Abstract
We have used activation tagging with T-DNA carrying cauliflower mosaic virus 35S enhancers to investigate the complex signaling networks underlying disease resistance in Arabidopsis. From a screen of ∼5000 lines, we identified constitutive disease resistance (CDR1) encoding an apoplastic aspartic protease, the overexpression of which causes dwarfing and resistance to virulent Pseudomonas syringae. These phenotypes reflect salicylic-acid-dependent activation of micro-oxidative bursts and various defense-related genes. Antisense CDR1 plants were compromised for resistance to avirulent P. syringae and more susceptible to virulent strains than wild type. CDR1 accumulates in intercellular fluid in response to pathogen attacks. Induction of CDR1 generates a small mobile signal, and CDR1 action is blocked by the protease inhibitor pepstatin and by mutations in the protease active sites. We propose that CDR1 mediates a peptide signal system involved in the activation of inducible resistance mechanisms.
Keywords: activation tagging, Arabidopsis thaliana, aspartic protease, CDR1, disease resistance
Introduction
Plants use complex recognition and response mechanisms to protect themselves from pathogen attack. Major resistance (R) genes specify recognition of pathogens carrying the corresponding avirulence genes leading to the rapid activation of a battery of inducible defenses, including reinforcements of the cell wall, synthesis of phytoalexin antibiotics, and deployment of pathogenesis-related proteins such as chitinases and glucanases (Yang et al, 1997; Dangl and Jones, 2001). This battery of induced defenses is often accompanied by the collapse of challenged plant cells in the hypersensitive response (HR), which results in a restricted lesion clearly delimited from surrounding healthy tissue. In addition, immunity to subsequent attack by a broad range of normally virulent pathogens develops throughout the plant (Dong, 2001; Glazebrook, 2001).
R-gene-mediated recognition triggers a number of rapid cellular responses, including perturbations in ion fluxes and the pattern of protein phosphorylation, which precede the accumulation of reactive oxygen intermediates (ROIs), nitric oxide (NO) and salicylic acid (SA) and the transcriptional activation of defense-related genes (Yang et al, 1997; McDowell and Dangl, 2000; Cohn et al, 2001). Interplay between ROIs, NO, and SA contributes to the establishment of the HR and to the potentiation of defense responses. Many of the defense signaling and effector mechanisms associated with localized expression of R-gene-mediated resistance are also subsequently activated throughout the plant during the establishment and expression of systemic acquired resistance (SAR) (Dong, 2001). Activation of these mechanisms also contributes to the basal resistance underlying attempted lesion limitation in the later stages of compatible interactions (Glazebrook et al, 1997; Tao et al, 2003), implying flexible deployment of conserved defense signals and effectors in different biological settings.
Mutational analysis in the model plant Arabidopsis thaliana has started to identify genes involved in the complex signal networks underlying expression of inducible resistance mechanisms. For example, NDR1 and PBS2 (AtRAR1) function downstream of a group of R genes encoding members of the coiled-coil subclass of nucleotide-binding site leucine-rich repeat proteins, while EDS1 and PAD4 are required for the action of a group of R genes encoding nucleotide-binding site leucine-rich repeat proteins with amino terminal domains similar to the Drosophila Toll and mammalian interleukin-1 receptors (Dangl and Jones, 2001; Glazebrook, 2001). Likewise, loss-of-function mutations in NPR1 compromise salicylic-acid-dependent responses (Dong, 2001).
While such loss-of-function mutagenesis is a powerful approach to the genetic dissection of signal pathways (Glazebrook, 2001), many genes cannot be uncovered in this way because of lethal effects or functional redundancy, especially in complex reiterative signal networks. Thus, typically ∼90% of mutants showing loss of resistance are r gene alleles with only a small minority in loci involved in downstream signaling (Dangl and Jones, 2001).
T-DNA activation tagging provides a means of generating dominant, gain-of-function mutations (Weigel et al, 2000). Ectopic expression of defense response signaling molecules can result in constitutively expressed resistance phenotypes (Verberne et al, 2000), and this type of screen might therefore be expected to uncover genes that quantitatively impact resistance even where there is considerable redundancy and crosstalk within and between signaling pathways. Using this approach, we have identified an Arabidopsis gene, constitutive disease resistance (CDR1), the overexpression of which leads to enhanced resistance to bacterial pathogens. CDR1 encodes an aspartic protease that releases an endogenous peptide elicitor of salicylic-acid-dependent inducible resistance responses.
Results
CDR1-D is a dominant mutation conferring resistance to P. syringae
About 5000 activation tagged T1 A. thaliana ecotype Columbia (Col-0) lines were screened for resistance to sprayed suspensions of virulent P. syringae pathovars tomato (Pst) or maculicola (Psm). constitutive disease resistance 1-Dominant (CDR1-D) was almost asymptomatic when challenged with virulent Pst, whereas neighboring plants were severely infected (Figure 1A). CDR1-D was of dwarf stature and its leaves were slightly curled and darker than wild type.
Figure 1.
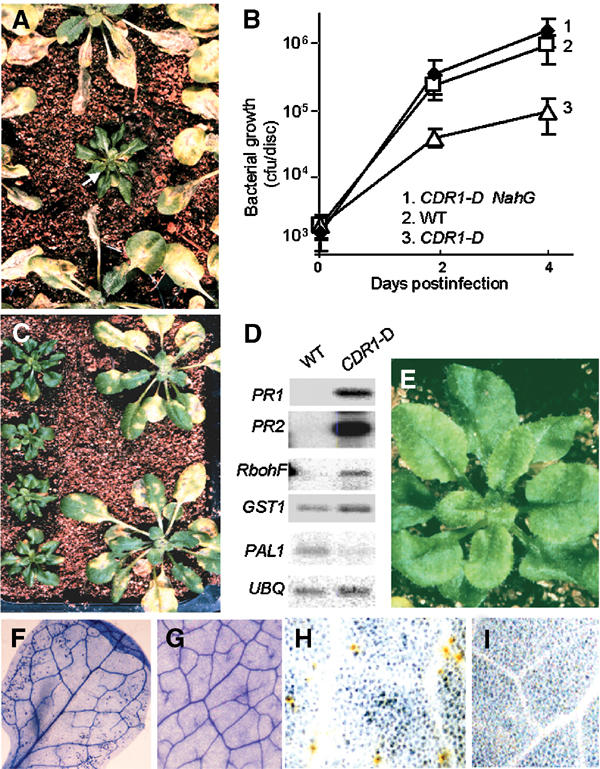
Visible and molecular phenotypes of the CDR1-D mutant. (A) Identification of the CDR1-D mutant in the T1 generation. T1 plants were infected by spraying with virulent Pst DC3000. (B) In planta growth of virulent Pst in CDR1-D, CDR1-D × NahG F1 plants and wild-type Col-0. Plants were infected by the dipping procedure. (C) CDR1-D exhibits enhanced resistance to Psm. CDR1-D T3 plants (left) and wild-type Col-0 plants (right) were infected by spraying with the virulent Psm strain M4. (D) Defense gene expression in uninfected CDR1-D plants compared to wild-type Col-0 plants. See text for details. UBQ is a ubiquitin gene used to control for RNA loading. (E) No obvious lesion formation is observed with the naked eye in CDR1-D. (F–I) Micro-oxidative bursts and micro-HRs in CDR1-D plants. (F) Trypan blue staining shows small clusters of dead cells throughout the leaf of an uninfected CDR1-D plant, but not in a wild-type plant (G). (H) DAB staining shows localized H2O2 production in uninfected CDR1-D plants, but not in control plants (I).
