Abstract
Interferon regulatory factor 5(IRF5) located on human chromosome 7q32 is associated with many chronic inflammatory disorders. IRF5 is the key regulator of proinflammatory cytokines and type I interferons. We surveyed two cohorts of inflammatory bowel disease (IBD) patients from a North American Consortium. Six single-nucleotide polymorphisms and a 5-base-pair (bp) insertion-deletion (CGGGG indel)polymorphism were investigated. Cytokine secretion was measured in primary lymphocytes after toll-like receptor 9 stimulation. Two-marker haplotypes containing the pairs (rs4728142-CGGGG indel) and (CGGGG indel-rs7808907) were associated with IBD protection (P = 2.89 × 10−6, P = 9.32 × 10−4 (non-Jewish ancestry) and P = 4.68 × 10−8, P = 2.50 × 10−8 (Jewish ancestry)) and IBD risk (P = 0.004, P = 0.003 (Jewish ancestry), respectively. IRF5 polymorphisms were risk factors for IBD in a single cohort. Interleukin-12-p70 cytokine production was higher (P = 0.04) in lymphocytes from controls with two alleles of the 5-bp insertion. IRF5 polymorphisms contribute to the risk profile for Crohn’s disease and ulcerative colitis along with ancestry and NOD2 genotypes.
Keywords: inflammatory bowel disease, Crohn’s disease, ulcerative colitis, interferon regulatory factor 5, polymorphisms, haplotype
INTRODUCTION
Crohn’s disease (CD) and ulcerative colitis (UC) are the two forms of the chronic gastrointestinal disorder, inflammatory bowel disease (IBD). The causes of IBD and its exact mechanisms are unknown. However, pathogenesis is clearly multifactorial and involves a genetic predisposition, antigenic stimuli and a dysregulation in the normal cytokine milieu of the mucosal immune system.1 The recurrent organ damage seen in IBD is characteristic of a group of immune-mediated inflammatory disorders (IMIDs) that have significant clinical and economic outcomes.2,3 Chronic inflammatory or autoimmune disorders affect over 5% of Americans and studies have shown that families with one IMID have higher rates of co-occurrence over other IMIDs.4 This is corroborated by genome-wide association studies that have reported shared loci or genes between multiple IMIDs.5 – 7
Interferon regulatory factor 5 (IRF5) is a transcription factor that forms one of the three major downstream inflammatory pathways that include the nuclear factor-kappa B (NF-κB), mitogen-activated protein kinase and IRFs8 (Figure 1). It is constitutively expressed in the cytoplasm of plasmacytoid dendritic cells and B cells, but is induced in most lymphocytes upon activation of the Toll-like receptor (TLR) 7 and 9 pathway. IRF5 regulates type I interferons and is crucial for activation of the pro-inflammatory cytokines interleukin (IL)-6, IL-12 and tumor necrosis factor alpha (TNF-α).9 – 12
Figure 1.
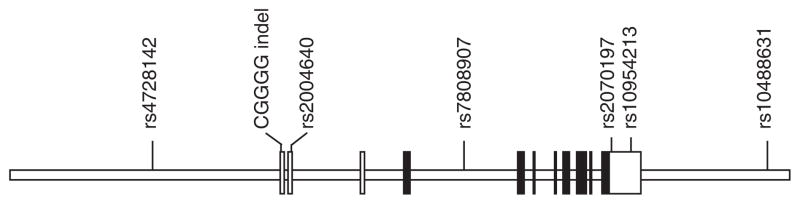
Interferon regulatory factor 5 gene: clear boxes are untranslated exons, shaded are translated.
