidium iodide and 100 µg of RNaseA for 30 min at 37°C. Samples were then analyzed for their DNA content by FACSCalibur (Becton-Dickinson, Mountain View, CA).
Isolation of RNA and microarray analysis
Total RNA was extracted from untreated or curcumin-treated H2373 and H2461 MPM cells. At the end of treatments, the untreated and treated cells were harvested and total RNA were isolated, and purified using the RNeasy Mini kit and RNase-free DNase Set (Qiagen, Valencia, CA) according to the manufacturer’s protocols.
Cucumin-dependent changes in gene expression in MPM cells were performed at the Genomic Core Facility, Karmanos Cancer Institute utilizing Illumina BeadChip® Arrays essentially according to manufacturer’s instruction (Illumina). In brief, 0.5 µg total RNA was biotin-labeled and hybridized with BeadChips. The signal was detected with streptovadin-Cy3 according to manufacturer’s instruction (Illumina). The imaging of the BeadChips was conducted using a Bead Array Reader in conjunction with Bead Studio software (Illumina). Normalization of the data was carried out using a quantile-based approach which transforms the raw data so that the resulting normalized expression values of each sample have the same distribution [29]. An unsupervised cluster analysis was performed to detect similarities among samples based on gene expression profiles. The genes retained to perform the clustering were those varying the most regardless their group membership as described elsewhere [30]. Significance of the differentially expressed genes among various groups was tested using a moderated t-test to allow for P-value computation for the significance of gene changes. The P-values were then adjusted using the False Discovery Rate method [30] to derive corrected P-values. The P-values of <0.5 were considered significant provided that the fold change in expression was also equal to or larger than 2.
Murine mesothelioma allograft experiments
Female BALB/c mice aged 5 weeks were purchased from Taconic Research Animal Services and housed in accordance with protocols approved by the Institutional Laboratory Animal Care and Use Committee of Wayne State University. On day 1, AB12 murine mesothelioma cells (0.5 × 106) were suspended in 0.1 ml serum-free DMEM medium and inoculated subcutaneously (s.c.) in the right flank of each mouse (n = 5 each group). When the tumors became palpable (on day 10 after inoculation), the mice were randomly assigned into two groups and given various treatments by gavage. The control group received the vehicle (DMSO) while the test group received 500 mg/kg curcumin daily by oral gavage. Tumor sizes were measured daily using calipers and their volumes calculated using a standard formula: width2 × length/2. Body weight was measured weekly. The mice were sacrificed after 16 day-treatment when control tumors reached to ~ 1,000 mm3. H&E staining confirmed the presence of tumor.
Immunohistochemical analysis
Apoptosis in tumor tissues was determined by TUNEL assay using in situ cell Death Detection kit from Roche Applied Science (Indianapolis, IN) according to the manufacturer’s instruction. The formalin-fixed tumor xenograft biopsies from untreated or curcumin-treated animals were paraffin embedded and processed essentially following our previously described procedures [31, 32]. The tumor tissue slides were stained for presence of CARP-1, cyclin-dependent kinase inhibitor p27, HSULF1, or p-p38 by utilizing respective antibodies, and were then photographed under different magnifications using Zeiss microscope with a 35-mm camera attached for recording the photomicrographs. H&E counter-staining of tumor tissues was performed following our previously described methods [32].
Results
Curcumin inhibits MPM cell growth
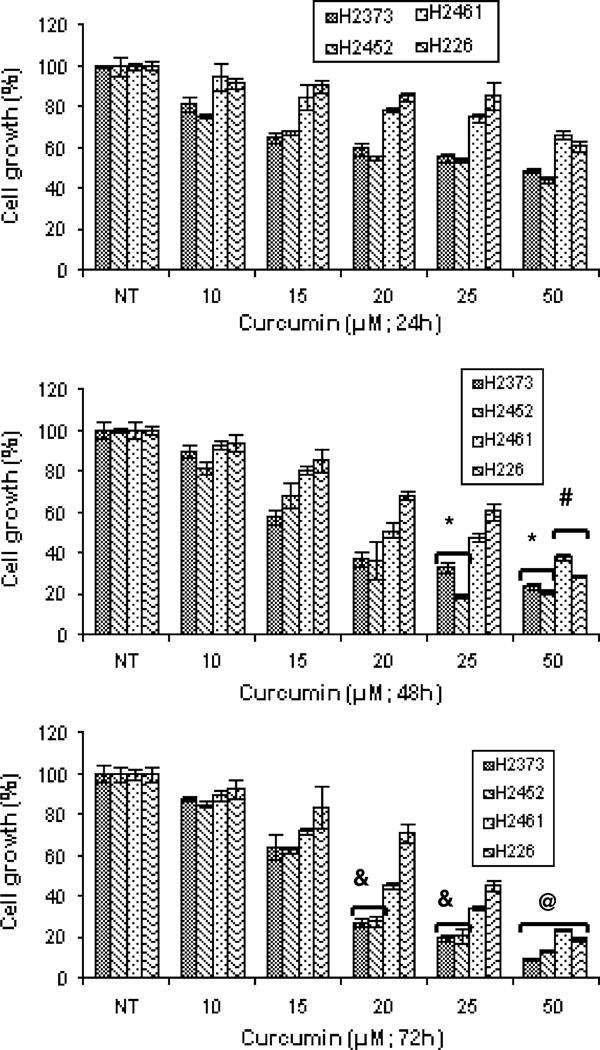
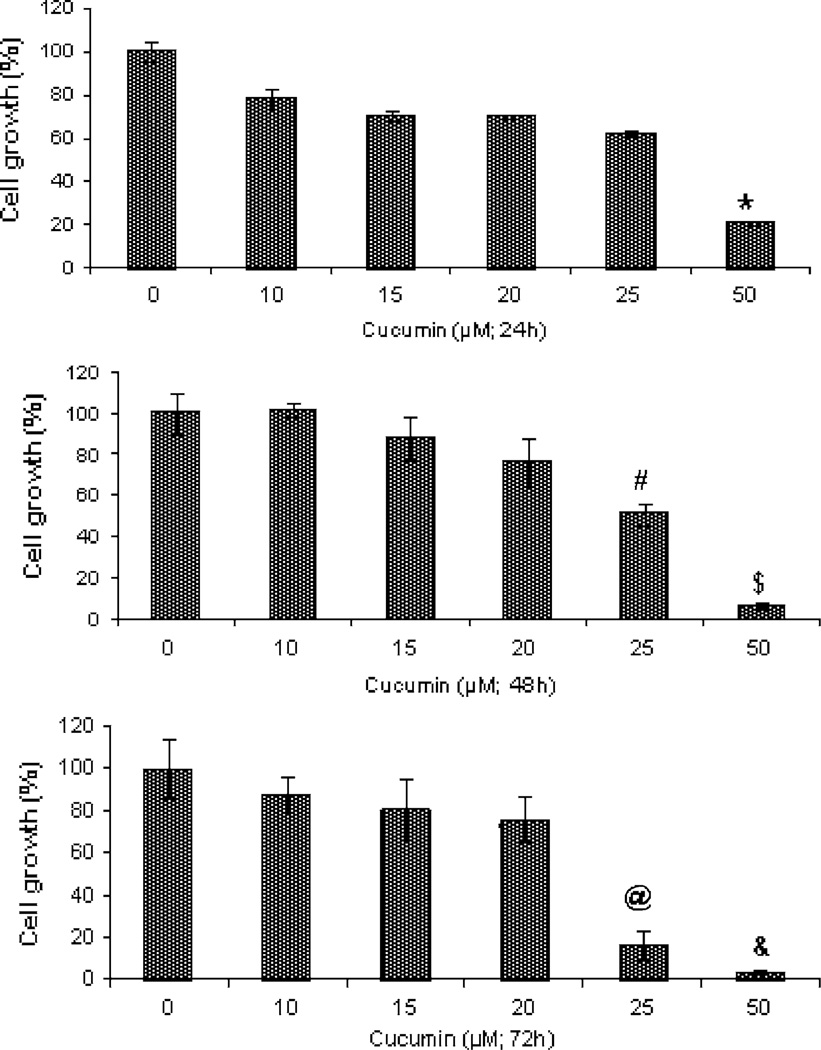
A number of studies have revealed chemo-preventive and anti-cancer properties of curcumin that are mainly due to its ability to cause cell cycle arrest and promote apoptosis in various cancer cells of diverse histological origins [21]. In this study, we utilized a number of MPM cells to investigate their growth inhibition by curcumin. As a first step, we determined the effects of curcumin on the growth of human MPM (H2373, H2452, H2461, and H226) as well as AB12 murine MPM cells. Each of the cells was separately treated with medium containing various doses of curcumin for periods of 24, 48, and 72 h followed by measurements of their viabilities by conducting MTT assays as detailed in “Materials and methods” section. Curcumin inhibited growth of all the MPM cells in a dose-dependent manner. Although, curcumin was cytotoxic to the MPM cells, the IC50 of ~ 20–25 µM were noted for the MPM cells (Fig. 1). Curcumin treatments also inhibited growth of the murine mesothelioma cells with an IC50 of ~ 25 µM (Fig. 2). Thus, curcumin inhibits growth of MPM cells in a dose-dependent manner.