T2 plants segregated 3:1 for the CDR1-D and wild-type phenotypes, and progeny from a back-cross between wild-type Col-0 and the heterozygous CDR1-D T1 plant segregated 1:1 for these phenotypes. Thus, the CDR1-D mutation is dominant over its wild-type allele. Among >80 T2 progeny scored, all CDR1-D plants were dwarfs and resistant to the herbicide Basta. Basta resistance is encoded by the BAR gene incorporated into the T-DNA as a selectable marker. These data indicate that the CDR1-D mutation is tightly linked to the T-DNA insertion and the dwarf stature is associated with the enhanced disease resistance.
In CDR1-D plants, the growth of virulent Pst, measured as bacterial numbers 4 days after inoculation, was over 12-fold less than in equivalent wild-type plants (Figure 1B). The growth of virulent Psm was also significantly reduced (data not shown), and CDR1-D plants exhibited much less severe symptoms than wild-type plants (Figure 1C).
CDR1-D plants constitutively overexpress defense-related genes
Expression of disease resistance is often accompanied by marked increases in the expression of batteries of defense genes (Dong et al, 1991). Therefore, we examined the expression of several of these genes in CDR1-D plants. These included genes encoding the pathogenesis-related proteins PR1 and PR2, phenylalanine ammonia-lyase (PAL), glutathione S-transferase (GST), and RbohF, a subunit of the NADPH oxidase that contributes to the oxidative burst induced by avirulent pathogens (Torres et al, 1998; Torres et al, 2002). The levels of PR1 and PR2 transcripts were approximately 200-fold higher in uninfected CDR1-D mutant plants than in equivalent healthy wild-type plants (Figure 1D). The levels of RbohF transcripts were at least 20-fold higher in CDR1-D than in wild-type plants, and GST transcripts were two- to three-fold elevated in the mutant. In contrast, the level of PAL transcripts was lower in CDR1-D plants. PDF1.2 transcripts, encoding a defensin protein that is induced in Arabidopsis by a salicylic-acid-independent pathway involving jasmonic acid (Penninckx et al, 1996), were not elevated in the CDR1-D mutant (data not shown).
CDR1-D develops spontaneous micro-lesions
Many constitutive disease resistance mutants develop visible spontaneous lesions (Glazebrook et al, 1997). While the CDR1-D mutant does not exhibit macroscopic lesions (Figure 1E), Trypan blue staining revealed small micro-lesions, comprising one or a few dead cells, in leaves of uninfected CDR1-D plants (Figure 1F) but not in leaves of wild-type plants (Figure 1G). These small lesions were reminiscent of the micro-HRs induced by systemic micro-oxidative bursts in the establishment of immunity underlying SAR (Alvarez et al, 1998) and, taken together with the high levels of RbohF transcripts, suggested that the CDR1-D mutant might generate high levels of ROIs. To examine this, diaminobenzidine (DAB) staining was used to detect H2O2 in situ. DAB polymerizes instantly and locally on contact with H2O2, giving reddish-brown polymers. Figure 1H shows that discrete cells in uninfected CDR1-D leaves exhibit DAB staining, indicating local generation of high levels of H2O2. This was not seen in uninfected wild-type Col-0 leaves (Figure 1I).
SA is required for CDR1-D-mediated disease resistance
Many plant defense responses are mediated via increases in the levels of the signal molecule SA and its glucoside (Dong, 2001; Glazebrook, 2001). We extracted SA from CDR1-D and wild-type plants, and found that the levels of SA and its glucoside were approximately 15- and 35-fold higher, respectively, in healthy CDR1-D plants than in wild-type plants.
To determine whether the CDR1-D mutant phenotypes were a result of activation of the SA signaling pathway, CDR1-D plants were crossed to plants expressing the bacterial NahG gene. NahG encodes a hydroxylase that converts SA to catechol, thereby suppressing SA accumulation (Gaffney et al, 1993). The dwarf stature of CDR1-D was suppressed in CDR1-D × NahG F1 plants (Figure 2A). Furthermore, the CDR1-D NahG plants no longer exhibited enhanced resistance to virulent Pst (Figure 2B) or Psm (data not shown). Measurement of bacterial growth in planta following hand infiltration demonstrated that CDR1-D-mediated resistance was completely suppressed in the CDR1-D NahG F1 plants (Figure 1C). In addition, the CDR1-D NahG plants did not develop spontaneous micro-oxidative bursts or micro-lesions as seen in the original CDR1-D line. Defense gene activation was also suppressed in the CDR1-D NahG plants; in contrast to the CDR1-D mutant, the CDR1-D NahG plants did not exhibit elevated levels of PR1 or PR2 transcripts (Figure 2C). However, CDR1 transcripts (see below) remained at elevated levels in the CDR1-D NahG plants.
Figure 2.
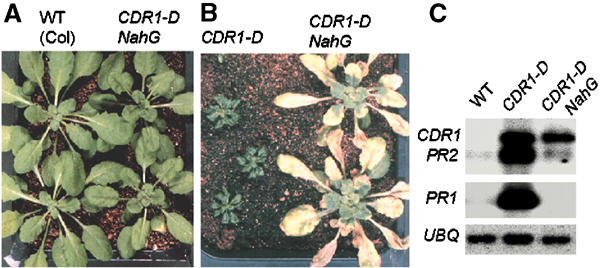
Involvement of SA in expression of CDR1-D phenotypes. (A) NahG suppresses the CDR1-D dwarf stature. (B) CDR1-D NahG plants no longer exhibit enhanced resistance to Pst. (C) CDR1-D NahG plants do not accumulate high levels of PR gene transcripts.
Cloning of CDR1
DNA gel blot hybridization of EcoRI-digested genomic DNA from pooled selfed progeny of the CDR1-D T1 plant revealed two fragments containing 35S enhancer sequences (Figure 3A), consistent with Basta resistance segregation data that indicated that the original CDR1-D T1 plant contained at least two T-DNA insertions. The 5 kb EcoRI fragment was present in both wild-type Basta resistance progeny and CDR1-D plants, whereas the 10 kb fragment cosegregated with the CDR1-D allele (Figure 3A and data not shown). Progeny CDR1-D lines that contained the 10 kb but not the 5 kb EcoRI fragment were recovered and studied in the subsequent experiments reported here. We isolated the 10 kb fragment by plasmid rescue. Nucleotide sequence and restriction digestion analysis of the rescued plasmid, pCDR1E, showed that T-DNA is inserted approximately 0.8 kb upstream of an open reading frame encoding a 437-amino-acid polypeptide (At5g33340) (Figure 3B). To examine whether this gene is hyperactivated by the 35Se insertion, total RNA prepared from leaves was probed with this gene in an RNA blot hybridization experiment. A single strong band corresponding to a 1.5 kb transcript was detected in CDR1-D mutant plants, but it was barely detectable in wild-type plants (Figure 3C). The massive elevation of the level of the 1.5 kb transcript in the mutant suggested that it was a good candidate for the CDR1 transcript.
