Abstract
Src/Yes tyrosine kinase signaling contributes to the regulation of bone homeostasis and inhibits osteoblast activity. Here we show that the endogenous Yes-associated protein (YAP), a mediator of Src/Yes signaling, interacts with the native Runx2 protein, an osteoblast-related transcription factor, and suppresses Runx2 transcriptional activity in a dose-dependent manner. Runx2, through its PY motif, recruits YAP to subnuclear domains in situ and to the osteocalcin (OC) gene promoter in vivo. Inhibition of Src/Yes kinase blocks tyrosine phosphorylation of YAP and dissociates endogenous Runx2–YAP complexes. Consequently, recruitment of the YAP co-repressor to subnuclear domains is abrogated and expression of the endogenous OC gene is induced. Our results suggest that Src/Yes signals are integrated through organization of Runx2–YAP transcriptional complexes at subnuclear sites to attenuate skeletal gene expression.
Keywords: Cbfa1, nuclear matrix, osteoblasts, osteocalcin, Src signaling
Introduction
The spatial distribution of binding sites for transcription factors, regulation of their binding to cognate sites by chromatin, and the distribution of regulatory factors and their co-regulators in distinct subnuclear domains together contribute to the control of gene expression (Berezney and Jeon, 1995; Stein et al, 2000). Transcription factors possess intrinsic nuclear import signals that localize these proteins to the nucleus. Importantly, several transcription factors have an additional protein module, the nuclear matrix-targeting signal (NMTS), that directs these factors to subnuclear sites where activation and repression take place (van Steensel et al, 1995a, 1995b; Zeng et al, 1997; Davie and Chadee, 1998; McNeil et al, 1998; Stenoien et al, 1998; Zeng et al, 1998; Mancini et al, 1999; DeFranco and Guerrero, 2000; Stenoien et al, 2000; Zaidi et al, 2001). Proteins that transduce signals to the nucleus contain both nuclear import and export signals, and their nucleo-cytoplasmic shuttling is controlled by physiological cues. Within the nucleus, these proteins interact with transcription factors and modulate their ability to regulate gene expression (Massague, 1998; Akiyama, 2000; Finidori, 2000; Ihle, 2001). Our studies focus on the mechanisms by which combinatorial activities of transcription factors and signaling molecules are integrated at specific subnuclear sites for physiological control of tissue formation.
Bone formation requires transcriptional mechanisms for sequential induction and repression of genes that support progressive osteoblast phenotype development. The Runx transcription factors and their co-regulators control cellular differentiation and lineage commitment (Ito, 1999; Westendorf and Hiebert, 1999; Li et al, 2002) by influencing the functional architecture of target gene promoters (Stein et al, 2000). Runx proteins are directed to subnuclear domains through the C-terminal NMTS and interact with DNA through the N-terminal runt homology domain (Ogawa et al, 1993; Zeng et al, 1997, 1998; Tang et al, 1999; Zaidi et al, 2001). The Runx2 family member is essential for osteoblast maturation in vivo and is associated with cleidocranial dysplasia (Komori et al, 1997; Mundlos et al, 1997; Otto et al, 1997). In vivo genetic evidence indicates that interference with subnuclear targeting and associated co-regulatory functions of Runx2 can account for this block in bone formation (Choi et al, 2001).
Runx2 is a target of several extracellular signals that regulate skeletal formation and homeostasis. The C-terminus of Runx2, which includes the NMTS, interacts with proteins involved in the TGFβ/BMP (i.e., Smads), the transducin-like enhancer (TLE)/groucho and the Src/Yes tyrosine kinase (e.g., the Yes-associated protein, YAP) signaling pathways (Hanai et al, 1999; Yagi et al, 1999; Zhang and Derynck, 1999; Javed et al, 2000; Zaidi et al, 2002). Src family tyrosine kinases are activated by a variety of extracellular stimuli, and broadly control cell cycle regulation, cell migration, cell metabolism and survival, as well as cell proliferation and differentiation (Thomas and Brugge, 1997; Schlessinger, 2000). Src homology (SH) domains present in these kinases mediate interactions with downstream signaling proteins such as YAP. YAP, in turn, contains a WW domain in the N-terminus (Sudol, 1994) that recognizes a proline-rich motif (PPxY) present in a broad range of proteins including Runx factors (Sudol et al, 1995; Sudol and Hunter, 2000).
The significance of Src/Yes tyrosine kinase signaling in bone development is suggested by the osteopetrotic phenotype of Src null mice (Soriano et al, 1991) and the accelerated differentiation of Src-deficient osteoblasts (Marzia et al, 2000). These results indicate that Src signaling inhibits osteoblast differentiation, but the underlying molecular mechanisms are not known. Here we show that Src signaling in osteoblasts function through YAP to inhibit Runx2 activity. In response to Src/Yes signaling, YAP is phosphorylated and recruited by Runx2 to subnuclear sites and chromatin of the bone-specific OC gene, resulting in its repression. Thus, Runx2–YAP interactions integrate Src signals at architecturally associated subnuclear sites, where regulatory complexes assemble to control osteoblast gene expression.
Results
Endogenous Runx2 and YAP proteins interact in vivo and associate in situ at subnuclear sites in osteoblasts
Direct interaction between YAP and Runx protein segments has been documented in a cell-free system (Yagi et al, 1999). We therefore determined whether endogenous Runx2 and YAP interact in osteoblasts. Co-immunoprecipitation assays show that endogenous YAP and Runx2 proteins form a complex in vivo in osteoblastic ROS 17/2.8 cells (Figure 1A). These findings indicate that Runx2 and YAP interact under physiological conditions.
Figure 1.
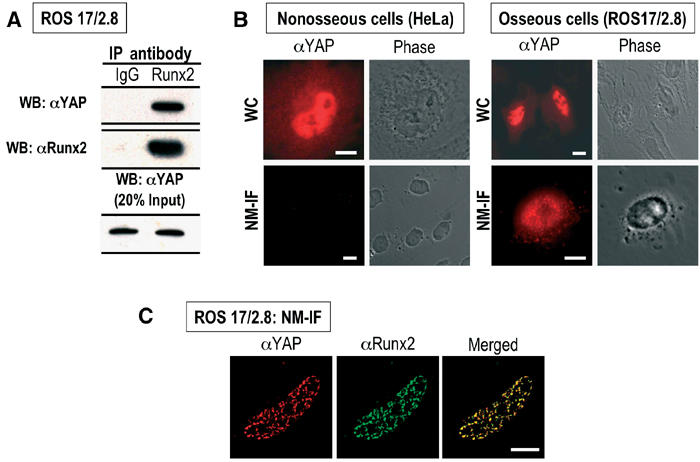
Endogenous YAP and Runx2 proteins interact in vivo and co-localize in situ in osseous cells. (A) Endogenous Runx2 was immunoprecipitated from ROS 17/2.8 cells with a rabbit polyclonal antibody (1:2000) raised against the Runx2 C-terminus (Zhang et al, 2000). A rabbit polyclonal antibody was used to detect endogenous YAP (top panel). Normal goat IgG was used as a control. The middle panel shows efficient immunoprecipitation of endogenous Runx2. The bottom panel shows the expression of endogenous YAP in ROS 17/2.8 cells (20% of total input). (B) In situ immunofluorescence of whole cell (WC) and nuclear matrix-intermediate filament (NM-IF) preparations was performed to assess the nucleo-cytoplasmic distribution and subnuclear localization of endogenous YAP in nonosseous (HeLa) and osseous (ROS 17/2.8) cells. YAP is predominantly nuclear in both HeLa and ROS 17/2.8 cells (top panels), but is only associated with the nuclear matrix ROS 17/2.8 cells (bottom panels). (C) Same as (B), using deconvoluted images. The merged image reveals that endogenous YAP resides in Runx2 containing subnuclear foci in ROS 17/2.8 cells (bar=10 μm).
