Abstract
Diabetic kidney disease is the leading cause of ESRD, but few biomarkers of diabetic kidney disease are available. This study used gas chromatography-mass spectrometry to quantify 94 urine metabolites in screening and validation cohorts of patients with diabetes mellitus (DM) and CKD(DM+CKD), in patients with DM without CKD (DM–CKD), and in healthy controls. Compared with levels in healthy controls, 13 metabolites were significantly reduced in the DM+CKD cohorts (P≤0.001), and 12 of the 13 remained significant when compared with the DM–CKD cohort. Many of the differentially expressed metabolites were water-soluble organic anions. Notably, organic anion transporter-1 (OAT1) knockout mice expressed a similar pattern of reduced levels of urinary organic acids, and human kidney tissue from patients with diabetic nephropathy demonstrated lower gene expression of OAT1 and OAT3. Analysis of bioinformatics data indicated that 12 of the 13 differentially expressed metabolites are linked to mitochondrial metabolism and suggested global suppression of mitochondrial activity in diabetic kidney disease. Supporting this analysis, human diabetic kidney sections expressed less mitochondrial protein, urine exosomes from patients with diabetes and CKD had less mitochondrial DNA, and kidney tissues from patients with diabetic kidney disease had lower gene expression of PGC1α (a master regulator of mitochondrial biogenesis). We conclude that urine metabolomics is a reliable source for biomarkers of diabetic complications, and our data suggest that renal organic ion transport and mitochondrial function are dysregulated in diabetic kidney disease.
Diabetic kidney disease continues to increase worldwide without much evidence of abating.1 Apart from driving increasing rates of ESRD, progression of kidney disease is a major sign of overall cardiovascular disease and all-cause mortality in patients with diabetes.2 The basis of the progression of diabetic kidney disease remains unknown despite numerous investigations using genomics, transcriptomics, and proteomics.3–5
Metabolomics is a systematic evaluation of small molecules and may provide fundamental biochemical insights into disease pathways. Prior studies have evaluated plasma metabolomics in diabetes6 and ESRD and revealed alterations in branched-chain and aromatic amino acid metabolism and accumulation of metabolites.7 However, with current methods, plasma presents a narrow subset of compounds related to intermediary metabolism. In contrast, urine metabolomics offers a wider range of measurable metabolites8–10 because the kidney is responsible for concentrating a variety of metabolites and excreting them in the urine. In addition, urine metabolomics may offer direct insights into biochemical pathways linked to kidney dysfunction. However, the relationship of urine metabolomics with diabetic kidney disease has not been comprehensively evaluated. To that end, in the present study we evaluated cohorts of patients with diabetes with and without overt kidney disease using a rigorous quantitative urine metabolomics approach. The studies reveal a novel and characteristic signature for diabetic kidney disease.
Results
Clinical Characteristics of Screening and Validation Cohorts
The healthy control group (n=23) consisted of patients primarily drawn from the San Diego region. They had a mean age of 37.7 years, 52% were white, and 26% were female. The clinical characteristics of the screening cohort with diabetes and CKD, the validation cohort with diabetes and CKD, and the cohorts of patients with type 1 (n=32) or type 2 (n=41) diabetes without CKD are shown in Table 1. The screening cohort (n=24) consisted exclusively of patients with type 2 diabetes with CKD from the San Diego region, who were predominantly male; 42% were white. The validation group included patients with type 1 (n=12) or type 2 (n=49) diabetes from different regions across the United States and Finland (characteristics of the validation group separated by diabetes type are listed in Supplemental Table 1). Although patients in the screening and validation groups differed with respect to ethnicity and geographic location, they were similar with respect to estimated GFR (eGFR) and overall medical management (Table 1).
Table 1.