Figure 3.

Cloning of CDR1. (A) Southern blot analysis showing the 10 kb EcoRI fragment that cosegregates with the CDR1-D mutant phenotype. (B) Structure of the CDR1-D allele. The rescued plasmid pCDR1E contains the 10 kb EcoRI fragment consisting of part of the T-DNA and the flanking 5 kb plant sequence. CDR1-like is likely a pseudogene that shares a sequence similarity (60% identify at the nucleotide level). (C) CDR1 expression is massively enhanced in the CDR1-D mutant. The bottom panel shows a portion of the RNA gel stained with ethidium bromide. (D) pBI/CDR1X construct containing the XbaI fragment of pCDR1E in the binary vector pBI101, which contains one copy of the 35S enhancer and the genomic fragment of CDR1. (E) Plants transformed with pBI/CDR1X exhibit the CDR1-D phenotypes of dwarf stature and resistance to Pst (wild-type Co-l plants are shown below). Only one of the transgenic lines was shown.
To confirm that the candidate gene is indeed the CDR1 gene whose hyperactivation causes the CDR1-D phenotype, the XbaI fragment from pCDR1E, which includes one copy of the 35S enhancer and the candidate CDR1 gene (Figure 3B), was subcloned into the binary vector pBI101 to generate the pBI/CDR1X construct (Figure 3D). The construct was used to transform wild-type Arabidopsis to generate transgenic lines that overexpress the candidate gene. Among >45 independent transgenic lines, approximately half exhibited the dwarfing and enhanced resistance to Pst (Figure 3E) and Psm (data not shown) observed in the original CDR1-D mutant, thus confirming that, when overexpressed, the candidate gene did indeed confer the CDR1-D phenotype.
Loss-of-function experiments confirm that CDR1 functions in disease resistance
To further investigate the role of CDR1 in disease resistance, a full-length CDR1 cDNA, isolated by screening a cDNA library (see Materials and methods), was fused to the CaMV 35S promoter in the antisense orientation, and the construct was introduced into wild-type Col-0 plants to generate CDR1 antisense lines (aCDR1). We expected that if CDR1 plays an important role in the defense response, antisense suppression of CDR1 could compromise disease resistance. To test this, 42 independent aCDR1 lines were screened by spaying with suspensions of avirulent Psm harboring the avrRpm1 avirulence gene (Psm avrRpm1), which does not cause obvious disease symptoms on wild-type Col-0 plants. Three of the aCDR1 lines showed compromised resistance. RNA blot hybridization revealed that CDR1 is suppressed in the above three lines but not in two phenotypically normal antisense lines that were examined (Figure 4A).
Figure 4.
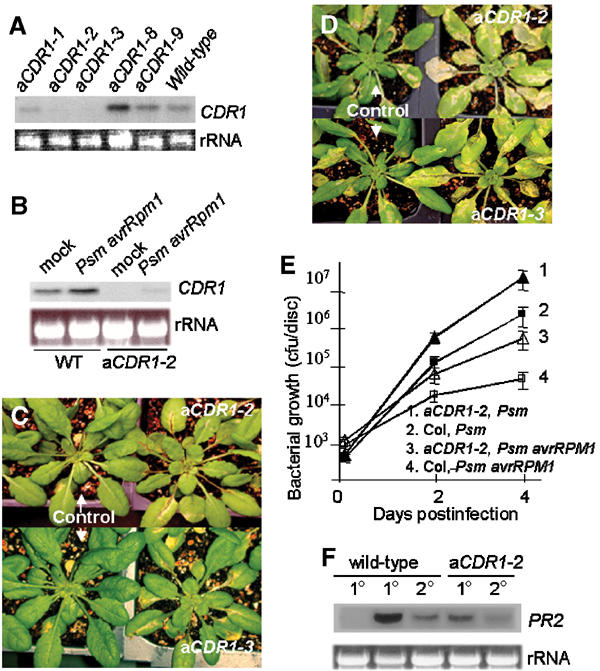
Phenotypes of the CDR1 antisense suppression lines. (A, B) Suppression of CDR1 expression in the three aCDR1 lines. RNA was isolated from uninfected wild type and the five aCDR1 lines (A) and from wild type and aCDR1-2 plants 8 h postinoculation with Psm avrRpm1 (B). Mock inoculation was used as a control. (C, D) aCDR1-2, aCDR1-3, and control plants containing an empty vector were infected by spraying with Psm avrRpm1 (C) and Psm (D). (E) In planta growth of Psm avrRpm1 and Psm in aCDR1-2 and control plants. (F) The antisense line is impaired in local and systemic induction of PR2 by inoculation of Psm avrRpm1. RNA was isolated from inoculated (1°, 8 h postinoculation) and systemic (2°, 48 h postinoculation) leaves of wild-type and the anti-CDR1 plants. The first lane is from mock-inoculated plants as a negative control.
In wild-type plants, CDR1 was expressed constitutively but at a low level, and its transcript level increased slightly after bacterial inoculation (Figure 4B). However, in the antisense plants, the CDR1 transcript was reduced to an undetectable level in the uninoculated leaves and was barely detectable in the inoculated leaves (Figure 4B). Examination of the CDR1 protein levels further confirmed that CDR1 is suppressed in the aCDR1-2 and 1-3 lines (Figure 5F, see below). The disease resistance phenotype of these two lines was further examined in progeny plants. These plants were not only compromised for resistance to avirulent Psm avrRpm1, but also exhibited enhanced susceptibility to virulent Psm (Figure 4C–E). In addition, the aCDR1-2 line was impaired in local and systemic induction of PR genes in response to inoculation with Psm avrRpm1 (Figure 4F). Genetic analysis indicated that the enhanced disease susceptibility phenotype in the aCDR1 lines was dominant and cosegregated with the transgene, further demonstrating that the phenotype is the consequence of the antisense-mediated suppression of CDR1.
Figure 5.
