Abstract
RNA silencing is an evolutionarily conserved surveillance system that occurs in a broad range of eukaryotic organisms. In plants, RNA silencing acts as an antiviral system; thus, successful virus infection requires suppression of gene silencing. A number of viral suppressors have been identified so far; however, the molecular bases of silencing suppression are still poorly understood. Here we show that p19 of Cymbidium ringspot virus (CymRSV) inhibits RNA silencing via its small RNA-binding activity in vivo. Small RNAs bound by p19 in planta are bona fide double-stranded siRNAs and they are silencing competent in the in vitro RNA-silencing system. p19 also suppresses RNA silencing in the heterologous Drosophila in vitro system by preventing siRNA incorporation into RISC. During CymRSV infection, p19 markedly diminishes the amount of free siRNA in cells by forming p19–siRNA complexes, thus making siRNAs inaccessible for effector complexes of RNA-silencing machinery. Furthermore, the obtained results also suggest that the p19-mediated sequestration of siRNAs in virus-infected cells blocks the spread of the mobile, systemic signal of RNA silencing.
Keywords: in vivo siRNA binding, silencing suppression, tombusvirus p19 suppressor, virus-induced RNA silencing
Introduction
RNA silencing is an evolutionarily conserved surveillance system that occurs in a broad range of eukaryotic organisms including fungi (quelling), animals (RNA interference (RNAi)) and plants (post-transcriptional gene silencing (PTGS)). Accumulation of double-stranded (ds) RNA in eukaryotic cells triggers RNA silencing and the dsRNAs are converted to small interfering (siRNAs) which guide the degradation of homologous single-stranded (ss) mRNAs (reviewed in Hannon, 2002). RNA silencing is an ancient self-defence mechanism acting against molecular parasites, including transposons and viruses (Voinnet, 2002). RNA silencing is likely involved in the maintenance of genome stability by regulating heterochromatin formation in the fission yeast (Provost et al, 2002; Volpe et al, 2002) and in plants (Hamilton et al, 2002; Zilberman et al, 2003). A conserved set of gene products is required for RNA silencing in plants, animals and fungi (reviewed in Bernstein et al, 2001b; Vance and Vaucheret, 2001; Cogoni, 2002; Mlotshwa et al, 2002).
The molecular mechanism of RNA silencing is also conserved in eukaryotes. Cytoplasmic dsRNAs could be derived from the expression of inverted repeats (Beclin et al, 2002) or production of complementary transcripts. Current models of RNA silencing suggest that dsRNAs generated by RNA-dependent RNA polymerases (RdRP) also act as RNA-silencing inducers both in plants and nematodes (Dalmay et al, 2000; Mourrain et al, 2000; Smardon et al, 2000; Sijen et al, 2001). Moreover, RNA silencing is efficiently triggered by dsRNA intermediates of cytoplasmically replicating viruses (Ahlquist, 2002).
The mechanism for RNA silencing involves an initial processing of the inducing dsRNA into siRNAs of 21–26 nucleotides by the RNase III-like enzyme DICER (Bernstein et al, 2001a; Billy et al, 2001; Grishok et al, 2001; Ketting et al, 2001; Knight and Bass, 2001). siRNAs guide a nuclease complex referred to as the RNA-induced silencing complex (RISC) to target RNAs for degradation (Hammond et al, 2000; Zamore et al, 2000; Zamore, 2001; Carmell et al, 2002). siRNAs could also provide sequence specificity for other effector complexes of RNA silencing such as RdRP, proteins involved in heterochromatin formation or complexes playing a role in systemic silencing (see below).
RNA silencing plays an antiviral role in plants, insects and perhaps in many other eukaryotes. Thus, virus invasion requires evasion or suppression of silencing. Indeed, both plant and insect viruses have evolved proteins that suppress RNA silencing (Li and Ding, 2001; Voinnet, 2001; Baulcombe, 2002; Li et al, 2002) by targeting different steps of silencing pathways (Li and Ding, 2001). However, the molecular mechanism of RNA-silencing suppression remains unravelled. A characteristic feature of plant RNA silencing is that it operates at both single-cell and whole-plant levels (reviewed in Matzke et al, 2001; Voinnet, 2001). Activation of RNA silencing in a plant cell leads to processing of dsRNAs to siRNAs and to the degradation of cognate RNAs (referred to as cell-autonomous silencing), and causes generation of a mobile signal, which spreads in the plant and results in sequence-specific RNA degradation in distant cells (systemic silencing) (Mlotshwa et al, 2002). Systemic silencing might play a substantial role in virus control. For instance, infection with nepo-, tobra- and caulimoviruses, which are not able to cope with RNA silencing, often leads to a recovery phenotype of infected plants. While the first systemically infected leaves of recovered plants are fully invaded and contain a high level of viruses, the upper leaves show mild symptoms and low virus titre. Presumably, the inoculated leaves serve as a source of mobile signal that triggers systemic silencing; thus plants can restrict virus spread in the upper leaves (Ratcliff et al, 1997, 1999).
Cymbidium ringspot virus (CymRSV) contains a positive-sense ssRNA genome with five open reading frames (ORFs). The 19 kDa protein (p19) product of ORF5 has been recently shown to be an RNA-silencing suppressor (Voinnet et al, 1999; Silhavy et al, 2002; Qiu et al, 2002; Qu and Morris, 2002). While CymRSV infects Nicotiana benthamiana systemically and kills the host within 2 weeks, infection with a mutant virus, in which p19 was inactivated (Cym19stop), results in a recovery-like phenotype showing mild symptoms and low virus levels in the upper leaves (Szittya et al, 2002).
Apparently, p19 does not affect virus-induced cell-autonomous silencing, as CymRSV and Cym19stop virus RNAs as well as siRNAs derived from these viruses accumulate to the same levels in transfected single cells (Silhavy et al, 2002). In addition, the p19 protein was shown to repress the accumulation of all size classes of siRNA produced in agroinfiltration assays (Hamilton et al, 2002; Silhavy et al, 2002). Furthermore, p19 is the only viral silencing suppressor for which the basis of silencing suppression was analysed at the molecular level. It has been shown recently that p19 specifically binds in vitro to synthetic and natural 21-nt ds siRNAs having 2-nt 3′ overhangs. Based on these observations, it was postulated that the RNA-silencing suppressor activity of p19 depends on binding and inactivation of silencing-generated ds siRNAs (Silhavy et al, 2002).
Indeed, we demonstrate here that p19 inhibits RNA silencing in planta by sequestering siRNAs. In CymRSV-infected plants, siRNAs are present in p19–siRNA complexes, while in plants infected with the p19 defective mutant Cym19stop siRNAs were found in free forms. Furthermore, we show that p19 suppresses RNA silencing in the heterologous Drosophila in vitro RNA-silencing system, preventing the formation of an active RISC complex. Based on the obtained results, we propose a model for the natural role of the virus-encoded p19 silencing suppressor.