Baseline characteristics by group
| Characteristic | Screening DM+CKD (n=24) | Validation DM+CKD (n=61) | Type 1 DM without CKD (n=32) | Type 2 DM without CKD (n=41) |
|---|---|---|---|---|
| Age (yr) | 64.2±7.8 | 60.6±11.9 | 45.7±10.4 | 59.1±6.8 |
| Race | ||||
| White | 10 (42) | 38 (63) | 32 (100) | 40 (98) |
| Nonwhite | 14 (58) | 22 (37) | 0 (0) | 1 (2) |
| Women | 3 (12) | 25 (41) | 15 (47) | 18 (44) |
| BMI (kg/m2) | 34.2±6.2 | 32.4±7.2 | 25.5±3.5 | 24.8±3.7 |
| Ever smoking | 14 (61) | 29 (49) | 18 (56) | 24 (59) |
| Systolic BP (mmHg) | 131.8±20.5 | 131±14.9 | 137.9±17.5 | 138.9±15.7 |
| Diastolic BP (mmHg) | 70.9±10.6 | 72.2±8.0 | 76.6±8.2 | 82.5±8.6 |
| Type 2 DM duration (yr) | 16.0 (10.0, 21.0) | 15.0 (9.5, 23.0) | — | 11.0 (8.0, 15.0) |
| Type 1 DM duration (yr) | — | 30.0 (24.2, 37.8) | 31.5 (25.0, 37.2) | — |
| HbA1c (%) | 7.2±1.1 | 7.4±1.3 | 7.9±1.0 | 8.4±1.3 |
| Serum creatinine (mg/dl) | 2.2±0.6 | 2.2±0.9 | 0.8±0.2 | 0.9±0.2 |
| Albumin-to-creatinine ratio | 0.81 (0.12, 1.28) | 0.21 (0.06, 1.09) | 0.08 (0.03, 0.81) | 0.09 (0.05, 0.28) |
| eGFR (ml/min per 1.73 m2) | 35.5±10.9 | 36.0±13.4 | 106±25.8 | 79.8±15.2 |
Values are expressed as n (%), mean ± SD, or median (quartile 1, quartile 3). DM, diabetes mellitus; BMI, body mass index; HbA1c, hemoglobin A1c.
Urine Metabolites in Healthy Controls and Diabetic Patients with CKD
Ninety-four separate metabolites were measured in the healthy control group (n=23) and the screening cohort (n=24). With use of the Benjamini-Hochberg step-down approach for false discovery rate correction, 17 metabolites were found to be significantly different in the screening cohort compared with the healthy controls (Table 2). In the validation cohort of patients with diabetic kidney disease, urine metabolomics were analyzed with the same platform. We observed a high concordance rate of the urine metabolites in the validation group, as urine concentrations of 13 of the 17 metabolites identified in the screening cohort were confirmed to be statistically significantly different from those in the healthy control sample (Table 2). Under the null hypothesis of no association between these markers and diabetic kidney disease, we would expect not more than 3 of the 17 markers to be replicated in the validation set just by chance.11 Hence, recapturing 13 of 17 suggests that this set of markers is likely associated with diabetic kidney disease. In addition, the direction of association was consistent, as all 13 validated urine metabolites had lower concentrations in both groups of patients with diabetic kidney disease compared with the healthy control sample (Table 2).
Table 2.