Multiple functional polymorphisms of the IRF5 gene are associated with systemic lupus erythematosus (SLE), rheumatoid arthritis, Sjogren’s syndrome, multiple sclerosis, psoriasis, and IBD. Dideberg et al.13 reported the first association of a 5-bp insertion-deletion (indel) polymorphism in the promoter region of IRF5 with IBD. They postulated that, since different combinations of IRF5 polymorphisms are correlated with different forms of pathophysiology, altered IRF5 gene expression could form part of the genetic background that predisposes to the development of chronic inflammation. IRF5 is a complex gene with multiple isoforms that may have varying functions.14 A recent study demonstrated that the late-phase secretion of TNF-α in plasmacytoid dendritic cells is dependent upon the interaction of IRF5 with the NF-κB protein RelA.15 The aberrant overexpression of the TNF-α cytokine in IMIDs has led to the development of anti-TNF-α therapies that improved the morbidity and quality of life in patients. We postulated that IRF5 polymorphisms alter gene regulation and result in a genetic predisposition to chronic inflammatory disorders. We therefore investigated whether IRF5 polymorphisms were associated with IBD in a cohort from the North American NIDDK IBD Genetics Consortium. We also examined the correlation of risk markers with cytokine production.
STUDY SUBJECTS
Our study cohorts consisted of a case-control cohort with 1601 controls and 2059 IBD patients, as well as a family cohort with 620 trio and tetrad families. Both cohorts were from the NIDDK IBD consortium. IBD cases previously diagnosed using standard criteria were included along with healthy controls. Patient phenotypes were classified according to the Montreal Classification. All research protocols for human subjects’ research were reviewed and approved by the individual institutional review boards at six institutions. A standardized clinical questionnaire was completed for each patient at the time of enrollment and entered into a database program. The questionnaire included date of birth, sex, age at diagnosis, ethnicity, disease location, disease behavior, family history, extraintestinal manifestations, surgeries, smoking status and therapeutic management.
PBMC ISOLATION AND CULTURE
Thirty-five healthy controls were recruited from the IBD registry. From each patient, 50–60 ml of whole blood was collected and peripheral blood mononuclear cells (PBMCs) were isolated by density gradient separation (Ficoll-Paque, GE Healthcare) and frozen at −80°. Approximately 10 million PBMCs were thawed and diluted in RPMI (25 mm HEPES and 2 mm L-glutamine) and washed twice. Cells were then suspended in complete medium (CM): sterile RPMI-1640 medium with 10% sterile heat-inactivated FBS (Sigma) and 1% sterile antibiotic/antimycotic (60 μg ml−1 penicillinand 100 μg ml−1 streptomycin) and incubated in 12-well plates for 72 h at a cell density of 2 × 106 cells per ml (2 ml). At 24 h, cells were stimulated with 2.5 μM of CpG ODN (Invivogen) and interferon alpha (100 IU μl−1). Samples were centrifuged and supernatants were frozen at −80° for later analysis of cytokines.
CYTOKINE MEASUREMENT
The cytokine analysis was performed using the Bio-Plex 200 (Luminex) system. This bead-based multiplex analysis system permits simultaneous analysis of up to 100 different biomolecules (proteins, peptides or nucleic acids) in a single-microplate well.
Assay kits were purchased from Millipore Corporation (Billerica, MA, USA). Product number TGFB-64K-01 was used to assay TGF-b1; number MPXHCYTO-60K-10 was used to assay G-CSF, GM-CSF, IL-1ra, IL-1b, IL-6, IL-10, IL-12(p40), IL-12(p70), MCP-1 and TNF-α. The procedures followed the manufacturer’s recommendations.
One-way analysis of variance was used to derive means for quantitative cytokine data using SPSS version 19.0 (SPSS Inc., IBM Corporation, NY, USA). Nominal P-values < 0.05 were considered significant.