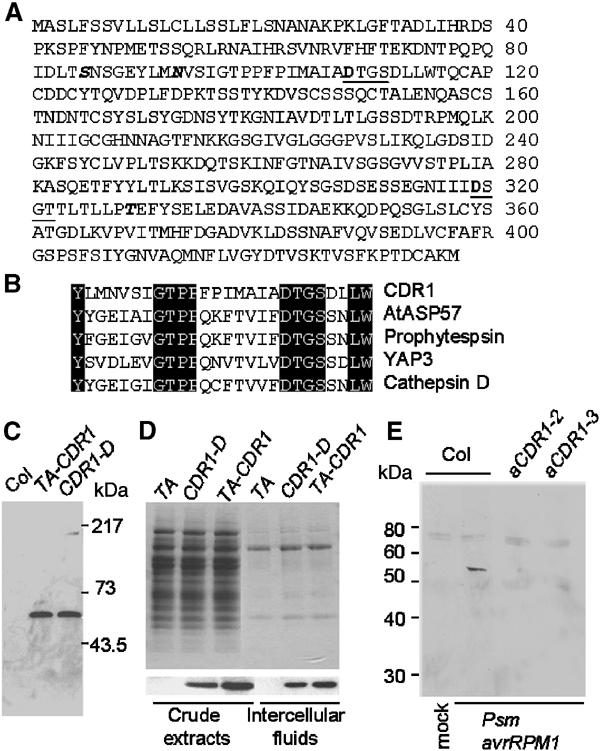
CDR1 is an extracellular protein and is induced in response to bacterial inoculation. (A) Deduced amino acid sequence of CDR1. The two active sites are underlined, and the active site aspartyl residues are in bold. One potential N-glycosylation (N93) and two potential O-glycosylation sites (S85 and T329) are in italic. (B) Partial sequence alignment of CDR1 and four other aspartic proteases in the regions surrounding the first active site. (C) Affinity-purified anti-CDR1 polyclonal antibodies detect an approximately 54 kDa polypeptide in extracts from leaves of the CDR1-D mutant and from TA-CDR1 plants 16 h after Dex application. (D) Coomassie staining of total protein extracts and IFs from wild-type Col-0 plants carrying the empty vector (TA) and plants overexpressing CDR1 (upper panel) and immunodetection of CDR1 protein in the samples (lower panel). (E) CDR1 protein accumulates in IFs of wild-type plants but not the aCDR1 lines in response to bacterial inoculation. IFs were isolated from leaves 8 h after inoculation. The 54 kDa band represents CDR1, and other weak bands likely resulted from cross-reactions of the antibody to other proteins.
CDR1 encodes an extracellular aspartic protease whose level is induced in response to pathogen infection
Sequence analysis of the CDR1 genomic and two cDNA clones indicated that the gene had no intron. The deduced 47 kDa CDR1 protein (Figure 5A) is predicted by the PSORT algorithm (Nakai and Kanehisa, 1992) to be extracellular, with a signal peptide cleaved at position 25. A BLAST (Altschul et al, 1990) search of the GenBank databases revealed that the CDR1 protein shares sequence identity with aspartic proteases, one of the five types of endopeptidases (Barrett, 1998). Like other eukaryotic aspartic proteases of the pepsin family, CDR1 contains two catalytic aspartic acid residues within two conserved active sites with the sequence motif Asp-Thr/Ser-Gly-Ser/Thr (Figure 5A). Figure 5B shows a sequence alignment of one of the active site regions of CDR1 with corresponding regions from cathepsin D (a human aspartic protease), phytepsin (an aspartic protease isolated from barley seeds), yeast aspartic protease 3 (YAP3), and AtASP57 (At1g62290; another putative aspartic protease from Arabidopsis). Overall, CDR1 shares approximately 22% identity to human cathepsin D with much higher sequence similarity around the two active sites. Unlike some plant aspartic proteases purified from seeds, CDR1 does not contain the plant-specific sequence (PSS) of about 100 amino acids located near the C-termini of these plant aspartic proteases that is believed to be involved in targeting the proteins to vacuoles (Mutlu and Gal, 1999).
Anti-CDR1 polyclonal antibodies were raised against a synthetic peptide derived from a sequence near the C-terminus of the deduced CDR1 protein. The affinity-purified antibodies detected a single polypeptide with an approximate size of 54 kDa in the CDR1-D mutant and the dexamethasone (Dex)-induced TA-CDR1 plant (Figure 5C). The TA-CDR1 plants carry a chemically inducible-CDR1 construct pTA-CDR1 (Figure 6A), generated by using the glucocorticoid-mediated transcriptional induction system (Aoyama and Chua, 1997). CDR1 was induced by spraying with Dex. The level of CDR1 protein in total extracts of wild-type plants is too low to be detected by the antibodies. The experimentally determined size of plant-expressed CDR1 is greater than the size calculated from its open reading frame (47 kDa), indicating that the mature CDR1 protein might be post-translationally modified. One potential N-glycosylation site (N93) and two potential O-glycosylation sites (S85 and T329) (Figure 5A) are predicted by the NetOGlyc and NetNGlyc algorithms (http://us.expasy.org/tools/).
Figure 6.

Induction of CDR1 leads to local and systemic activation of defense responses. (A) pTA-CDR1: construct for Dex inducible CDR1 expression. (B) Induction of TA-CDR1 by hand infiltration with Dex induces local and systemic PR gene expression. Two or three leaves of each plant were hand infiltrated with 5 μM Dex. The infiltrated (1°) and uninfiltrated (2°) leaves were collected separately. The TA plants carry the empty vector. (C) Grafting experiment showing induction of PR2 expression in wild-type scions grafted onto TA-CDR1 rootstocks following CDR1 induction. CDR1 was induced by watering with 10 μM Dex, and leaves were collected for RNA isolation 24 h after Dex application. Scions grafted onto TA rootstocks were used as a control. Lane 1: without Dex application; lanes 2 and 3: the samples from two sets of grafting experiments. (D) IFs isolated from Dex-treated TA-CDR1 plants induce PR gene expression. RNA was isolated from the wild-type leaves 10 h after hand infiltration with the IFs. IFs were isolated from TA transgenics (control) (16 h after spraying with 2 μM Dex, lane 1) and from TA-CDR1 transgenics either before (lane 3) or 16 h after (lane 2) Dex treatment. (E) Induction of PR2 by the IF was compromised in NahG and eds5-1 mutant. (F) Local and systemic induction of PR gene expression by HMW and LMW fractions of the IFs. (G) Immunodetection of CDR1 in the crude IF and two fractions of the IF. IF from TA transgenics was used as a negative control. (H) The elicitor in the LMW fraction has a molecular weight of 3–10 kDa. Only PR2 induction in the secondary leaves is shown. (I) The elicitor activity in the 3–10 kDa fraction is sensitive to heating and Pronase treatments.
Based on the presence of an N-terminal signal sequence, CDR1 may be extracellular. To test this, intercellular fluids (IFs) were isolated from CDR1-overexpressing plants and control plants. Protein gel-blot analysis indicated that the amount of CDR1 in the IF preparation was about the same as in the cellular protein extract, whereas the latter contained ∼30-fold more total protein (Figure 5D). This substantial enrichment of CDR1 in the IF indicates that CDR1 indeed accumulates in the apoplast. Furthermore, fusion of the CDR1 signal peptide to the N-terminus of green fluorescent protein (GFP) was sufficient to result in secretion of GFP to the external culture medium when the CDR1–GFP construct was expressed in tobacco BY2 cells (data not shown).
