Abstract
The effects of the PufX polypeptide on membrane architecture were investigated by comparing the composition and structures of photosynthetic membranes from PufX+ and PufX− strains of Rhodobacter sphaeroides. We show that this single polypeptide profoundly affects membrane morphology, leading to highly elongated cells containing extended tubular membranes. Purified tubular membranes contain helical arrays composed solely of dimeric RC–LH1–PufX (RC, reaction centre; LH, light harvesting) complexes with apparently open LH1 rings. PufX− cells contain crystalline membranes with a pseudo-hexagonal packing of monomeric core complexes. Analysis of purified complexes by electron microscopy and atomic force microscopy shows that LH1 and PufX form a continuous ring of protein around each RC. A model of the tubular membrane is presented with PufX located adjacent to the stained region created by a vacant LH1β. This arrangement, coupled with a flexible ring, would give the RC QB site transient access to the interstices in the lattice, which might be of functional importance. We discuss the implications of our data for the export of quinol from the RC, for eventual reduction of the cytochrome bc1 complex.
Keywords: 2D crystals, cytochrome bc 1 complex, light harvesting complex, reaction centre, photosynthesis
Introduction
The detailed visualisation of the intact machinery of a biological membrane presents a major challenge in structural biology. Exploration of the machinery involved in the conversion of light energy to proton-motive force can be undertaken because the purple photosynthetic bacteria offer a unique system for obtaining and integrating structural and functional information. These bacteria, exemplified by Rhodobacter sphaeroides, require only four protein–pigment complexes for the transduction of light energy: a peripheral light-harvesting complex (LH2), a central light-harvesting apparatus (LH1), which complexes with the photochemical reaction centre (RC), and a proton translocating cytochrome bc1 complex.
The work described in this paper builds upon models for the individual components of this machinery obtained from the application of electron cryomicroscopy (cryoEM) and X-ray crystallography. The RC was the first membrane protein structure determined by X-ray crystallography (Deisenhofer et al, 1985; Allen et al, 1987). A similar level of structural detail is available for the LH2 complexes from Rhodopseudomonas acidophila and Rhodospirillum molischianum (McDermott et al, 1995; Koepke et al, 1996), as is a 6 Å resolution projection map of the Rb. sphaeroides complex (Walz et al, 1998). An ∼8.5 Å cryoEM projection of the RC–LH1 complex shows that it consists of a 115 Å LH1 ring completely enclosing the RC, and is composed of 16 subunits in Rhodospirillum rubrum (Karrasch et al, 1995; Jamieson et al, 2002; Qian et al, 2003). Each subunit of LH1 consists of one α and one β polypeptide, and two bacteriochlorophylls (Bchl). This complete encirclement of the RC raises the question of how quinol molecules are exported from the RC to the quinone pool, prior to reducing the cytochrome bc1 complex. The observation that the LH1 ring can adopt both circular and elliptical forms was taken as evidence of its flexibility; this might allow ‘breathing' motions to open the ring transiently and release quinol (Karrasch et al, 1995; Jamieson et al, 2002; Fotiadis et al, 2004).
The discovery that bacteria such as Rb. sphaeroides and Rhodobacter capsulatus possess an extra polypeptide, PufX, adds another layer of complexity to the issue of quinol export from the RC. This protein is required for photosynthetic growth in these species (Farchaus et al, 1992; Lilburn et al, 1992; McGlynn et al, 1994; Barz et al, 1995a, 1995b) and was proposed to form part of the LH1 ring, acting as a portal for quinol export (Cogdell et al, 1996). PufX is proposed to have a single transmembrane span, and has a cytoplasmically exposed N-terminus (Pugh et al, 1998). It associates with the LH1 α polypeptide in vitro (Recchia et al, 1998), although its association with the RC appears to precede encirclement of the RC by LH1 (Pugh et al, 1998). Very recently, an X-ray crystal structure of an RC–LH1 complex from Rhodopseudomonas palustris at 4.8 Å resolution has shown that the RC is surrounded by 15 LH1 subunits, as well as a PufX-like polypeptide (Roszak et al, 2003).
This wealth of structural data on the individual complexes invites investigation into the next level of complexity – the supramolecular organisation of the individual complexes within a functional photosynthetic membrane. Fortunately, there are several examples of the natural crystallinity of such membranes in the photosynthetic bacteria, which allow investigation by electron microscopy (EM) and atomic force microscopy (AFM) (Miller, 1982; Stark et al, 1984; Jungas et al, 1999; Scheuring et al, 2003). In each case, the peripheral LH2 complex was absent, either naturally or as a consequence of mutation. It has been known for a long time that although bacteria such as Rb. sphaeroides normally synthesise spherical intracytoplasmic membranes (ICMs), tubular membranes are formed in the absence of LH2 (Hunter et al, 1988; Golecki and Heinrich, 1991; Golecki et al, 1991). These membranes contain an ordered array of proteins when analysed by EM (Sabaty et al, 1994; Jungas et al, 1999; Westerhuis et al, 2002). The data of Jungas et al were obtained on membranes containing RC–LH1–PufX complexes and were therefore especially intriguing, because this was the first structural information on any complex containing PufX. This analysis offered an explanation for how quinones could pass from the RC to the cytochrome bc1 complex, by suggesting that the RC is not surrounded by a complete ring of protein and that core complexes may be organised in dimeric complexes consisting of a partially open ‘S'-shaped LH1 assembly, with an intervening cytochrome bc1 complex. Linear dichroism (LD) showed that PufX was responsible for the ordering of RC–LH1–PufX complexes in the membrane, and that in the absence of PufX the natural crystallinity of the membrane was abolished (Frese et al, 2000).
These remarkable effects of PufX now require further investigation. Specifically within tubular membranes, it is important to establish whether the cytochrome bc1 complex is really associated with the RC–LH1–PufX complex as proposed (Jungas et al, 1999) and whether the RC is only partially enclosed by LH1 and PufX. Secondly, it is important to establish the significance of these ordered membranes in the photosynthetic mechanism. In this paper, we describe the separation of a variety of natural membrane types including the tubular vesicles, and show that specifically for these tubular vesicles there are no detectable levels of cytochrome bc1 complex. Moreover, we use AFM to show that purified and reconstituted RC–LH1–PufX complexes form a continuous ring of polypeptides around the RC, inconsistent with the notion of a permanently open ring, at least in vitro. In addition, this study was augmented by the preparation of membranes lacking PufX. Again, a proportion of these membranes is highly ordered, but they form flat sheets containing monomeric RC–LH1 units, highlighting a role for PufX in promoting the formation of long-range helical arrays of dimeric core complexes.
Results
Cell ultrastructure of PufX+ and PufX−strains
The membrane ultrastructure of LH2− mutants of Rb. sphaeroides was observed in thin cell sections examined by EM. Abundant highly elongated membrane-bound structures were observed in all cells of the PufX+ strain, along with a remarkable elongation of the cells themselves (Figure 1A). The membranes appeared similar to those observed previously (Hunter et al, 1988; Golecki et al, 1991; Sabaty et al, 1994) and were up to ∼2 μm in length and ∼0.1 μm in diameter. PufX− cells contained large membranous structures up to 0.4 μm in diameter, but these were never elongated (Figure 1B). The PufX polypeptide therefore appears to be an absolute requirement for the formation of elongated membranes in LH2− Rb. sphaeroides.
Figure 1.
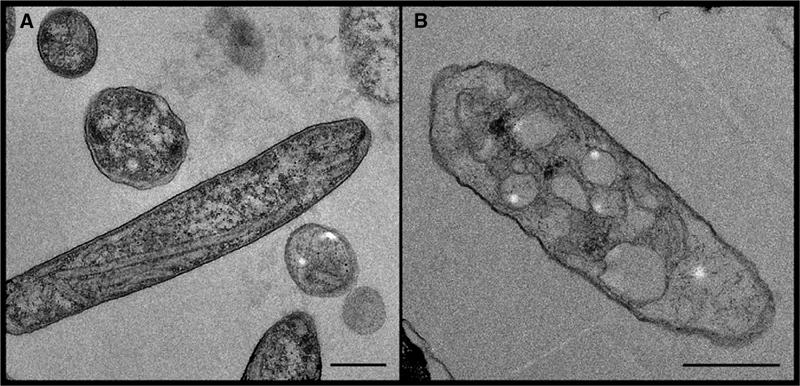
Electron micrographs of sections of (A) PufX+ (strain DD13/G1[RKEH]) and (B) PufX− (DD13/G1[RKEHX−]) cells of Rb. sphaeroides, both lacking the peripheral LH2 complex. Scale bars: 0.5 μm.