YAP interacts with Src family kinases at the plasma membrane, with 14-3-3 proteins in the cytoplasm and with transcription factors in the nucleus (Mohler et al, 1999; Yagi et al, 1999; Strano et al, 2001; Vassilev et al, 2001; Basu et al, 2003). We therefore assessed the subcellular localization of YAP in osseous (ROS 17/2.8) and nonosseous (HeLa) cells. Endogenous YAP is present in the cytoplasm, but is predominantly nuclear in both HeLa and ROS 17/2.8 cells (Figure 1B, top panels). YAP associates with the nuclear matrix-intermediate filament (NM-IF) fraction in osseous cells that express endogenous Runx proteins (Figure 2B, bottom right panel); however, it is not detected in the NM-IF preparations of HeLa cells, which do not express any of the three Runx proteins (Armesilla et al, 1996) (Figure 1B, bottom left panel). We have previously shown that Runx proteins contain a conserved NMTS that targets Runx to punctate subnuclear sites (Zeng et al, 1997, 1998; Tang et al, 1999; Zaidi et al, 2001). As our data with nonosseous cells suggest that YAP lacks an intrinsic subnuclear targeting signal, we immunostained NM-IF preparations of ROS 17/2.8 cells for both endogenous YAP and Runx2. As shown in Figure 1C, YAP and Runx2 colocalize at punctate foci in the NM-IF preparations of osseous cells. Hence, YAP and Runx2 proteins interact in vivo and associate in situ at subnuclear sites in osteoblasts. Importantly, YAP association with the nuclear matrix in osseous but not nonosseous cells may depend on the presence of Runx2.
Figure 2.
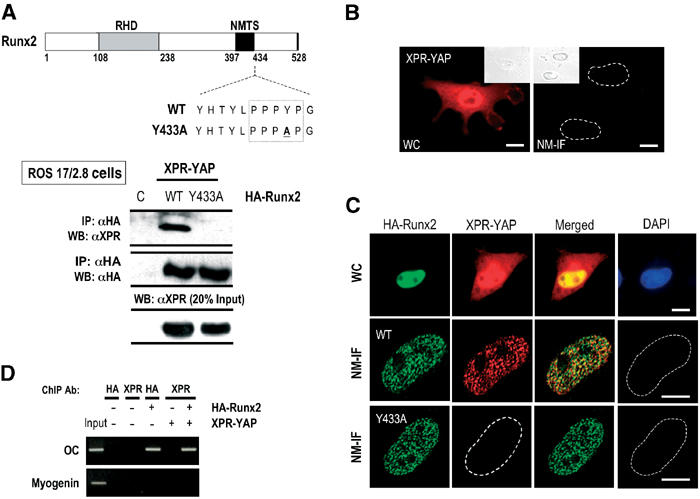
The PY motif of Runx2 is required for interaction with YAP and its recruitment to subnuclear sites as well as target gene promoters in vivo. (A) ROS 17/2.8 cells were cotransfected with Xpress-YAP- and HA-tagged wild-type or the Y433A mutant of Runx2 (numbering according to the mouse MASNS/Runx2 isoform). After 24 h of transfection, proteins were precipitated using a monoclonal antibody against the HA tag (2 μg) and separated by SDS–PAGE. A mouse monoclonal antibody (1:5000) was used to detect XPR-YAP. The blots were stripped and were incubated with HA antibody (1:3000) to assess the expression of Runx2 (wild type or mutant) proteins or with XPR antibody to assess the expression of exogenous YAP. (B) HeLa cells, transfected with XPR-YAP (0.5 μg), were processed for WC and NM-IF preparations. YAP is present both in the cytoplasm and nucleus (right panel), but neither exogenous nor endogenous YAP is found in the nuclear matrix of HeLa cells (left panel). (C) Runx2 does not alter the subcellular localization of YAP in HeLa cells cotransfected with XPR-YAP and HA-Runx2 (wild type and Y433A) (WC, top panels). YAP is associated with the nuclear matrix when co-expressed with wild-type Runx2 (middle panel), but not if the interaction of Runx2 with YAP is disrupted (Y433A mutant, bottom panel) (bar=10 μm). (D) Chromatin immunoprecipitation assay was performed using ROS 17/2.8 cells transfected with indicated tagged expression constructs. Purified immunoprecipitated DNA was amplified with primers spanning the Runx-binding sites B and C in rat osteocalcin promoter as described. OC-derived DNA was observed in chromatin immunoprecipitations with the HA and Xpress antibodies from cells expressing both HA-tagged Runx2 and Xpress-tagged YAP after 21 cycles of PCR amplification. Control lanes (−) from untransfected cells show that antibodies against HA or XPR tags do not nonspecifically precipitate chromatin (lanes 2 and 3). A low level of OC-derived DNA was detected from cells expressing YAP alone after 25 cycles of PCR amplification due to endogenous Runx2 (data not shown). The unrelated myogenin promoter does not exhibit signals (bottom panel).
The PY motif of Runx2 is required for targeting of YAP to subnuclear domains in situ and its recruitment to chromatin in vivo
The absence of YAP from the nuclear matrix of nonosseous cells suggests that the interaction of YAP with Runx2 is required for its subnuclear targeting. Therefore, we introduced a point mutation into the PY motif of Runx2 (Runx2 Y433A; Figure 2A, top panel), a mutation that has been reported to disrupt Runx2–YAP interaction in vitro (Yagi et al, 1999). As seen for endogenous proteins (Figure 1), exogenously expressed YAP and Runx2 form precipitable complexes in osseous cells (Figure 2A, bottom panel), but the Y433A mutation abrogates this interaction (Figure 2A). Thus, the interaction of YAP with Runx2 inside the cell requires the PY motif of Runx2.
As YAP lacks an intrinsic subnuclear targeting signal, it may require Runx2 for subnuclear trafficking in osseous cells (Figure 1B). We directly tested whether Runx2 mediates YAP intranuclear targeting. Runx2 (wild-type or the Y433A mutant) and YAP were expressed in nonosseous cells that lack endogenous Runx proteins, and their subnuclear localization was assessed using immunofluorescence microscopy (Figure 2B and C). Wild-type Runx2 and the Y433A mutant are exclusively nuclear and exhibit a punctate subnuclear distribution (Figure 2C, left panels). Exogenously expressed YAP is present in both the cytoplasm and the nucleus, and is not detected in nuclear matrix preparations (Figure 2B; see also Figure 1). When co-expressed with wild-type Runx2, YAP shows a discrete subnuclear distribution and the two proteins are colocalized (Figure 2C, middle panels). However, co-expression of the Runx2 Y433A mutant with YAP fails to target YAP to the nuclear matrix (Figure 2C, bottom panels). Similarly, mutations that compromise subnuclear targeting of Runx2 also fail to target YAP to the nuclear matrix (Zaidi et al, 2002). Although Runx2 recruits YAP to nuclear matrix-associated subnuclear foci, it does not alter the nucleo-cytoplasmic distribution of YAP (Figure 2C, top panels). Collectively, these results demonstrate that while the nuclear import of YAP is independent of Runx2, targeting of the Runx2–YAP complex to subnuclear domains requires an intact PY motif and the functional NMTS of Runx2.
As YAP lacks DNA-binding activity, we postulated that Runx2 may recruit YAP to promoters of target genes. Chromatin immunoprecipitation (ChIP) assays were performed to assess the in vivo association of YAP with the osteocalcin promoter. YAP alone is unable to bind to the promoter region of the OC gene (Figure 2D). However, when co-expressed with Runx2, YAP is specifically recruited to the OC promoter (see control lanes in upper panel) and not to the myogenin gene promoter, which is not a Runx-responsive gene (Figure 2D, bottom panel). Hence, the interaction with Runx2 results in targeting of YAP to subnuclear domains in situ and its recruitment to a target gene promoter in vivo.