IFs were then used to examine whether native CDR1 protein accumulates in wild-type plants in response to pathogen attack. We isolated IFs from leaves of wild-type plants and two aCDR1 lines after inoculation with Psm avrRpm1. The IF proteins were concentrated 10-fold through ultrafiltration using 10 kDa molecular weight cut-off filters and were subjected to Western blotting analysis using the anti-CDR1 antibodies. CDR1 protein was detected in the IF of the Psm avrRpm1-inoculated wild-type leaves 8 h postinoculation, but not in those of aCDR1 leaves or the mock-inoculated leaves (Figure 5E).
Induction of CDR1 leads to activation of local and systemic defense responses
We used the TA-CDR1 plants to further study the effect of CDR1 on induction of the defense response. To induce CDR1 expression, TA-CDR1 plants were hand infiltrated with Dex. Accumulation of CDR1 transcripts was detected within 3 h of Dex application and was maximal after 1–2 days (Figure 6B). Dex induction of CDR1 was followed by expression of PR2 (Figure 6B) and PR1 (data not shown) genes with about a 5 h lag time. Moreover, local induction of CDR1 by hand infiltration of Dex was found to induce PR2 (Figure 6B) and PR1 (data not shown) expression in distant leaves. However, no CDR1 transcripts were detected in the systemic leaves following local Dex application (Figure 6B), suggesting that the systemic induction of PR1 and PR2 reflected the action of an endogenous mobile signal rather than the movement of Dex through the plant.
A micrografting experiment, based on the technique previously described by Turnbull et al (2002), confirmed that CDR1 activation leads to systemic induction of defense responses. Wild-type Col-0 seedlings were used as scion and grafted onto rootstocks of TA-CDR1 and TA (carrying the empty vector and used as a negative control) seedlings, respectively. At 2 weeks after grafting, the plants were watered with Dex to induce CDR1 expression in the rootstocks. As shown in Figure 6C, following the Dex treatment, PR2 was activated in the wild-type plants grafted onto the TA-CDR1 rootstocks, but not in those on the TA rootstocks.
IFs from leaves of CDR1-D and Dex-induced TA-CDR1 plants were found to induce marked accumulation of PR1 and PR2 transcripts when hand infiltrated into leaves of wild-type plants (Figure 6D and data not shown). In line with the NahG CDR1-D data showing that CDR1 action is SA dependent, induction of PR2 expression by the IF was significantly compromised not only in NahG leaves but also in the eds5-1 mutant, which is defective in SA-dependent signaling (Nawrath et al, 2002) (Figure 6E). No micro-lesion or necrosis was observed in the leaves infiltrated with the IF (data not shown), suggesting that micro-lesion formation in the CDR1-D mutant might not be the cause of activation of the defense response.
To further characterize this elicitor activity, the IFs were size fractionated using a 10 kDa molecular weight cut-off filter. The high-molecular-weight (HMW) fraction (>10 kDa), which contained CDR1 protein (Figure 6G), accounted for most of the elicitor activity seen in the crude IFs (Figure 6F). This fraction induced PR transcripts in both primary and systemic secondary leaves. The low-molecular-weight (LMW) fraction (<10 kDa), which does not contain CDR1 protein (Figure 6G), also induced PR transcripts in both the primary and secondary leaves (Figure 6F). The accumulation of PR transcripts in inoculated tissues was relatively weak compared to that observed in tissues directly inoculated with the HMW fraction. However, the response in distant tissues to local inoculation with the LMW fraction was comparable to the distant effects of local inoculation with the HMW fraction, suggesting that the elicitor activity in the LMW fraction makes a substantial contribution to the systemic effects of CDR1 activity. Further size fractionation using a filter with a 3 kDa cut-off demonstrated that the elicitor activity in the LMW fraction is associated with molecules in the 3–10 kDa range (Figure 6(H)). When this fraction was subjected to heating (boiling for 5 min) or Pronase treatments, the elicitor activity was severely reduced (Figure 6I).
Protease activity is required for CDR1 function
CDR1 expressed in Escherichia coli as a fusion protein to GST exhibited protease activity against the model substrate FITC-BSA (Figure 7A). In this in vitro assay, CDR1 was approximately 30% as active as cathepsin D. To determine whether the aspartic protease activity of CDR1 was essential for its biological activity, various mutations were made by site-directed mutagenesis, and these were reintroduced into wild-type Col-0. The mutations were D108N and D319N (which alter the aspartate residues in each of the two active sites), the double mutant D108N/D319N, and D174N (which alters an aspartate residue not in the active site). Unlike plants overexpressing wild-type CDR1, plants expressing the mutant forms of CDR1 in one or both active site aspartate residues did not show the CDR1-D-like phenotype (Figure 7B and data not shown). In contrast, plants expressing D174N showed the typical CDR1-D phenotype. Likewise, Dex inducible expression of CDR1 genes encoding proteins with active site mutations fails to stimulate PR expression (data not shown).
Figure 7.
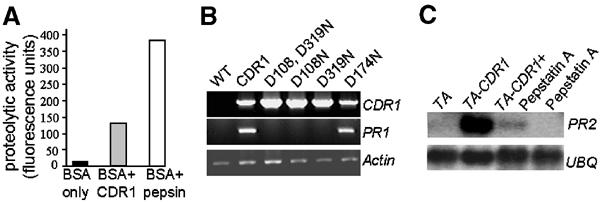
CDR1 function requires aspartic protease activity. (A) In vitro proteolytic activity assay of E. coli-expressed CDR1. FITC-BSA was used as the substrate, and pepsin was used as a positive control. (B) RT–PCR analysis of PR gene expression in Arabidopsis transformed with CDR1 or various site-directed mutants in which active site or nonactive site aspartate residues are mutated. (C) Influence of pepstatin A on PR gene expression induced by IFs from CDR1 overexpressing Arabidopsis.
To determine whether aspartic protease activity is required for the CDR1-mediated induction of PR genes by components of IFs from CDR1-overexpressing Arabidopsis, the HMW fraction was coinfiltrated with pepstatin A, a reversible aspartic protease inhibitor. Pepstatin A greatly reduced induction of PR2 expression by the IF fraction (Figure 7C).
Discussion
Using activation tagging, which generates gain-of-function mutations (Weigel et al, 2000), we have identified the Arabidopsis CDR1 gene, whose hyperactivation induces an SA-dependent disease resistance response. CDR1-D exhibited a phenotype that mimics constitutive activation of SAR, including the accumulation of high levels of SA and transcripts of SAR marker genes such as PR1 and PR2. The mutant also exhibited localized cell death and oxidative bursts, which have been previously shown to be associated with the establishment of systemic immunity underlying SAR (Alvarez et al, 1998). The CDR1-D phenotype is blocked almost completely by NahG and is partially blocked by the npr1 mutation that affects signaling downstream of SA action (Cao et al, 1997) (data not shown). Consistent with the phenotype caused by the CDR1-D gain-of-function mutation, suppression of CDR1 impairs induction of PR genes by bacterial inoculation, and causes not only enhanced susceptibility to infection by virulent bacterial pathogens but also compromised gene-for-gene-mediated resistance.