RESULTS AND DISCUSSION
The case–control (CC) cohort consisted of 1601 controls, 1158 CD patients and 901 UC patients. The family cohort had 620 trio and tetrad families that comprised 1098 controls, 712 CD patients, and 331 UC patients. Forty five and twenty eight samples from each cohort, respectively, came from patients with indeterminate colitis. The clinical characteristics of the IBD patients in both cohorts are shown in Table 1. Phenotypic characteristics in both cohorts were compared by Fisher’s exact test, Pearson’s χ2-test or independent-sample T-tests for continuous variables. The proportion of males and females were significantly different between cohorts (P = 0.007). Other variables with significantly different proportions between the cohorts included the age at diagnosis and disease location. Disease behavior, smoking, appendectomy and surgery information were available for the CC cohort (Supplementary Table 5). The combined cohorts consisted of European-ancestry individuals, but included a subset of individuals of Jewish ancestry (Supplementary Table 4). To account for this difference that could introduce population admixture, we completed the analysis in subsets of non-Jewish and Jewish ancestry separately. IRF5 has been well studied in the autoimmune disorder SLE. The markers selected for our study are based on the polymorphisms most significantly associated with SLE and includes the CGGGG indel that was previously associated with IBD. The single-nucleotide polymorphism (SNP) rs2004640 failed the Hardy-Weinberg equilibrium test and this SNP was removed from further analysis. The SNP rs2070197 was also removed due to missing data. The single-marker association analysis for the remaining five polymorphisms is shown in Table 2. The four repeats of the CGGGG indel (rs77571059) demonstrate a significant association (P = 0.025) with CD in the Jewish Ancestry CD subgroup. In Table 3, haplotypes comprising 2 SNP windows and involving rs4728142, CGGGG and rs7808907 were significantly associated with risk and protection of IBD in our combined analysis. The overall haplotype association signals are P = 4.704 × 10−7in nonJewish European ancestry individuals and P = 9.856 × 10−9in Jewish ancestry individuals for haplotype A (rs4728142-CGGGG indel); P = 0.00212 in non-Jewish European ancestry individuals and P = 2.12 × 10−8 in Jewish ancestry individuals for haplotype B (CGGGG indel-rs7808907).
Table 1.
Phenotypic characteristics of the IBD patients
| Group | Case/control | Family |
|---|---|---|
| CD | 1158 | 712 |
| Male/female (%) | 634/643 (49.6/50.4) | 309/403 (43.4/56.6) |
| Age, mean±s.d. (range) | 41.9±15.7 (7–101) | |
| Age at diagnosis, mean±s.d. (range) | 23±11.1 (2–80) | 22.3±9.8 (0–64) |
| UC | 901 | 331 |
| Male/female (%) | 476/425 (52.8/47.2) | 155/176 (46.8/53.2) |
| Age, mean±s.d. (range) | 43.5±16.3 (5–91) | |
| Age at diagnosis, mean±s.d. (range) | 30.1±14.8 (2–81) | 26±13.0 (2–75) |
| Age at diagnosis of CD (Montreal A) | ||
| A1<16 years | 327 (25.6) | 187 (26.3) |
| A2 = 17–40 years | 686 (53.7) | 434 (61) |
| A>440 years | 83 (6.5) | 29 (4.1) |
| Age at diagnosis of UC (Montreal A) | ||
| A1<16 years | 57 (17.2) | 162 (18) |
| A2 = 17–40 years | 180 (54.4) | 532 (59) |
| A>440 years | 33 (10) | 201 (22.3) |
| Disease behavior, CD (Montreal B) | ||
| B1 non-stricturing, non-penetrating | 437 (37.7) | |
| B2 stricturing | 264 (22.8) | |
| B3 penetrating excludes perianal | 344 (29.7) | |
| Disease location, CD (Montreal L) | ||
| L1 ileal | 418 (36.1) | 279 (39.2) |
| L2 colonic | 65 (5.6) | 91 (12.8) |
| L3 ileocolonic | 635 (54.8) | 284 (39.9) |
| Perianal only | 3 | 4 |
| Disease location, UC (Montreal E) | ||
| E1 ulcerative proctitis | 94 (10.4) | 32 (9.7) |
| E2 left-sided UC (distal UC) | 261 (29.0) | 90 (27.2) |
| E3 extensive UC (pancolitis) | 537 (59.6) | 156 (47.1) |
Abbreviations: CD, Crohn’s disease; IBD, inflammatory bowel disease; UC, ulcerative colitis.
Table 2.