YAP suppression of Runx2-mediated osteocalcin activation is relieved by a dominant-negative (DN) inhibitor of Src tyrosine kinase
The functional consequences of the Runx2–YAP interaction at subnuclear sites were examined by monitoring YAP- and Runx2-mediated OC promoter activity. Runx2 activates the OC promoter from three- to four-fold in nonosseous cells, while YAP alone has no effect on the basal promoter activity (Figure 3A). YAP suppresses Runx2-enhanced OC promoter activity in a dose-dependent manner (Figure 3A, center lanes). Strikingly, expression of the Runx2 Y433A mutant that does not interact with YAP dramatically increases activation of the OC promoter (15-fold) compared to wild-type Runx2 (Figure 3A, last group). As expected, increasing concentrations of YAP do not alter the activity of the Y433A mutant, which cannot bind YAP (Figure 2A). Thus, disruption of the PY motif allows Runx2 to function unabated by the presence of endogenous YAP in HeLa cells. In ROS 17/2.8 osteoblastic cells, exogenous YAP expression slightly increases OC promoter activity and suppresses Runx2-mediated OC activation (Figure 3B). In addition, the Runx2 Y433A mutant shows a relatively moderate activation of the OC promoter in ROS 17/2.8 compared to HeLa cells, presumably due to the presence of endogenous Runx2. Similar results were obtained in mouse osteoblastic MC3T3 cells, premyoblast C2C12 cells and NIH3T3 fibroblasts (Figure 3C), demonstrating that the activity of the Runx2–YAP complex is independent of cell type. Interestingly, when YAP is expressed as a Gal4 fusion protein in a heterologous Gal4 system, we observe strong transcriptional activation, as has been previously reported (data not shown; Yagi et al, 1999]. However, exogenously expressed YAP suppresses Runx2-mediated OC gene transcription on the native OC gene promoter (Figure 3), suggesting that the activity of YAP depends on the promoter context.
Figure 3.
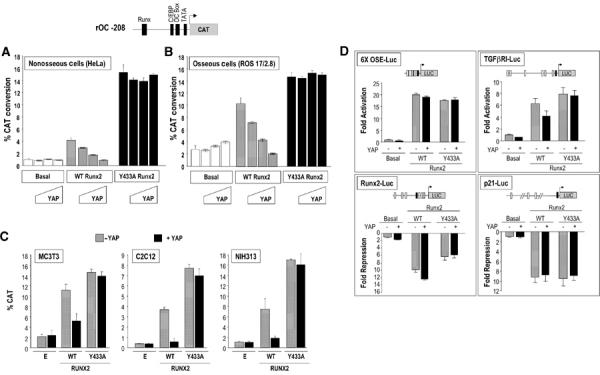
YAP suppresses Runx2-mediated activation of the rat osteocalcin promoter. (A) The rOC −208 CAT reporter was transfected in HeLa cells along with the indicated Runx2 constructs (250 ng) and increasing concentrations (0 (first bar of each group), 250, 500 ng and 1 μg) of full-length YAP. Expression of YAP alone does not affect basal promoter activity. Runx2 activates osteocalcin promoter activity, which is suppressed in a dose-dependent manner by increasing concentrations of YAP. The Runx2 Y433A mutant, that does not interact with YAP, shows a 3–4-fold higher activity compared to wild-type Runx2. (B) YAP suppresses the activity of Runx2 in osteoblastic ROS 17/2.8 cells. In contrast to HeLa cells, wild-type YAP slightly enhances basal promoter activity (first group), and the Runx2 Y433A mutant moderately activates rOC −208 due to endogenous Runx2 in ROS 17/2.8 cells. The graphs represent at least three independent experiments (n=6 each); error bars=standard error of mean. (C) The rOC −208/CAT reporter was transfected with 250 ng each of Runx2 and YAP. Runx2 activates osteocalcin promoter activity, which is suppressed by YAP in osteoblastic MC3T3 cells, premyoblast C2C12 cells and fibroblast NIH3T3 cells. The Runx2 Y433A mutant which does not interact with YAP shows higher activity compared to wild-type Runx2 in C2C12 and NIH 3T3 cells. This effect, however, is not apparent in MC3T3 cells, probably because of higher levels of endogenous Runx2 protein. (D) Runx2–YAP complex regulates the activity of multiple Runx target promoters. ROS 17/2.8 cells were transfected with the indicated reporter and expression constructs. Cells were harvested 30 h after the transfection and subjected to luciferase reporter assay. The luciferase activity is expressed as fold activation (6 × OSE-Luc and TGFβRI-Luc promoter) or fold suppression (in case of Runx2-Luc and p21-Luc promoters). The open squares in promoter constructs represent functional Runx sites.
We tested whether the activity of the Runx2–YAP complex is promoter context dependent, by examining the regulation of various Runx target gene promoters by the Runx2–YAP complex. We used luciferase reporters driven by multimerized Runx-binding sites (6X OSE-Luc; activated by Runx2), the TGFβ receptor type I promoter (TGFβRI-Luc; activated by Runx2), the Runx2 promoter (Runx2-Luc; suppressed by Runx2) and the p21 promoter (p21-Luc; suppressed by Runx2). Our results show that the co-expression of YAP does not alter Runx2 activity on the 6X OSE-Luc; however, it moderately suppresses Runx2 activity on a natural TGFβRI promoter (Figure 3D, upper panels). YAP co-expression results in increased suppression of the Runx2 promoter by Runx2, but does not affect Runx2 suppression of the p21 promoter (Figure 3D, bottom panels), which is known to involve HDAC6 (Westendorf et al, 2002). We conclude that the activity of the Runx2–YAP complex is promoter context dependent.
YAP is a substrate of the Src/Yes tyrosine kinases (Sudol, 1994). To investigate whether YAP-mediated suppression of Runx2 activity involves Src signaling, we used a DN inhibitor of Src tyrosine kinase (Src K295R/Y527F or Src DN), which is kinase inactive (Mukhopadhyay et al, 1995). This variant constitutively binds to cellular proteins that interact with activated Src under physiological conditions. Expression of Src DN increases the basal activity of the osteocalcin gene promoter in ROS 17/2.8 cells in a dose-dependent manner (Figure 4). The Src DN has a similar effect when co-expressed with Runx2 in nonosseous cells such as HeLa, which express high endogenous levels of YAP and Src family members (data not shown). This increased basal OC transcription in the presence of the Src DN may result from inhibition of interaction between endogenous YAP and Src proteins. In addition, YAP-mediated suppression of Runx2 activity is relieved in the presence of the Src DN (Figure 4). Thus, YAP strongly blocks Runx2-mediated transcriptional activation of OC, and this suppression may involve Src tyrosine kinase activity. We propose that the PY motif of Runx2 is an essential protein module that mediates the suppression of OC promoter activity in response to Src signaling.
Figure 4.
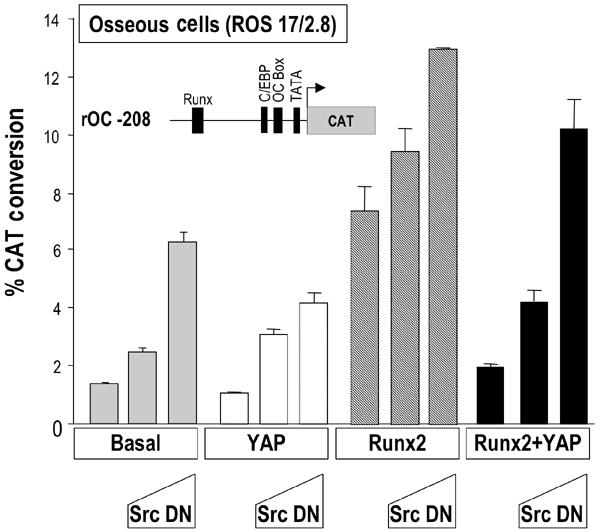
A DN inhibitor of Src tyrosine kinase relieves YAP-mediated suppression of Runx2 activity. ROS 17/2.8 cells were transfected with 1 μg rOC −208/CAT reporter along with 250 ng of Runx2 and/or YAP expression constructs. Two different concentrations of Src DN construct (200 and 800 ng) were used (bars 2 and 3, respectively, of each group). Cells were harvested 24–30 h after transfection and were subjected to CAT reporter assay. The graph represents results obtained from three independent experiments (n=6 each).