CDR1 encodes an apoplastic protein that shares significant sequence similarity to aspartic proteases. Like other eukaryotic aspartic proteases, CDR1 contains two active sites with the conserved motifs Asp-Thr-Gly-Ser and Asp-Ser-Gly-Thr, respectively. High-level expression in Arabidopsis of mutant versions of CDR1, in which the active site aspartyl residues were altered, failed to recapitulate the CDR1-D-like phenotype. In addition, the induction of defense responses by the HMW fraction from IFs, which contain CDR1, was strongly inhibited by pepstatin. Taken together with the in vitro proteolytic activity of E. coli-expressed CDR1, these data indicate that CDR1 encodes an aspartic protease that functions biologically by the proteolytic cleavage of its endogenous target.
Aspartic proteases have been implicated in regulating a variety of biological processes through limited proteolytic processing of peptide hormones, receptors, and other regulatory proteins. CDR1 protein is accumulated in the apoplast in response to inoculation of avirulent bacterial pathogens. CDR1 might process a cell surface protein that could be a component of the basal host defense complex (Dangl and Jones, 2001). Alternatively, CDR1 activation might lead to the generation of an endogenous extracellular peptide elicitor. Local induction of CDR1 was found to induce the systemic defense response, and a mobile elicitor activity was detected in the IFs. The hypothesized CDR1-released elicitor can rapidly activate basal defense responses in local and systemic leaves, suggesting its high capacity of mobility. It is tempting to speculate that the putative CDR1-released peptide elicitor could function as a mobile SAR signal in a manner analogous to that of the proteinaceous wound response mediator systemin (Pearce et al, 1991). Plant cells are equipped with sensitive and specific receptors for recognizing complex signals generated from both pathogen and host plant cells, such as recognition of race-specific elicitors as well as ‘general' peptide elicitors serving as pathogen-associated molecular patterns (Nürnberger et al, 1994; Keller et al, 1996; Gomez-Gomez and Boller, 2000). Several molecules, such as SA, ROIs, NO, and some lipids and lipid derivatives, have been suggested to be putative short- or long-distance mobile signals mediating the development of a variety of defense mechanisms, underlying the complexity of the plant innate immunity response. An endogenous peptide elicitor is an attractive candidate as a mobile SAR signal because plant cells appear to contain receptors for perceiving peptide signals and machinery for translocating such peptides, as demonstrated for the wound signal systemin (Bergey et al, 1996; Scheer and Ryan, 2002).
Recently, several proteases have been found to play important roles in plant–microbe interactions. Several avirulence genes encode different classes of proteases (Jia et al, 2000; Orth et al, 2000; Shao et al, 2002). In addition, the tomato RCR3 gene encoding a secreted cysteine protease is required for Cf-2-mediated gene-for-gene resistance against the fungus Cladosporium fulvum expressing the avr2 avirulence gene (Kruger et al, 2002). It has been speculated that such proteases mediate defense responses either by directly processing R proteins, or by generating elicitors that could potentially be recognized by R proteins (Bonas and Lahaye, 2002; Kruger et al, 2002; Shao et al, 2002). Identification of CDR1 reveals a host protease that might function by generating an endogenous peptide elicitor to induce local and systemic defense responses. Future identification of the endogenous elicitor will enable us to examine whether it is generated directly or indirectly by CDR1.
Materials and methods
Plant materials
A. thaliana ecotype Columbia (Col-0) plants were grown at 23°C under short-day conditions (9 h 150 μE light and 15 h dark) for 5 weeks, followed by growth under long-day conditions (16 h light and 8 h dark) until mature.
Activation tagging
Plants were transformed by a dipping procedure (Clough and Bent, 1998) using Agrobacterium strain GV3101 pMP90RK, harboring the activation tagging vector pSKI15 (Kardailsky et al, 1999). After transformation, seeds were harvested from T0 plants, pooled and sown on soil. Basta-resistant seedlings (T1 plants) were transplanted individually and grown for 4–5 weeks before mutant screening.
Infection
Pseudomonas syringae pv. tomato (Pst) and P. syringae pv. maculicola (Psm) were grown as described (Cameron et al, 1994). For the spraying and dipping infection procedures (Whalen et al, 1991), 1–3 × 108 cfu/ml bacterial suspensions containing 0.02% of the detergent Silwet-L77 were used. Plants were sprayed with the bacterial suspension using a hand spray bottle. Inoculated plants were covered for 24 h to maintain high humidity. The hand-infiltration inoculation procedure was as previously described (Cameron et al, 1994).
Salicylic acid
SA was extracted according to published procedures (Meuwly and Metraux, 1993). Separation and determination was performed on an HP1100 (Agilent) HPLC, Waters Spherisorb ODS2 5 μm C18 reverse phase column (250 × 4.6 mm), flow rate 0.8 ml/min, with a diode array detector (Agilent) and fluorescence detector (Jasco Model FP920) in tandem. Samples were monitored at 230 and 300 nm using the diode array detector, and the fluorescence detector was set at an excitation wavelength of 305 nm and emission of 407 nm. SA levels in the samples were calculated based on fluorescence compared to a standard curve using authentic SA.
Histochemistry
DAB and Trypan blue staining were performed as described (Alvarez et al, 1998).
Nucleic acid analysis
Arabidopsis genomic DNA was isolated using a modified cetyltrimethylammonium bromide (CTAB) procedure (Saghai-Maroof et al, 1984). Total RNA was isolated from aerial parts of 4–5-week-old plants that had not started to bolt using the Trizol reagent according to the manufacturer's instructions (GIBCO-BRL). Labeling of DNA fragments with 32P, electrophoresis, blotting of nucleic acids, and hybridization were conducted according to standard procedures (Sambrook et al, 1989).
Reverse transcriptase–polymerase chain reaction (RT–PCR) analysis of transcript levels was carried out using total leaf RNA from 4–5-week-old plants. First-strand cDNA was synthesized from 4 μg total RNA using Superscript II reverse transcriptase (GIBCO) and oligo dT primer. PCR was carried out for 25 cycles using ExTaq DNA polymerase (Takara) with gene-specific primers for CDR1, PR1 and actin as follows: 5′-TACGAACGATAGCCATGGCCTCTC-3′ and 5′-GGATCCTACATCTTTGCACAATCTGTTGGC-3′ for CDR1; 5′-ATGAATGAAATGTCGTTCTTTGGTTATAG-3′ and 5′-CCAATCACTAATATGGACGTTGACCGATG-3′ for PR1; 5′-GATATGGAAAAGATCTGGCATCAC-3′ and 5′-TCATACTCGGCCTTGGAGATCCAC-3′ for actin.
The plant genomic DNA flanking the T-DNA borders was cloned by plasmid rescue (Borevitz et al, 2000).