Fig. 1.
Antiproliferative effect of curcumin on human MPM cells. Cells were treated with vehicle (Control, denoted as NT) or indicated doses of curcumin for noted times. Determination of viable/live cells was carried out by MTT assay. Columns in each of the histograms represent means of 3–4 independent experiments; bars, S.E. *, #, &, and @ significantly different from NT, P = <0.01, 0.015, 0.017 and 0.006, respectively
Fig. 2.
Antiproliferative effect of curcumin on murine MPM cells. Cells were treated with vehicle (Control, denoted as 0) or indicated doses of curcumin for noted times. Determination of viable/live cells was carried out by MTT assay. Columns in each of the histograms represent means of three independent experiments; bars, S.E. *, #, $, @, and & significantly different from untreated controls noted as 0, P = 0.002, 0.0005, 0.0005, 0.008, and 0.0009, respectively
Curcumin enhances cisplatin-induced MPM cell growth inhibition
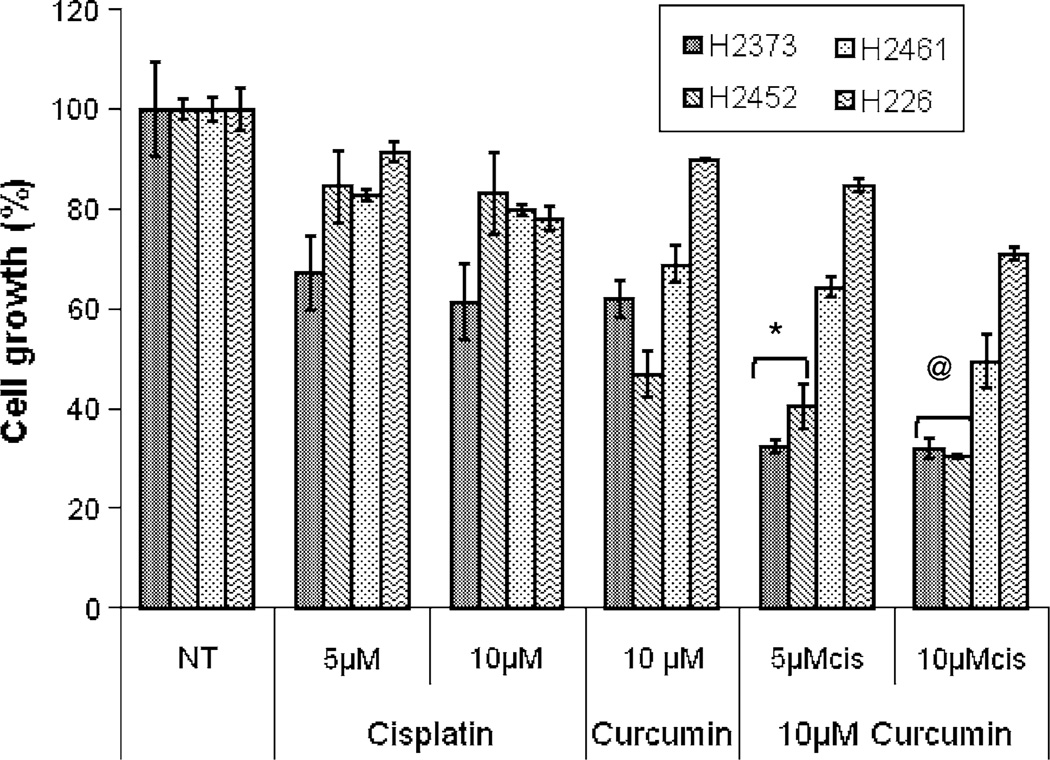
We have previously performed survival studies to establish a baseline cytotoxicity profile for MPM cell lines exposed to cisplatin alone. The MPM cells were relatively resistant to cisplatin with the IC50 values around 80 µM [33]. These IC50 values were approximately three-to-four fold higher than those for curcumin noted in Figs. 1 and 2. Since 20 µM dose of curcumin for 48-h treatment period caused ~ 60%, ~60%, ~50%, and ~30% inhibition of H2373, H2452, H2461, and H226 MPM cell growth, respectively, (Fig. 1), we utilized sub-optimal doses of this agent to examine its potential to enhance activity of cisplatin. MPM cells were pretreated with either 10 µM curcumin for 24 h followed by treatments with 5 or 10 µM cisplatin for additional 72 h. Of note is the fact that all the MPM cells were treated with different doses of cisplatin alone for 24-h periods. MPM cell viabilities were measured by MTT assay as above. As shown in Fig. 3, curcumin pre-treatments enhanced the growth inhibitory activity of cisplatin in H2373 and H2452 MPM cells when compared with their untreated, curcumin-or cisplatin-treated counterparts. A combination of the 10 µM doses of curcumin and cisplatin each, however, elicited modest growth inhibition of H2461 cells.
Fig. 3.