Comparison of metabolites in validation and screening groups versus healthy control group
| Metabolite | Screeninga versus Control (95% CI) (%)b | P Valuec | Validationd versus Control (95% CI) (%)b | P Value c |
|---|---|---|---|---|
| 3-methyl adipic acid | −85.82 (−93.86 to −67.25) | 2.72 E−05 | −89.07 (−92.62 to −83.8) | 6.51 E-18 |
| 2-methyl acetoacetate | −75.26 (−90.36 to −36.51) | 0.004641 | −88.56 (−93.09 to −81.04) | 8.10 E-13 |
| 3-methyl crotonyl glycine | −66.72 (−81.29 to −40.8) | 0.0003909 | −74.71 (−82.24 to −63.98) | 3.01 E-11 |
| 2-ethyl 3-OH propionate | −46.13 (−61.01 to −25.55) | 0.000429 | −51.48 (−59.92 to −41.27) | 8.83 E-11 |
| 3-hydroxy isovalerate | −77.98 (−86.6 to −63.8) | 2.47 E−07 | −71.73 (−80.24 to −59.54) | 7.28 E-10 |
| Glycolic acid | −67.6 (−77.25 to −53.88) | 9.29 E−08 | −54.76 (−65.06 to −41.41) | 3.69 E-08 |
| Tiglylglycine | −44.07 (−58.47 to −24.67) | 0.0003034 | −54.59 (−65.56 to −40.12) | 2.19 E-07 |
| 3-hydroxy propionate | −59.87 (−74.07 to −37.91) | 0.0001275 | −62.44 (−73.42 to −46.93) | 2.64 E-07 |
| Citric acid | −72.55 (−82.03 to −58.05) | 2.40 E−07 | −73.67 (−83.57 to −57.79) | 2.71 E-07 |
| Homovanillic acid | −51.89 (−70.95 to −20.33) | 0.005496 | −57.59 (−69.88 to −40.29) | 3.59 E-06 |
| Aconitic acid | −46.81 (−60.41 to −28.52) | 9.59 E−05 | −56.68 (−69.16 to −39.14) | 5.13 E-06 |
| 3-hydroxy isobutyrate | −61.61 (−75.59 to −39.62) | 0.0001104 | −63.69 (−77.82 to −40.58) | 0.0001027 |
| Uracil | −67.2 (−82.44 to −38.75) | 0.0008285 | −46.47 (−64.16 to −20.07) | 0.002667 |
| 4-hydroxy hippurate | −5.88 (−9.63 to −1.97) | 0.00444 | −3.95 (−7.63 to −0.12) | 0.04333 |
| N-acetyl aspartate | −72.57 (−82.58 to −56.82) | 9.57 E−07 | 80.97 (−3.88 to 240.72) | 0.06574 |
| Succinic acid | −63.74 (−81.89 to −27.43) | 0.005167 | −22.64 (−50.97 to 22.05) | 0.27 |
| 2-hydroxy butyrate | −35.34 (−52.78 to −11.47) | 0.007674 | 8.58 (−17.66 to 43.19) | 0.56 |
Values are adjusted for age, race, and sex, and metabolites are natural log transformed. CI, confidence interval.
Significant in screening cohort after false discovery rate correction.
Percentage difference between screening or validation groups versus healthy controls.
P values are based on analysis of covariance comparison of validation or screening groups to the healthy controls group.
First 13 rows are significant metabolites in validation cohort after Bonferroni correction (0.05/17=0.0029).
Differentiating Diabetes versus Diabetic Kidney Disease with Urine Metabolomics
To determine whether diabetes itself contributed to the observed mean differences in urine metabolites, patients with diabetes alone (eGFR >60 ml/min per 1.73 m2) were compared with patients with diabetes and CKD (Table 3). After adjustment for potential confounders, including age, sex, body mass index, mean arterial pressure, hemoglobin A1c, and duration of diabetes mellitus, 12 of the 13 metabolites that significantly differed between the validation cohort and healthy controls were also significantly different between patients with diabetes and CKD (n=85) and those with diabetes alone (n=73) using a Bonferroni-adjusted cutoff P value of 0.0038 (0.05/13) to account for 13 metabolite comparisons. The raw geometric means of the 13 metabolites for each group is presented in Supplemental Table 2. In further exploratory analysis, the groups were also separated by type of diabetes, and there remained an overall similar pattern of regulation of urine metabolites in both patients with type 1 and those with type 2 diabetes and CKD compared with their respective control groups (Supplemental Table 3). Eight of 13 urine metabolites remained statistically significant after Bonferroni correction with each type of diabetes with CKD, and an additional three metabolites were reduced in both groups (homovanillic acid, 3-methyl crotonyl glycine, and tiglylglycine) but did not achieve significance at the stringent Bonferroni P value cutoff of 0.0038. One metabolite was significantly different only in the type 1 diabetic group (2-methyl acetoacetate) and not in the type 2 diabetic group, and two metabolites were significantly reduced in the type 2 diabetic group and not with type 1 diabetes with CKD (3-methyl adipic acid and uracil).