Association of IRF5 polymorphisms with IBD
| Group | SNP | Risk allele | F cases | F controls | P value | OR (95% CI) |
|---|---|---|---|---|---|---|
| Non-Jewish IBD | Rs4728142 | A | 0.437 | 0.433 | 0.541 | 0.99 (0.91–1.07) |
| CGGGG indel | G | 0.550 | 0.548 | 0.715 | 1.01 (0.93–1.11) | |
| rs7808907 | C | 0.496 | 0.498 | 0.263 | 0.90 (0.83–0.98) | |
| rs10954213 | A | 0.594 | 0.598 | 0.879 | 1.01 (0.91–1.12) | |
| rs10488631 | T | 0.893 | 0.872 | 0.0569 | 1.04 (0.89–1.23) | |
| Non-Jewish CD | rs4728142 | A | 0.416 | 0.433 | 0.194 | 0.93 (0.84–1.04) |
| CGGGG indel | G | 0.433 | 0.453 | 0.169 | 0.92 (0.83–1.04) | |
| rs7808907 | C | 0.484 | 0.498 | 0.316 | 0.95 (0.87–1.08) | |
| rs10954213 | A | 0.410 | 0.402 | 0.594 | 1.03 (0.92–1.15) | |
| rs10488631 | T | 0.892 | 0.876 | 0.114 | 0.84 (0.72–1.01) | |
| Non-Jewish UC | rs4728142 | A | 0.462 | 0.433 | 0.0634 | 1.09 (0.99–1.18) |
| CGGGG indel | G | 0.470 | 0.453 | 0.252 | 1.07 (0.96–1.17) | |
| rs7808907 | C | 0.489 | 0.503 | 0.317 | 0.95 (0.84–1.04) | |
| rs10954213 | A | 0.398 | 0.402 | 0.841 | 0.98 (0.79–1.42) | |
| rs10488631 | T | 0.893 | 0.872 | 0.086 | 0.84 (0.72–1.02) | |
| Jewish IBD | rs4728142 | A | 0.493 | 0.48 | 0.34 | 1.06 (0.91–1.24) |
| CGGGG indel | G | 0.504 | 0.47 | 0.027 | 1.16 (0.99–1.37) | |
| rs7808907 | C | 0.587 | 0.607 | 0.34 | 0.93 (0.79–1.08) | |
| rs10954213 | A | 0.31 | 0.31 | 0.909 | 0.95 (0.78–1.15) | |
| rs10488631 | T | 0.895 | 0.904 | 0.429 | 0.89 (0.67–1.17) | |
| Jewish CD | rs4728142 | A | 0.494 | 0.481 | 0.387 | 1.10 (0.93–1.29) |
| CGGGG indel | G | 0.514 | 0.47 | 0.025 | 1.22 (1.02–1.46) | |
| rs7808907 | C | 0.592 | 0.607 | 0.464 | 0.95 (0.80–1.12) | |
| rs10954213 | A | 0.31 | 0.31 | 0.952 | 0.95 (0.78–1.17) | |
| rs10488631 | T | 0.891 | 0.905 | 0.273 | 0.84 (0.63–1.13) | |
| Jewish UC | rs4728142 | A | 0.485 | 0.48 | 0.968 | 0.95 (0.72–1.26) |
| CGGGG indel | G | 0.466 | 0.47 | 0.848 | 1.00 (0.75–1.32) | |
| rs7808907 | C | 0.573 | 0.607 | 0.248 | 0.85 (0.64–1.12) | |
| rs10954213 | A | 0.293 | 0.31 | 0.837 | 0.90 (0.52–1.56) | |
| rs10488631 | T | - | - | - |
Abbreviations: CD, Crohn’s disease; CI, confidence interval; IBD, inflammatory bowel disease; IRF, interferon regulatory factor; OR, odds ratio; UC, ulcerative colitis.
Table 3.
IRF5 gene-significant two-marker-haplotype association results
| Non-Jewish European ancestry Individuals
|
Jewish ancestry individuals
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Haplotype freq.
|
P value | OR (95% CI) | Haplotype freq.