Isolation of CDR1 cDNA clones
A cDNA library derived from Arabidopsis ecotype Col-0 (Kieber et al, 1993) was screened according to standard procedures (Sambrook et al, 1989). The two cDNA clones isolated were named pCDR1C1 and pCDR1C2.
Expression of CDR1 fusion protein in E. coli
The open reading frame from pCDR1C1 was amplified by PCR and subcloned into pGEX-4T-2 expression vector (Pharmacia) to generate pGEX-CDR1 in which CDR1 is fused at the C-terminus of GST. E. coli strain BL21 cells harboring the expression construct were grown to an OD600 of 0.6, and expression was induced by the addition of isopropyl 1-thio-β-D-galactopyranoside (IPTG) to a final concentration of 1 mM, with further incubation at 16°C for 9 h. Purification of the CDR1 fusion protein was performed on a glutathione sepharose 4B column according to the manufacturer's instructions. Aspartic protease was assayed using FITC-BSA as substrate (Murakami et al, 2000).
Immunoblot analysis
A peptide of 14 amino acids, corresponding to the sequence DTVSKTVSFKPTDC adjacent to the CDR1 C-terminus, was synthesized and purified by HPLC. The peptides were conjugated to keyhole limpet hemocyanin and injected into rabbits. The resulting antisera were purified using the CDR1 fusion protein immobilized on a nitrocellulose filter and immunoblotting was performed according to standard protocols (Sambrook et al, 1989).
Intercellular fluids
IFs were isolated based on the procedure described by De Wit and Spikman (1982). For fractionation, IFs were loaded into a 10 kDa cut-off Ambion centrifuge filter and centrifuged at 8000 g for approximately 3–6 h until completion. The run-through and the retentate were labeled as the LMW and HMW fractions, respectively. The LMW fraction was further fractionated using a 3 kDa cut-off filter.
For the Pronase treatment of the 3–10 kDa IF fraction, 1 M Tris–HCl (pH 7.5) was added to the IF to a final concentration of 10 mM. Pronase E (Sigma) (5 mg/ml stock in 10 mM Tris–HCl, pH 7.5) was added to the IF to a final concentration of 10 μg/ml, and the IF was incubated at 37°C for 1.5 h. The digested IF was then filtered through a 10 kDa cut-off filter, and the run-through was used to infiltrate Arabidopsis leaves.
Induction of PR genes by IFs
Approximately 10 μl of IFs were pressure infiltrated using a syringe into leaves of 4–5-week-old Arabidopsis Col-0 plants. For the pepstatin A inhibition experiments, pepstatin A was dissolved in 100% ethanol and added to the intercellular fractions to a final concentration of 1 mM immediately prior to infiltration. The same volume of 100% ethanol was added to controls. Total RNA was isolated from the infiltrated and distal leaves and subjected to RNA gel blot analysis.
Construction of pBI/CDR1X, pTA-CDR1, and pAnti-CDR1
The XbaI fragment of the rescued plasmid pCDR1E that contains one copy of the 35S enhancer sequence and the candidate CDR1 gene was subcloned into the XbaI site of the binary vector pBI101.3 (Clontech) to generate pBI/CDR1X. To construct pTA-CDR1, the cDNA clone pCDR1C1 was completely digested with SpeI and partially digested with XhoI. The 1.5 kb fragment containing the entire cDNA insert was subcloned into the XhoI/SpeI site of pTA7002 (Aoyama and Chua, 1997). To make the antisense construct pAnti-CDR1, the insert from pCDR1C1 was excised with BamHI/EcoRV and ligated into the BamHI/SmaI sites of pBI121.
CDR1 site-directed mutants
For the construction of CDR1 mutants, the full-length cDNA encoding CDR1 was used as a template for PCR. For D108N, in which the aspartate residue at position 108 was replaced by asparagine, mutation was carried out using the NcoI upstream primer 5′-CGATGCCATGGCCTCTCTATTCTCTTCAGTTCTCT-3′ and the BamHI downstream primer 5′-CGCGGATCCTACATCTTTGCACAATCTGTTGGCTT-3′ (primer set 1), and the internal forward primer 5′-TGGCCATCGCCAACACCGGAAGTGATCTCC-3′ and the internal reverse primer 5′-CACTTCCGGTGTTGGCGATGGCCATGATC-3′ (primer set 2). The double underlined nucleotides indicate the positions of mutagenesis (GAC codon for aspartate replaced by AAC codon for asparagines). The first amplification was performed using the two sets of primers in separate reaction tubes. The D108N fragment was then generated from a mixture of the two PCR-amplified fragments, the NcoI upstream primer and BamHI downstream primer. The PCR product was purified and cloned into pGEMTeasy vector (Promega), and subcloned between the NcoI and BamHI sites of pRTL2 (Restrepo et al, 1990) under the CaMV 35S promoter. The mutated CDR1 expression cassette was excised with HindIII and cloned into the binary vector pGA482 (An, 1986). The other site-directed mutations of CDR1 were also introduced by PCR using the following combinations of primers: the internal forward primer 5′-TTGTCTTACGGGAATAACTCATACACAAAGGG-3′ and the internal reverse primer 5′-TGTATGAGTTATTCCCGTAAGACAATGAG-3′ for generation of the D174N mutation; the internal forward primer 5′-CATCATCATCAATTCAGGCACAACTTTAACG-3′ and the internal reverse primer 5′-AGTTGTGCCTGAATTGATGATGATGTTTCCC-3′ for the D319N mutation. Constructs were mobilized into the A. tumefaciens strain.
Micro-grafting
Micro-grafting was conducted according to the technique described by Turnbull et al (2002). Sterile seeds were germinated on the medium containing MS basal salts, 1% sucrose, and 2% agar. Seedlings were grown on plates vertically for 3–5 days. Seedlings were transverse cut at hypocotyls and butt aligned using collar. At 6 days after grafting, seedlings were examined and successfully grafted ones were transplanted into soil.
Accession number
The sequence data of CDR1 have been submitted to the GenBank database under accession number AV243479.