Inhibition of the Src tyrosine kinase family abrogates Runx2–YAP interaction and induces expression of the endogenous osteocalcin gene
As YAP is an in vitro substrate of the Src/Yes tyrosine kinases (Sudol, 1994), we examined whether YAP interacts with Yes and Src tyrosine kinases in vivo in osteoblasts. Endogenous Src or Yes proteins were immunoprecipitated from ROS 17/2.8 cells expressing XPR-YAP. We find that YAP is indeed associated with both Yes and Src tyrosine kinases in osteoblasts (Figure 5A). We therefore assessed whether the suppressor function of YAP on the OC promoter may involve Src/Yes signaling using the inhibitor PP2 (Hanke et al, 1996). PP2 is a broad inhibitor of the Src tyrosine kinase family, and has been reported to inhibit the activity of at least four Src family members, including Src, Yes, Fyn and Hck (Hanke et al, 1996). Endogenous Runx2 was immunoprecipitated from untreated (control) or PP2-treated (5 μM for 1 h) ROS 17/2.8 cells expressing XPR-YAP. Runx2 is associated with exogenously expressed YAP in control cells (Figure 5B, lane 3), but not in the presence of PP2 (Figure 5B, lane 4). To address more specifically the involvement of Src tyrosine kinase in regulating interaction between Runx2 and YAP, we examined the Runx2–YAP interaction in the presence of Src DN (Src K295R/Y527F). Forced expression of Src DN significantly reduces (2–3-fold) interaction between Runx2 and YAP (compare lanes 3 and 4; Figure 5C); residual interactions between Runx2 and YAP may occur through signaling pathways involving other Src family members. Taken together, the findings with PP2 and Src DN indicate that the Src family tyrosine kinase signaling regulates the Runx2–YAP interaction.
Figure 5.
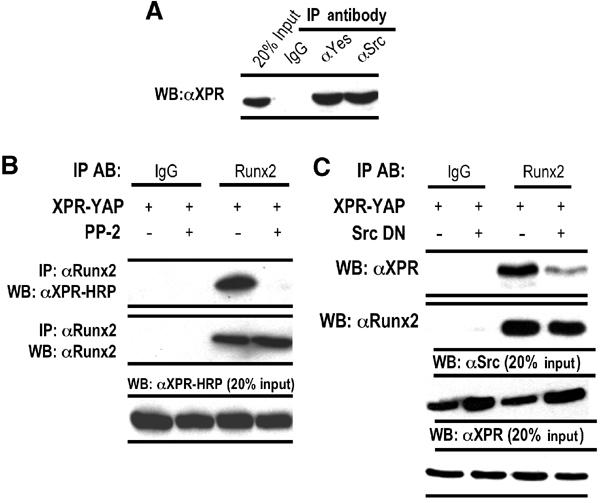
Activated Src family kinases contribute to the YAP–Runx2 interaction. (A) ROS 17/2.8 cells, transiently transfected with XPR-YAP and antibodies, were used to immunoprecipitate endogenous Yes or Src tyrosine kinases. Association of YAP with these kinases was assessed by western blotting using HRP-conjugated anti-XPR antibody. (B) ROS 17/2.8 cells, expressing indicated constructs, were treated with 5 μM of PP2 for 1 h and processed for immunoprecipitation. The Runx2–YAP interaction is completely abrogated in the presence of PP2 (panel 1). Panel 2 shows the efficiency of Runx2 immunoprecipitation, while panel 3 shows comparable levels of YAP overexpression in all lanes. (C) As in (B), but antibodies were used to immunoprecipitate endogenous Runx2. The effect of Src DN on the YAP–Runx2 interaction was assessed by western blotting using the HRP-conjugated anti-XPR antibody (panel 1). A mouse monoclonal antibody against Runx2 was used to assess the efficiency of immunoprecipitation of the endogenous Runx2 protein (panel 2). Expression of Src DN and XPR-YAP was confirmed with Src and HRP-Xpress antibodies, respectively (panels 3 and 4). In each immunoprecipitation, appropriate normal IgG was used as a control.
Elevated osteocalcin mRNA in Src-deficient osteoblasts (Marzia et al, 2000) suggests an inhibitory role for Src tyrosine kinase in OC gene expression. We therefore addressed the involvement of the Src-YAP signaling pathway in the regulation of the endogenous osteocalcin gene in osteoblast-like ROS 17/2.8 cells using PP2 and Src DN, as well as a previously characterized DN inhibitor of YAP (Yagi et al, 1999). Inhibition of Src family members with various concentrations of PP2 results in a dose-dependent increase in endogenous OC expression as measured by mRNA levels (Figure 6A; quantitated in Figure 6C). More importantly, endogenous OC gene expression was also upregulated 2–3-fold when the activity of endogenous Src tyrosine kinase (Figure 6B, Src DN lanes) or YAP (Figure 6B, YAP DN lanes; also see Figure 6C for quantitation) was blocked. Our results demonstrate that YAP suppresses Runx2-mediated expression of the endogenous OC gene in response to Src tyrosine kinases.
Figure 6.
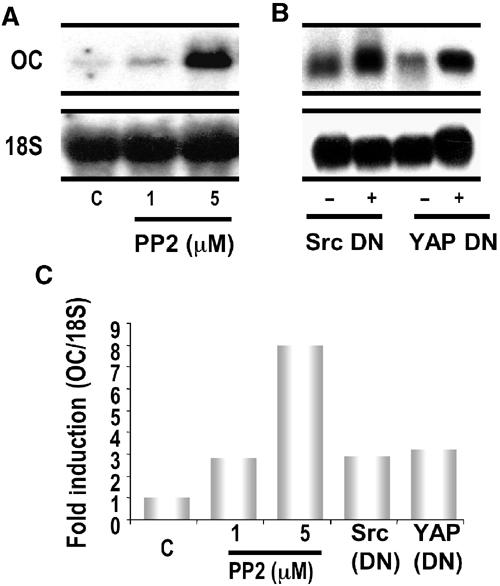
Inhibition of endogenous Src signaling or YAP activity induces endogenous osteocalcin gene transcription. (A) Total cellular RNA from ROS 17/2.8 cells was analyzed by northern blotting and treated with indicated concentrations of PP2 for 1 h. Endogenous osteocalcin gene transcription is induced in a dose-dependent manner when the activity of Src family members is inhibited by PP2 (upper panel). 18S RNA was used as a loading control (bottom panel). (B) Activity of Src tyrosine kinase or YAP was selectively inhibited by overexpressing DN inhibitors (Src DN for Src tyrosine kinase and YAP DN for YAP). Total cellular RNA was subjected to northern blot analysis from ROS 17/2.8 cells overexpressing the indicated plasmids. As shown in the top panel, the DN inhibitor of Src (lane 2) or YAP (lane 4) induces endogenous osteocalcin gene transcription. (C) Densitometric analysis reveals the extent to which endogenous osteocalcin is induced under each experimental condition analyzed. (The bar graph represents the ratio of densitometric units of osteocalcin and 18S transcripts.)