Curcumin enhances cisplatin efficacy. MPM cells were either untreated (denoted as NT), treated with different concentrations of cisplatin (cis) for 24 h, or pretreated with noted doses of curcumin for 24 h followed by addition of indicated concentrations of cisplatin for further 24 h. Determination of viable/live cells was carried out by MTT assay as detailed in methods. Columns in each of the histograms represent means of 3–4 independent experiments; bars, S.E. * and @ significantly different from NT, P = <0.03 and <0.002, respectively
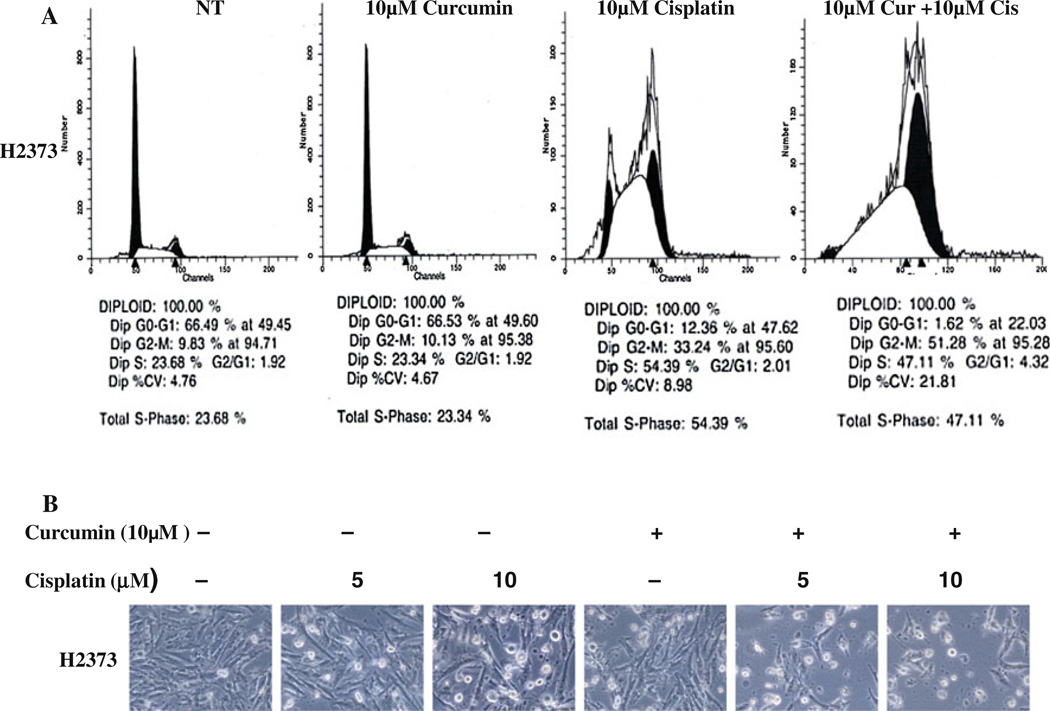
To investigate the growth inhibitory mechanisms utilized by curcumin, we conducted cell cycle analyses of untreated, curcumin-, cisplatin-, and curcumin-plus-cisplatin-treated H2373 MPM cells using propidium iodide staining and flow cytometry. Consistent with our earlier studies [34], 10 µM cisplatin treatment caused significant accumulation of cells in S and G2 M phases (Fig. 4a). Treatments with curcumin alone, however, failed to alter accumulation of these cells in any of the cell cycle phases when compared with the cell cycle distribution of their untreated counterparts. Moreover, MPM cells treated with a combination of curcumin and cisplatin had significant accumulation in the apoptotic (sub G0) fractions while undergoing a drastic reduction in cell numbers in the G0/ G1 phase (Fig. 4a). Together, these data support targeting of cell cycle progression in MPM cells by cisplatin alone or in combination with curcumin that involve their accumulation in S and G2M phases. We next determined whether growth inhibition of MPM cells by these agents involved induction of apoptotic cell death. In the first instance, we measured cellular apoptosis-associated morphological changes (such as nuclear condensation and fragmentation) in cells treated with curcumin, cisplatin, or a combination of both the agents. Cellular morphology changes (i.e., spherical and detached changes) were visualized by phase-contrast microscopy. The apoptotic cellular changes (Fig. 4b) were observed in the H2373 cells treated with curcumin, cisplatin, or a combination of both, but not in the cells that were treated with DMSO.
Fig. 4.
The effect of curcumin on the cell cycle distribution of MPM cell lines, a H2373 cells were treated with 10 µM curcumin for 94 h, 10 µM cisplatin for 72 h, or pretreated with 10 µM of curcumin for 24 h followed by 10 µM cisplatin for 72 h, followed by FACS analysis. DMSO was used as vehicle control, b H2373 cells were treated with 10 µM of curcumin for 94 h, or 5, 10 µM cisplatin for 72 h, or pretreated with 10 uM of curcumin for 24 h followed by 5, 10 µM cisplatin for 72 h, followed by photography of cellular morphologic changes. DMSO was used as vehicle control
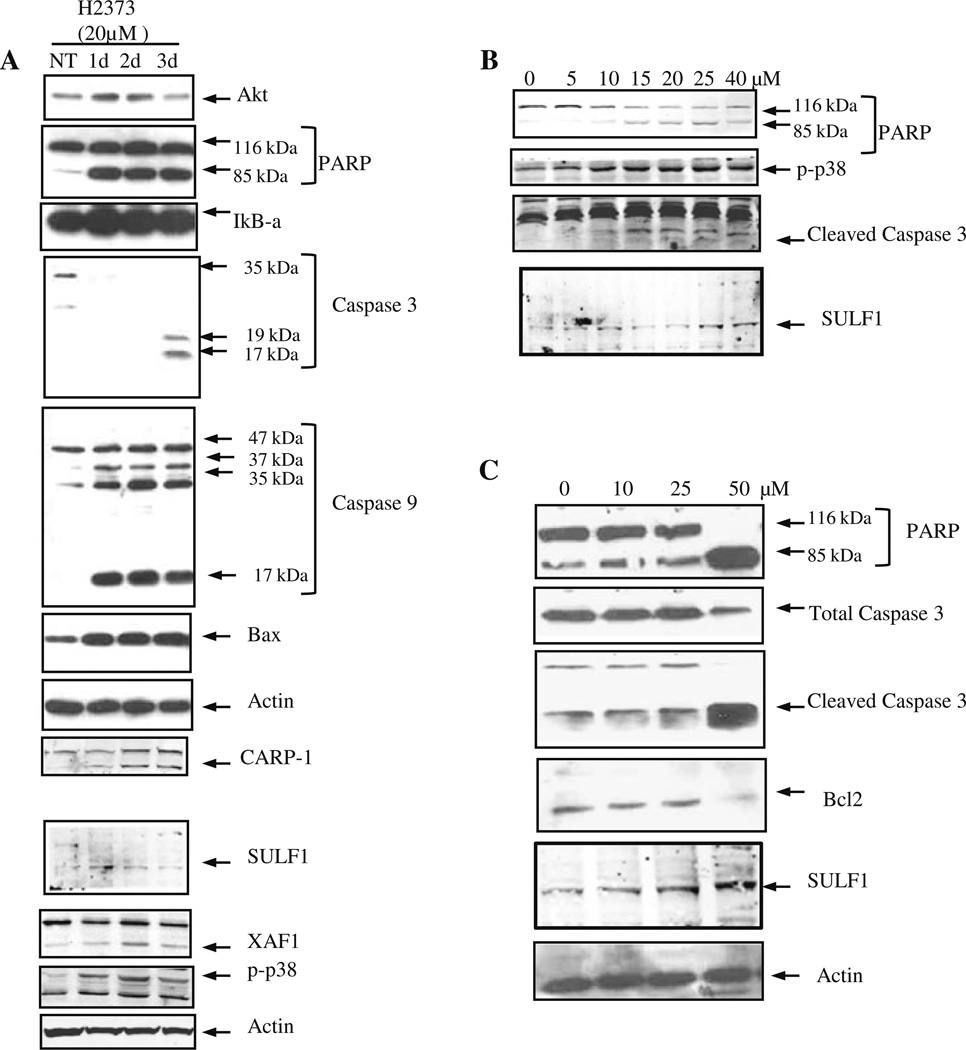
In an attempt to elucidate the mechanism of enhanced apoptosis of the MPM cells by curcumin, we assessed the levels of PARP, caspase 3, caspase 9, Bax, and Bcl2 proteins, which are known to be involved in diverse apoptosis-signaling pathways. H2373 cells were either untreated or treated with 20 µM curcumin for 24, 48, and 72 h, followed by western blot analysis of the cell lysates in conjunction with respective antibodies for the above noted proteins. Treatment with curcumin caused significant increase in expression of pro-apoptotic protein Bax, cleaved caspase 3, caspase 9, and PARP proteins (Fig. 5a). In addition, the H2373 human MPM and AB12 murine MPM cells were treated with various doses of curcumin for 72 h and levels of the apoptosis-signaling associated proteins p38 SAPK, PARP, caspase 3, and Bcl2 were determined by western blot analysis. In the human H2373 MPM cells, a 10 µM dose of curcumin induced activation of p38, caspase 3, and cleavage of PARP (Fig. 5b). Since curcumin also inhibited growth of the murine MPM cells, the western blot analysis revealed activation of caspase 3, cleavage of PARP, and loss of Bcl2 in the cells that were exposed to 50 µM dose of curcumin (Fig. 5c). Of note is the fact that 48-h treatments with 20 µM curcumin elicited ~60 and ~20% inhibition of H2373 and AB12 cells (Figs. 1, 2), respectively, suggest that H2373 cells are likely more sensitive to inhibition by curcumin when compared with the murine AB12 cells. This is also consistent with the western blot data in Fig. 5 where 10 µM dose of curcumin activated apoptosis signaling in H2373 cells, while a higher, 50 µ dose of curcumin was found to elicit activation of apoptosis in AB12 cells. Taken together, our in vitro data thus far suggest that curcumin suppresses growth of the MPM cells in part by promoting apoptosis.
Fig. 5.
Curcumin induces apoptosis in MPM cells. H2373 cells were treated with 20 µM of curcumin for 1–3 days in (a), or with different concentrations of curcumin for 72 h in (b). In panel c, AB12 cells were treated with different concentrations of curcumin for 72 h. Cell lysates were prepared from DMSO-treated vehicle control and curcumin-treated cells in each panel, and analyzed by SDS-PAGE, followed by western immunoblotting of the membranes with antibodies for indicated proteins as described in methods. All the membranes were probed with anti-actin antibodies for actin protein as a loading control