Results
p19 binds RNA-silencing-generated siRNAs in vivo
Although the symptoms of CymRSV- and Cym19stop-infected plants are markedly different (see Introduction), high levels of virus-derived 21-nt siRNAs could be detected in both CymRSV- and Cym19stop-inoculated plants (Szittya et al, 2002) (see also Figure 1A, lanes 1 and 4). It was hypothesised that in CymRSV-infected plants p19 binds RNA-silencing-generated siRNAs, and thus inhibits antiviral response and leads to lethal systemic infection (Silhavy et al, 2002). To test whether p19 binds ds siRNAs in vivo, p19 was immunoprecipitated from the leaf extracts of CymRSV- and Cym19stop-infected N. benthamiana plants using antisera raised against purified p19 protein (Havelda et al, 1998). The protein content of recovered immunoprecipitations (IPs) was analysed by Western blotting, while the RNAs extracted from the same IPs were detected by Northern analysis. Analysis of α-p19 IP derived from CymRSV-infected plants showed a high level of p19 protein and virus-specific 21-nt RNAs (Figure 1A and B, lane 2). As expected, neither p19 protein nor 21-nt RNAs were detected in the eluates of the α-p19 IP of extracts derived from Cym19stop-infected plants (Figure 1A and B, lane 5). Similarly, no short RNAs were detected in the eluates of control IP using antisera raised against CymRSV coat protein (CP) (Figure 1A, lanes 3 and 6). Interestingly, a part of siRNAs derived from α-p19 IP migrated slightly faster in denaturing gel than 21-nt siRNAs in the input RNA sample (Figure 1A, lane 2). To better understand the molecular nature of p19-bound siRNAs, Northern analysis was performed from native nondenaturing gel-separated siRNAs derived from α-p19 IP. The extracted siRNAs were run alongside ss and ds synthetic RNA oligonucleotides of 19 and 21 nt in length, respectively. As expected from a previous in vitro binding experiment (Silhavy et al, 2002), siRNAs from α-p19 IP comigrated with ds siRNAs in a size range of 19–21 nt (Figure 1D, compare lane 1 with lanes 3–6). The observed siRNAs shorter than 21 nt are probably generated artificially during the IP, because RNA extracts directly from virus-infected plant tissue (Szittya et al, 2003) or from extracts prior to IP contained only 21-nt siRNAs (Figure 1A, lane 1).
Figure 1.
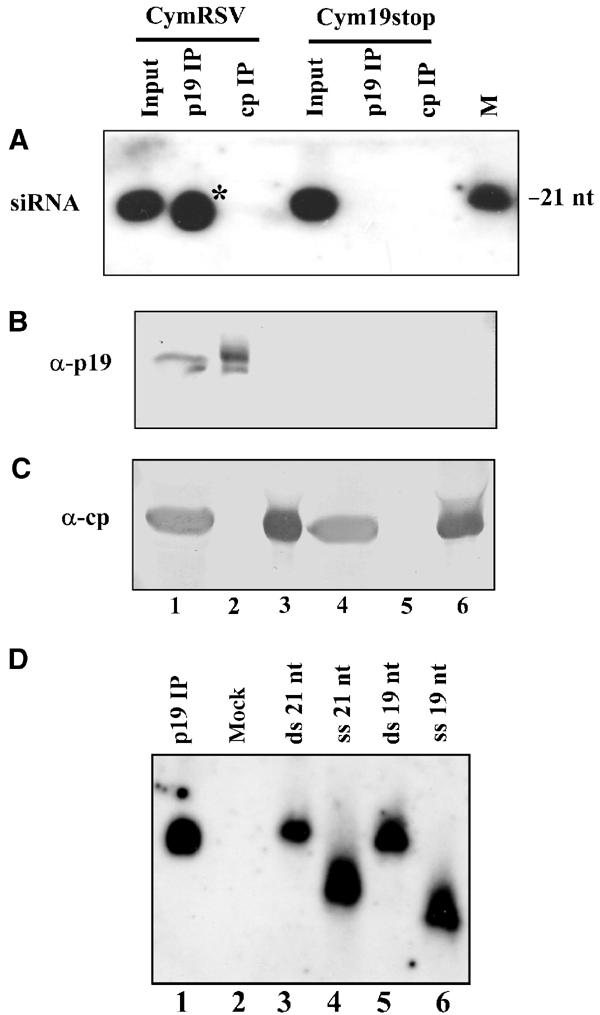
p19 binds silencing-generated 21-nt RNAs in planta. Extracts prepared from upper systemically infected leaves of CymRSV- or Cym19stop-inoculated plants at 6 dpi were immunoprecipitated with either α-p19 or α-CP (control) antibody. Inputs and eluates of IPs were analysed with Northern (A) and Western blotting (B, C). In vitro transcribed internally labelled positive-strand RNA of the CymRSV CP ORF was used as a probe for Northern blot analyses. As a size marker, γ-32P-ATP-labelled synthetic 21-nt ssRNA was applied. Protein blots were probed with α-p19 (B) and α-CP (C) antibodies, respectively. * indicates p19-bound RNAs shorter than 21 nt, which were generated artificially during the IP. (D) p19-bound 21-nt RNAs are double stranded. Extract of leaves of CymRSV-infected plants first separated on a gel-filtration column at 4°C to reduce nonspecific degradation of p19-bound RNA. p19 containing peak fractions were used to perform IP as in (A). Eluates of IPs were loaded onto a 15% polyacrylamide containing native 1 × TBE gel and analysed with Northern blotting. In vitro transcribed internally labelled positive-strand RNA of CymRSV was used as a probe. Duplexes of 21-nt (synthetic siRNA) and 19-nt (‘blunt' duplex) RNA oligonucleotides and single-stranded 21- and 19-nt synthetic RNA oligonucleotides were used as size markers. Oligonucleotides were labelled with γ-32P-ATP. The complementary strands of the duplexes were phosphorylated with ATP before annealing.
This result confirms our hypothesis that p19 RNA-silencing suppressor forms stable complexes with silencing-produced 21-nt siRNAs in vivo (Silhavy et al, 2002).
p19 binds bona fide siRNAs in planta
As in CymRSV-infected plants siRNAs accumulate to high levels and p19 forms complexes with siRNAs, we hypothesised that p19 suppresses silencing by preventing the siRNA incorporation into effector complexes. Hypothetically, p19 could prevent the formation of the active silencing complex in two distinct ways. p19 binds silencing-generated ds siRNAs and catalyses their modification into silencing-incompetent molecules. Alternatively, p19 does not modify siRNAs, instead prevents active RISC complex formation by sequestering ds siRNAs. To distinguish between these possibilities, we first tested whether the p19-bound small RNAs are silencing-competent siRNAs.