Tyrosine phosphorylation of YAP in response to the activity of Src family members regulates its interaction with Runx2 and subsequent subnuclear targeting
The molecular mechanism(s) underlying Src-YAP-mediated suppression of Runx2 activity on the endogenous OC promoter was further examined by in situ immunofluorescence microscopy of nonosseous cells co-expressing Runx2 and YAP in the presence of PP2 or Src DN. Inhibition of tyrosine kinase activity by the Src DN or PP2 does not alter the nucleo-cytoplasmic distribution of YAP. However, Runx2-mediated subnuclear targeting of YAP is severely compromised in the presence of the Src DN, that is, only 20–25% of cells in the NM-IF preparation are positive for YAP signal (Figure 7A, middle panel). As YAP and Runx2 do not interact in the presence of PP2 (Figure 5B), YAP is absent in the NM-IF preparation of PP2-treated cells, even though Runx2 association with the nuclear matrix is unaltered (Figure 7A, right panel and data not shown). Thus, subnuclear targeting, but not nucleo-cytoplasmic distribution, of YAP requires activated Src tyrosine kinases.
Figure 7.
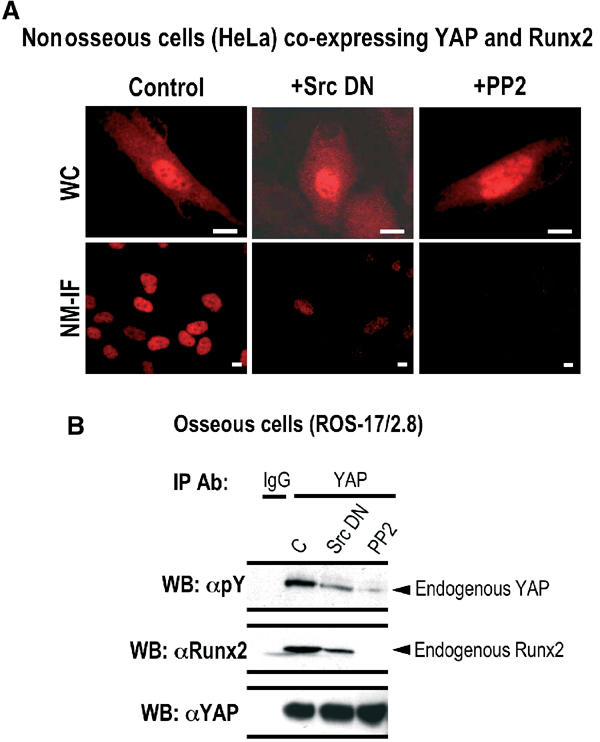
Tyrosine phosphorylation of YAP regulates its interaction with Runx2 and subsequent subnuclear trafficking. (A) HeLa cells, co-expressing YAP and Runx2, were transfected with Src DN or treated with PP2 (5 μM) for 1 h and WC or NM-IF preparations and in situ immunofluorescence microscopy. Inhibition of either Src tyrosine kinase (+Src DN) or its family members (+PP2) does not alter subcellular localization of YAP. Subnuclear trafficking of YAP is severely compromised when the kinase activity of Src alone or its family members is inhibited. (B) To directly assess the effects of Src DN or PP2 on tyrosine phosphorylation of endogenous YAP and its interaction with native Runx2, endogenous YAP was immunoprecipitated from ROS 17/2.8 cells. The immunoprecipitates were resolved by SDS–PAGE and subjected to western blot analysis. The tyrosine phosphorylation of YAP was determined by a mouse monoclonal phospho-specific antibody (Py; 1:2000) raised against phospho-tyrosine (top panel). The blot was stripped and re-probed with a mouse monoclonal antibody against Runx2 to assess the presence of endogenous Runx2 in the immunoprecipitates (middle panel). The efficiency of YAP immunoprecipitation was assessed by a YAP rabbit polyclonal antibody (bottom panel).
To further explore the role of Src signaling in the Runx2–YAP interaction and control of OC gene expression, we assessed the tyrosine phosphorylation status of endogenous YAP in osteoblasts upon inhibition of Src signaling. Expression of Src DN significantly decreases tyrosine phosphorylation of YAP (Figure 7B, upper panel) and reduces its interaction with the native Runx2 protein (Figure 7B, middle panel). In addition, treatment of cells with PP2 abrogates YAP tyrosine phosphorylation and its interaction with Runx2 (Figure 7B, right lanes). Taken together, our results indicate that Src-related tyrosine phosphorylation of YAP is required for its interaction with Runx2, and its targeting to Runx subnuclear sites.
Discussion
In this study, we have shown that YAP, a downstream target of Src tyrosine kinases, functions to suppress the activity of Runx2, a transcription factor required for osteoblast maturation. We find that YAP interacts with both Src and Yes tyrosine kinases in osteoblasts. Tyrosine phosphorylation of endogenous YAP promotes its interaction with Runx2. Runx2 then recruits YAP to the bone-specific osteocalcin promoter and to subnuclear sites where Runx2 resides to suppress osteocalcin promoter activity. Interference with the Src-YAP–Runx2 pathway at any level results in inhibition of YAP tyrosine phosphorylation, disruption of the Runx2–YAP interaction, block in recruitment of YAP to subnuclear sites and induction of osteocalcin gene expression. We conclude that Runx2–YAP interactions are obligatory for integration of Src signals at architecturally associated sites where regulatory complexes assemble to control osteoblast gene expression.
Physiological significance of Src/Yes signaling to Runx2 activity
Negative regulation of Runx2 by Src/Yes kinases may be important for maintenance of bone homeostasis. Bone remodeling is a dynamic process that requires resorption and formation of bone tissue to ensure the continuous renewal of bone throughout life. This process is regulated by the physiological balance between the activities of bone-forming osteoblasts and bone-resorbing osteoclasts in response to several physiological cues. Like vitamin D and bone morphogenetic proteins (BMPs), Src/Yes signaling coordinates both osteoblast and osteoclast activities for bone turnover (Katagiri et al, 1990; Soriano et al, 1991; Boyce et al, 1992; Horne et al, 1992; Suda et al, 1992; Van Leeuwen et al, 2001). The molecular mechanisms involving Src/Yes tyrosine kinases in osteoclast differentiation are well studied, but a role for this pathway and its downstream signaling molecules such as YAP in osteoblasts has only recently been suggested. Osteoblasts isolated from Src−/− mice exhibit accelerated differentiation and elevated levels of OC mRNA and other osteoblast markers, suggesting that c-Src negativity regulates osteoblast maturation (Thomas and Lafage-Proust, 1999; Marzia et al, 2000). We find that several osteoblast-related genes are regulated by the Runx2–YAP complex. Thus, YAP modulation of Runx2-mediated gene expression provides a molecular mechanism for Src signaling in the control of osteoblast differentiation.
Our studies provide evidence for the novel concept that YAP can function as a suppressor of gene activation. The carboxy-terminus of YAP contains a strong activation domain as assessed by heterologous gene transcription assays (Yagi et al, 1999; Strano et al, 2001; Vassilev et al, 2001), and YAP enhances the Runx-mediated activation of the IgαC promoter. However, the effect of YAP on the OC promoter has not been examined previously (Yagi et al, 1999). Our studies on the native OC gene promoter establish that YAP suppresses Runx2-dependent transcriptional activation. Of interest, a multimerized Runx2 element does not respond to YAP repression, supporting the concept that the formation of multimeric regulatory complexes at Runx sites in native promoters is influenced by surrounding sequences (Canon and Banerjee, 2003). As Runx2–YAP may be part of a larger regulatory complex, we propose that the functional outcome of YAP activity depends on specific protein–protein interactions that are promoter context dependent, as is the case for other co-regulators (Lemon and Tjian, 2000).