Curcumin targets novel apoptosis transducers
The molecular complexity of cancers and therapy-associated side effects often limit effectiveness of many anticancer modalities, and necessitate identification of novel cancer cell growth inhibitory targets/pathways for potential exploitation in devising efficacious therapeutic strategies. To further investigate mechanisms of MPM cell growth regulation by curcumin, a gene-array-based strategy was employed. H2373 and H2461 MPM cells were either untreated or separately treated with two doses of curcumin as described in methods. The RNAs from each group were hybridized with gene-array chips, and the data computed to identify genes that had a significant two-fold or higher altered expression following curcumin treatments of both the MPM cells. Next, a subset of genes that had similar curcumin-dependent altered expression in both the MPM cells were derived from this data and a select subset of these genes are indicated in Table 1. Of note is the fact that curcumin treatment caused down-regulation of several growth-promoting genes. These included receptor type protein tyrosine phosphatase R (PTPRR), RAS oncogene family member RAB8A, inhibitor of DNA binding (ID) 1, 2, 3, and regulator of G-protein signaling (RGS) 4. On the other hand, curcumin caused elevated expression of several growth inhibitory genes. These group of genes included XIAP-associated factor 1 (XAF1), osteoblast specific factor periostin, decorin, myosin light chain kinase, sulfatase (SULF) 1, damage-regulated autophagy modulator, filamin A interacting protein 1-like protein, growth arrest-specific (GAS) 6 protein, insulin-like growth factor binding proteins (IGFBP) 3, 7, Sparc/osteonectin, and interleukin (IL) 6β. Our western blot analyses further demonstrate that treatments of the 2373 MPM cells with curcumin results in elevated expression of XAF1 and SULF1 proteins (Fig. 5a).
Table 1.
List of select curcumin regulated genes in MPM cells
| ID | Adj.P.val | Fold change | Direction | SYMBOL | Name | ENTREZ |
|---|---|---|---|---|---|---|
| 5220451 | 0.999951362 | 4.216387218 | UP | POSTN | Periostin, osteoblast specific factor | 10631 |
| 6980296 | 0.999951362 | 3.893999579 | UP | DCN | Decorin | 1634 |
| 6350608 | 0.999951362 | 2.931475288 | UP | MYLK | Myosin light chain kinase | 4638 |
| 2640519 | 0.999951362 | 2.495608056 | UP | COL4A1 | Collagen, type IV, alpha 1 | 1282 |
| 7050703 | 0.999951362 | 3.756081427 | UP | SULF1 | Sulfatase 1 | 23213 |
| 3460278 | 0.999951362 | 2.051210568 | DOWN | PTPRR | Protein tyrosine phosphatase, receptor type, R | 5801 |
| 840537 | 0.999951362 | 2.728935376 | UP | PDGFRB | Platelet-derived growth factor receptor, beta polypeptide | 5159 |
| 5810471 | 0.999951362 | 2.289118934 | UP | DRAM | Damage-regulated autophagy modulator | 55332 |
| 4570377 | 0.999951362 | 2.956642534 | UP | ABCG1 | ATP-binding cassette, sub-family G (WHITE), member 1 | 9619 |
| 940220 | 0.999951362 | 9.987995292 | DOWN | RAB8A | RAB8A, member RAS oncogene family | 4218 |
| 7380300 | 0.999951362 | 2.204011716 | UP | FILIP1L | Filamin A interacting protein 1-like | 11259 |
| 3310368 | 0.999951362 | 2.407088299 | DOWN | ID3 | Inhibitor of DNA binding 3, dominant negative helixloophelix protein | 3399 |
| 4780008 | 0.999951362 | 2.523453734 | UP | IGFBP7 | Insulin-like growth factor binding protein 7 | 3490 |
| 1070403 | 0.999951362 | 2.489312792 | UP | GAS6 | Growth arrest-specific 6 | 2621 |
| 4670092 | 0.999951362 | 2.086346517 | DOWN | ID2 | Inhibitor of DNA binding 2, dominant negative helixloop-helix protein | 3398 |
| 5570538 | 0.999951362 | 2.125908257 | DOWN | ID1 | Inhibitor of DNA binding 1, dominant negative helixloop-helix protein | 3397 |
| 7050682 | 0.999951362 | 2.616469369 | UP | CXCR7 | Chemokine (C-X-C motif) receptor 7 | 57007 |
| 1740341 | 0.999951362 | 2.139974705 | UP | XAF1 | XIAP-associated factor 1 | 54739 |
| 3440500 | 0.999951362 | 2.705678862 | UP | IL6 | Interleukin 6 (interferon, beta 2) | 3569 |
| 1400709 | 0.999951362 | 2.198712979 | DOWN | RGS4 | Regulator of G-protein signaling 4 | 5999 |
| 2480450 | 0.999951362 | 2.064923366 | UP | SPOCK1 | Sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 | 6695 |
| 4540551 | 0.999951362 | 2.044121375 | UP | IGFBP3 | Insulin-like growth factor binding protein 3 | 3486 |
Genes that are down regulated by curcumin are made italic
CARP-1/CCAR1, a novel signaling transducer, was previously identified as a target of select chemotherapy such as adriamycin in the human breast cancer and lymphoma cells [23, 31, 32]. We further observed that treatments of MPM cells with proteasome inhibitor velcade also caused elevated levels of CARP-1 [33]. In this study, we investigated whether curcumin-dependent MPM cell growth inhibition involved elevated CARP-1 expression. H2373 Cells were treated with 20 µM curcumin for various time periods, and the cell lysates were analyzed by western blotting for CARP-1 expression. Our data in Fig. 5a show that curcumin treatments caused increased expression of CARP-1 in MPM cells. Together, data in Fig. 5 and Table 1 strongly suggest that curcumin-dependent inhibition of MPM cancer cells is accomplished, in part, by stimulating novel transducers of apoptosis signaling such as CARP-1/CCAR1 and XAF1.