As a plant-derived in vitro RNA-silencing system that can be programmed by exogenous siRNAs is not available, we used the heterologous Drosophila in vitro RNAi system (referred to as an in vitro RNA-silencing system in this paper) to test the silencing competency of p19-bound siRNAs. Drosophila in vitro RNA-silencing system can be efficiently programmed only with a silencing-competent, bona fide siRNA characterised as ∼21-nt dsRNAs having 2-nt 3′ overhangs (Tuschl et al, 1999; Zamore et al, 2000), although a high molar excess of ss 21 siRNA can also trigger RNA degradation (Martinez et al, 2002; Schwarz et al, 2002). To test the silencing competence of p19-bound siRNAs, IP using p19 antibody was performed to pull down p19-small RNA complexes from CymRSV-infected plant extracts. To reduce the artificial degradation of p19-bound siRNAs observed in Figure 1A (lane 2), plant extracts were first separated on a Superdex-200 HR 10/30 column at 4°C, and p19 protein containing peak fractions were collected and used to perform IPs. Indeed, the obtained RNA samples derived from α-p19 IP contained ds siRNAs (Figure 1D), mostly 21 nt in length, containing CymRSV sequences (Figure 2A, lane 1). The concentration of CymRSV-specific siRNA derived from α-p19 IP was quantified (Figure 2A and B) before testing to initiate target RNA degradation in the Drosophila in vitro RNA-silencing system (Tuschl et al, 1999; Zamore et al, 2000; Elbashir et al, 2001). Figure 2C lanes 3–7 show that the CymRSV-derived ds 21-nt RNAs extracted from p19-small RNA complexes were able to programme the in vitro RNA-silencing system to degrade the 32P-labelled CymRSV RNA target. The results shown in Figure 2C also indicate that the target degradation was dependent on the amount of applied siRNAs. The nearly complete degradation of target viral RNA was observed in relatively high siRNA concentration (262 nM), which presumably reflects that some degradation of siRNAs occurs during the extraction. However, the p19-bound siRNA-guided target degradation was clearly specific, because the unrelated control GFP target RNAs were not degraded (Figure 2D, lanes 3–7).
Figure 2.
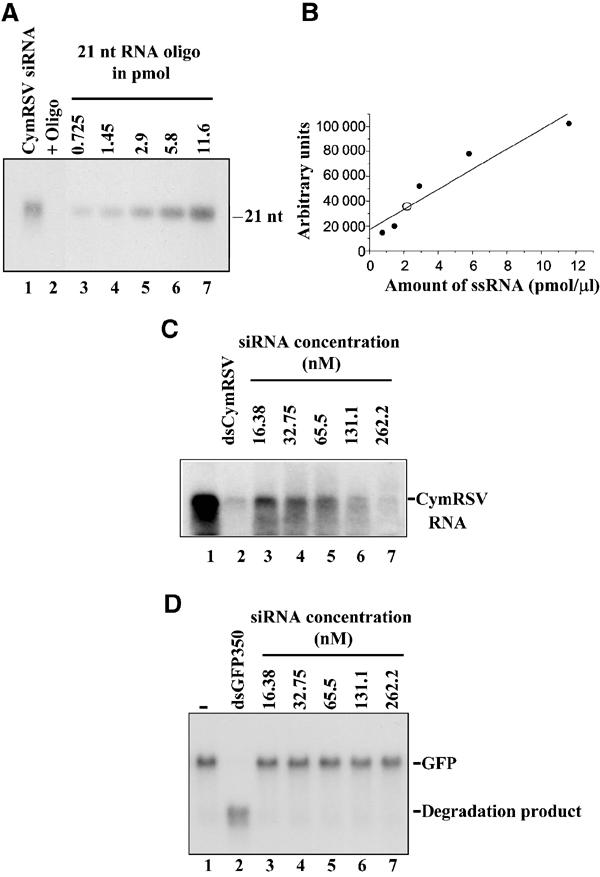
Characterisation of p19-bound 21-nt dsRNAs. (A) Analyses of p19-bound 21-nt RNAs by quantitative Northern blotting using increasing amounts of a synthetic 21-nt oligonucleotide complementer to the positive strand of CymRSV and RNA prepared from α-p19 IP (Figure 1D). An internally labelled positive strand of CymRSV was used as a probe. (B) Quantification of data obtained in (A). Closed circles, concentration standards; open circle, RNA isolated from α-p19 IP. The line shows the linear fit of the standards calculated with the computer program Microcal Origin 5.00. (C) 21-nt dsRNA from α-p19 IP driving the degradation of the cognate target RNA in the in vitro RNA-silencing system. A final concentration of 16.38–262.2 nMp19-bound 21-nt dsRNAs was added to the reactions. For target RNA, we used the full-length CymRSV1−4733 transcript at 100 pM concentration. dsRNA (5 nM) corresponding to the full-length CymRSV was used as a positive control. For negative control, the same system except GFP target RNA at 200 pM and dsGFP350 at 3 nM in lane 2 was used.
We demonstrated that p19-bounded siRNAs are double stranded, which were able to programme the Drosophila in vitro RNA-silencing system, confirming that at least a part of them are silencing-competent bona fide siRNAs.
p19 inhibits RNA silencing in the Drosophila in vitro system in a dose-dependent manner
Findings that p19 inhibits RNA silencing in planta (Figure 1D) and specifically binds 21-nt ds siRNAs in vitro (Silhavy et al, 2002) suggest that no other factors are required for the silencing suppressor activity of p19. If it is so, p19 is expected to inhibit RNA silencing also in the heterologous Drosophila in vitro RNA-silencing system, which offers an excellent possibility to analyse the mechanism of p19-mediated silencing suppression. Before testing in vitro the suppressor activity-purified p19-GST recombinant protein (Silhavy et al, 2002), we quantified the ds siRNA-binding activity of the recombinant p19-GST fusion protein. Gel mobility shift assay was carried out with 0.17–43.75 nM of p19-GST and a constant amount of (0.144 nM final concentration) radioactively labelled synthetic siRNA (Figure 3A). Based on the obtained binding data, the apparent dissociation constant (Ka) for p19-GST was 2.50±0.3 nM (Figure 3B). Considering the quantified binding activity of p19-GST, we set up the Drosophila in vitro RNA-silencing system to test the suppressor activity of p19. In our standard silencing assay, 32P-labelled 725-nt GFP ssRNA as a target and 350-bp dsRNA GFP (3 nM) as inducer molecules were added to Drosophila embryo extract. During the in vitro silencing reaction, a part of the 725-nt GFP target RNA—corresponding to the 350-bp GFP dsRNA—was completely degraded and lower molecular weight RNAs representing the cleaved target accumulated (Figure 4A, lane 2). However, addition of an increasing amount of recombinant p19-GST to the silencing reaction resulted in increasing inhibition of target RNA degradation (Figure 4A, lanes 3–8). Adding 1400 nM p19-GST resulted in the complete inhibition of target RNA degradation. In contrast, addition of the same amount of GST did not interfere with target RNA degradation (Figure 4A, lanes 9–14). Note that in further in vitro experiments p19-GST will be referred to as p19.
Figure 3.