Regulatory aspects of Runx2-mediated recruitment of YAP to subnuclear domains and the OC gene promoter
Runx transcription factors play a pivotal role in the maintenance of different levels of nuclear organization (Stein et al, 2000, 2003). Intact Runx-binding sites are required for transcription, steroid hormone responsiveness and for the maintenance of open chromatin structure of the rat OC gene promoter (Javed et al, 1999). In addition, Runx factors possess a specific intranuclear targeting signal (the NMTS) that directs these factors to distinct subnuclear domains and supports transcriptional control of target genes (Zeng et al, 1997, 1998; Tang et al, 1999; Stein et al, 2000; Choi et al, 2001; Zaidi et al, 2001). The NMTS overlaps with interacting domains of several coregulators of Runx function, including YAP and Smads (Hanai et al, 1999; Yagi et al, 1999). Smads, which transduce BMP signals, require the subnuclear targeting signal of Runx transcription factors to assemble as distinct subnuclear complexes at sites of active transcription to regulate gene expression (Zaidi et al, 2002). Here, we show that YAP must bind to Runx2 to associate with nuclear substructures and the promoter regions of target genes. Taken together, these studies suggest that Runx2 functions as a multifunctional organizer of regulatory complexes at subnuclear sites.
Our studies also suggest differences in the importance of serine and tyrosine phosphorylation in defining subcellular and subnuclear distribution of YAP. The Akt-kinase-mediated serine phosphorylation of YAP disrupts its interaction with 14-3-3 proteins, thus resulting in cytosolic localization of YAP (Basu et al, 2003). We find that tyrosine phosphorylation of YAP in response to Src/Yes signals does not alter its nucleo-cytoplasmic distribution, but is required for interaction of YAP with Runx2 and its consequent subnuclear trafficking. Taken together, these observations support a novel mechanism by which regulatory complexes spatially and temporally assemble within the nucleus in response to extracellular signals, leading to activation or suppression of target genes. We propose that the dynamic presence of coregulatory proteins in Runx subnuclear domains provides a mechanism for positive and negative transcriptional control of Runx-dependent promoters within the three-dimensional context of nuclear architecture in response to physiological signals to ensure proper bone formation.
Materials and methods
Reporter assays
Rat osteosarcoma ROS 17/2.8 and human cervical carcinoma HeLa cells were plated at a density of 0.04 × 106 cells/ml. Transient transfections were initiated 24 h later using either Superfect (Qiagen Inc., Valencia, CA, USA) or Fugene (Roche Applied Science, Indianapolis, IN, USA) with the indicated expression constructs and 1 μg of −208 OC-CAT (chloramphenicol acetyl transferase), spanning one Runx-binding site (Site C) of the rat osteocalcin promoter. RSV-Luc was included as a control for transfection efficiency. After 24 h, cells were processed for CAT assay and quantitated using a Storm PhosphorImager (Molecular Dynamics, Sunnyvale, CA, USA) as described (Javed et al, 1999). Luciferase activity was assessed with a Luciferase Assay Kit (Promega, Madison, WI, USA) in the same cell lysate and used to normalize the CAT values. Graphs are representative of three independent experiments (n=6 in each experiment) with different DNA preparations.
Expression constructs
The following constructs have been previously reported: HA-Runx2 (Zaidi et al, 2001), Xpress (XPR)-tagged full-length YAP (1–472) and its DN inhibitor (1–301; YAP DN hereafter) (Yagi et al, 1999), and rat osteocalcin with one Runx-binding site (rOC −208-CAT; Heinrichs et al, 1993). The Y433A mutant of Runx2 was generated by PCR-based site-directed mutagenesis. Src DN (Src K295R Y527F) was purchased from Upstate Biotechnology (Lake Placid, NY, USA; Mukhopadhyay et al, 1995).
In situ immunofluorescence and digital microscopy
HeLa or ROS 17/2.8 cells (untransfected or transfected with 0.5 μg of indicated expression constructs) were extracted for in situ nuclear matrix preparation 24 h after transfection and were processed for digital microscopy as described before (Javed et al, 2000). HA-Runx2 was detected by a rabbit polyclonal antibody against HA-tag at a dilution of 1:3000 (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA). XPR-YAP was detected with mouse monoclonal antibodies at a dilution of 1:1000. Rabbit polyclonal antibodies raised against Runx2 (Oncogene Research Products, La Jolla, CA, USA) or YAP (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) were used to detect endogenous proteins at a dilution of 1:200. The secondary antibodies used were either Alexa 488 anti-rabbit or Alexa 568 anti-mouse (Molecular Probes, Eugene, OR, USA) at a dilution of 1:800.
Co-immunoprecipitation and western blotting
ROS 17/2.8 cells were transfected with expression constructs for HA-tagged wild-type or mutant Runx2 proteins and XPR-YAP. Cells were prepared 36 h after transfection in a buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.5% Triton X-100, 1% aprotinin and 1 mM phenylmethylsulfonyl fluoride (PMSF). Lysates were incubated with specific antibodies for 1 h, followed by incubation with protein A/G plus-sepharose beads. Immunoprecipitates were boiled in the SDS sample buffer (100 mM Tris–HCl (pH 6.8), 5% glycerol, 2% SDS, 100 mM DTT and 0.002% bromophenol blue) and subjected to western blot analysis. The following antibodies (2 μg in each case) were used for immunoprecipitation: rabbit polyclonal antibodies against Runx2, YAP and Src tyrosine kinase, and mouse monoclonal antibody against Yes tyrosine kinase (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Proteins were separated on 10% SDS–polyacrylamide gel and processed for western blotting using appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2000). The signal was visualized using the enhanced chemiluminescence kit (Amersham Pharmacia Biotech Inc., Piscataway, NJ, USA) according to the manufacturer's instructions.
Chromatin immunoprecipitation
ROS 17/2.8 cells transfected with HA-Runx2 and/or XPR-YAP by Fugene transfection reagent were crosslinked with 1.1% formaldehyde in 1 × PBS at room temperature for 10 min. Cells were scraped in 500 μl of lysis buffer (150 mM NaCl, 50 mM Tris–HCl (pH 8.0), 1% NP-40, 25 μM MG132, 1 × Complete™ protease inhibitor cocktail). The cell lysate was then sonicated to generate 500–1000 bp long fragments. Cell lysate was microfuged at 14 000 r.p.m. for 15 min at 4°C and the supernatant was precleared with 40 μl protein A/G plus agarose beads, 2 μg of sheared salmon sperm DNA and 2 μg appropriate normal IgG for 1 h at 20 r.p.m. The precleared supernatant was incubated with 2 μg antibody for 1 h at 4°C with rotation. The immuno-complexes were collected by adding 40 μl of protein A/G plus agarose beads and rotating at 4°C at 30 r.p.m. for another 1 h, following centrifugation by extensive washing in multiple buffers as described in Shen et al (2002). Upon reversal of crosslinking, DNA was purified using the QIAGEN PCR Purification kit (Qiagen Inc., Valencia, CA, USA), stored in 50 μl, and 1 μl was used for PCR. The following primer sets were used to amplify promoter regions: OC promoter 5′-GGT GAA TGA GGA CAT TAC TGA CCG CTCC and 5′-CCA AAG GAT GCT GTG GTT GGT GAC, and myogenin promoter 5′-ACC CCT TTC TTG TTC CCT TCC′ and 5′-CTC CCG CAG CCC CTC AC. The PCR program used was: 2 min 95°C denaturing step, then 30 s 95°C, 30 s 60°C, 45 s 72°C for 21 cycles, and finally 5 min 72°C elongation step.
Northern blotting
Total cellular RNA was isolated by the Trizol method (Invitrogen Corporation, Carlsbad, CA, USA) from ROS 17/2.8 cells (untreated or treated for 1 h with PP2 (4-amino-5-(4-chlorophenyl)-7-(t-butyl) pyrazolo [3,4-d] pyrimidine); Calbiochem-Novabiochem Corp., La Jolla, CA, USA). The probes used to detect OC mRNA and 18S rRNA have been described (Gutierrez et al, 2002). To determine the role of Src tyrosine kinase and YAP, ROS 17/2.8 cells were transfected with 10 μg of the Src DN or the YAP DN 36 h prior to RNA isolation using Fugene transfection reagent.