|
P value | OR (95% CI) | |||
| IBD | Controls | IBD | Controls | |||||
| Haplotype A | ||||||||
| A-A | 0.0187 | 0.0349 | 0.00450 | 0.53 (0.36–0.79) | 0.014 | 0.052 | 4.68 × 10308 | 0.26 (0.14–0.50) |
| G-A | 0.536 | 0.517 | 0.372 | 1.08 (0.95–1.23) | 0.480 | 0.476 | 0.612 | 1.02 (0.85–1.23) |
| A-G | 0.419 | 0.400 | 0.181 | 1.10 (0.96–1.25) | 0.482 | 0.428 | 0.004 | 1.24 (1.03–1.51) |
| G-G | 0.0261 | 0.0510 | 2.89 × 10306 | 0.50 (0.36–0.70) | 0.024 | 0.043 | 0.033 | 0.55 (0.33–0.94) |
| Haplotype B | ||||||||
| A-C | 0.105 | 0.140 | 0.337 | 0.72 (0.59–0.87) | 0.124 | 0.215 | 2.50 × 10308 | 0.51 (0.40–0.66) |
| G-C | 0.393 | 0.359 | 0.0938 | 1.17 (1.02–1.32) | 0.463 | 0.392 | 0.003 | 1.34 (1.11 -1.61) |
| A-T | 0.447 | 0.410 | 0.749 | 1.16 (1.02–1.32) | 0.368 | 0.319 | 0.154 | 1.24 (1.03–1.51) |
| G-T | 0.0558 | 0.0914 | 0.000932 | 0.59 (0.46–0.75) | 0.044 | 0.073 | 0.280 | 0.58 (0.39–0.88) |
Abbreviations: CI, confidence interval; freq., frequency; IBD, inflammatory bowel disease; IRF, interferon regulatory factor; OR, odds ratio; SNP, single-nucleotide polymorphism. SNPs include haplotype A: rs4728142 (A/G) - CGGGG indel (A/G) and haplotype B: CGGGG indel (A/G) - rs7808907 (C/T). Overall haplotype A (rs4728142 - CGGGG indel) association P value = 4.704E-07 in non-Jewish European ancestry individuals, and P value = 9.856E-09 in Jewish ancestry individuals. Overall haplotype B (CGGGG indel - rs7808907) association P value = 0.00212 in non-Jewish European ancestry individuals, and P value = 2.125E-08 in Jewish ancestry individuals. Seven IRF5 variants were tested for their association with IBD. The analysis was limited to Caucasian patients and controls from the NIDDK consortium. SNP genotyping was completed by several methods. Three polymorphisms (rs2004640, rs10954213 and rs2070197) were sequenced by Sequenom, a technology that uses primer extension chemistry and mass spectrometric analysis24. Additional SNPs (rs10488631, rs4728142, and rs7808907) were determined using TaqMan MGB technology from Applied Biosystems and following the manufacturer’s recommendations. One additional polymorphism, the CGGGG indel, was characterized by fragment analysis of fluorescently labeled PCR products using the Applied Biosystems Genemapper 3730. As the samples contain both unrelated case/control as well as family samples, single-marker and haplotype association are performed in IQLS (quasi-likelihood scoring test).25 IQLS is a case - control association testing method between a binary trait and single or multiple SNPs. It is designed for samples with combined known family pedigree structure and unrelated individuals. By incorporating parental genotype information into an individual’s haplotype, it has higher power than the standard association test, assuming no family structure. Association testing is performed in non-Jewish European ancestry cohort and Jewish ancestry cohorts. The indel CGGGG is coded as a biallelic SNP (A/G), assuming an additive genetic inheritance model. Haplotype frequencies are estimated by IQLS (the IQL_b estimator). The OR is calculated from the estimated haplotype frequencies from the IQLS program; it is more accurate than the frequency calculated from founder samples only.
Genotype-to-phenotype associations were also determined by univariate and multivariate logistic regression analysis using SPSS Statistics software (version 19, SPSS Inc., IBM Corporation, NY, USA). This approach enabled us to take into account a dose-response effect (wild type, heterozygote or homozygote) and allowed for the inclusion of additional variables including other genetic polymorphisms with potential confounding effects. Using data from the CC cohort (Supplementary Table 6), the presence of one or more risk markers for the rs4728142 alleles corresponded with a decreased risk of an IBD diagnosis while presence of one or more risk alleles for rs7808907 was associated with an increased risk of IBD. Sample sizes were too small to determine a disease association for any additional haplotype permutation. Additional factors that corresponded to increased odds for CD included one or more of the three NOD2 polymorphisms (R702W, G908R and 1007 fsinsC) and ancestry. The smoking and appendectomy data were incomplete for the controls and were not included in this analysis.