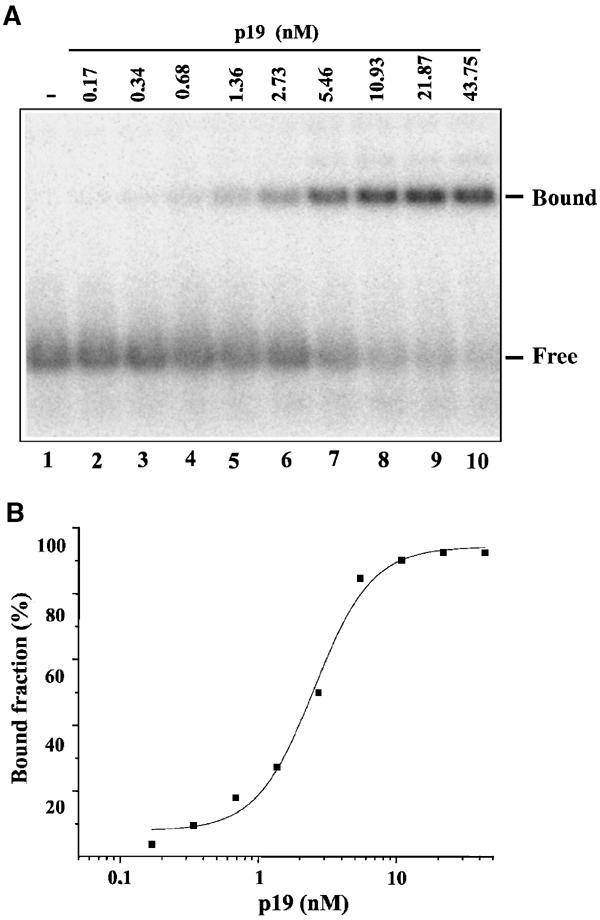
Determination of the apparent dissociation constant (Ka) of p19-GST recombinant protein. (A) Representative gel mobility shift assay carried out with 0.17–43.75 nM of p19-GST and a constant amount of (0.144 nM) radioactively labelled synthetic siRNA. (B) Representative plot of direct binding of p19-GST to siRNAs. The curve is best fitted to the indicated sets of data with the computer program Microcal Origin 5.00. The apparent dissociation constant (Ka) is estimated as the concentration of the protein required to give 50% saturation. Note that we do not know the percent of active p19-GST in our protein extract; therefore, we can calculate the apparent dissociation constant (Ka).
Figure 4.
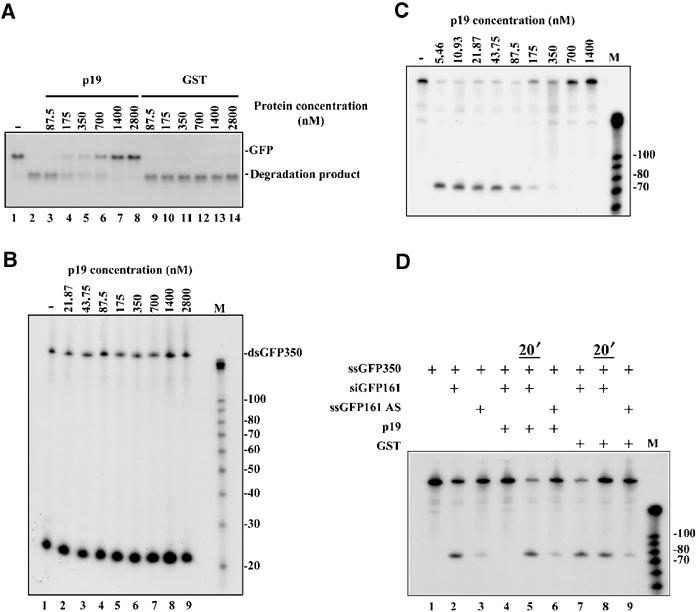
p19 suppresses RNA silencing in the heterologous in vitro RNA-silencing system. (A) p19 inhibits RNA silencing in the Drosophila in vitro system in a dose-dependent manner. In the RNA-silencing reaction, dsGFP350 dsRNA was used as inducer at 3 nM concentration and radioactively labelled full-length (725 nt) GFP transcript (200 pM) was added as a target RNA to the Drosophila embryo lysate. In vitro reactions were supplemented with various concentrations of CymRSV p19 expressed as a GST fusion protein (p19) or GST (87.5–2800 nM). Target degradation was monitored on a 1.2% agarose gel. RNA-silencing activity was indicated by loss of target RNA and appearance of the shorter degradation products (degraded target). (B) Effect of p19 on DICER activity in vitro. Radioactively labelled dsGFP350 was added to Drosophila embryo lysate at 3 nM concentration and p19 was used at various concentrations (21.87–2800 nM). dsRNA processing to siRNA was analysed on an 8% polyacrylamide gel. As a size marker, the Decade Marker System (Ambion) was used. (C) p19 inhibits RNA silencing by sequestering siRNAs in vitro. siRNA duplex (siGFP161) at 20 nM was used to induce the degradation of labelled ssGFP350 target RNA (100 pM) in the in vitro RNA-silencing system. Reactions were supplemented with various amounts of p19 (5.46–1400 nM final concentration). Inhibition of RNA-silencing activity is indicated by loss of the 74 base in the size 5′ end cleavage product. As a size marker, the Decade Marker System (Ambion) was used. (D) p19 cannot counteract with active RISC complex. Degradation of the ssGFP350 target RNA (lane 1) was induced by 20 nM siRNA (siGFP161, lane 2), or 500 mM ss siRNA (GFP161AS, lane 3). As indicated by the ±sign, 700 nM of p19 or GST was added simultaneously with siGFP161 or siGFP161AS to inhibit RNA silencing (lanes 4, 6, 7, 9). In lanes 5 and 8, p19 or GST was added 20 min after the addition of siGFP161. Inhibition of RNA silencing activity is indicated by loss of the 74 base in the size 5′ end cleavage product.
These results demonstrate that p19 could act as an efficient RNA-silencing suppressor in the heterologous system and show that the silencing inhibition is dose dependent, which is consistent with our suggestion that p19 inhibits RNA silencing by ds siRNA sequestering.
p19 inhibits the formation of active RISC complex in vitro
Since p19 inhibits RNA silencing in the Drosophila embryo extract as well as in plants, the well-established Drosophila in vitro system provides the possibility of assessing which step of RNA silencing is targeted by p19. According to the current model, DICER processes dsRNAs into siRNAs (Bernstein et al, 2001a), which guide RISC to their cognate substrates and degrade them (Hammond et al, 2000; Zamore et al, 2000). To unravel the molecular bases of p19-mediated silencing suppression, we first tested whether p19 could interfere with the DICER-mediated dsRNA processing. As expected, the Drosophila embryo extract rapidly processed the 32P-labelled 350-bp GFP dsRNA into siRNAs (Figure 4B, lane 1), and the presence of p19 in the concentrations which were inhibitory to RNA silencing (Figure 4A) did not interfere significantly with DICER activity (Figure 4B, lanes 2–9). In very high p19 concentration, a partial inhibition of DICER activity was also observed (not shown); however, it is unlikely that this inhibition reflects the natural role of the p19-silencing suppressor.
To analyse whether p19 targets steps of RNA silencing downstream of dsRNA processing, we tested the effect of p19 on siRNA-induced RNA silencing in the Drosophila in vitro system. In our experiment, we used GFP-specific synthetic siRNAs at 20 nM, which directed the cleavage of the 350-nt GFP RNA, yielding a 74-nt 5′ end product in the Drosophila RNA-silencing system. The obtained data show that the presence of p19 significantly inhibits the ds siRNA-triggered target RNA degradation (Figure 4C), indicating that p19 interfered with RNA silencing downstream of siRNA generation.