Table 3.
Comparison of candidate urine metabolites between patients with diabetes and CKD (n=85) and patients with diabetes without CKD (n=73)
| Metabolite | DM+CKD versus DM–CKD (95% CI) (%)a | P Valueb |
|---|---|---|
| 3-hydroxy isovalerate | −61.11 (−69.81 to −49.91) | 1.11E-11 |
| Aconitic acid | −45.4 (−54.18 to −34.93) | 2.22E-10 |
| Citric acid | −65.96 (−74.31 to −54.9) | 3.82E-12 |
| 2-ethyl 3-OH propionate | −43.82 (−52.26 to −33.89) | 8.80E-11 |
| Glycolic acid | −54.19 (−62.54 to −43.99) | 2.18E-12 |
| Homovanillic acid | −38.72 (−53.04 to −20.04) | 0.0003812 |
| 3-hydroxy isobutyrate | −58.35 (−68.88 to −44.25) | 2.00E-08 |
| 2-methyl acetoacetate | −49.74 (−68.29 to −20.35) | 0.00367 |
| 3-methyl adipic acid | −48.63 (−58.74 to −36.05) | 1.42E-08 |
| 3-methyl crotonyl glycine | −41.52 (−57.72 to −19.1) | 0.001349 |
| 3-hydroxy propionate | −22.54 (−42.29 to 3.97) | 0.08851 |
| Tiglylglycine | −35.71 (−50.53 to −16.44) | 0.001097 |
| Uracil | −50.1 (−63.93 to −30.97) | 4.04E-05 |
Values are adjusted for age, race, and sex; mean arterial pressure, body mass index, hemoglobin A1c, and diabetes duration; metabolites are natural log transformed. DM, diabetes mellitus; CI, confidence interval.
Percentage difference between DM+CKD versus DM–CKD groups.
P values based on based on analysis of covariate comparison of DM+CKD versus DM–CKD; Bonferroni-adjusted P value cutoff is 0.0038 to adjust for 13 significant metabolites from validation cohort.
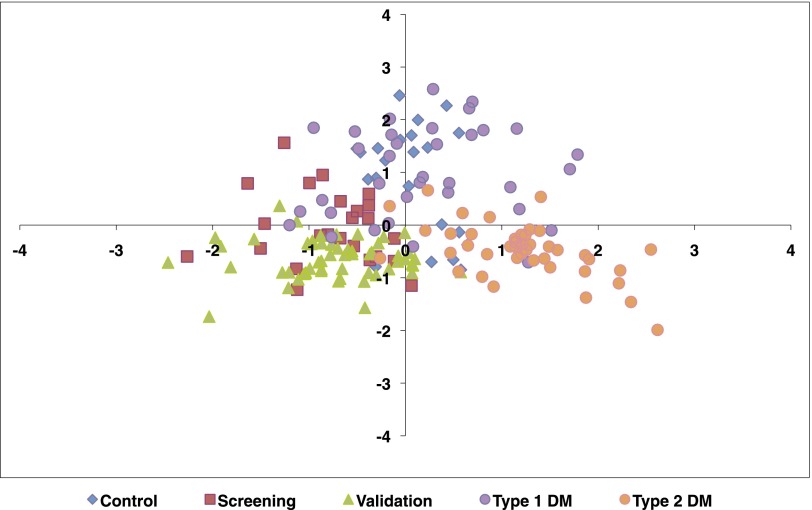
Principal components (PC) analysis of the 13 metabolites demonstrated that the first two components (PC1 and PC2) separated the groups reasonably well (Figure 1). The patients with type 1 and type 2 diabetes with CKD co-migrated along the left horizontal axis. The patients with type 2 diabetes without CKD were isolated on the right horizontal axis. Interestingly, the healthy control group and the type 1 diabetic group without CKD co-migrated along the positive vertical axis. The component loadings of PC1 and PC2 are listed in Supplementary Table 4.