After TLR9 ligand stimulation, cytokine production in primary lymphocytes was quantified and analyzed by one-way analysis of variance after stratification by the alleles for each polymorphism. Each assay was measured in duplicate and 35 independent samples were included. Individuals with two risk markers for the CGGGG indel (P = 0.04) had elevated IL-12-p70 cytokine levels (Figure 2).
Figure 2.
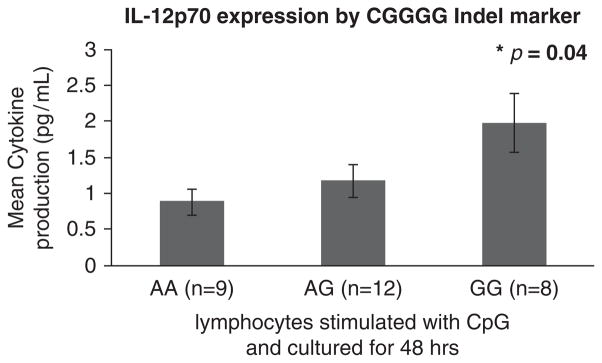
Cytokine production in primary lymphocytes after culture & TLR9 ligand stimulation.
We have identified two previously unreported risk and protective haplotypes of the IRF5 gene for the immune-mediated disease IBD. IRF5 is a complex gene with multiple isoforms and many functional polymorphisms. The CGGGG indel is located in the promoter region 64bp upstream of the first untranslated exon (exon 1A) of IRF5.16 This indel is diallelic and is part of a repetitive DNA stretch that consists of either three or four CGGGG units. The minor allele has four repeats and is predicted to have three so-called GCboxes ‘GGGCGGG’, which are binding sites for the transcription factor SP1.16 The shorter (3 × CGGGG) allele is predicted to have just two of the same binding sites. In a recent study of SLE patients, electrophoretic mobility shift assays (EMSA) demonstrated a higher level of binding of the SP1 protein to the 4 × CGGGG allele of IRF when compared with the 3 × CGGGG allele. The rs10954213 (A/G) allele is located in the 3′ untranslated regions of exon 9. Sequence analysis around this variant illustrates that the SNP lies in a potential polyadenylation site (AAT (G/A) AA).17 In SLE studies the overtransmitted A allele creates a novel polyadenylation site (AATAAA). Samples that are homozygous for the overtransmitted A allele are predicted to preferentially express shorter and more stable transcripts, whereas G/G homozygotes will produce mRNAs with a longer 3′ untranslated region. The remaining three SNPs (rs10488631, rs4728142 and rs7808907) have no known function and fall within the intronic and untranslated regions that may have a role in gene regulation.
In the present study, single-marker analysis showed a significant association of IBD with the CGGGG indel among Jewish ancestry individuals (P = 0.025). Further, several recent publications propose that haplotype-based association has increased power in detection of disease risk.18,19 In our study, two-marker haplotypes comprising the rs4728142, CGGGG indel and rs7808907 polymorphisms were significantly associated with the risk and protection of IBD (Table 3). The overall association was mainly attributable to the two haplotypes that demonstrated an increased frequency in controls compared with the other cases. Importantly, two of the polymorphisms involved span the regulatory region that encompasses the start sites of three alternate and untranslated first exons in IRF5, while the third polymorphism is found within the intronic region between exon 2 and 3. In a multivariate binary logistic regression analysis, we determined the contribution of all the three polymorphisms to the risk of IBD or to CD and UC independently. We found that each polymorphism had a distinct pattern of association (Supplementary Table 2), suggesting a more complex relationship of the polymorphisms. Better characterization of the involved region by genetic sequencing will be pursued. NOD2/CARD15 on chromosome 16 has been identified as a susceptibility gene for CD.20,21 Three major mutations (R702W, G908R, 1007fs) have been identified. In agreement with previous reports, the presence of one or more of the three NOD2 polymorphisms investigated (66% of the CD data set) was significantly associated with an increased risk of developing CD. This was independent of the IRF5 polymorphisms.