In Drosophila extract, active RISC complex formation occurs in two steps (Caplen et al, 2001; Elbashir et al, 2001; Martinez et al, 2002; Schwarz et al, 2002). siRNA duplexes are first incorporated into an inactive RISC complex; subsequently, an RNA helicase unwinds ds siRNA, and thus converts the inactive complex into a silencing-competent active complex (Nykanen et al, 2001). We asked whether p19 inhibits the incorporation of ds siRNAs into an inactive RISC and/or blocks the activated RISC complexes. The effect of p19 on activated complexes was tested by ordered addition of siRNA and p19 into the RNA-silencing reaction. siRNAs were added first to the extract and allowed for 20 min to form active RISC complexes, and then target RNA and p19 were provided. As Figure 4D shows, p19 failed to inhibit the already activated RISC-mediated target RNA cleavage (Figure 4D, lane 5), while simultaneous addition of p19 and siRNAs into the silencing reaction resulted in the inhibition of target degradation (Figure 4D, lane 4). These results demonstrate that p19 interferes with RNA silencing downstream of siRNA generation, but fails to inhibit the target RNA degradation by the active RISC complexes. However, it has been reported that a high amount (∼500 nM) of ss siRNAs can also activate RISC as efficiently as 20 nM ds siRNAs (Martinez et al, 2002; Schwarz et al, 2002). To test the specificity of p19 in RNA cleavage reaction, as much as 500 nM of GFP ss siRNA was also applied (Figure 4D, lane 3). Consistent with the binding experiments in which p19 failed to bind ss siRNAs (Silhavy et al, 2002), the presence of p19 did not inhibit target cleavage programmed by ss siRNA (Figure 4D, lane 6).
In conclusion, the combined data further support our hypothesis that p19 inhibits RNA silencing by specifically binding ds siRNA, thus preventing the assembly of active RISC complexes.
p19 sequesters siRNAs in CymRSV-infected cells
The finding that p19-bound siRNAs derived from virus-infected plants are silencing competent (Figure 2C) suggests that p19 prevents the active silencing effector complex formation by sequestering siRNAs instead of catalytically inactivating small RNAs. To test whether the p19-mediated silencing suppression is based on the sequestering of siRNAs, we carried out gel-filtration experiments to establish the distribution of siRNAs and p19 in leaves of N. benthamiana plants infected with CymRSV or Cym19stop. Extracts of the first leaves showing systemic viral symptoms were subjected to gel filtration on a Superdex-200 chromatography column. Then the gel-filtration fractions were divided into two parts and tested for the presence of siRNAs and p19 by Northern and Western blotting. In fractions derived from CymRSV-infected plants, siRNAs were almost exclusively found in a complex migrating in fractions 31–35 (Figure 5A). Western analysis of a second aliquot demonstrated that p19 was present in the same fractions in which virus-derived siRNAs were identified (Figure 5B). Importantly, p19 was not detectable in other fractions. In a control experiment, we examined the distribution of siRNAs in the first leaves showing systemic viral symptoms of Cym19stop-infected plants. As Figure 5C shows, Cym19stop siRNAs were exclusively found in fractions 43–45. These siRNAs are likely free molecules, since siRNAs of Proteinase K-treated extracts from CymRSV-infected plants were also found in fractions 43–45 (data not shown). Surprisingly, we did not observe a higher molecular weight siRNA–protein complex (RISC), as it was found in the Drosophila systems (Hammond et al, 2001; Nykanen et al, 2001). This can be a difference between the animal and plant systems, or more likely only small fractions of siRNAs are incorporated into large complexes; thus they could not be detected by Northern analysis of the gel filtrations. Indeed, approximately 1–3% of total virus specific siRNAs were found in large protein complexes in partially purified extracts derived from Cym19stop-infected plants (L Lakatos, unpublished results).
Figure 5.
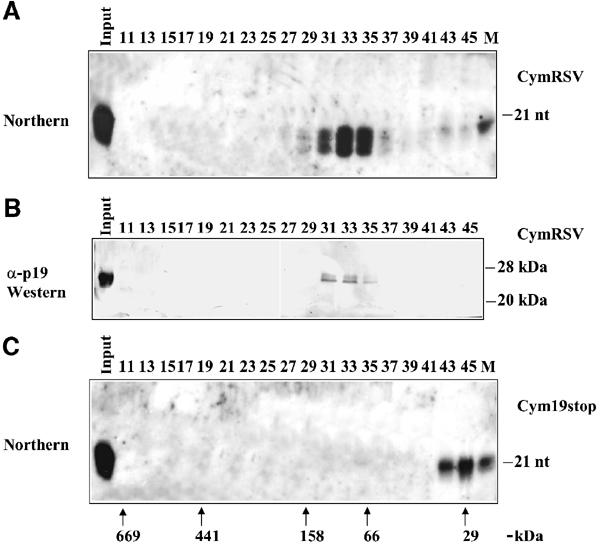
siRNAs and p19 are present in the same high molecular weight chromatography fractions of virus-infected plant extracts. The extracts prepared from systemic leaves of CymRSV- or Cym19stop-infected N. benthamiana plants were size separated by the Superdex-200 gel-filtration column, and then fractions were tested for the presence of siRNAs and for p19. (A) Northern blot of RNAs isolated from a half volume of gel-filtration fractions of extract from CymRSV-infected plants. (B) Western blot of the other half of the same fractions shown in panel (A) were probed with an α-p19 antibody. (C) Northern blot of RNA isolated from gel-filtration fractions of Cym19stop-infected plants. RNA gel blots were probed with radioactively labelled in vitro CymRSV transcript. γ-32P-ATP-labelled 21-nt synthetic RNA oligo was used as a size marker (M) for RNA gel blots. The elution position of protein molecular weight markers for all panels is shown in (C). 669 kDa, thyroglobulin; 440 kDa, ferritin; 150 kDa, aldolase; 66 kDa, bovine serum albumin; 29 kDa, carbonic anhydrase.
Discussion
In this study, we have analysed the molecular mechanism of RNA-silencing suppression by p19 using both virus-infected plant and the Drosophila in vitro RNA-silencing system. We demonstrate that p19 silencing suppressor—previously shown to bind ds siRNA—inhibits RNA silencing also in the Drosophila in vitro system by targeting ds siRNAs, the most conserved element of silencing machinery. Combining our in vitro and in vivo results, we found that p19-mediated silencing suppression was similar in many aspects in the Drosophila in vitro system and in virus-infected plant; therefore, the results derived from in vitro studies could help to understand the p19-mediated RNA-silencing inhibition in plants.
p19-mediated RNA-silencing suppression in vitro and in vivo
Accumulation of siRNAs was observed in virus-infected plant, regardless of the presence or absence of p19 suppressor (Silhavy et al, 2002; Szittya et al, 2002), suggesting that p19 does not impair the siRNA generation from ds replicative intermediates (Ahlquist, 2002) or highly structured regions of viral RNAs (Szittya et al, 2002). Similarly, p19 failed to inhibit the dsRNA processing to siRNAs in the Drosophila in vitro system (the present study).
Furthermore, we demonstrated that p19 inhibited RNA silencing in a dose-dependent manner in the Drosophila system, as was shown for both virus-infected (Szittya et al, 2003) and agroinfiltrated plants (Qiu et al, 2002; Silhavy et al, 2002).