Purification of ICMs
Membrane fragments were initially separated on 15/40/50% w/w sucrose gradients. For both strains, major coloured membrane bands were found at the 15/40% interface (bands 1) and 40/50% interface (bands 2) (Figures 2A and D). Electron micrographs of band 1 from each strain revealed small vesicles (Figures 2B and E). These ICM vesicles had a maximum diameter of ∼0.4 μm. Similar ICM vesicles were found in bands 2 (Figures 2C and F). However, in addition, for band 2 from PufX+ cells, we observed a high proportion of elongated tubules (Figure 2C). For band 2 from PufX− cells, we found that the average size of the vesicles was higher than for band 1, up to a maximum of ∼1 μm in diameter, with many of these having the appearance of ordered 2D arrays of protein and lipid (Figure 2F). These large ordered arrays were often opened up into single-layered sheets. No tubular vesicles were found in the PufX− strain.
Figure 2.
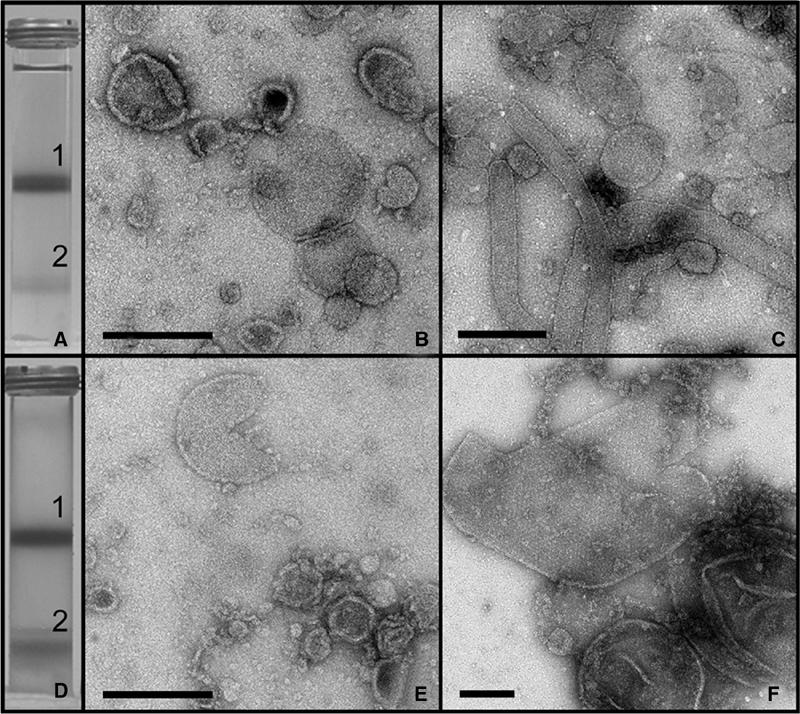
Sucrose density gradient fractionation of photosynthetic membranes. (A) Result of centrifugation of cell extract from the PufX+ strain on a 15/40/50% w/w sucrose gradient. (B) Negatively stained sample from band 1 recovered from the 15/40% interface in (A). Scale bar: 0.2 μm. (C) Negatively stained sample from band 2 recovered from the 40/50% interface in (A). Scale bar: 0.5 μm. (D) Centrifugation of cell extract from the PufX− strain on a 15/40/50% w/w sucrose gradient. (E) Negatively stained sample from band 1 recovered from the 15/40% interface in (D). Scale bar: 0.2 μm. (F) Negatively stained sample from band 2 recovered from the 40/50% interface in (D). Scale bar: 0.2 μm.
The results of SDS–PAGE analysis of bands 1 and 2 from the PufX+ gradient are shown in Figure 3A. This silver-stained gel reveals proteins running close to the molecular weights expected for the RC L, M and H subunits (∼22, ∼25 and ∼30 kDa, respectively). We probed these fractions with antibodies generated against a preparation of subunit IV of the cytochrome bc1 complex. This preparation contained small amounts of other components of the complex and these are also detectable in Western blots as shown in Figure 3B. These blots show that PufX+ membranes from bands 1 and 2 contain the cytochrome bc1 complex (subunit IV at ∼15 kDa, the Rieske Fe–S protein at ∼23 kDa, and cytochrome c1 at ∼31 kDa). Similar silver staining and Western blot data were obtained for bands 1 and 2 from PufX− cells (data not shown).
Figure 3.
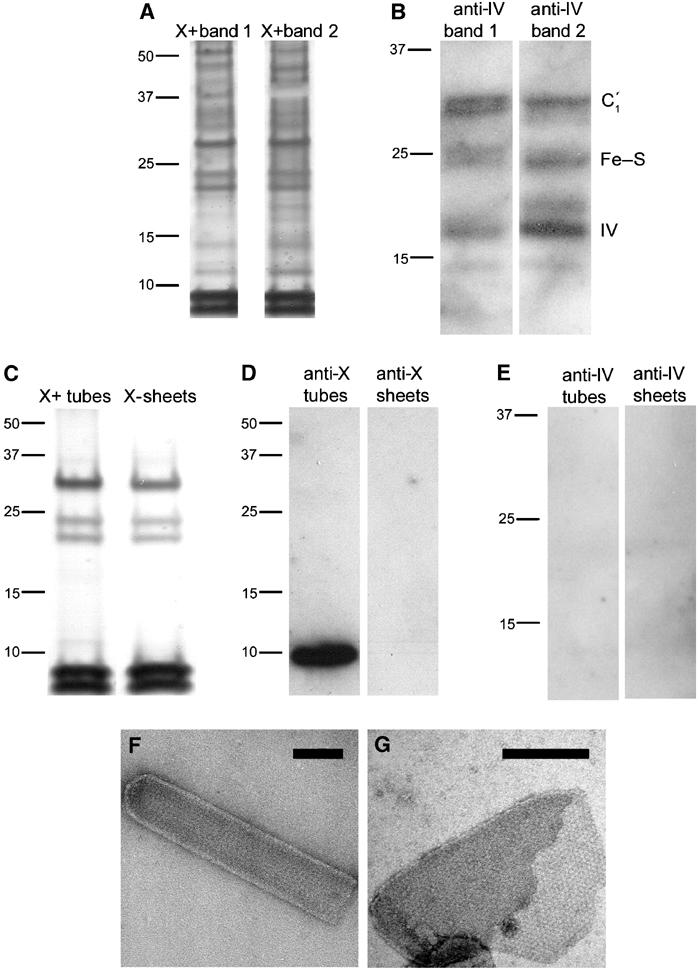
(A) Silver-stained SDS–PAGE of crude PufX+ membrane fractions from the first centrifugation step (see Figure 2A). Left lane, band 1; right lane, band 2. Molecular weights are marked in kDa. (B) Western blot analysis of membrane fractions using antibodies directed against subunit IV of the cytochrome bc1 complex. Left lane, band 1; right lane, band 2 from the first centrifugation of PufX+ membranes. (C) Silver-stained SDS gel electrophoresis of crystalline membrane fractions isolated in the second centrifugation step. Left lane, PufX+ tubes; right lane, PufX− sheets. (D) Corresponding Western blot analysis using antibodies directed against PufX. Left lane, PufX+ tubes; right lane, PufX− sheets. (E) Western blot analysis of PufX+ tubes (left lane) and PufX− sheets (right lane) using antibodies directed against subunit IV of the cytochrome bc1 complex. (F) Negatively stained PufX+ tube isolated in the second centrifugation step. Scale bar: 0.1 μm. (G) Negatively stained PufX− sheet isolated in the second centrifugation step. Scale bar: 0.2 μm.