Acknowledgments
This work was supported by NIH grants DE12528, POI AR48818 and AR39588 POI CA82834. The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. We thank Judy Rask and Karen Concaugh for editorial assistance, Daniel Young for helpful comments on the manuscript and Dr William Horne (Department of Cell Biology, Yale University School of Medicine, New Haven, CT, USA) for providing reagents and for critical suggestions.
References
- Akiyama T (2000) Wnt/beta-catenin signaling. Cytokine Growth Factor Rev 11: 273–282 [DOI] [PubMed] [Google Scholar]
- Armesilla AL, Calvo D, Vega MA (1996) Structural and functional characterization of the human CD36 gene promoter: identification of a proximal PEBP2/CBF site. J Biol Chem 271: 7781–7787 [DOI] [PubMed] [Google Scholar]
- Basu S, Totty NF, Irwin MS, Sudol M, Downward J (2003) Akt phosphorylates the Yes-associated protein, YAP, to induce interaction with 14-3-3 and attenuation of p73-mediated apoptosis. Mol Cell 11: 11–23 [DOI] [PubMed] [Google Scholar]
- Berezney R, Jeon KW (1995) Structural and Functional Organization of the Nuclear Matrix. New York: Academic Press [Google Scholar]
- Boyce BF, Yoneda T, Lowe C, Soriano P, Mundy GR (1992) Requirement of pp60c-src expression for osteoclasts to form ruffled borders and resorb bone in mice. J Clin Invest 90: 1622–1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canon J, Banerjee U (2003) In vivo analysis of a developmental circuit for direct transcriptional activation and repression in the same cell by a Runx protein. Genes Dev 17: 838–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J-Y, Pratap J, Javed A, Zaidi SK, Xing L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, Stein JL, Jones SN, Stein GS (2001) Subnuclear targeting of Runx/Cbfa/AML factors is essential for tissue-specific differentiation during embryonic development. Proc Natl Acad Sci USA 98: 8650–8655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie JR, Chadee DN (1998) Regulation and regulatory parameters of histone modifications. J Cell Biochem Suppl 30–31: 203–213 [PubMed] [Google Scholar]
- DeFranco DB, Guerrero J (2000) Nuclear matrix targeting of steroid receptors: specific signal sequences and acceptor proteins. Crit Rev Eukaryot Gene Expr 10: 39–44 [PubMed] [Google Scholar]
- Finidori J (2000) Regulators of growth hormone signaling. Vitam Horm 59: 71–97 [DOI] [PubMed] [Google Scholar]
- Gutierrez S, Javed A, Tennant D, van Rees M, Montecino M, Stein GS, Stein JL, Lian JB (2002) CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J Biol Chem 277: 1316–1323 [DOI] [PubMed] [Google Scholar]
- Hanai J, Chen LF, Kanno T, Ohtani-Fujita N, Kim WY, Guo WH, Imamura T, Ishidou Y, Fukuchi M, Shi MJ, Stavnezer J, Kawabata M, Miyazono K, Ito Y (1999) Interaction and functional cooperation of PEBP2/CBF with Smads. Synergistic induction of the immunoglobulin germline Calpha promoter. J Biol Chem 274: 31577–31582 [DOI] [PubMed] [Google Scholar]
- Hanke JH, Gardner JP, Dow RL, Changelian PS, Brissette WH, Weringer EJ, Pollok BA, Connelly PA (1996) Discovery of a novel, potent, and Src family-selective tyrosine kinase inhibitor. Study of Lck- and FynT-dependent T cell activation. J Biol Chem 271: 695–701 [DOI] [PubMed] [Google Scholar]
- Heinrichs AAJ, Banerjee C, Bortell R, Owen TA, Stein JL, Stein GS, Lian JB (1993) Identification and characterization of two proximal elements in the rat osteocalcin gene promoter that may confer species-specific regulation. J Cell Biochem 53: 240–250 [DOI] [PubMed] [Google Scholar]
- Horne WC, Neff L, Chatterjee D, Lomri A, Levy JB, Baron R (1992) Osteoclasts express high levels of pp60c-src in association with intracellular membranes. J Cell Biol 119: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihle JN (2001) The Stat family in cytokine signaling. Curr Opin Cell Biol 13: 211–217 [DOI] [PubMed] [Google Scholar]
- Ito Y (1999) Molecular basis of tissue-specific gene expression mediated by the runt domain transcription factor PEBP2/CBF. Genes Cells 4: 685–696 [DOI] [PubMed] [Google Scholar]
- Javed A, Guo B, Hiebert S, Choi J-Y, Green J, Zhao S-C, Osborne MA, Stifani S, Stein JL, Lian JB, van Wijnen AJ, Stein GS (2000) Groucho/TLE/R-Esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J Cell Sci 113: 2221–2231 [DOI] [PubMed] [Google Scholar]
- Javed A, Gutierrez S, Montecino M, van Wijnen AJ, Stein JL, Stein GS, Lian JB (1999) Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D responsive transcription and contribute to chromatin organization. Mol Cell Biol 19: 7491–7500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Yamaguchi A, Ikeda T, Yoshiki S, Wozney JM, Rosen V, Wang EA, Tanaka H, Omura S, Suda T (1990) The non-osteogenic mouse pluripotent cell line, C3H10T1/2, is induced to differentiate into osteoblastic cells by recombinant human bone morphogenetic protein-2. Biochem Biophys Res Commun 172: 295–299 [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao Y-H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T (1997) Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89: 755–764 [DOI] [PubMed] [Google Scholar]
- Lemon B, Tjian R (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev 14: 2551–2569 [DOI] [PubMed] [Google Scholar]
- Li QL, Ito K, Sakakura C, Fukamachi H, Inoue K, Chi XZ, Lee KY, Nomura S, Lee CW, Han SB, Kim HM, Kim WJ, Yamamoto H, Yamashita N, Yano T, Ikeda T, Itohara S, Inazawa J, Abe T, Hagiwara A, Yamagishi H, Ooe A, Kaneda A, Sugimura T, Ushijima T, Bae SC, Ito Y (2002) Causal relationship between the loss of RUNX3 expression and gastric cancer. Cell 109: 113–124 [DOI] [PubMed] [Google Scholar]
- Mancini MG, Liu B, Sharp ZD, Mancini MA (1999) Subnuclear partitioning and functional regulation of the Pit-1 transcription factor. J Cell Biochem 72: 322–338 [PubMed] [Google Scholar]
- Marzia M, Sims NA, Voit S, Migliaccio S, Taranta A, Bernardini S, Faraggiana T, Yoneda T, Mundy GR, Boyce BF, Baron R, Teti A (2000) Decreased c-Src expression enhances osteoblast differentiation and bone formation. J Cell Biol 151: 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J (1998) TGF-beta signal transduction. Annu Rev Biochem 67: 753–791 [DOI] [PubMed] [Google Scholar]
- McNeil S, Guo B, Stein JL, Lian JB, Bushmeyer S, Seto E, Atchison ML, Penman S, van Wijnen AJ, Stein GS (1998) Targeting of the YY1 transcription factor to the nucleolus and the nuclear matrix in situ: the C-terminus is a principal determinant for nuclear trafficking. J Cell Biochem 68: 500–510 [PubMed] [Google Scholar]
- Mohler PJ, Kreda SM, Boucher RC, Sudol M, Stutts MJ, Milgram SL (1999) Yes-associated protein 65 localizes p62(c-Yes) to the apical compartment of airway epithelia by association with EBP50. J Cell Biol 147: 879–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay D, Tsiokas L, Zhou XM, Foster D, Brugge JS, Sukhatme VP (1995) Hypoxic induction of human vascular endothelial growth factor expression through c-Src activation. Nature 375: 577–581 [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JHM, Owen MJ, Mertelsmann R, Zabel BU, Olsen BR (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89: 773–779 [DOI] [PubMed] [Google Scholar]