Our results obtained in the Drosophila in vitro system are in line with the previous suggestion (Silhavy et al, 2002) that p19 prevents the ds siRNA-mediated activation of silencing-competent RISC complexes. However, in line with p19-binding specificity, p19 failed to inhibit the activation of RISC by ss siRNA. Similarly, p19 was not able to suppress the target cleavage of the already activated RISC (Figure 4D). Consistent with the latter finding, CymRSV infection was not able to revert the already established GFP RNA silencing (Silhavy et al, 2002), contrary to other silencing suppressors such as HC-Pro that likely interferes with active RISC complexes (Kasschau et al, 2003) reverting RNA silencing (Brigneti et al, 1998). Although it has not been experimentally proven, our observation suggests that p19 also inhibits the assembly of active RISC (or other siRNA-guided effector complexes) in plants (see below).
In conclusion, these results suggest that p19-mediated silencing suppression could be similar at least partly in plants and in heterologous systems. Thus, we suggest that heterologous systems could be used for analysing the molecular mechanism of suppressors that target conserved elements of silencing machinery. More strikingly, we expect that p19 could emerge as a powerful tool to inhibit RNA silencing in other organisms if RNA silencing is triggered by dsRNAs or siRNA, but can be circumvented by using ss siRNA for induction (Martinez et al, 2002).
However, the data obtained in the Drosophila in vitro system may not reflect directly the operating RNA-silencing mechanism in plants, since there are remarkable differences between the two systems, such as lack of a cellular RdRP and intramolecular transitivity in the RNA-silencing system of Drosophila (Schwarz et al, 2002).
Mechanism of virus-induced RNA silencing suppression mediated by p19
The results obtained from Drosophila in vitro experiments suggest that p19 does not interfere with siRNA generation from long dsRNA (Figure 4B), supporting the previous findings that virus-derived siRNAs accumulate to similar levels in both CymRSV- and Cym19stop-infected plants and protoplasts (Silhavy et al, 2002; Szittya et al, 2002).
Moreover, in CymRSV-infected plants, virus-specific siRNAs are complexed with p19 and these p19-bound ds siRNAs are at least partly silencing-competent molecules. These findings indicate that p19 binds and sequesters silencing-generated siRNAs, and thus depletes the specificity determinants of silencing effector complexes. Consequently, RNA silencing effector complexes including RISC will not be activated. Previously, it was suggested that p19 does not affect cell-autonomous silencing (Silhavy et al, 2002). Now we propose that p19 inhibits RISC activation also along with cell-autonomous silencing. However, in tombusvirus infection this effect is negligible, since cell-autonomous silencing cannot compete with the rapidly replicating CymRSV or Cym19stop; therefore, the suppressor-less mutant virus accumulates to wild-type levels in protoplasts (Silhavy et al, 2002). By contrast, cell-autonomous silencing might be important in the control of slowly replicating viruses as potyviruses, since Tobacco etch potyvirus bearing a certain mutation in HC-Pro failed to accumulate in protoplasts (Kasschau et al, 1997).
Unlike cell-autonomous silencing, systemic RNA silencing can protect plants from quickly replicating viruses. In the lack of p19, siRNAs are accumulating in free forms and systemic silencing could restrict the virus extent in and around the veins of upper leaves of Cym19stop-infected recovered plants (Havelda et al, 2003; Szittya et al, 2003). In contrast, in CymRSV-infected plants siRNAs are bound by p19 and the upper leaves are fully invaded by the pathogen, indicating that sequestering of siRNAs prevents the development of systemic silencing. Importantly, p19 could only be detected in fractions together with siRNAs in the extracts of CymRSV-infected plants, suggesting that under these conditions a majority of p19 proteins are used for siRNA sequestering.
Recently, we showed that RNA silencing is temperature dependent and that at high temperature (27°C) virus-derived siRNAs accumulate to very high levels, enhancing systemic silencing that restricts CymRSV spreading in the upper leaves (Szittya et al, 2003). We propose that at high temperature p19 molecules are overloaded by the extreme amount of siRNAs; thus, virus-derived free siRNAs could accumulate and consequently virus-induced systemic silencing develops. A similar phenomenon was also observed recently in the presence of defective interfering (DI) RNAs in CymRSV-infected plants. It was shown that DI RNAs significantly enhanced the level of siRNA with respect to the level of p19, leading to the accumulation of free siRNAs that resulted in the symptom attenuation of virus-infected plants (Z Havelda, unpublished results).
The exact role of siRNAs in the generation and spreading of a mobile signal is still not known. It is possible that a systemic signal is generated by siRNA-guided RdRP in the aberrant RNA-silencing pathway (Voinnet, 2001); thus, depletion of siRNAs by p19 results in the inactivation of a signal-generating pathway. However, we prefer the more obvious explanation that free siRNA (or complexed with endogenous proteins) is the mobile signal itself. siRNAs might be small enough to spread cell to cell along with or ahead of the viral front (Hamilton and Baulcombe, 1999), and can be transported through the phloem (Wu et al, 2002). Consistently, it was suggested recently that primary siRNAs from locally initiated silencing can move to 10–15 adjacent cells and activate RNA silencing (Himber et al, 2003).
Taken together, we can establish that p19 suppresses RNA silencing via depletion of all siRNAs, thus either eliminating the mobile signal or preventing the formation of the mobile signal in vivo.
In eukaryotes—including plants—RNA silencing plays an important role in the developmental timing by micro (mi) RNA-controlled endogenous gene regulation (reviewed in Bartel and Bartel, 2003), and viruses can interact with this miRNA pathway (Kasschau et al, 2003). A virus-encoded silencing suppressor—P1/HC-Pro of Turnip mosaic virus—has been shown to interfere with conserved miRNA-controlled RNA-silencing pathways and the virus symptoms are likely to be a consequence of this interference (Kasschau et al, 2003). Moreover, it was suggested that P1/HC-Pro interacts with RISC, inhibiting the target mRNA cleavage (Kasschau et al, 2003); however, the molecular bases of the RISC inhibition by HC-Pro are not known.
The unique nature of p19 to bind specifically ds siRNAs both in plants and the heterologous system provides a potential for p19 to interfere any RNA-silencing pathways having ds siRNA intermediates. Recent reports suggesting short-lived ds intermediates of matured ss miRNAs (Bartel and Bartel, 2003; Schwarz et al, 2003) and the observation that p19 expressing transgenic Arabidopsis displayed altered phenotype and miRNA accumulation (Papp et al, 2003) might indicate that p19 also interferes with miRNA-mediated pathways. In addition, the recently identified endogenous siRNA-like molecules (Ambros et al, 2003) are also potential targets of p19.
In conclusion, we suggest that p19 could emerge as a versatile tool to study these recently discovered gene regulation pathways, which involve ds siRNA-like molecules.
Materials and methods
Plant materials
N. benthamiana plants grown in soil under normal growth conditions were used for virus inoculation with in vitro transcripts of CymRSV and Cym19stop, as described previously (Dalmay et al, 1993). Virus-infected plants were grown in a test chamber (Versatile Enviromental Test Chambers (Sanyo, Tokyo Japan)) under 14 h light (50 μE m−2 s−1) and 10 h dark regime at 22°C.