Purification of native crystalline vesicles and tubes
In view of the demonstration of different membrane types in the band 2 samples (Figures 2C and F), we subjected these to a second centrifugation step on a continuous sucrose gradient of 30–50%. The PufX+ and PufX− samples each yielded two distinct, coloured membrane bands (data not shown). Silver-stained SDS–PAGE indicated that the upper bands in the second gradient contained several proteins, including those of the RC, LH1 and cytochrome bc1 complexes (data not shown). Figure 3C (left) shows that, in contrast, the silver stain detected only five polypeptides in membranes from the lower PufX+ band, arising from RC and LH1 complexes; PufX is not detected by silver staining. EM confirmed that this lower band consisted only of tubular membranes; an example is shown in Figure 3F. The analogous lower band from the PufX− experiment gave an almost identical silver-staining pattern (Figure 3C, right). EM showed this band to contain only large crystalline vesicles and sheets as displayed in Figure 3G. The presence of PufX in the tubes from the PufX+ strain and its absence in sheets from the PufX− strain was confirmed by Western blotting using an anti-PufX antibody (Figure 3D). We also probed the pure PufX+ tubes and PufX− sheets for the cytochrome bc1 complex in a Western blot analysis. Figure 3E shows that purified tubes and sheets from the second sucrose gradients contain no detectable cytochrome bc1 complex.
The membranes in this study were prepared from cells grown semiaerobically in the dark, to permit a direct comparison between PufX+ and PufX− cells, given that the latter strain is not capable of photosynthetic growth. PufX+ cells were also grown photosynthetically, and their tubular membranes were analysed by silver-stain SDS–PAGE and EM. These membranes were essentially identical to those from semiaerobically grown cells, and they had no detectable cytochrome bc1 complex (data not shown).
Absorbance spectra were recorded in order to provide further evidence for the enrichment of RC–LH1 complexes and the lack of the cytochrome bc1 complex in the purified PufX+ tube and PufX− sheet fractions. Figure 4A shows spectra comparing membranes from the first sucrose gradient in Figures 2A and D and from the second gradient, normalised according to protein concentration. The effect of purification of the membrane is clearly seen, with the tubes and sheets in Figure 4A(a) and (b) enriched in LH1 complexes, which absorb at ∼875 nm. The spectra in Figure 4B are reduced minus oxidised difference spectra, which display peaks at ∼550 and ∼560 nm, arising from c- and b-type cytochromes, respectively. These spectra do not discriminate between the many types of cytochrome that exist in these cells, but they do provide a qualitative guide to the amount of cytochrome bc1 complex in each fraction. It is clear that the relatively crude membrane fractions corresponding to bands 1 and 2 from the first sucrose gradients contain the highest levels of cytochrome (Figure 4B, spectra c, e), and that the purified PufX+ tubes (after the second centrifugation) (spectrum a) and PufX− sheets (spectrum b) contain no detectable cytochrome, consistent with the Western blotting data.
Figure 4.
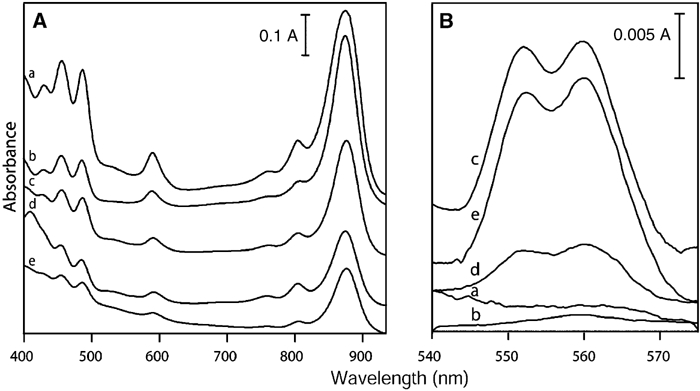
Absorbance spectra of membrane fractions. (A) Absorbance spectra normalised to equivalent total protein concentration. (B) Difference spectra of reduced minus oxidised samples, normalised to equivalent sample absorbance at 875 nm. (a) Purified PufX+ tubes; (b) purified PufX− sheets; (c) band 1 of first PufX+ centrifugation; (d) upper band of second PufX+ centrifugation step; (e) band 2 of first PufX+ centrifugation.
Electron crystallography of naturally crystalline PufX+ tubes and PufX− sheets
The natural tubular membranes (Figure 3F) isolated from semiaerobically grown cells of the PufX+ strain and embedded in negative stain were suitable for standard crystallographic image processing. Tube diameters varied from 80 to 115 nm with a mean of ∼95 nm. Lengths varied from 0.5 to 2 μm. Fourier analysis (Figure 5C) indicated upper and lower crystalline layers of the tubes, with mean unit cell parameters of a=125±3 Å, b=195±5 Å, γ=106±1°. Similar unit cell parameters were determined for tubular membranes from photosynthetically grown cells. Comparison of phases of predicted symmetry-related Fourier terms for the semiaerobic membrane sample (Valpuesta et al, 1994) showed the data at this resolution to be consistent with p2 symmetry (Table I). A projection map of the PufX+ crystal averaged from five images is shown in Figures 5A and B with p2 symmetry imposed. The significant features are the dimeric ‘S'-shaped stain-excluding density, enclosing two central densities. Each ‘C'-shaped unit within this ‘S' shape forms an apparent ellipse. Given that even the highly sensitive silver-stain method (Figure 3C) detects only the three RC subunits and the LH1 α and β polypeptides (PufX is not detected by this stain), we conclude that the densities in the EM projection map can be attributed solely to the RC–LH1–PufX complex.
Figure 5.
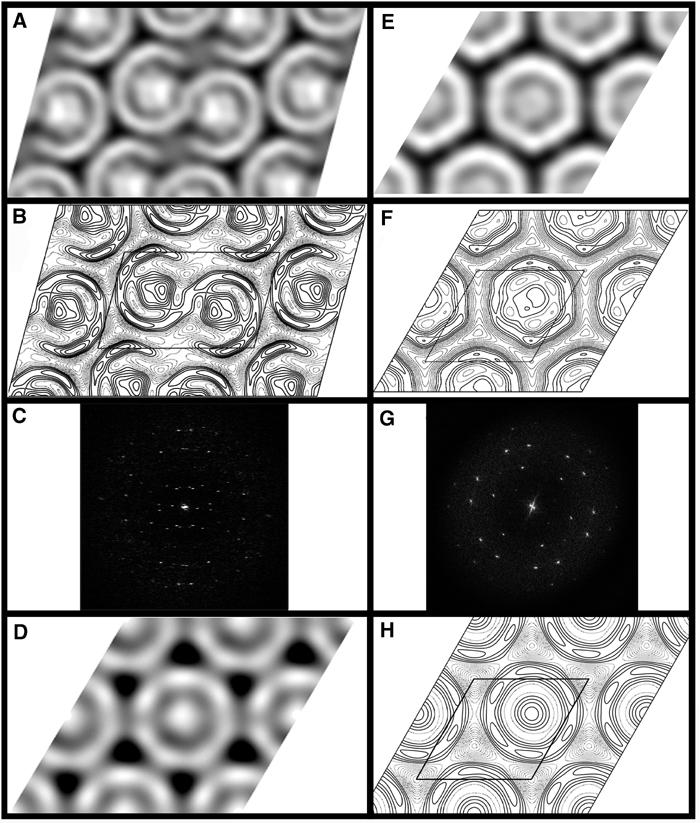
(A) Grey-level representation of five merged images of PufX+ tubes with p2 symmetry. Black represents the densest staining. (B) Contour map of the density in (A). Dashed contours represent density above the mean, and solid contours represent density below the mean. Contour interval is at one-third of the root-mean-square density. One unit cell is outlined, a=125 Å, b=195 Å, γ=106°. (C) Fourier transform from a PufX+ tubular crystal, showing lattices arising from both surfaces of the collapsed tubules. The edge of the box corresponds to 0.04 Å−1. (D) Grey-level representation of a Fourier-filtered image of a 2D crystal grown from purified RC–LH1–PufX complexes. (E) Grey-level representation of four merged images of PufX− crystalline sheets with p1 symmetry. (F) Contour map of the density in (E). One unit cell is outlined, a=130 Å, b=132 Å, γ=120°. (G) Fourier transform from a PufX− crystalline sheet. The edge of the box corresponds to 0.04 Å−1. (H) Contour map of the density in (D). One unit cell is outlined, a≈b≈142 Å, γ=120°.