In a study first described by Graham et al.14 the rs2004640 T allele creates a 5′ donor splice site allowing for the expression of several unique IRF5 isoforms. These isoforms were different in their transactivation properties. While they are not examined here, some IRF5 haplotypes may correspond to a differential isoform expression that predisposes to risk or protection in IBD. In a study by Krausgruber et al.15 IRF5 was shown to be critical for the late-phase secretion of TNF-α. This role may be critical for the sustained inflammation that results in the organ damage seen in IMIDs. Unmethylated CpG is a ligand that stimulates the activation and nuclear translocation of IRF5 via the TLR9-MyD88 pathway. In mouse bone marrow-derived dendritic cells, IRF5 was required for Fas-induced apoptosis after CpG stimulation.12,22,23 Increased IRF5 levels can result in enhanced cell death that can potentially promote the release of nucleic acids and the development of autoantigens.16
We examined the production of key proinflammatory and anti-inflammatory cytokines in primary lymphocytes from healthy controls after stimulation with CpG-ODN, a TLR9 ligand. We found that homozygous carriage of the CGGG insertion corresponded with significantly higher levels of the IL12-p70 cytokine.
In summary, the IRF5 gene is a risk marker for both CD and UC. In addition, increased IRF5 expression may lead to the overproduction of proinflammatory cytokines along with enhanced cellular apoptosis. Further studies are needed to determine the functional role of the highly expressed isoforms, and the relationship between IRF5 expression and the inflammatory response, particularly at the intestinal level. IRF5 is a putative therapeutic target for IMIDs.
Supplementary Material
Acknowledgments
We thank Evelyn Ng for her skillful technical assistance. We are also greatful to Yashoda Sharma and other members of the NIDDK IBD genetics consortium for assistance with the clinical database. This work was supported by the following grants from the National Institutes of Health: T32 DK 07017 30 and U01 DK 062422-08 S1.
References
- 1.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev. 2008;8:458– 466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 2.Gibson TB, Ng E, Ozminkowski RJ, Wang S, Burton WN, Goetzel RZ, et al. The direct and indirect cost burden of Crohn’s disease and ulcerative colitis. J Occup Environ Med. 2008;50:1261– 1272. doi: 10.1097/JOM.0b013e318181b8ca. [DOI] [PubMed] [Google Scholar]
- 3.Kappelman MD, Rifas-Shiman SL, Porter CQ, Ollendorf DA, Sandler RS, Galanko JA, et al. Direct health care costs of Crohn’s disease and ulcerative colitis in US children and adults. Gastroenterology. 2008;135:1907– 1913. doi: 10.1053/j.gastro.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weng X, Liu L, Barcellos LF, Allison JE, Herrinton LJ. Clustering of inflammatory bowel disease with immune mediated diseases among members of a Northern California-managed care organization. Am J Gastroenterol. 2007;102:1429– 1435. doi: 10.1111/j.1572-0241.2007.01215.x. [DOI] [PubMed] [Google Scholar]
- 5.Franke A, Balschun T, Karlsen TH, Hedderich J, May S, Lu T, et al. Replication of signals from recent studies of Crohn’s disease identifies previously unknown disease loci for ulcerative colitis. Nat Genet. 2008;40:713– 715. doi: 10.1038/ng.148. [DOI] [PubMed] [Google Scholar]
- 6.Liu Y, Helms C, Liao W, Zaba LC, Duan S, Gardner J, et al. A genome-wide association study of psoriasis and psoriatic arthritis identifies new disease loci. PLoS Genet. 2008;4:e1000041. doi: 10.1371/journal.pgen.1000041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rueda B, Orozco G, Raya E, Fernandez-Sueiro JL, Mulero J, Blanco FJ, et al. The IL23R Arg381Gln non-synonymous polymorphism confers susceptibility to ankylosing spondylitis. Ann Rheum Dis. 2008;67:1451– 1454. doi: 10.1136/ard.2007.080283. [DOI] [PubMed] [Google Scholar]
- 8.Kozyrev SV, Lewen S, Reddy PM, Pons-Estel B, Witte T, Junker P, et al. Structural insertion/deletion variation in IRF5 is associated with a risk haplotype and defines the precise IRF5 isoforms expressed in systemic lupus erythematosus. Arthritis Rheum. 2007;56:1234– 1241. doi: 10.1002/art.22497. [DOI] [PubMed] [Google Scholar]