Curcumin inhibits MPM cell-derived xenograft growth
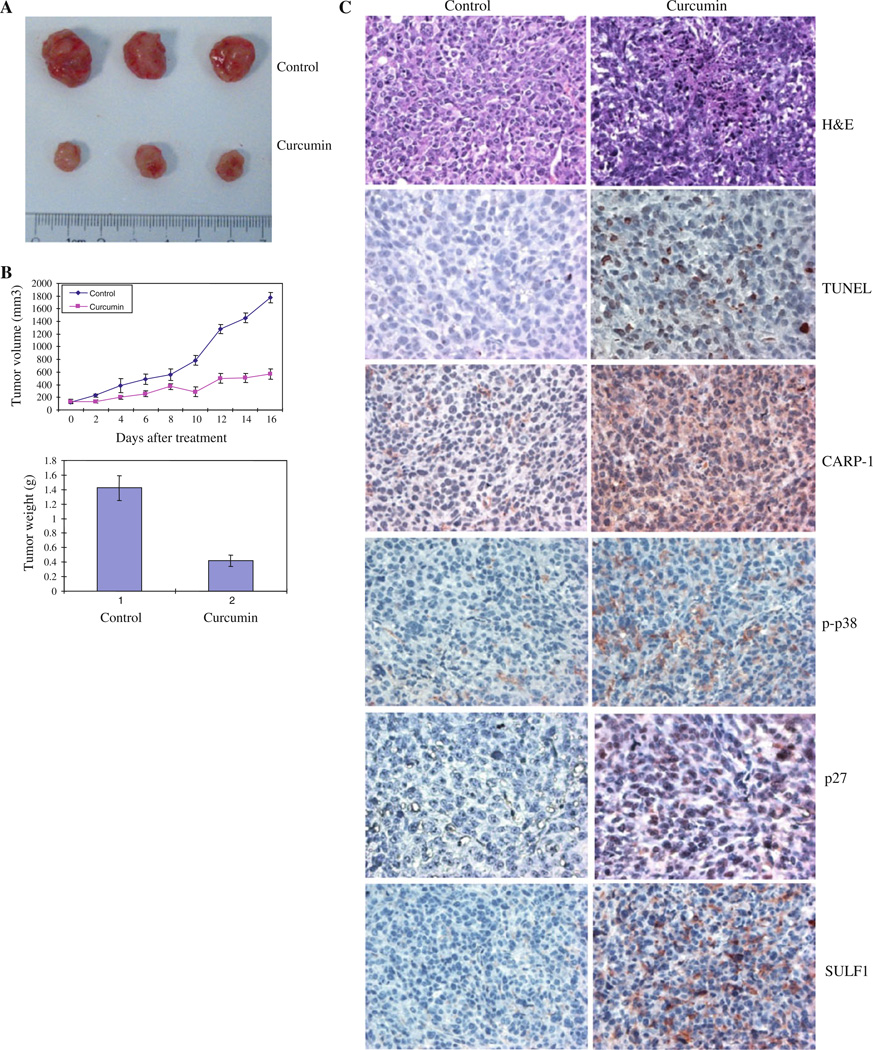
The Balb/c mice in each group were injected with AB12 murine MPM cells as in methods. After the tumors became palpable, the mice were randomly assigned to control (untreated) or treated groups. The mice were gavage treated with curcumin as detailed in methods. The animals were observed throughout the treatment period and did not show any visible toxicity including symptoms of diarrhea, dehydration, weight loss, hair loss, or any other discomfort. Mice were sacrificed on day 18, and tumor size and appearance from one flank for three mice each under the control and treatment condition is shown in Fig. 6a. Subcutaneous tumor volume in each flank was measured on regular intervals, and tumor progression under each treatment condition is shown in Fig. 6b. In addition, the bar histogram in Fig. 6b shows tumor weight from control and treated animals following calculations using the formula indicated in methods. As is evident from Fig. 6a, b, curcumin administration caused profound suppression of the MPM cell-derived xenograft growth in vivo. Whether curcumin treatments suppressed murine MPM xenograft growth by stimulating apoptosis was determined next by conducting immunohistochemical analyses of the xenografted tumor biopsies from the control and treated animals for levels of apoptosis. Consistent with our data in Figs. 4 and 5, immuno- staining showed elevated levels of pro-apoptotic phos-phorylated p38 MAPK and CARP-1 proteins in the xenograft biopsies from the curcumin-treated animals when compared with their untreated control counterparts (Fig. 6c). Moreover, levels of cyclin-dependent kinase inhibitor p27 protein, SULF1, and the number of apoptotic, TUNEL-positive cells were elevated in xenograft biopsies from curcumin-treated animals (Fig. 6c). The data in Fig. 6 therefore strongly suggest that curcumin suppresses MPM tumor growth in vivo in part by stimulating apoptosis.
Fig. 6.