Table 1.
Mean p2 phase residuals for five merged images of PufX+ crystals in resolution shells, for all spots with IQ>9
| Resolution range (Å) | No. of spots | Mean phase residual (°) (random=45°) | Standard error of mean (°) |
|---|---|---|---|
| ∞-32 | 39 | 17.8 | 3.4 |
| 32-22 | 34 | 15.9 | 3.2 |
| ∞-22 | 73 | 16.9 | 2.3 |
It is essential to compare this density map with one obtained from naturally crystalline membranes isolated from PufX− cells, in order to assign the features and organisation imposed by PufX. Figure 5G shows a Fourier transform from one image of a PufX− sheet. At the resolution attained, we have not been able to distinguish between an oblique primitive or a larger unit cell (e.g. orthorhombic), the RC–LH1 complexes simply appearing as stain-embedded quasi-hexagonally close-packed rings. Figures 5E and F show a map averaged with p1 symmetry from four independent images (a=130±1 Å, b=132±1 Å, γ=120±0.3°). The stain-excluding densities appear as independent intact ring structures, each enclosing a central RC. It should be noted that at this resolution no significance should be attributed to density variations of one or two contour levels or to small deviations from the circular symmetry of the LH1 ring. Individual subunits would not be distinguishable at this resolution.
Purification, reconstitution and crystallisation of RC–LH1 complexes
The S-shaped density observed in the projection map of the natural tubes in Figures 5A and B appears to show that there is a gap in the LH1 structure. In order to examine this in a different way, we purified RC–LH1–PufX complexes and reconstituted them into 2D crystals as described previously (Qian et al, 2003). Figures 5D and H show a projection map calculated from the negatively stained 2D crystals. The crystals appear to contain monomeric complexes where the central RC density is fully encircled by the LH1 complex. Western blot analysis confirmed the presence of PufX in these crystals (data not shown). Crystals were tightly packed with complexes and we found no evidence for separate clusters of PufX. Given the very limited resolution, we have assigned a primitive oblique unit cell (a≈b≈142 Å, γ=120°) containing one RC–LH1 unit. Fourier filtered images from these reconstituted crystals showed no evidence of the apparent dimeric units found in the natural PufX+ tubes, nor the gaps in the LH1 ring.
AFM analysis of individual RC–LH1–PufX complexes
The reconstituted 2D crystals were somewhat disordered and we could not exclude the possibility that Fourier filtering of EM images averaged out gaps in the LH1–PufX ring. In order to circumvent this problem, we undertook an AFM analysis of the crystals. This was intended to show the arrangement of the LH1 complex in individual molecules. The advantage of the reconstituted crystals over the natural tubes is that they form large flat areas more amenable to AFM analysis. We have not obtained molecular resolution AFM images of native membrane tubes to date. Figure 6 shows a progression in terms of scanning sequence and magnification. Figure 6A shows the results of an initial scan of the 2D crystal, where little detail is visible, and most notable is the large number of bright features, which correspond to the RC-H subunit, by analogy with data obtained on 2D crystals of the RC–LH1 complex of Rs. rubrum (Fotiadis et al, 2004). This study also highlighted the difficulties of obtaining detailed information on the LH1 complex when the large cytoplasmic surface of the RC-H subunit is present. We found that in contrast to the relatively stable Rs. rubrum complex, the RC-H subunit of the Rb. sphaeroides RC–LH1–PufX complex is easily removed by nanodissection and so successive scans were used to remove this interfering subunit, producing the field of images first in Figure 6B and then in Figure 6C. This final image clearly shows that all the LH1–PufX rings are complete. Other interesting features have been revealed for the first time, including the presence of a major population of LH1 complexes of ∼113 Å diameter and some larger ones of up to ∼134 Å, perhaps corresponding to rings of up to 18 αβBchl2 units.
Figure 6.
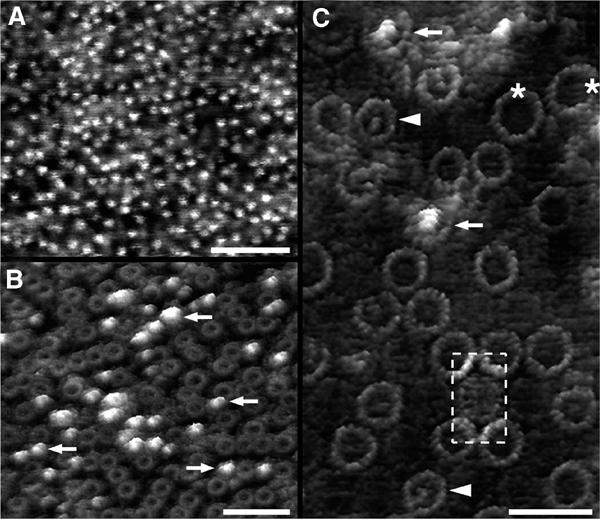
AFM of reconstituted RC–LH1–PufX 2D crystals. (A) Low-magnification topograph of a crystal. The bright spots represent the RC-H subunits protruding from the cytoplasmic side of the complexes. (B) After several scans over the same area, the number of RC-H subunits (arrows) decreased significantly due to displacement by the AFM tip. (C) RC–LH1–PufX complexes imaged at high resolution. Different types of complexes exposing the cytoplasmic side can be discerned: intact complexes with (arrows) and without (arrowheads) the RC-H subunit. The LH1 rings of such complexes had a diameter of ∼113 Å. Occasionally, complexes with larger LH1 rings and depleted of the RC-H subunit (asterisks) could be visualised (LH1 ring diameter: ∼134 Å). The periplasmic surface of the RC–LH1–PufX complex protruded less from the bilayer than the cytoplasmic surface. For better visualisation, the contrast of an RC–LH1 complex exposing the periplasmic side was enhanced (broken box). The topographs in (B) and (C) are shown in relief, tilted by 15°. Scale bars: 50 nm (A and B) and 25 nm (C). Vertical brightness ranges: 3.0 nm (A), 3.5 nm (B) and 4.0 nm (C).
Discussion
It is well established that the PufX polypeptide plays a crucial role in the function and organisation of the components of bacterial photosynthesis (Farchaus et al, 1990; Lilburn et al, 1992; McGlynn et al, 1994; Barz et al, 1995a, 1995b; McGlynn et al, 1996; Frese et al, 2000). However, there have been no detailed reports of a direct visual comparison of the effects of PufX on membrane architecture at a molecular level. In this paper, we have shown that PufX has a dramatic influence on the global organisation of the membranes in which it is inserted, with important implications for the way in which these RC–LH1 complexes interact with each other and how they export quinol.
Effect of PufX on membrane morphology and on the organisation of RC–LH1–PufX complexes
EM of cell sections of LH2− mutants (Figure 1) provides the first clue to the crucial role of PufX in determining the morphology of photosynthetic membranes and the shape of the cell. In the presence of PufX highly elongated tubular membranes are synthesised, whereas in its absence membranes with the appearance of large vesicles are assembled. For both strains, membrane fractionation has yielded two populations of membranes: small ICM vesicles (Figures 2B and E) and large crystalline vesicles (Figures 2C and F). The small ICM vesicles are typical of the ‘chromatophores' commonly described for several species of wild-type purple bacteria (e.g. Schachman et al, 1952; Drews and Golecki, 1995). They contain RC, LH1, cytochrome bc1 complex and a number of other proteins; they were not suitable for detailed microscopic analysis and we are not yet able to analyse the effect of PufX on their structure. However, for the ordered tubular and sheet-like membranes, the EM data in Figures 2C and F show that the presence or absence of PufX is a critical determinant of membrane morphology. This provides a direct visual confirmation that PufX enforces a long-range ordering of at least a proportion of the photosynthetic membrane in vivo, as originally detected by LD (Frese et al, 2000). In the presence of PufX, it is likely that tubes are formed as a natural consequence of a slight tilt of one dimer in relation to the next one along the longitudinal plane. In the absence of PufX, membranes composed of monomeric RC–LH1 complexes form vesicles often opening out to extended flat sheets, reminiscent of those naturally found in Blastochloris viridis, which also lacks a PufX analogue. The characteristics of these membranes are a pseudo-hexagonal close packing of core complexes, each made up of an intact ring of LH1 density surrounding a single RC (Miller, 1982; Stark et al, 1984; Walz and Ghosh, 1997; Stahlberg et al, 1998; Walz et al, 1998; Jamieson et al, 2002; Scheuring et al, 2003).