Figure 1.
Principal components analysis reveals separation of diabetic CKD from healthy controls and diabetes without CKD. The figure shows the plot of principal component 1 (x-axis) versus principal component 2 (y-axis). Blue diamonds represent the control group, red squares represent the screening group, green triangles represent the validation group, purple circles represent the type 1 diabetes group without CKD, and orange circles represent the type 2 diabetes group without CKD.
A cohort of patients (n=12) with biopsy-proven FSGS who were refractory to standard immunotherapies and had a similar degree of CKD (mean eGFR, 43.2±25 ml/min per 1.73 m2) were also evaluated by the same metabolomics platform to determine whether the signature of diabetic kidney disease varies from another type of CKD. As shown in Supplemental Table 5, 5 of 13 metabolites remained statistically significant, demonstrating that several metabolites appear to be specific for diabetic CKD. There is also overlap in the urine metabolomic profile in patients with nondiabetic CKD as 8 of the 13 urine metabolites did not significantly differ between diabetic CKD and FSGS-CKD.
Regulation of Urinary Metabolite Differences
To examine dose-response associations between each of these 13 metabolites and common markers of kidney disease in diabetes, we evaluated whether the urine concentrations of the 13 metabolites were statistically related to eGFR or albuminuria (albumin-to-creatinine ratio) levels, in linear regression models adjusted for potential confounders, (Supplemental Table 6). In this exploratory analysis, 5 of the metabolites showed a significant (P<0.05) correlation with eGFR (3-hydroxy isovalerate, aconitic acid, glycolic acid, uracil and citric acid). A separate set of three metabolites were associated (P<0.05) with albuminuria (2-methyl acetoacetate, 3-methyl crotonyl glycine, and 3-methyl adipic acid). Thus, many of the metabolites were not simply correlated with reduced levels of eGFR and/or increased albuminuria and may be independently regulated. The 13 metabolites were also measured in the plasma of a subset of the healthy controls and the group with diabetic kidney disease to determine whether circulating levels may be informative. As noted in Table 4, 7 of the 13 metabolites could be reliably measured in the plasma; of these, 4 were increased in patients with diabetic kidney disease, 2 were unchanged, and 1 was significantly reduced in the circulation compared with healthy controls.
Table 4.
Plasma levels of metabolites in patients with diabetes and CKD and healthy controls
| Metabolite | DM+CKD (n=16) versus Healthy Controls (n=16) (95% CI) (%)a | P Valueb |
|---|---|---|
| 3-hydroxy isovalerate | 185.1 (74.13 to 366.78) | 0.0001621 |
| Aconitic acid | 127.34 (39.6 to 270.22) | 0.001796 |
| Citric acid | 150.14 (25.21 to 399.7) | 0.01125 |
| 2-ethyl 3-OH propionate | 43.38 (8.43 to 89.6) | 0.01335 |
| Glycolic acid | −3.66 (−17.11 to 11.97) | 0.62 |
| Homovanillic acid | ND | – |
| 3-hydroxy isobutyrate | −95.63 (−96.71 to −94.18) | 1.90E-19 |
| 2-methyl acetoacetate | ND | – |
| 3-methyl adipic acid | ND | – |
| 3-methyl crotonyl glycine | ND | – |
| 3-hydroxy propionate | 7.38 (−28.58 to 61.46) | 0.72 |
| Tiglylglycine | ND | – |
| Uracil | ND | – |
DM, diabetes mellitus; CI, confidence interval; ND, not detected.
Percentage difference for DM+CKD patients versus healthy controls.
P values based on analysis of covariance comparison of DM+CKD versus healthy controls.