Curcumin inhibits growth of MPM xenografts. The efficacy studies were carried out as in methods, and data were analyzed essentially as described before [33]. Animals were either untreated or treated with curcumin as in methods. a Tumor volumes were monitored every other day in each group and tumor weights were determined by the end of the experiment. b Points, mean tumor volume in each experimental group containing four mice; columns, mean tumor weights in vehicle or curcumin-treated group. Bars, SD. In panel c, the formalin-fixed tumor xenograft biopsies from panel (a) were paraffin embedded, processed, and subsequently subjected to immunohistochemical staining as detailed in methods. Representative photomicrographs (200× magnification) in the first row show hematoxylin and eosin (H&E) staining of the tissue sections. Representative photomicrographs (200× magnification) are also presented demonstrating apoptosis in xenografted tumors following their staining using TUNEL assay (second row), with anti-CARP-1 (third row), anti-phospho-p38 (fourth row), anti-p27 antibodies (fifth row), or anti sulfatasel antibodies (sixth row)
Discussion
A large number of cell biological and animal studies have suggested potential therapeutic or preventive effects associated with curcumin in a variety of malignancies including cancer. It is thought to have antitumor, antioxidant, antiarthritic, anti-amyloid, anti-ischemic, and antiinflammatory properties [34–36]. With regard to the human pathologies, recent and current clinical trials have further elaborated potential utility of curcumin for multiple myeloma, pancreatic cancer, myelodysplastic syndromes, colon cancer, psoriasis, and Alzheimer’s disease. The anticancer effects of curcumin however are due largely to its ability to inhibit the NF-κB- and Akt-dependent survival pathways as well as induce apoptosis in cancer cells with minimal cytotoxicity toward the healthy cells [21]. Moreover, synergistic, pro-apoptotic effects of curcumin and various chemotherapies have also been described [22]. Despite a wealth of knowledge with regard to anti-cancer and other beneficial properties of curcumin, the molecular mechanisms of its action are yet to be fully elucidated. In this investigation, we conducted in vitro and in vivo studies by utilizing human and murine MPM cells to investigate anti-MPM effects of curcumin and the molecular mechanisms involved. Consistent with observations in other models, we found that curcumin suppressed MPM growth in vitro and in vivo and enhanced efficacy of the first line anti-MPM therapeutic cisplatin.
Targeting of cancer cell growth and survival pathways remains an attractive anti-cancer strategy. Our current studies indicating involvement of apoptosis in curcumin-dependent inhibition of MPM growth highlights anti-MPM potential for this safe and non-toxic agent. In this context, prior micro-array analyses of the curcumin-treated human breast cancer cells have revealed a subset of apoptosis-associated genes that were altered by curcumin treatment [22]. Because alterations in apoptosis-signaling pathways often contribute to MPM growth and survival [38], we undertook an in vitro and gene-array-based profiling to identify novel transducers of apoptosis signaling that may be activated/induced by curcumin. Our studies revealed that while curcumin suppressed levels of several growth-promoting genes, it also caused elevated expression of multiple novel cell growth inhibitory and apoptosis transducers such as CCAR1/CARP-1, XAF1, and SULF1.
Our current data and several previous studies [21, 22] amply demonstrate that curcumin treatments induced apoptosis in a variety of cancer cells types, including MPM. Although, a robust cleavage of PARP, caspase 3, and induction of Bax was noted in H2373 MPM cells that were treated with 10–20 µM curcumin, a somewhat higher dose of curcumin was required for robust activation of apoptosis transducers in murine MPM cells (Figs. 5b, c). Whether this difference in the IC50 of curcumin is due to additional cell-selective signaling leading to increased sensitivity of human MPM cells or is a consequence of differences in the cellular uptake and metabolism of curcumin by the MPM cells remains to be clarified. These data nonetheless suggest that curcumin inhibits MPM growth by inducing growth arrest and apoptosis, and its effects are likely transduced in part by targeting specific mediator(s). Our gene-array-based analyses (Table 1) together with western blot data in Figs. 5 and 6 convincingly reveal a new group of apoptosis signal-transducing genes that are activated by curcumin in MPM cells in vitro and in vivo. Since, CCAR1/CARP-1 is an emerging and novel target of diverse cell growth and apoptosis-signaling pathways [23, 32, 33, 38–40] and is not involved in cisplatin-dependent HBC growth inhibition [23], identification of CCAR1/CARP-1 as a downstream effector of curcumin signaling is anticipated to allow for design of effective strategies for optimizing curcumin potential as suppressor of MPM and its drug (Cisplatin)-resistant phe-notype. Moreover, curcumin also stimulated XAF1 expression (Fig. 5; Table 1). XAF1 is an inhibitor of XIAP, a transducer of cell survival signaling. Together with our earlier studies demonstrating increased levels of XAF1 in MPM and breast cancer cells in the presence of velcade [33], loss of XIAP expression in velcade-treated MPM cells further underscores an important role for XIAP-dependent cell survival pathway by velcade and curcumin.
The gene-array data also revealed a number of interesting genes that may be involved in regulating MPM cell growth and survival in the presence of curcumin. Of note is the down-regulation of three members of the inhibitor of DNA binding (ID) protein family (ID1, 2, 3; Table 1). ID proteins are a group of helix-loop-helix (HLH) proteins that lack a basic DNA-binding domain but are able to form hetero-dimers with other HLH proteins, thereby inhibiting DNA binding and transcription functions. ID proteins that are thought to be necessary for G1 progression as well as transduction of p5 3-dependent DNA damage response [41, 42]. Whether and to what extent curcumin interferes with cell cycle progression of the MPM cells in part by diminishing levels of ID proteins remains to be clarified. Curcumin treatments however resulted in up-regulation of several proteins that are known to function as inhibitors of cell growth. In addition to XAF1, curcumin interestingly stimulated expression of SULF1 (Table 1; Figs. 5, 6). Recent studies have highlighted a potential tumor suppressor function of human SULF1 in carcinogenesis. SULF1 inhibits the co-receptor function of cellular heparin sulfate proteoglycans (HSPGs) and thus impacts multiple receptor tyrosine kinase (RTK)-signaling pathways regulated by the heparin-binding growth factors such as FGF, HB-EGF, VEGF, PDGF, and HGF [25]. SULF1 is down-regulated by epigenetic mechanisms in many cancers and its forced expression interferes with cancer cell proliferation, migration, and invasion processes. Although, curcumin induces apoptosis in MPM cells, increased levels of SULF1 in the presence of curcumin likely attenuates RTK-dependent MPM growth and survival signaling. If proven correct, SULF-1 -dependent signaling in the curcumin-treated MPM cells will not only support a novel mechanism of action of this agent, but will underscore a broader utility for this compound as a future anti-cancer and perhaps chemo-preventive agent for MPM and possibly other cancers.
Acknowledgments
This study was supported by the department of Veterans Affairs Merit Review grants to AKR and AW, Susan G Komen for the Cure grant to AKR, and MARF support to AW.
Contributor Information
Ying Wang, John D. Dingell VA Medical Center, Karmanos Cancer Institute, Wayne State University, VAMC, 4646 John R., Detroit, MI 48201, USA.
Arun K. Rishi, Email: Rishia@Karmanos.org, John D. Dingell VA Medical Center, Departments of Oncology and Internal Medicine, Karmanos Cancer Institute, Wayne State University, Room B4334, VAMC, 4646 John R., Detroit, MI 48201, USA.
Wenjuan Wu, John D. Dingell VA Medical Center, Karmanos Cancer Institute, Wayne State University, Room B4325, VAMC, 4646 John R., Detroit, MI 48201, USA.
Lisa Polin, Departments of Oncology and Internal Medicine, Karmanos Cancer Institute, Wayne State University, 110 E. Warren Street, Detroit, MI 48201, USA.