Projection structure of the RC–LH1–PufX complex in vivo – an open ‘C'-shaped LH1 ring?
The apparent opening of the LH1 ring in the tubes containing RC–LH1–PufX complexes to form a ‘C' shape (Figures 5A and B) must be reconciled with the negative stain EM projection map for the purified RC–LH1–PufX complex in Figures 5D and H and with the AFM data (Figure 6), both of which show a continuous ring of density surrounding the RC. EM and AFM report on different features: projected density averaged over multiple complexes and surface topography of individual complexes, respectively. In our AFM images we are not able to resolve the surfaces of separate α and β polypeptides, and so if just one α was replaced by PufX, it is unlikely that the vacant β position would be visible; the ring would appear intact. By contrast, negative stain EM reports on projected density of stain that has penetrated to varying depths throughout the sample, and not just on upper or lower surfaces. Any ‘notch' left by a vacant β subunit on the outer part of the ring would very likely fill with stain. This could account for the stained ‘gap' (dashed contours) in the LH1 in Figure 5B. AFM shows that reconstituted 2D crystals have individual complexes with cytoplasmic faces in both ‘up' and ‘down' orientations, distributed in a quasi-random fashion. In contrast, these two orientations would have been averaged in our EM analysis, making the LH1 ring appear continuous (Figures 5D and H). We note that an X-ray crystal structure of the RC–LH1 complex of the closely related Rps. palustris indeed shows a closed ring of 15 αβ subunits, one PufX type polypeptide and a single vacant β subunit (Roszak et al, 2003). We have modelled the projection of such a structure at 30 Å resolution and find indeed that a large apparent ‘gap' is seen, despite the RC being fully encircled with protein (data not shown). Moreover, a continuous ring containing PufX (Cogdell et al, 1996) is consistent with the binding of PufX to the LH1α polypeptide (McDermott et al, 1995; Recchia et al, 1998) and its copurification with the RC–LH1 complex (Farchaus et al, 1992; Pugh et al, 1998; Francia et al, 1999). An additional factor that could account for a variation in contrast around the ring in dimers is that the monomers within a pair of RC–LH1–PufX complexes could be mutually tilted towards one another. The capacity of LH and RC complexes to tilt has previously been demonstrated (Walz et al, 1998; Katona et al, 2003).
PufX promotes the formation of dimeric RC–LH1 core complexes in vivo
The projection map of PufX+ tubes in Figures 5A and B appears to consist of pairs of fused, elongated ‘C' shapes, each arising from an LH1 complex. At the resolution achievable, the Fourier phases are fully consistent with p2 symmetry (Table I), with one of the two-fold symmetry axes located at the point of fusion of the two ‘C' shapes, this being the most likely dimer interface. We found no extra protein density lying at this two-fold axis that could be attributed to PufX, although this interface has been proposed as the PufX location for reconstituted RC–LH1–PufX crystals (Scheuring et al, 2004). The striking difference between the projection maps in Figures 5A and E is attributable solely to PufX; this is an in vivo counterpart to the results on detergent-solubilised complexes, which showed that PufX promotes the formation of dimeric core complexes (Francia et al, 1999). Our data are also consistent with EM and AFM studies of membranes from the naturally PufX− bacterium B. viridis, none of which has demonstrated any dimeric particles (Miller, 1982; Scheuring et al, 2003).
Similar membranes were analysed by LD (Frese et al, 2000) to show that PufX induces a specific orientation of the RC in the LH1 ring, as well as the formation of a long-range regular array with the Qy transition of the special pair Bchls of the RC lying at an angle of ∼80° with respect to the long axis of the tubular membrane. The diagram in Figure 7 shows a hypothetical model for the arrangement of two monomeric RC–LH1–PufX core complexes, positioned according to the LD data (Frese et al, 2000), with the Qy transition dipoles (red lines) lying at ∼80° to the tube long axis (arrow). We have chosen one of four possible ways of achieving this orientation relative to the tube axis. Firstly, we note that the long axis of the RC lies in the same direction as the long axis of the LH1 ellipse, consistent with the analyses of elliptical core complexes by EM (Jamieson et al, 2002) and AFM (Fotiadis et al, 2004). Secondly, although we have already stated that there is complete continuity in protein surrounding the RC, since PufX binds to LH1α (Recchia et al, 1998), it is likely that there would be a corresponding discontinuity in the outer LH1β ring (Roszak et al, 2003). We suggest that PufX is located adjacent to the apparent opening in each ‘C' shape where there is heavier staining in place of the vacant LH1β. This arrangement, coupled with a flexible ring, would give the RC QB site transient access to the interstices in the lattice, which might be functionally important.
Figure 7.
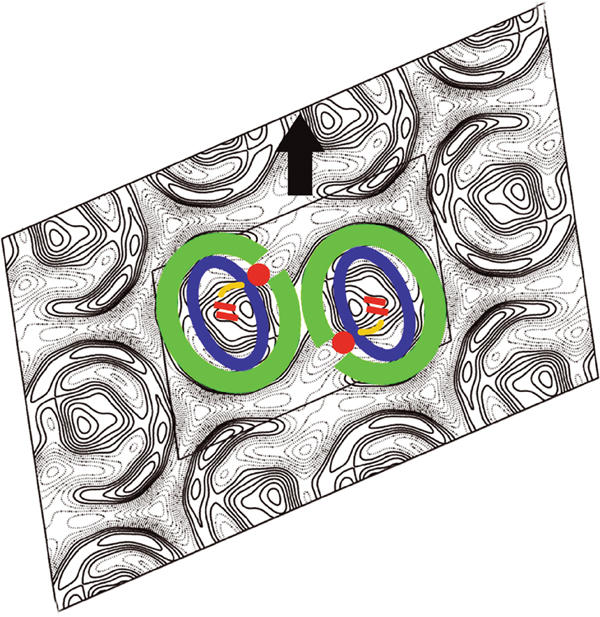
Hypothetical schematic representation of the components of the RC–LH1–PufX dimer in PufX+ tubes. The arrow shows the typical direction of the longitudinal tube axis relative to the unit cell. Green represents an elliptical ring of LH1αβ dimers. The location of the PufX polypeptide is suggested by the red circle, interacting with both the RC (approximated by the blue ellipse) and the inner ring of LH1α subunits. The Qy transition dipoles of the special pair bacteriochlorophylls are indicated in red, lying at 80° to the tube axis (Frese et al, 2000). The approximate location of the QB site is indicated in yellow.
Existence of an RC–LH1–PufX–cytochrome bc1 supercomplex within tubular membranes
It has been proposed that the cytochrome bc1 complex forms a stoichiometric association with the RC–LH1 dimer in tubular membranes (Jungas et al, 1999; Verméglio and Joliot, 2002). Our refined tube purification procedure gave us the opportunity to test rigorously for the presence of the cytochrome bc1 complex without any significant contaminating material. We tested for the cytochrome complex using silver-stain SDS–PAGE (Figure 3C), reduced–oxidised difference absorbance spectroscopy (Figure 4B) and Western blot analysis (Figure 3B), and found no detectable amounts, although the cytochrome bc1 complex was found in other membrane fractions, as expected. Our data point to tubes containing only RC–LH1–PufX complexes, irrespective of whether the cells were grown in the dark under oxygen limitation, or grown photosynthetically in the light. This raises the question of the location of the cytochrome bc1 complex; even though it is not present in the tubular membranes, abundant quantities of this complex are found in both membrane fractions in the first sucrose gradient (e.g. Figures 3B and 4B). It should be emphasised that our data do not prove that RC–LH1–PufX–cytochrome bc1 complexes do not exist, only that they are not to be found in tubular membranes.