- Ogawa E, Maruyama M, Kagoshima H, Inuzuka M, Lu J, Satake M, Shigesada K, Ito Y (1993) PEBP2/PEA2 represents a family of transcription factors homologous to the products of the Drosophila runt gene and the human AML1 gene. Proc Natl Acad Sci USA 90: 6859–6863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GWH, Beddington RSP, Mundlos S, Olsen BR, Selby PB, Owen MJ (1997) Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89: 765–771 [DOI] [PubMed] [Google Scholar]
- Schlessinger J (2000) New roles for Src kinases in control of cell survival and angiogenesis. Cell 100: 293–296 [DOI] [PubMed] [Google Scholar]
- Shen J, Montecino MA, Lian JB, Stein GS, van Wijnen AJ, Stein JL (2002) Histone acetylation in vivo at the osteocalcin locus is functionally linked to vitamin D dependent, bone tissue-specific transcription. J Biol Chem 277: 20284–20292 [DOI] [PubMed] [Google Scholar]
- Soriano P, Montgomery C, Geske R, Bradley A (1991) Targeted disruption of the c-src proto-oncogene leads to osteopetrosis in mice. Cell 64: 693–702 [DOI] [PubMed] [Google Scholar]
- Stein GS, van Wijnen AJ, Stein JL, Lian JB, Montecino M, Choi J-Y, Zaidi K, Javed A (2000) Intranuclear trafficking of transcription factors: implications for biological control. J Cell Sci 113: 2527–2533 [DOI] [PubMed] [Google Scholar]
- Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi J-Y, Stein JL, Lian JB, Javed A (2003) Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol 13: 584–592 [DOI] [PubMed] [Google Scholar]
- Stenoien D, Sharp ZD, Smith CL, Mancini MA (1998) Functional subnuclear partitioning of transcription factors. J Cell Biochem 70: 213–221 [PubMed] [Google Scholar]
- Stenoien DL, Mancini MG, Patel K, Allegretto EA, Smith CL, Mancini MA (2000) Subnuclear trafficking of estrogen receptor-alpha and steroid receptor coactivator-1. Mol Endocrinol 14: 518–534 [DOI] [PubMed] [Google Scholar]
- Strano S, Munarriz E, Rossi M, Castagnoli L, Shaul Y, Sacchi A, Oren M, Sudol M, Cesareni G, Blandino G (2001) Physical interaction with Yes-associated protein enhances p73 transcriptional activity. J Biol Chem 276: 15164–15173 [DOI] [PubMed] [Google Scholar]
- Suda T, Takahashi N, Etsuko A (1992) Role of vitamin D in bone resorption. J Cell Biochem 49: 53–58 [DOI] [PubMed] [Google Scholar]
- Sudol M (1994) Yes-associated protein (YAP65) is a proline-rich phosphoprotein that binds to the SH3 domain of the Yes proto-oncogene product. Oncogene 9: 2145–2152 [PubMed] [Google Scholar]
- Sudol M, Bork P, Einbond A, Kastury K, Druck T, Negrini M, Huebner K, Lehman D (1995) Characterization of the mammalian YAP (Yes-associated protein) gene and its role in defining a novel protein module, the WW domain. J Biol Chem 270: 14733–14741 [DOI] [PubMed] [Google Scholar]
- Sudol M, Hunter T (2000) NeW wrinkles for an old domain. Cell 103: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Tang L, Guo B, Javed A, Choi J-Y, Hiebert S, Lian JB, van Wijnen AJ, Stein JL, Stein GS, Zhou GW (1999) Crystal structure of the nuclear matrix targeting signal of the transcription factor AML-1/PEBP2αB/CBFα2. J Biol Chem 274: 33580–33586 [DOI] [PubMed] [Google Scholar]
- Thomas SM, Brugge JS (1997) Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol 13: 513–609 [DOI] [PubMed] [Google Scholar]
- Thomas T, Lafage-Proust MH (1999) Contribution of genetically modified mouse models to the elucidation of bone physiology. Rev Rhum Engl Ed 66: 728–735 [PubMed] [Google Scholar]
- Van Leeuwen JP, van Driel M, van den Bemd GJ, Pols HA (2001) Vitamin D control of osteoblast function and bone extracellular matrix mineralization. Crit Rev Eukaryot Gene Expr 11: 199–226 [PubMed] [Google Scholar]
- van Steensel B, Brink M, van der MK, van Binnendijk EP, Wansink DG, de Jong L, de Kloet ER, van Driel R (1995a) Localization of the glucocorticoid receptor in discrete clusters in the cell nucleus. J Cell Sci 108 (Part 9): 3003–3011 [DOI] [PubMed] [Google Scholar]
- van Steensel B, Jenster G, Damm K, Brinkmann AO, van Driel R (1995b) Domains of the human androgen receptor and glucocorticoid receptor involved in binding to the nuclear matrix. J Cell Biochem 57: 465–478 [DOI] [PubMed] [Google Scholar]
- Vassilev A, Kaneko KJ, Shu H, Zhao Y, DePamphilis ML (2001) TEAD/TEF transcription factors utilize the activation domain of YAP65, an Src/Yes-associated protein localized in the cytoplasm. Genes Dev 15: 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westendorf JJ, Hiebert SW (1999) Mammalian runt-domain proteins and their roles in hematopoiesis, osteogenesis, and leukemia. J Cell Biochem Suppl 32–33: 51–58 [DOI] [PubMed] [Google Scholar]
- Westendorf JJ, Zaidi SK, Cascino JE, Kahler R, van Wijnen AJ, Lian JB, Yoshida M, Stein GS, Li X (2002) Runx2 (Cbfa1, AML-3) interacts with histone deacetylase 6 and represses the p21(CIP1/WAF1) promoter. Mol Cell Biol 22: 7982–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi R, Chen LF, Shigesada K, Murakami Y, Ito Y (1999) A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J 18: 2551–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Javed A, Choi J-Y, van Wijnen AJ, Stein JL, Lian JB, Stein GS (2001) A specific targeting signal directs Runx2/Cbfa1 to subnuclear domains and contributes to transactivation of the osteocalcin gene. J Cell Sci 114: 3093–3102 [DOI] [PubMed] [Google Scholar]
- Zaidi SK, Sullivan AJ, van Wijnen AJ, Stein JL, Stein GS, Lian JB (2002) Integration of Runx and Smad regulatory signals at transcriptionally active subnuclear sites. Proc Natl Acad Sci USA 99: 8048–8053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, McNeil S, Pockwinse S, Nickerson JA, Shopland L, Lawrence JB, Penman S, Hiebert SW, Lian JB, van Wijnen AJ, Stein JL, Stein GS (1998) Intranuclear targeting of AML/CBFα regulatory factors to nuclear matrix-associated transcriptional domains. Proc Natl Acad Sci USA 95: 1585–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng C, van Wijnen AJ, Stein JL, Meyers S, Sun W, Shopland L, Lawrence JB, Penman S, Lian JB, Stein GS, Hiebert SW (1997) Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc Natl Acad Sci USA 94: 6746–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Derynck R (1999) Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol 9: 274–279 [DOI] [PubMed] [Google Scholar]
- Zhang YW, Yasui N, Ito K, Huang G, Fujii M, Hanai J, Nogami H, Ochi T, Miyazono K, Ito Y (2000) A RUNX2/PEBP2αA/CBFA1 mutation displaying impaired transactivation and Smad interaction in cleidocranial dysplasia. Proc Natl Acad Sci USA 97: 10549–10554 [DOI] [PMC free article] [PubMed] [Google Scholar]