We found that several metabolites affected by diabetic kidney disease were water-soluble organic anions and may therefore be regulated by members of the organic anion transporter (OAT) subfamily of SLC22 transporters. Indeed, on the basis of in vitro studies of microinjected oocytes and/or transfected cell lines (Supplemental Table 7; see Km and Ki values), several of these organic anions were found to be substrates for OAT1 or OAT3.12,13 For example, homovanillic acid is a substrate for OAT1 (SLC22A6, also known as NKT) and OAT3.14 Furthermore, because of the reduction in the urine of a similar set of molecules in the Oat1 knockout13,15,16 (Supplemental Table 7), we analyzed the expression of OAT1 and OAT3 in microdissected tubular segments from kidney biopsy specimens from patients with biopsy-proven diabetic nephropathy (mean values ± SD: age, 60±10.5 years; serum creatinine, 2.99±0.49 mg/dl; eGFR, 26±4.2 ml/min per 1.73 m2; and overt proteinuria) and nondiseased kidneys. This nephron segment expressed less than half the normal levels of OAT1 and OAT3 (Table 5). Taken together, these data suggest that the altered metabolite profiles seen in patients with diabetic kidney disease may be related to decreased organic anion elimination due to diminished OAT1 and OAT3 expression in the cortical tubule.
Table 5.
Regulation of OAT1 and OAT3 gene expression in diabetic kidneys
| Probe Set ID | Entrez Gene ID | Gene Symbol | Representative Public ID | Gene Title | Fold Change | Q Value (%) |
|---|---|---|---|---|---|---|
| 210343_s_at | 9356 | SLC22A6 | AF124373 | Solute carrier family 22 (organic anion transporter 1), member 6 | 0.556 | 0.029 |
| 216599_x_at | 9356 | SLC22A6 | AJ271205 | Solute carrier family 22 (organic anion transporter 1), member 6 | 0.591 | 0.088 |
| 221298_s_at | 9376 | SLC22A8 | NM_004254 | Solute carrier family 22 (organic anion transporter 3), member 8 | 0.519 | 0.029 |
| 231352_at | 9376 | SLC22A8 | AW025165 | Solute carrier family 22 (organic anion transporter 3), member 8 | 0.533 | 0.029 |
Biochemical Implications of Urinary Metabolite Reduction in Diabetic CKD
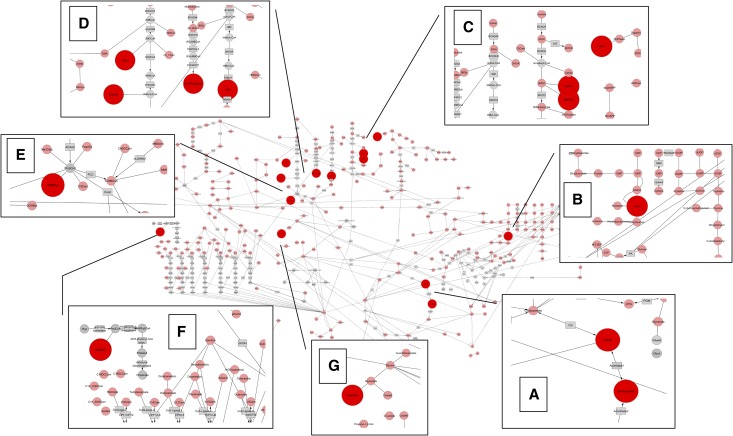
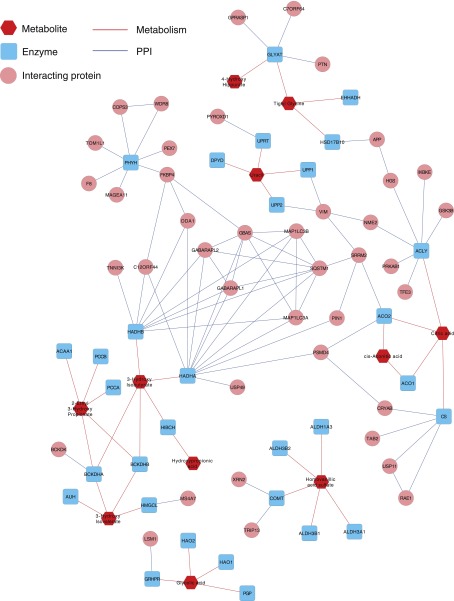
The data separating the diabetic CKD group from the other groups indicated a panel of metabolites (n=13) that provides a metabolomic signature of diabetic kidney disease and may provide biochemical insight. As shown in Figure 2, metabolites from the Krebs cycle, pyrimidine metabolism, amino acid, propionate, fatty acid, and oxalate metabolism were all significantly reduced in the urine of patients with diabetic kidney disease. Eleven of 13 metabolites were connected by network analysis using a protein-protein interaction network linking the enzymes involved in production of the metabolites (Figure 3). Two of the 13 metabolites (glycolic acid and homovanillic acid) were out of network. A classification table based on organelle localization of the metabolites and the enzymes producing the metabolites identified that 12 of 13 were associated with mitochondria, with 10 exclusively produced in mitochondria (Table 6). Because of a reduction in all of the mitochondrial metabolite markers, the combined biochemical and systems analysis suggests that there may be a generalized reduction in several aspects of mitochondrial function in patients with diabetic kidney disease.