Acknowledgments
This work was supported by the Samuel Roberts Noble Foundation and the Biotechnology and Biological Sciences Research Council.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Alvarez ME, Pennell R, Meijer P-J, Ishikawa A, Dixon RA, Lamb C (1998) Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 92: 773–784 [DOI] [PubMed] [Google Scholar]
- An G (1986) Development of plant promoter expression vectors and their use for analysis of differential activity of nopaline synthase promoter in transformed tobacco cells. Plant Physiol 81: 86–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyama T, Chua N-H (1997) A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Barrett AJ (1998) Proteolytic enzymes: nomenclature and classification. In Handbook of Proteolytic Enzymes, Woessner JF (ed) London: Academic Press pp 1–20 [Google Scholar]
- Bergey DR, Howe GA, Ryan CA (1996) Polypeptide signaling for plant defensive genes exhibits analogies to defense signaling in animals. Proc Natl Acad Sci USA 93: 12053–12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonas U, Lahaye T (2002) Plant disease resistance triggered by pathogen-derived molecules: refined models of specific recognition. Curr Opin Microbiol 5: 44–50 [DOI] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron RK, Dixon RA, Lamb CJ (1994) Biologically induced systemic acquired resistance in Arabidopsis thaliana. Plant J 5: 715–725 [Google Scholar]
- Cao H, Glazebrook J, Clarke JD, Volko S, Dong X (1997) The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88: 57–63 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cohn J, Sessa G, Martin GB (2001) Innate immunity in plants. Curr Opin Immunol 13: 55–62 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- De Wit PJGM, Spikman G (1982) Evidence for the occurrence of race and cultivar-specific elicitors of necrosis in intercellular fluids of compatible interactions of Cladosporium fulvum and tomato. Physiol Plant Pathol 21: 1–11 [Google Scholar]
- Dong X (2001) Genetic dissection of systemic acquired resistance. Curr Opin Plant Biol 4: 309–314 [DOI] [PubMed] [Google Scholar]
- Dong X, Mindrinos M, Davis KR, Ausubel FM (1991) Induction of Arabidopsis defense genes by virulent and avirulent Pseudomonas syringae strains and by a cloned avirulence gene. Plant Cell 3: 61–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaffney T, Friedrich B, Vernooij D, Negrotto G, Nye S, Uknes E, Ward E, Kessmann H, Ryals J (1993) Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261: 754–756 [DOI] [PubMed] [Google Scholar]
- Glazebrook J (2001) Genes controlling expression of defense responses in Arabidopsis—2001 status. Curr Opin Plant Biol 4: 301–308 [DOI] [PubMed] [Google Scholar]
- Glazebrook J, Rogers EE, Ausubel FM (1997) Use of Arabidopsis for genetic dissection of plant defense responses. Annu Rev Genet 31: 547–569 [DOI] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T (2000) FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B (2000) Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J 19: 4004–4014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky I, Shukla VK, Ahn JH, Dagenais N, Christensen SK, Nguyen JT, Chory J, Harrison MJ, Weigel D (1999) Activation tagging of the floral inducer FT. Science 286: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Keller H, Blein JP, Bonnet P, Ricci P (1996) Physiological and molecular characteristics of elicitin-induced systemic acquired resistance in tobacco. Plant Physiol 110: 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber JJ, Rothenberg M, Roman G, Feldmann KA, Ecker JR (1993) CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72: 427–441 [DOI] [PubMed] [Google Scholar]
- Kruger J, Thomas CM, Golstein C, Dixon MS, Smoker M, Tang S, Mulder L, Jones JD (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science 296: 744–747 [DOI] [PubMed] [Google Scholar]
- McDowell JM, Dangl JL (2000) Signal transduction in the plant immune response. Trends Plant Sci 25: 79–82 [DOI] [PubMed] [Google Scholar]
- Meuwly P, Metraux JP (1993) Ortho-anisic acid as internal standard for the simultaneous quantitation of salicylic acid and its putative biosynthetic precursors in cucumber leaves. Anal Biochem 214: 500–505 [DOI] [PubMed] [Google Scholar]
- Murakami S, Kondo Y, Nakano T, Sato F (2000) Protease activity of CND41, a chloroplast nucleoid DNA-binding protein, isolated from cultured tobacco cells. FEBS Lett 468: 15–18 [DOI] [PubMed] [Google Scholar]
- Mutlu A, Gal S (1999) Plant aspartic proteinases: enzymes on the way to a function. Physiol Plant 105: 569–576 [Google Scholar]
- Nakai K, Kanehisa M (1992) A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14: 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawrath C, Heck S, Parinthawong N, Metraux JP (2002) EDS5, an essential component of salicylic acid-dependent signaling for disease resistance in Arabidopsis, is a member of the MATE transporter family. Plant Cell 14: 275–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nürnberger T, Nennstiel D, Jabs T, Sacks WR, Hahlbrock K, Scheel D (1994) High affinity binding of a fungal oligopeptide elicitor to parsley plasma membranes triggers multiple defense responses. Cell 78: 449–460 [DOI] [PubMed] [Google Scholar]
- Orth K, Xu Z, Mudgett MB, Bao ZQ, Palmer LE, Bliska JB, Mangel WF, Staskawicz B, Dixon JE (2000) Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science 290: 1594–1597 [DOI] [PubMed] [Google Scholar]
- Pearce G, Strydom D, Johnson S, Ryan CA (1991) A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–898 [DOI] [PubMed] [Google Scholar]
- Penninckx IAMA, Eggermont K, Terras FRG, Thomma BPHJ, De Samblanx GW, Buchala A, Métraux J-P, Manners JM, Broekaert WF (1996) Pathogen-induced systemic activation of a plant defensin gene in Arabidopsis follows a salicylic acid-independent pathway. Plant Cell 8: 2309–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Restrepo MA, Freed DD, Carrington JC (1990) Nuclear transport of plant potyviral proteins. Plant Cell 2: 987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW (1984) Ribosomal DNA spacer-length polyporphism in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc Natl Acad Sci USA 81: 8014–8018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning. A Laboratory Manual, 2nd edn New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Scheer JM, Ryan CA (2002) The systemin receptor SR160 from Lycopersicon peruvianum is a member of the LRR receptor kinase family. Proc Natl Acad Sci USA 99: 9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao F, Merritt PM, Bao Z, Innes RW, Dixon JE (2002) A Yersinia effector and a Pseudomonas avirulence protein define a family of cysteine proteases functioning in bacterial pathogenesis. Cell 109: 575–588 [DOI] [PubMed] [Google Scholar]
- Tao Y, Xie Z, Chen W, Glazebrook J, Chang HS, Han B, Zhu T, Zou G, Katagiri F (2003) Quantitative nature of Arabidopsis responses during compatible and incompatible interactions with the bacterial pathogen Pseudomonas syringae. Plant Cell 15: 317–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, Jones JDG (2002) Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci USA 99: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Onouchi H, Hamada S, Machida C, Hammond-Kosack KE, Jones JD (1998) Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J 14: 365–370 [DOI] [PubMed] [Google Scholar]
- Turnbull CGN, Booker JP, Leyser HMO (2002) Micrografting techniques for testing long-distance signaling in Arabidopsis. Plant J 32: 255–262 [DOI] [PubMed] [Google Scholar]
- Verberne MC, Verpoorte R, Bol JF, Mercado-Blanco J, Linthorst HJM (2000) Overproduction of salicylic acid in plants by bacterial transgenes enhances pathogen resistance. Nat Biotechnol 18: 779–783 [DOI] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borewitz J, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malanchaurvil EJ, Neff MM et al. (2000) Activation tagging in Arabidopsis. Plant Physiol 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whalen MC, Innes RW, Bent AF, Staskawicz BJ (1991) Identification of Pseudomonas syringae pathogens of Arabidopsis and a bacterial locus determining avirulence on both Arabidopsis and soybean. Plant Cell 3: 49–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Shah J, Klessig DF (1997) Signal perception and transduction in plant defense responses. Genes Dev 11: 1621–1639 [DOI] [PubMed] [Google Scholar]