RNA isolation and Northern blotting and native Northern blotting
RNA was isolated from extracts made in IP or 1 × lysis buffer from leaves, by adding an equal volume of 2 × PK buffer (Nykanen et al, 2001) with 50 μg Proteinase K (80 ng/μl final concentration) and then incubating at 65°C for 15 min. For isolation of siRNAs under native conditions, Proteinase K digestion was carried out at room temperature for 30 min. After phenol–chloroform extraction, the RNA was precipitated by 2.5 vol of ethanol. Northern blots were prepared as described in Szittya et al (2002).
Polyacrylamide gel (15% native) was run in 1 × TBE buffer and blotted onto a Hybond N+ (Pharmacia-Biotech) membrane in solution containing 50 mM NaOH. Hybridisation was carried out as described in Szittya et al (2002).
Protein separation and Western analysis
Proteins were separated in a 12% volume of SDS–polyacrylamide gel, and then transferred onto a Hybond C Extra filter (Pharmacia-Biotech). Western analysis was performed using α-p19 (Havelda et al, 1998) and α-CP (Havelda et al, 2003) polyclonal antibodies.
Immunoprecipitation
For IPs, 2 g of CymRSV- or Cym19stop-infected N. benthamiana leaves showing systemic symptoms were collected at 6 days postinoculation (dpi) and used to prepare extracts in 1 × IP buffer containing 30 mM HEPES-KOH (pH 7.5), 100 mM NaCl, 66 mM KCl, 1 mM MgCl2, 1 mM DTT, 0.2 mM PMSF, and centrifuged at 15 000 g for 10 min. IPs were carried out at 4°C for 3–5 h, and then beads were centrifuged and washed in 1 × IP buffer two times. Input extracts and eluates of IPs were used for Western analysis and RNA isolation. RNA molecules were separated on a sequencing gel containing 12% polyacrylamide and 8 M urea.
For isolation of a high amount of virus-specific siRNA, extracts of systemic leaves of CymRSV-infected N. benthamiana prepared in a buffer containing 30 mM HEPES-KOH (pH 7.5), 100 mM KCl, 2 mM MgCl2, 1 mM DTT and 5% glycerol were separated on a Superdex-200 HR 10/30 column. To reduce degradation of p19-bound siRNA, the column was run at 4°C. p19 protein containing peak fractions were collected and used to perform the IPs.
Electrophoretic mobility shift assay
In all, 30 pmol of RNA oligonucleotides was phosphorylated in the presence of γ-32P ATP in a 20 μl reaction using 20 U of T4 polynucleotide kinase (Fermentas). The 5′-phosphorylated complementary strand was added in 10 times molar excess, and then the reaction was heated to 90°C for 1 min and cooled slowly to anneal the strands. Duplexes were purified by PAGE on a native 15% polyacrylamide containing TBE gel. The labelled duplex was cut out of the gel and eluted in a solution containing 0.3 M NaCl, and then precipitated with ethanol. Binding reactions were performed with a 0.144 nM final concentration of labelled siRNA and 0.173–43.75 nM of p19 protein in the presence of 1 × lysis buffer (Zamore et al, 2000) supplemented with 0.02% of Tween 20 for 30 min at room temperature. Complexes were resolved on a 6% polyacrylamide-containing 0.5 × TBE gel. The gels were then dried, exposed to a storage phosphor screen (Molecular Dynamics Typhoon Phosphorimager, Amersham Biosciences), and bands were quantified with a Genius Image Analyser (Syngene).
In vitro RNA silencing
Drosophila embryo lysate and inducer dsRNA preparation were as described in Zamore et al (2000). Primers used to synthesise the 350 bp T7 promoter containing PCR product for in vitro transcription of target ssRNA and inducer dsRNA were T7gfp5′ 5′-GTA ATA CGA CTC ACT ATA GGG AGA GGG TGA AGG TGA TGC AAC, GFP5′ GGG AGA GGG TGA AGG TGA TGC AAC-3′, T7gfp3′ 5′-GTA ATA CGA CTC ACT ATA GGG CTG CCA TGA TGT ATA CAT TGT GT-3′ and Gfp3′ 5′-GGG CTG CCA TGA TGT ATA CAT TGT GT-3′. For preparation of the full-length GFP target RNA, primers T7Gfp5′ 5′-GTA ATA CGA CTC ACT ATA GGG ATG AGT AAA GGA GAA GAA CTT TT-3′, GFPatg 5′-ATG AGT AAA GGA GAA GAA CTT TT-3′, T7GFP3′ 5′-GTA ATA CGA CTC ACT ATA GGG TCG ACA TTT ATT TGT ATA GTT CAT-3′ and GFPtga 5′-TCG ACA TTT ATT TGT ATA GTT CAT-3′ were used. For CymRSV target RNA preparation, the full-length cDNA clone described by Dalmay et al (1993) was used. Target RNA was internally labelled with radioactive α-32P UTP in a 10 μl in vitro transcription reaction driven by T7 RNA polymerase. Template DNA was digested by RQ1 DNase (Promega) and residual nucleotides were removed by a Sephadex G-50 spin-column chromatography, and then the target RNA was quantified by measuring the absorbance at 260 nM. Typically, we used target RNA at 100–200 pM. p19-GST purification was essentially as described by Havelda et al. (1998). After purification, RNA was loaded onto an 8% polyacrylamide sequencing gel or a 1.2% agarose gel containing 1 × TBE.
Assay for siRNA distribution by gel filtration
Extracts were prepared from 0.5 to 1 g of systemic leaves of CymRSV- or Cym19stop-infected N. benthamiana in 0.5–1 ml buffer containing 30 mM HEPES-KOH (pH 7.5), 100 mM KCl, 2 mM MgCl2, 1 mM DTT and 5% glycerol, and 200 μl of the extracts were chromatographed at room temperature on a Superdex-200 HR 10/30 column (Pharmacia) at 0.4 ml/min in a column buffer containing 30 mM HEPES-KOH (pH 7.5), 100 mM KCl, 2 mM MgCl2, 1 mM DTT and 5% glycerol. In all, 60 200 μl fractions were collected and used for Western analysis and for RNA isolation. RNA molecules were separated on a 12% polyacrylamide and 8 M urea containing sequencing gel.
Acknowledgments
We are grateful to Tracy T Hall for helpful comments during the preparation of the manuscript and Miklós Erdélyi for kindly providing the Drosophila culture. We thank Marjori A Matzke for providing a preprint of her manuscript. We also thank Szilárd Bodó for help in preparing the figures. This research was supported by grants from the Hungarian Ministry of Education (NKFP 4/023/2001) and ‘VIS' EU project (QLG2-CT-2002-01673).