Figure 2.
Biochemical pathway analysis reveals prominent role of Krebs cycle and amino acid metabolites in urine signature of diabetic kidney disease. Pink nodes represent chemicals that are measured, and large red nodes represent metabolites that are altered in diabetic renal disease. Gray nodes represent compounds that are not measured and gray rectangles represent enzymes. Compounds whose concentrations are significantly altered are shown in the magnified inserts, falling in the following areas of metabolism: (A) Krebs cycle (citrate, aconitate). (B) Pyrimidine metabolism (uridine). (C) Leucine catabolism (3-hydroxyisovaleric acid [3HIVA], 3-methylcrotonylglycine [3MCGly]) and tyrosine metabolism (vanillylmandelic acid [VMA]). (D) Valine catabolism (3-hydroxyisobutyric acid [HIBA]) and the isoleucine catabolism L-pathway (tiglylglycine [TigGly], 2-methylacetoacetic acid [2MAcAc]) and R-pathway (2-ethyl-3-hydroxypropionate [2E3Hpropionate]). (E) Propionate metabolism (3-hydroxypropionate [3OHProp]). (F) Branched-chain fatty acid metabolism (3-methyladipic acid [3MAdipic]). (G) Oxalate metabolism (glycolic acid).
Figure 3.
Network analysis of metabolites with enzymes reveals connectivity of 11/13 metabolites in large network. Proteins that interact with these enzymes are also shown (i.e., first neighbors of the enzymes on protein-protein interaction [PPI] network). The map was drawn using Cytoscape.
Table 6.
Subcellular localization of enzymes producing the metabolites associated with diabetic kidney disease
| Metabolite Decreased in Diabetic Kidney Disease | HMDB ID (Molecular Weight) | Function in Intermediary Metabolism | Enzyme(s) Producing the Metabolite | Subcellular Location |
|---|---|---|---|---|
| 3-hydroxy isovalerate (3-OH 3-methyl butyric acid) | HMDB00754 (118.131) | Leucine metabolite | 3-methyl glutaconyl CoA hydratasea | Mitochondria |
| Glycolic acid | HMDB00115 (76.051) | Glycine (peroxisomes) and 4OH-proline (mitochondria) | NADPH-glyoxylate reductase | Peroxisomesb Mitochondria |
| Citric acid | HMDB00094 (192.124) | Krebs cycle and lipid synthesis | Citrate synthase | Mitochondria |