Sunita Sharma, John D. Dingell VA Medical Center, Department of Surgery, Karmanos Cancer Institute, Wayne State University, VAMC, 4646 John R., Detroit, MI 48201, USA.
Edi Levi, John D. Dingell VA Medical Center, Departments of Pathology and Oncology, Wayne State University, VAMC, 4646 John R., Detroit, MI 48201, USA.
Steven Albelda, Pulmonary, Allergy & Critical Care Division, Abramson Research center, University of Pennsylvania Medical Center, Philadelphia, PA, USA.
Harvey I. Pass, Division of Cardiothoracic Surgery, New York University Cancer Center, New York, USA
Anil Wali, John D. Dingell VA Medical Center, Department of Surgery, Karmanos Cancer Institute, Wayne State University, Room B4325, VAMC, 4646 John R., Detroit, MI 48201, USA.
References
- 1.Belli C, Fennell D, Giovannini M, Gaudino G, Mutti L. Malignant pleural mesothelioma: current treatments and emerging drugs. Expert Opin Emerg Drugs. 2009;14:423–437. doi: 10.1517/14728210903074563. [DOI] [PubMed] [Google Scholar]
- 2.Fennell DA, Gaudino G, O’Byrne KJ, Mutti L, van Meerbeeck J. Advances in the systemic therapy of malignant pleural mesothelioma. Nat Clin Pract Oncol. 2008;5:136–147. doi: 10.1038/ncponc1039. [DOI] [PubMed] [Google Scholar]
- 3.Janne PA. Chemotherapy for malignant pleural mesothelioma. Clin Lung Cancer. 2003;5:98–106. doi: 10.3816/CLC.2003.n.023. [DOI] [PubMed] [Google Scholar]
- 4.Goudar RK. New therapeutic options for mesothelioma. Curr Oncol Rep. 2005;7:260–265. doi: 10.1007/s11912-005-0048-3. [DOI] [PubMed] [Google Scholar]
- 5.Krug LM. An overview of chemotherapy for mesothelioma. Hematol Oncol Clin North Am. 2005;19:1117–1136. doi: 10.1016/j.hoc.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 6.De Rienzo A, Balsara BR, Apostolou S, Jhanwar SC, Testa JR. Loss of heterozygosity analysis defines a 3-cM region of 15q commonly deleted in human malignant mesothelioma. Oncogene. 2001;20:6245–6249. doi: 10.1038/sj.onc.1204828. [DOI] [PubMed] [Google Scholar]
- 7.Pylkkanen L, Sainio M, Ollikainen T, Mattson K, Nordling S, Carpen O, Linnainmaa K, Husgafvel-Pursiainen K. Concurrent LOH at multiple loci in human malignant mesothelioma with preferential loss of NF2 gene region. Oncol Rep. 2002;9:955–959. [PubMed] [Google Scholar]
- 8.Musti M, Kettunen E, Dragonieri S, Lindholm P, Cavone D, Serio G, Knuutila S. Cytogenetic and molecular genetic changes in malignant mesothelioma. Cancer Genet Cytogenet. 2006;170:9–15. doi: 10.1016/j.cancergencyto.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 9.Langerak AW, Williamson KA, Miyagawa K, Hagemeijer A, Versnel MA, Hastie ND. Expression of the Wilm’s tumor gene WT1 in human malignant mesothelioma cell lines and relationship to platelet-derived growth factor A and insulin-like growth factor 2 expression. Genes Chromosom Cancer. 1995;12:87–96. doi: 10.1002/gcc.2870120203. [DOI] [PubMed] [Google Scholar]
- 10.Adachi Y, Yoshio-Hoshino N, Aoki C, Nishimoto N. VEGF targeting in mesotheliomas using an interleukin-6 signal inhibitor based on adenovirus gene delivery. Anticancer Res. 2010;30:1947–1952. [PubMed] [Google Scholar]
- 11.Ohta Y, Shridhar V, Kalemkerian GP, Bright RK, Watanabe Y, Pass HI. Thrombospondin-1 expression and clinical implications in malignant pleural mesothelioma. Cancer. 1999;85:2570–2576. [PubMed] [Google Scholar]
- 12.Whitson BA, Jacobson BA, Frizelle S, Patel MR, Yee D, Mad-daus MA, Kratzke RA. Effects of insulin-like growth factor-1 receptor inhibition in mesothelioma. Ann Thorac Surg. 2006;82:996–1002. doi: 10.1016/j.athoracsur.2006.04.013. [DOI] [PubMed] [Google Scholar]
- 13.Pass HI, Bocchetta M, carbone M. Evidence of an important role for SV40 in mesothelioma. Thorac Surg Clin. 2004;14:489–195. doi: 10.1016/j.thorsurg.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Xia G, Kumar SR, Masood R, Koss M, Templeman C, Quinn D, Zhu S, Reddy R, Krasnoperov V, Gill PS. Up-regulation of EphB4 in mesothelioma and its biological significance. Clin Cancer Res. 2005;11:4305–1315. doi: 10.1158/1078-0432.CCR-04-2109. [DOI] [PubMed] [Google Scholar]
- 15.Fennell DA, Rudd RM. Defective core-apoptosis signalling in diffuse malignant pleural mesothelioma: opportunities for effective drug development. Lancet Oncol. 2004;5:354–362. doi: 10.1016/S1470-2045(04)01492-5. [DOI] [PubMed] [Google Scholar]
- 16.Surh YJ. Cancer chemoprevention with dietary phyto-chemicals. Nat Rev Cancer. 2003;3:768–780. doi: 10.1038/nrc1189. [DOI] [PubMed] [Google Scholar]
- 17.Scott EN, Gescher AJ, Steward WP, Brown K. Development of dietary phytochemical chemopreventive agents: bio-markers and choice of dose for early clinical trials. Cancer Prev Res. 2009;2:525–530. doi: 10.1158/1940-6207.CAPR-08-0223. [DOI] [PubMed] [Google Scholar]
- 18.Huang MT, Wang ZY, Georgiadis CA, Laskin JD, Conney AH. Inhibitory effects of curcumin on tumor initiation by benzo[a]pyrene and 7,12-dimethylbenz[a]anthracene. Carcinogenesis. 1992;13:2183–2186. doi: 10.1093/carcin/13.11.2183. [DOI] [PubMed] [Google Scholar]
- 19.Conney AH, Lysz T, Ferraro T, Abidi TF, Manchand PS, La-skin JD, Huang MT. Inhibitory effect of curcumin and some related dietary compounds on tumor promotion and ara-chidonic acid metabolism in mouse skin. Adv Enzyme Regul. 1991;31:385–396. doi: 10.1016/0065-2571(91)90025-h. [DOI] [PubMed] [Google Scholar]
- 20.Huang MT, Smart RC, Wong CQ, Conney AH. Inhibitory effect of curcumin, chlorogenic acid, caffeic acid, and ferulic acid on tumor promotion in mouse skin by 12-0 tetra-decanoylphorbol-13-acetate. Cancer Res. 1988;48:5941–5946. [PubMed] [Google Scholar]
- 21.Reuter S, Eifes S, Dicato M, Aggarwal BB, Diederich M. Modulation of anti-apoptotic and survival pathways by curcumin as a strategy to induce apoptosis in cancer cells. Biochem Pharmacol. 2008;76:1340–1351. doi: 10.1016/j.bcp.2008.07.031. [DOI] [PubMed] [Google Scholar]