- 9.Barnes BJ, Moore PA, Pitha PM. Virus-specific activation of a novel interferon regulatory factor, IRF-5, results in the induction of distinct interferon alpha genes. J Biol Chem. 2001;276:23382– 23390. doi: 10.1074/jbc.M101216200. [DOI] [PubMed] [Google Scholar]
- 10.Barnes BJ, Kellum MJ, Field AE, Pitha PM. Multiple regulatory domains of IRF-5 control activation, cellular localization, and induction of chemokines that mediate recruitment of T lymphocytes. Mol Cell Biol. 2002;22:5721– 5740. doi: 10.1128/MCB.22.16.5721-5740.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu G, Mancl ME, Barnes BJ. Signaling through IFN regulatory factor-5 sensitizes p53-deficient tumors to DNA damage-induced apoptosis and cell death. Cancer Res. 2005;65:7403– 7412. doi: 10.1158/0008-5472.CAN-05-0583. [DOI] [PubMed] [Google Scholar]
- 12.Takaoka A, Yanai H, Kondo S, Duncan G, Negishi H, Mizutani T, et al. Integral role of IRF-5 in the gene induction programme activated by Toll-like receptors. Nature. 2005;434:243– 249. doi: 10.1038/nature03308. [DOI] [PubMed] [Google Scholar]
- 13.Dideberg V, Kristjansdottir G, Milani L, Libioulle C, Sigurdsson S, Louis E, et al. An insertion-deletion polymorphism in the interferon regulatory factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Mol Genet. 2007;16:3008– 3016. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 14.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, et al. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550– 555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 15.Krausgruber T, Saliba D, Ryzhakov G, Lanfrancotti A, Blazek K, Udalova IA. IRF5 is required for late-phase TNF secretion by human dendritic cells. Blood. 2010;115:4421– 4430. doi: 10.1182/blood-2010-01-263020. [DOI] [PubMed] [Google Scholar]
- 16.Sigurdsson S, Goring HH, Kristjansdottir G, Milani L, Nordmark G, Sandling JK, et al. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Hum Mol Genet. 2008;17:872– 881. doi: 10.1093/hmg/ddm359. [DOI] [PubMed] [Google Scholar]
- 17.Cunninghame Graham DS, Manku H, Wagner S, Reid J, Timms K, Gutin A, et al. Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Hum Mol Genet. 2007;16:579– 591. doi: 10.1093/hmg/ddl469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shim H, Chun H, Engelman CD, Payseur BA. Genome-wide association studies using single-nucleotide polymorphisms versus haplotypes: an empirical comparison with data from the North American Rheumatoid Arthritis Consortium. BMC Proc. 2009;3(Suppl 7):S35. doi: 10.1186/1753-6561-3-s7-s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kang J, Kugathasan S, Georges M, Zhao H, Cho JH. Improved risk prediction for Crohn’s disease with a multi-locus approach. Hum Mol Genet. 2011;20:2435– 2442. doi: 10.1093/hmg/ddr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hugot JP, Chamaillard M, Zouali H, Lesage S, Cezard JP, Belaiche J, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn’s disease. Nature. 2001;411:599– 603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 21.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, Ramos R, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn’s disease. Nature. 2001;411:603– 606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 22.Barnes BJ, Kellum MJ, Pinder KE, Frisancho JA, Pitha PM. Interferon regulatory factor 5, a novel mediator of cell cycle arrest and cell death. Cancer Res. 2003;63:6424– 6431. [PubMed] [Google Scholar]
- 23.Couzinet A, Tamura K, Chen HM, Nishimura K, Wang Z, Morishita Y, et al. A cell-type-specific requirement for IFN regulatory factor 5 (IRF5) in Fas-induced apoptosis. Proc Natl Acad Sci USA. 2008;105:2556– 2561. doi: 10.1073/pnas.0712295105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jurinke C, Oeth P, van den Boom D. MALDI-TOF mass spectrometry: a versatile tool for high-performance DNA analysis. Mol Biotechnol. 2004;26:147– 164. doi: 10.1385/MB:26:2:147. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, McPeek MS. Incomplete-data quasi-likelihood approach to haplotype-based genetic association studies on related individuals. J Am Stat Assoc. 2009;104:1251– 1260. doi: 10.1198/jasa.2009.tm08507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