References
- Ahlquist P (2002) RNA-dependent RNA polymerases, viruses, and RNA silencing. Science 296: 1270–1273 [DOI] [PubMed] [Google Scholar]
- Ambros V, Lee RC, Lavanway A, Williams PT, Jewell D (2003) MicroRNAs and other tiny endogenous RNAs in C. elegans. Curr Biol 13: 807–818 [DOI] [PubMed] [Google Scholar]
- Bartel B, Bartel DP (2003) MicroRNAs: at the root of plant development? Plant Physiol 132: 709–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe D (2002) Viral suppression of systemic silencing. Trends Microbiol 10: 306–308 [DOI] [PubMed] [Google Scholar]
- Beclin C, Boutet S, Waterhouse P, Vaucheret H (2002) A branched pathway for transgene-induced RNA silencing in plants. Curr Biol 12: 684–688 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001a) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Denli AM, Hannon GJ (2001b) The rest is silence. RNA 7: 1509–1521 [PMC free article] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W (2001) Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA 98: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brigneti G, Voinnet O, Li WX, Ji LH, Ding SW, Baulcombe DC (1998) Viral pathogenicity determinants are suppressors of transgene silencing in Nicotiana benthamiana. EMBO J 17: 6739–6746 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Caplen NJ, Parrish S, Imani F, Fire A, Morgan RA (2001) Specific inhibition of gene expression by small double-stranded RNAs in invertebrate and vertebrate systems. Proc Natl Acad Sci USA 98: 9742–9747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Cogoni C (2002) Unifying homology effects. Nat Genet 30: 245–246 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Hamilton A, Rudd S, Angell S, Baulcombe DC (2000) An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101: 543–553 [DOI] [PubMed] [Google Scholar]
- Dalmay T, Rubino L, Burgyan J, Kollar A, Russo M (1993) Functional analysis of cymbidium ringspot virus genome. Virology 194: 697–704 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T (2001) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC (2001) Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D (2002) Two classes of short interfering RNA in RNA silencing. EMBO J 21: 4671–4679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286: 950–952 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Havelda Z, Hornyik C, Crescenzi A, Burgyan J (2003) In situ characterization of cymbidium ringspot tombusvirus infection-induced posttranscriptional gene silencing in Nicotiana benthamiana. J Virol 77: 6082–6086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havelda Z, Szittya G, Burgyan J (1998) Characterization of the molecular mechanism of defective interfering RNA-mediated symptom attenuation in tombusvirus-infected plants. J Virol 72: 6251–6256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O (2003) Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J 22: 4523–4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasschau KD, Cronin S, Carrington JC (1997) Genome amplification and long-distance movement functions associated with the central domain of tobacco etch potyvirus helper component-proteinase. Virology 228: 251–262 [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA, Carrington JC (2003) P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Dev Cell 4: 205–217 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight SW, Bass BL (2001) A role for the RNase III enzyme DCR-1 in RNA interference and germ line development in Caenorhabditis elegans. Science 293: 2269–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li WX, Ding SW (2002) Induction and suppression of RNA silencing by an animal virus. Science 296: 1319–1321 [DOI] [PubMed] [Google Scholar]
- Li WX, Ding SW (2001) Viral suppressors of RNA silencing. Curr Opin Biotechnol 12: 150–154 [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T (2002) Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Matzke MA, Matzke AJ, Pruss GJ, Vance VB (2001) RNA-based silencing strategies in plants. Curr Opin Genet Dev 11: 221–227 [DOI] [PubMed] [Google Scholar]
- Mlotshwa S, Voinnet O, Mette MF, Matzke M, Vaucheret H, Ding SW, Pruss G, Vance VB (2002) RNA silencing and the mobile silencing signal. Plant Cell 14 (Suppl): S289–S301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourrain P, Beclin C, Elmayan T, Feuerbach F, Godon C, Morel JB, Jouette D, Lacombe AM, Nikic S, Picault N, Remoue K, Sanial M, Vo TA, Vaucheret H (2000) Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101: 533–542 [DOI] [PubMed] [Google Scholar]
- Nykanen A, Haley B, Zamore PD (2001) ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell 107: 309–321 [DOI] [PubMed] [Google Scholar]
- Papp I, Mette MF, Aufsatz W, Daxinger L, Schauer SE, Ray A, van der Winden J, Matzke M, Matzke AJ (2003) Evidence for nuclear processing of plant micro RNA and short interfering RNA precursors. Plant Physiol 132: 1382–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Silverstein RA, Dishart D, Walfridsson J, Djupedal I, Kniola B, Wright A, Samuelsson B, Radmark O, Ekwall K (2002) Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc Natl Acad Sci USA 99: 16648–16653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu W, Park JW, Scholthof HB (2002) Tombusvirus P19-mediated suppression of virus-induced gene silencing is controlled by genetic and dosage features that influence pathogenicity. Mol Plant Microbe Interact 15: 269–280 [DOI] [PubMed] [Google Scholar]
- Qu F, Morris TJ (2002) Efficient infection of Nicotiana benthamiana by tomato bushy stunt virus is facilitated by the coat protein and maintained by p19 through suppression of gene silencing. Mol Plant Microbe Interact 15: 193–202 [DOI] [PubMed] [Google Scholar]
- Ratcliff F, Harrison BD, Baulcombe DC (1997) A similarity between viral defense and gene silencing in plants. Science 276: 1558–1560 [DOI] [PubMed] [Google Scholar]
- Ratcliff FG, MacFarlane SA, Baulcombe DC (1999) Gene silencing without DNA. RNA-mediated cross-protection between viruses. Plant Cell 11: 1207–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Haley B, Zamore PD (2002) Evidence that siRNAs function as guides, not primers, in the Drosophila and human RNAi pathways. Mol Cell 10: 537–548 [DOI] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD (2003) Asymmetry in the assembly of the RNAi enzyme complex. Cell 115: 199–208 [DOI] [PubMed] [Google Scholar]
- Sijen T, Fleenor J, Simmer F, Thijssen KL, Parrish S, Timmons L, Plasterk RH, Fire A (2001) On the role of RNA amplification in dsRNA-triggered gene silencing. Cell 107: 465–476 [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnar A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyan J (2002) A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. EMBO J 21: 3070–3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smardon A, Spoerke JM, Stacey SC, Klein ME, Mackin N, Maine EM (2000) EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr Biol 10: 169–178 [DOI] [PubMed] [Google Scholar]
- Szittya G, Molnar A, Silhavy D, Hornyik C, Burgyan J (2002) Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell 14: 359–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szittya G, Silhavy D, Molnar A, Havelda Z, Lovas A, Lakatos L, Banfalvi Z, Burgyan J (2003) Low temperature inhibits RNA silencing-mediated defence by the control of siRNA generation. EMBO J 22: 633–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuschl T, Zamore PD, Lehmann R, Bartel DP, Sharp PA (1999) Targeted mRNA degradation by double-stranded RNA in vitro. Genes Dev 13: 3191–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance V, Vaucheret H (2001) RNA silencing in plants—defense and counterdefense. Science 292: 2277–2280 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2001) RNA silencing as a plant immune system against viruses. Trends Genet 17: 449–459 [DOI] [PubMed] [Google Scholar]
- Voinnet O (2002) RNA silencing: small RNAs as ubiquitous regulators of gene expression. Curr Opin Plant Biol 5: 444. [DOI] [PubMed] [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC (1999) Suppression of gene silencing: a general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA 96: 14147–14152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA (2002) Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science 297: 1833–1837 [DOI] [PubMed] [Google Scholar]
- Wu X, Weigel D, Wigge PA (2002) Signaling in plants by intercellular RNA and protein movement. Genes Dev 16: 151–158 [DOI] [PubMed] [Google Scholar]
- Zamore PD (2001) RNA interference: listening to the sound of silence. Nat Struct Biol 8: 746–750 [DOI] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Cao X, Jacobsen SE (2003) ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299: 716–719 [DOI] [PubMed] [Google Scholar]