Implications for the growth and physiology of purple bacteria
Species such as Rs. rubrum and B. viridis lack a PufX analogue that facilitates quinol export from the RC. In such cases, it has been suggested that ‘breathing' motions in the LH1 ring allow quinol passage because the ring is flexible (Karrasch et al, 1995; Jamieson et al, 2002; Fotiadis et al, 2004). One critical factor for electron transfer is the state of reduction of the quinone pool, and its consequent ability to drive the Q-cycle within the cytochrome bc1 complex (Crofts et al, 1983). Under steady-state conditions, the redox balance of the quinone pool will depend on the provision of quinol by the RC and on its depletion by oxidases and other complexes downstream of the cytochrome bc1 complex (Verméglio et al, 1995). Perhaps, in comparison with bacteria such as Rs. rubrum, Rb. sphaeroides requires an enhanced provision of quinol for the Q-pool because it is also depleted more readily, which in turn necessitates PufX.
The presence of at least two distinct in vivo membrane architectures – crystalline membranes with long-range order, and smaller non-crystalline vesicles – raises a number of questions. For example, what is the physiological role of each membrane type? How is it possible for arrays of RC–LH1–PufX complexes in tubes to reduce the Q-pool, and eventually the cytochrome bc1 complexes, in a remote location? Given that measurements of electron transfer kinetics have been carried out on relatively crude membrane fractions without purification on successive rounds of sucrose gradients, how can differing kinetic models of electron transfer be reconciled with each other and with our structural data? The fact that many kinetic measurements have been carried out on LH2-containing membranes, which have a different morphology, complicates matters still further.
A supramolecular organisation has been proposed for the Rb. sphaeroides photosynthetic membrane (Verméglio and Joliot, 2002), based on kinetic evidence (Joliot et al, 1989, 1993, 1996) and the ordering of complexes containing two RC–LH1 units in LH2− strains (Jungas et al, 1999). Notably, one ‘supercomplex' model proposes a strict stoichiometry of two RCs to one cytochrome bc1 complex, but Crofts and co-workers note rapid electron transfer between RC and the cytochrome bc1 complex in strains presenting different stoichiometries (Crofts et al, 1998; Crofts, 2000a, 2000b). Crofts et al explain their kinetic data with a model in which local domains of photosynthetic complexes are distributed between small chromatophore vesicles. The concept of ‘supercomplexes' remains a matter of controversy (Verméglio and Joliot, 1999; Crofts, 2000a, 2000b; Verméglio and Joliot, 2000). Nevertheless, the present work dispels the idea that LH2− tubes contain RC–LH1–cytochrome bc1 supercomplexes, but does not rule out supercomplexes in the other membrane fractions. It should also be borne in mind that present notions of supercomplexes are largely confined to extrapolations from electron transfer kinetics, with limitations imposed by the preparation of uncharacterised membranes, or by comparisons of data from membranes and whole cells (Joliot et al, 1997; Crofts et al, 1998).
The absence of cytochrome bc1 complexes from tubular regions of invaginated membranes within the cell could preclude the transfer of quinol from these particular RC–LH1–PufX complexes to cytochrome bc1 complexes in other membrane types or locations. An analysis of a related problem, the diffusion of plastoquinone from Photosystem II in plants to the cytochrome b6f complex, concludes that quinone diffusion can be greatly limited within the highly protein obstructed thylakoid membrane (Tremmel et al, 2003). It can be calculated that the diffusion time for a quinone in a lipid-only tubular membrane 50 × 1000 nm in size is 0.5 ms, and that this time could be increased by several orders of magnitude in a protein-rich membrane. Given the known time of photoreduction of the cytochrome bc1 complex (∼0.5 ms; Crofts and Wraight, 1983), it seems unlikely that RC–LH1–PufX complexes in tubes are able to photoreduce cytochrome bc1 complexes on physiologically relevant timescales.
Conclusion
We have shown for the first time that PufX causes dimerisation of RC–LH1 complexes in vivo. The association of RC–LH1–PufX complexes into dimers has a profound effect on membrane morphology, leading to the synthesis of long helical tubes consisting entirely of dimeric RC–LH1–PufX complexes. We have been able to propose a location for the QB site within the dimer that could allow access to interstices in packed membranes at a point where PufX is inserted in the LH1 ring. The other membrane fractions remain more challenging for structural analysis, and future experiments should address the organisation of RC–LH1–PufX complexes in these membranes and their interactions with other proteins such as the cytochrome bc1 complex.
Materials and methods
Mutant preparation
The DD13/G1 double deletion strain (LH2− LH1− RC−), which synthesises the green carotenoid neurosporene (Jones et al, 1992), was complemented with RC–LH1+/−PufX genes as described (McGlynn et al, 1996). Transconjugants were grown semiaerobically in the dark on M22+ medium supplemented with neomycin (20 μg/ml) and tetracycline (1 μg/ml).
Cell growth and sectioning
Selected mutants were grown under oxygen-limited conditions in conical flasks in the dark, by shaking at 80% capacity at 150 rpm at 30°C. Cultures were subjected to an extra 48 h growth period upon reaching a cell density of ∼1.4 absorbance units at 680 nm. Photosynthetic cultures were grown photoheterotrophically in M22+ medium supplemented with neomycin (20 μg/ml) at low light intensity (2 × 60 W tungsten bulbs, ∼40 μE/m2/s1) for 120 h at 30°C. Cells were harvested by centrifugation at 4000 g for 25 min, washed twice in 1 mM Tris, 2 mM EDTA, pH 7.5, resuspended and frozen in membrane buffer (10 mM Tris, 20 mM EDTA, pH 7.5). Detailed methods for cell sectioning are described in the Supplementary material.
Membrane fractionation
Cells were supplemented with 500 μl of protease inhibitor cocktail per 25 ml of cells along with a few crystals of DNAse I prior to passage through a French press thrice at 3000 psi. Whole cells and cell wall material were pelleted at 10 000 g for 10 min. The supernatant was placed onto a 15/40/50% w/w sucrose/membrane buffer density gradient and spun for 4 h at 100 000 g. Band 2 (Figures 2A and D) was diluted 10-fold in membrane buffer and centrifuged for 4 h at 100 000 g. The pellet was resuspended overnight in 50 mM HEPES at pH 8 containing 0.03% α-dodecyl maltoside (DDM), gently homogenised and loaded onto a 30–50% w/w continuous sucrose gradient in 50 mM HEPES at pH 8 containing 0.03% DDM. Samples were centrifuged for 20 h at 200 000 g. Pigmented bands were harvested and frozen.
Purification of RC–LH1–PufX complexes
DHPC (1,2-diheptanoyl-sn-glycero-3-phosphocholine) was added to membranes in the dark (absorbance at 880 nm of 60, 1 cm path length) with gentle stirring at 4°C for 30 min. The solubilised material was applied to a continuous sucrose gradient (15–40% sucrose in buffer A) and spun at 150 000 g for 15 h at 4°C. The RC–LH1–PufX band was harvested and applied to a DEAE anion exchange column (20 × 70 mm). This was eluted with a gradient of buffer A (10 mM Tris, 1 mM EDTA, 3 mM DHPC, pH 7.9) and buffer B (300 mM NaCl in buffer A). The main peak, which contains the RC–LH1–PufX complex, corresponds to 250 mM NaCl. The best fractions judged from the 880:280 nm absorbance ratio (>1.8) were concentrated and loaded on a gel filtration column (Hiload 16/60, Superdex 200 preparative grade). The column was washed with buffer C (50 mM NaCl in buffer A). Fractions with an 880:280 absorbance ratio of more than 1.9 were pooled and used for crystallisation. 2D crystals were reconstituted with Escherichia coli lipids at a lipid/protein ratio from 0.6 to 1.0 (w/w) as described previously (Qian et al, 2003). Samples were applied to glow discharged carbon-coated grids and stained with 0.75% w/v uranyl formate. Micrographs were recorded at 100 kV on a Philips CM100 EM at ∼52 000 magnification on Kodak SO-163 film at an underfocus of ∼5000 Å. Films were digitised on a ZEISS SCAI scanner at a step size corresponding to 7 Å at the specimen.