- 22.Ramachandran C, Rodriguez S, Ramachandran R, Raveendran Nair PK, Fonseca H, Khatib Z, Escalon E, Melnick SJ. Expression profiles of apoptotic genes induced by curcumin in human breast cancer and mammary epithelial cell lines. Anticancer Res. 2005;25:3293–3302. [PubMed] [Google Scholar]
- 23.Rishi AK, Zhang L, Boyanapalli M, Wali A, Mohammad RM, Yu Y, Fontana JA, Hatfield JS, Dawson MI, Majumdar APN, Reichert U. Identification and characterization of a cell-cycle and apoptosis regulatory protein (CARP)-1 as a novel mediator of apoptosis signaling by retinoid CD437. J Biol Chem. 2003;278:33422–33435. doi: 10.1074/jbc.M303173200. [DOI] [PubMed] [Google Scholar]
- 24.Listen P, Fong WG, Kelly NL, Toji S, Miyazaki T, Conte D, Tamai K, Craig CG, McBurney MW, Korneluk RG. Identification of XAF1 as an antagonist of XIAP anti-caspase activity. Nat Cell Biol. 2001;3:128–133. doi: 10.1038/35055027. [DOI] [PubMed] [Google Scholar]
- 25.Lai J-P, Sandhu DS, Shire AM, Roberts LR. The tumor suppressor function of human sulfatase 1 (SULF1) in carcinogenesis. J Gastrointest Cancer. 2008;39:149–158. doi: 10.1007/s12029-009-9058-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pass HI, Lott D, Lonardo F, Harbut M, Liu Z, Tang N, Carbone M, Webb C, Wali A. Asbestos exposure, pleural mesothelioma, and serum osteopontin levels. N Engl J Med. 2005;353:1564–1573. doi: 10.1056/NEJMoa051185. [DOI] [PubMed] [Google Scholar]
- 27.Suzuki E, Kim S, Cheung HK, Corbley MJ, Zhang X, Sun L, Shan F, Singh J, Lee WC, Albelda SM, Ling LE. A novel small-molecule inhibitor of transforming growth factor beta type I receptor kinase (SM16) inhibits murine mesothelioma tumor growth in vivo and prevents tumor recurrence after surgical resection. Cancer Res. 2007;67:2351–2359. doi: 10.1158/0008-5472.CAN-06-2389. [DOI] [PubMed] [Google Scholar]
- 28.Odaka M, Sterman D, Wiewrodt R, Zhang Y, Kiefer M, Amin KM, Gao G-P, Wilson JM, Barsoum J, Kaiser LR, Albelda SM. Eradication of intraperitoneal and distant tumor by ade-novirus-mediated interferon-b gene therapy due to induction of systemic immunity. Cancer Res. 2001;61:6201–6212. [PubMed] [Google Scholar]
- 29.Smyth GK, Limma L. Liner models for microarray data. In: Gentleman R, Carey V, Duoit S, Irizarry R, Huber W, editors. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 30.Tarca AL, Carey VJ, Chen XW, Romero R, Draghici S. Machine learning and its applications to biology. PLoS Comput Biol. 2007;3 doi: 10.1371/journal.pcbi.0030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L, Levi E, Majumder P, et al. Transactivator of transcription tagged cell cycle and apoptosis regulatory protein-1 peptides suppress growth of human breast cancer cells in vitro and in vivo. Mol Cancer Ther. 2007;6:1661–1672. doi: 10.1158/1535-7163.MCT-06-0653. [DOI] [PubMed] [Google Scholar]
- 32.Levi E, Zhang L, Aboukameel A, Rishi S, Mohammad RM, Polin L, Hatfield JS, Rishi AK. Cell cycle and apoptosis regulatory protein (CARP)-l is a novel, adriamycin-inducible, diffuse large B-cell lymphoma (DLBL) growth suppressor. Cancer Chemother Pharmacol. in press doi: 10.1007/s00280-010-1442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Y, Rishi AK, Puliyappadamba VT, Sharma S, Yang H, Tarca A, Dou QP, Lonardo F, Ruckdeschel JC, Pass HI, Wali A. Targeted proteasome inhibition by velcade induces apoptosis in human mesothelioma and breast cancer cell lines. Cancer Chemother Pharmacol. 2010;66:455–466. doi: 10.1007/s00280-009-1181-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hatcher H, Planalp R, Cho J, Torti FM, Torti SV. Cur-cumin: from ancient medicine to current clinical trials. Cell Mol Life Sci. 2008;65:1631–1652. doi: 10.1007/s00018-008-7452-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aggarwal BB, Shishodia S. Molecular targets of dietary agents for prevention and therapy of cancer. Biochem Pharmacol. 2006;71:1397–1421. doi: 10.1016/j.bcp.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 36.Shukla PK, Khanna VK, Ali MM, Khan MY, Srimal RC. Anti-ischemic effect of curcumin in rat brain. Neurochem Res. 2008;33:1036–1043. doi: 10.1007/s11064-007-9547-y. [DOI] [PubMed] [Google Scholar]
- 37.Gordon GJ, Mani M, Mukhopadhyay L, Dong L, Yeap BY, Sugarbaker DJ, Bueno R. Inhibitor of apoptosis proteins are regulated by tumour necrosis factor-alpha in malignant pleural mesothelioma. J Pathol. 2007;211:439–446. doi: 10.1002/path.2120. [DOI] [PubMed] [Google Scholar]
- 38.Rishi AK, Zhang L, Yu Y, Jiang Y, Nautiyal J, Wali A, Fontana JA, Levi E, Majumdar APN. Cell cycle and apoptosis regulatory protein (CARP)-1 is involved in apoptosis signaling by epidermal growth factor receptor. J Biol Chem. 2006;281:13188–13198. doi: 10.1074/jbc.M512279200. [DOI] [PubMed] [Google Scholar]
- 39.Kim JH, Yang CK, Heo K, Roeder RG, An W, Stallcup MR. CCAR1, a key regulator of mediator complex recruitment to nuclear receptor transcription complexes. Mol Cell. 2008;31:510–519. doi: 10.1016/j.molcel.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ou CY, Kim JH, Yang CK, Stallcup MR. Requirement of cell cycle and apoptosis regulator 1 for target gene activation by Wnt and {beta}-catenin and for anchorage-independent growth of human colon carcinoma cells. J Biol Chem. 2009;284:20629–20637. doi: 10.1074/jbc.M109.014332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian Y, Chen X. ID1, inhibitor of differentiation/DNA binding, is an effector of the p53-dependent DNA damage response pathway. J Biol Chem. 2008;283:22410–22416. doi: 10.1074/jbc.M800643200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hara E, Yamaguchi T, Nojima H, Ide T, Campisi J, Okayama H, Oda K. Id-related genes encoding helix-loop-helix proteins are required for G1 progression and are repressed in senescent human fibroblasts. J Biol Chem. 1994;269:2139–2145. [PubMed] [Google Scholar]