Image processing
Images were processed using the MRC software (Crowther et al, 1996). Image areas were extracted and floated into square arrays of 1024 pixels on a side. In general, reflections in Fourier transforms were indexed on two separate lattices from the bottom and top layers of the collapsed tubules. For vesicular crystals from PufX− strains, Fourier transforms generally only showed one lattice. Structure factors were not corrected for the effects of the phase and amplitude contrast transfer functions.
Atomic force microscopy
A stock solution of 2D RC–LH1–PufX crystals (0.5 mg/ml protein in 10 mM Tris–HCl, 1 mM EDTA, pH 7.9) was diluted 30-fold in 20 mM Tris–HCl, 150 mM KCl, 25 mM MgCl2, pH 7.8 (imaging buffer), and adsorbed for 20–30 min on freshly cleaved muscovite mica. The sample was gently washed with imaging buffer. Images were recorded with a Nanoscope Multimode microscope equipped with an infrared laser head, fluid cell, and oxide-sharpened silicon nitride cantilevers of 100 and 200 μm length, and nominal spring constants of 0.08 and 0.06 N/m from Olympus Optical Co. and Digital Instruments, respectively. Topographs were acquired in contact mode at minimal loading forces (⩽100 pN). Trace and retrace signals were recorded simultaneously at line frequencies ranging between 4.1 and 5.1 Hz. Perspective views of raw data were prepared using the SXM program (University of Liverpool, UK).
Supplementary Material
Materials and methods
Acknowledgments
We would like to thank Dr R Frese for stimulating discussions, J Procter for assistance with thin-section EM, and Professor R Niederman for the subunit IV antibodies. AE acknowledges support by the Swiss National Research Foundation, the ME Müller Foundation, the Swiss National Center of Competence in Research (NCCR) ‘Structural Biology' and the NCCR ‘Nanoscale Science'. This work was funded by the UK Biotechnology and Biological Sciences Research Council. The Krebs Institute Structural Studies Group is a member of the BBSRC North of England Structural Biology Centre.
References
- Allen JP, Feher G, Yeates TO, Komiya H, Rees DC (1987) Structure of the reaction center from Rhodobacter sphaeroides R-26: the cofactors. Proc Natl Acad Sci USA 84: 5730–5734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barz WP, Francia F, Venturoli G, Melandri BA, Verméglio A, Oesterhelt D (1995a) Role of PufX protein in photosynthetic growth of Rhodobacter sphaeroides. 1. PufX is required for efficient light-driven electron-transfer and photophosphorylation under anaerobic conditions. Biochemistry 34: 15235–15247 [DOI] [PubMed] [Google Scholar]
- Barz WP, Verméglio A, Francia F, Venturoli G, Melandri BA, Oesterhelt D (1995b) Role of the PufX protein in photosynthetic growth of Rhodobacter sphaeroides. 2. PufX is required for efficient ubiquinone/ubiquinol exchange between the reaction center QB site and the cytochrome bc1 complex. Biochemistry 34: 15248–15258 [DOI] [PubMed] [Google Scholar]
- Cogdell RJ, Fyfe PK, Barrett SJ, Prince SM, Freer AA, Isaacs NW, McGlynn P, Hunter CN (1996) The purple bacterial photosynthetic unit. Photosynth Res 48: 55–63 [DOI] [PubMed] [Google Scholar]
- Crofts A, Guergova-Kuras M, Hong SJ (1998) Chromatophore heterogeneity explains phenomena seen in Rhodobacter sphaeroides previously attributed to supercomplexes. Photosynth Res 55: 357–362 [Google Scholar]
- Crofts AR (2000a) Photosynthesis in Rhodobacter sphaeroides. Trends Microbiol 8: 105–106 [DOI] [PubMed] [Google Scholar]
- Crofts AR (2000b) Photosynthesis in Rhodobacter sphaeroides—response. Trends Microbiol 8: 107–108 [DOI] [PubMed] [Google Scholar]
- Crofts AR, Meinhardt SW, Jones KR, Snozzi M (1983) The role of the quinone pool in the cyclic electron transfer chain of Rhodopseudomonas sphaeroides—a modified Q-cycle mechanism. Biochim Biophys Acta 723: 202–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crofts AR, Wraight CA (1983) The electrochemical domain of photosynthesis. Biochim Biophys Acta 726: 149–185 [Google Scholar]
- Crowther RA, Henderson R, Smith JM (1996) MRC image processing programs. J Struct Biol 116: 9–16 [DOI] [PubMed] [Google Scholar]
- Deisenhofer J, Epp O, Miki K, Huber R, Michel H (1985) X-ray structure analysis at 3 Å resolution of a membrane protein complex: folding of the protein subunits in the photosynthetic reaction centre from Rhodopseudomonas viridis. Nature 318: 618–624 [DOI] [PubMed] [Google Scholar]
- Drews G, Golecki JR (1995) Structure, molecular organization and biosynthesis of membranes of purple bacteria. In Anoxygenic Photosynthetic Bacteria, Bauer, CE (ed.), Vol. 2, pp 231–257. Dordrecht, Boston, London: Kluwer Academic Publishers [Google Scholar]
- Farchaus JW, Barz WP, Grunberg H, Oesterhelt D (1992) Studies on the expression of the pufX polypeptide and its requirement for photoheterotrophic growth in Rhodobacter sphaeroides. EMBO J 11: 2779–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farchaus JW, Gruenberg H, Oesterhelt D (1990) Complementation of a reaction center-deficient Rhodobacter sphaeroides pufLMX deletion strain in trans with pufBALM does not restore the photosynthesis-positive phenotype. J Bacteriol 172: 977–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotiadis D, Qian P, Philippsen A, Bullough PA, Engel A, Hunter CN (2004) Structural analysis of the RC-LH1 photosynthetic core complex of Rhodospirillum rubrum using atomic force microscopy. J Biol Chem 279: 2063–2068 [DOI] [PubMed] [Google Scholar]
- Francia F, Wang J, Venturoli G, Melandri BA, Barz WP, Oesterhelt D (1999) The reaction center-LH1 antenna complex of Rhodobacter sphaeroides contains one PufX molecule which is involved in dimerization of this complex. Biochemistry 38: 6834–6845 [DOI] [PubMed] [Google Scholar]
- Frese RN, Olsen JD, Branvall R, Westerhuis WH, Hunter CN, van Grondelle R (2000) The long-range supraorganization of the bacterial photosynthetic unit: a key role for PufX. Proc Natl Acad Sci USA 97: 5197–5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golecki JR, Heinrich UR (1991) Ultrastructural and electron spectroscopic analyses of cyanobacteria and bacteria. J Microsc 162 (Part 1): 147–154 [DOI] [PubMed] [Google Scholar]
- Golecki JR, Ventura S, Oelze J (1991) The architecture of unusual membrane tubes in the B800-850 light-harvesting bacteriochlorophyll-deficient mutant 19 of Rhodobacter sphaeroides. FEMS Microbiol Lett 77: 335–340 [Google Scholar]
- Hunter CN, Pennoyer JD, Sturgis JN, Farrelly D, Niederman RA (1988) Oligomerization states and associations of light-harvesting pigment protein complexes of Rhodobacter sphaeroides as analyzed by lithium dodecyl-sulfate polyacrylamide-gel electrophoresis. Biochemistry 27: 3459–3467 [Google Scholar]
- Jamieson SJ, Wang P, Qian P, Kirkland JY, Conroy MJ, Hunter CN, Bullough PA (2002) Projection structure of the photosynthetic reaction centre-antenna complex of Rhodospirillum rubrum at 8.5 Å resolution. EMBO J 21: 3927–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joliot P, Joliot A, Verméglio A (1997) Photo-induced cyclic electron transfer in frozen cells of Rhodobacter sphaeroides. Biochim Biophys Acta 1318: 374–384 [Google Scholar]
- Joliot P, Verméglio A, Joliot A (1989) Evidence for supercomplexes between reaction centers, cytochrome-c2 and cytochrome-bc1 complex in Rhodobacter sphaeroides whole cells. Biochim Biophys Acta 975: 336–345 [Google Scholar]
- Joliot P, Verméglio A, Joliot A (1993) Supramolecular membrane protein assemblies in photosynthesis and respiration. Biochim Biophys Acta 1141: 151–174 [Google Scholar]
- Joliot P, Verméglio A, Joliot A (1996) Supramolecular organization of the photosynthetic chain in chromatophores and cells of Rhodobacter sphaeroides. Photosynth Res 48: 291–299 [DOI] [PubMed] [Google Scholar]
- Jones MR, Fowler GJ, Gibson LC, Grief GG, Olsen JD, Crielaard W, Hunter CN (1992) Mutants of Rhodobacter sphaeroides lacking one or more pigment–protein complexes and complementation with reaction-centre, LH1, and LH2 genes. Mol Microbiol 6: 1173–1184 [DOI] [PubMed] [Google Scholar]
- Jungas C, Ranck JL, Rigaud JL, Joliot P, Verméglio A (1999) Supramolecular organization of the photosynthetic apparatus of Rhodobacter sphaeroides. EMBO J 18: 534–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karrasch S, Bullough PA, Ghosh R (1995) The 8.5 Å projection map of the light-harvesting complex I from Rhodospirillum rubrum reveals a ring composed of 16 subunits. EMBO J 14: 631–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katona G, Andreasson U, Landau EM, Andreasson LE, Neutze R (2003) Lipidic cubic phase crystal structure of the photosynthetic reaction centre from Rhodobacter sphaeroides at 2.35 Å resolution. J Mol Biol 331: 681–692 [DOI] [PubMed] [Google Scholar]
- Koepke J, Hu X, Muenke C, Schulten K, Michel H (1996) The crystal structure of the light-harvesting complex II (B800-850) from Rhodospirillum molischianum. Structure 4: 581–597 [DOI] [PubMed] [Google Scholar]
- Lilburn TG, Haith CE, Prince RC, Beatty JT (1992) Pleiotropic effects of pufX gene deletion on the structure and function of the photosynthetic apparatus of Rhodobacter capsulatus. Biochim Biophys Acta 1100: 160–170 [DOI] [PubMed] [Google Scholar]
- McDermott G, Prince SM, Freer AA, Hawthornthwaite-Lawless AM, Papiz MZ, Cogdell RJ, Isaacs NW (1995) Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature 374: 517–525 [Google Scholar]
- McGlynn P, Hunter CN, Jones MR (1994) The Rhodobacter sphaeroides PufX protein is not required for photosynthetic competence in the absence of a light harvesting system. FEBS Lett 349: 349–353 [DOI] [PubMed] [Google Scholar]
- McGlynn P, Westerhuis WH, Jones MR, Hunter CN (1996) Consequences for the organization of reaction center-light harvesting antenna 1 (LH1) core complexes of Rhodobacter sphaeroides arising from deletion of amino acid residues from the C terminus of the LH1 alpha polypeptide. J Biol Chem 271: 3285–3292 [DOI] [PubMed] [Google Scholar]
- Miller KR (1982) Three-dimensional structure of a photosynthetic membrane. Nature 300: 53–55 [Google Scholar]
- Pugh RJ, McGlynn P, Jones MR, Hunter CN (1998) The LH1-RC core complex of Rhodobacter sphaeroides: interaction between components, time-dependent assembly, and topology of the PufX protein. Biochim Biophys Acta 1366: 301–316 [DOI] [PubMed] [Google Scholar]
- Qian P, Addlesee HA, Ruban AV, Wang P, Bullough PA, Hunter CN (2003) A reaction center-light-harvesting 1 complex (RC-LH1) from a Rhodospirillum rubrum mutant with altered esterifying pigments: characterization by optical spectroscopy and cryo-electron microscopy. J Biol Chem 278: 23678–23685 [DOI] [PubMed] [Google Scholar]
- Recchia PA, Davis CM, Lilburn TG, Beatty JT, Parkes-Loach PS, Hunter CN, Loach PA (1998) Isolation of the PufX protein from Rhodobacter capsulatus and Rhodobacter sphaeroides: evidence for its interaction with the alpha-polypeptide of the core light-harvesting complex. Biochemistry 37: 11055–11063 [DOI] [PubMed] [Google Scholar]
- Roszak AW, Howard TD, Southall J, Gardiner AT, Law CJ, Isaacs NW, Cogdell RJ (2003) Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris. Science 302: 1969–1972 [DOI] [PubMed] [Google Scholar]
- Sabaty M, Jappé J, Olive J, Verméglio A (1994) Organization of electron-transfer components in Rhodobacter sphaeroides forma sp. denitrificans whole cells. Biochim Biophys Acta 1187: 313–323 [Google Scholar]
- Schachman HK, Pardee AB, Stanier RY (1952) Studies on the macromolecular organization of microbial cells. Arch Biochem Biophys 38: 245–260 [DOI] [PubMed] [Google Scholar]
- Scheuring S, Francia F, Busselez J, Melandri B, Rigaud J-L, Levy D (2004) Structural role of PufX in the dimerization of the photosynthetic core-complex of Rhodobacter sphaeroides. J Biol Chem, in press [DOI] [PubMed] [Google Scholar]
- Scheuring S, Seguin J, Marco S, Levy D, Robert B, Rigaud JL (2003) Nanodissection and high-resolution imaging of the Rhodopseudomonas viridis photosynthetic core complex in native membranes by AFM. Proc Natl Acad Sci USA 100: 1690–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg H, Dubochet J, Vogel H, Ghosh R (1998) Are the light-harvesting I complexes from Rhodospirillum rubrum arranged around the reaction centre in a square geometry? J Mol Biol 282: 819–831 [DOI] [PubMed] [Google Scholar]
- Stark W, Kühlbrandt W, Wildhaber H, Wehrli E, Mühlethaler K (1984) The structure of the photoreceptor unit of Rhodopseudomonas viridis. EMBO J 3: 777–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremmel IG, Kirchhoff H, Weis E, Farquhar GD (2003) Dependence of plastoquinol diffusion on the shape, size, and density of integral thylakoid proteins. Biochim Biophys Acta 1607: 97–109 [DOI] [PubMed] [Google Scholar]
- Valpuesta JM, Carrascosa JL, Henderson R (1994) Analysis of electron microscope images and electron diffraction patterns of thin crystals of phi 29 connectors in ice. J Mol Biol 240: 281–287 [DOI] [PubMed] [Google Scholar]
- Verméglio A, Joliot P, Joliot A (1995) Organization of electron transfer components and supercomplexes. In Anoxygenic Photosynthetic Bacteria, Bauer, CE et al. (ed.), Vol. 2, pp 279–295. Dordrecht, Boston, London: Kluwer Academic Publishers [Google Scholar]
- Verméglio A, Joliot P (1999) The photosynthetic apparatus of Rhodobacter sphaeroides. Trends Microbiol 7: 435–440 [DOI] [PubMed] [Google Scholar]
- Verméglio A, Joliot A (2000) Response from Verméglio and Joliot. Trends Microbiol 8: 106–107 [DOI] [PubMed] [Google Scholar]
- Verméglio A, Joliot P (2002) Supramolecular organisation of the photosynthetic chain in anoxygenic bacteria. Biochim Biophys Acta 1555: 60–64 [DOI] [PubMed] [Google Scholar]
- Walz T, Ghosh R (1997) Two-dimensional crystallization of the light-harvesting I-reaction centre photounit from Rhodospirillum rubrum. J Mol Biol 265: 107–111 [DOI] [PubMed] [Google Scholar]
- Walz T, Jamieson SJ, Bowers CM, Bullough PA, Hunter CN (1998) Projection structures of three photosynthetic complexes from Rhodobacter sphaeroides: LH2 at 6 Å, LH1 and RC-LH1 at 25 Å. J Mol Biol 282: 833–845 [DOI] [PubMed] [Google Scholar]
- Westerhuis WH, Sturgis JN, Ratcliffe EC, Hunter CN, Niederman RA (2002) Isolation, size estimates, and spectral heterogeneity of an oligomeric series of light-harvesting 1 complexes from Rhodobacter sphaeroides. Biochemistry 41: 8698–8707 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and methods