| 2-ethyl 3-OH propionate (2-ethyl hydracrylic acid) | HMDB00396 (118.131) | Isoleucine metabolite | From R-pathway of isoleucine metabolism (when 2MBDH is deficient) | Mitochondria |
| Uracil (uridine) | HMDB00300 (112.087) | Pyrimidine synthesis | Coenzyme Q10:dihdyroorotate dehydrogenase, uridine monophosphate synthetase (UMPS) | Mitochondria |
| 3-hydroxy isobutyrate | HMDB00023 (104.104) | Valine metabolite | 3HIBA CoA hydratase | Mitochondria |
| Aconitic acid | HMDB00072 (174.108) | Krebs cycle | Aconitase | Mitochondria |
| 3-methyl adipic acid | HMDB00555 (160.168) | Indicates incomplete branched-chain fatty acid oxidation | From decreased intake of phytanic acid or increased alpha oxidation of branch chain fatty acids | Peroxisome |
| Tiglylglycine | HMDB00959 (157.167) | Isoleucine metabolite | FAD+ 2-methylbutyryl-CoA dehydrogenase (2MBD) | Mitochondria |
| 3-methyl-crotonyl glycine | HMDB00459 (157.167) | Leucine metabolite | FAD+ isovaleryl-CoA dehydrogenase | Mitochondria |
| 2-Methyl Acetoacetate | HMDB03771 (116.115) | Isoleucine metabolite | NAD+ 2-methyl-3-hydroxy butyryl CoA dehydrogenase | Mitochondria |
| Homovanillic acid | HMDB00118 (182.173) | Dopamine metabolite | Catechol-O-methyl transferase (COMT) and monoamine oxidase (MAO) | Cytosolb Mitochondria |
| 3-hydroxy propionate (hydracrylic acid) | HMDB00700 (90.078) | Isoleucine, valine, threonine, and methionine metabolite | 2-methylacetoacetyl CoA thiolase (Ile); NAD+-methylmalonate semialdehyde dehydrogenase (Val); NAD+ 2-ketobutyrate dehydrogenase (Thr and Met) | Mitochondria |
2MBD, 2-methylbutyryl-CoA dehydrogenase in isoleucine metabolism.
Via alternative substrate utilization: 3-methylbutyryl-CoA versus 3-methylglutaconyl-CoA.
As an alternative substrate: 2-ethyl-3-hydroxypropionyl-CoA versus 2-methyl-3-hydroxybutyryl-CoA.
Validation of Biochemical Pathways
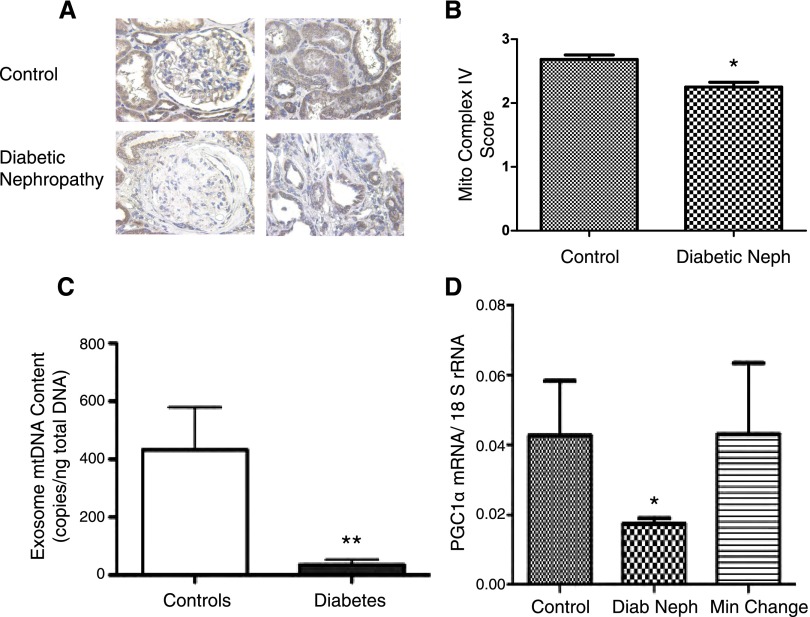
To determine whether there was indeed a reduction in mitochondria in patients with diabetic kidney disease, several approaches were pursued. We performed immunostaining with an antibody for cytochrome C oxidase (complex IV) on archived paraffin-embedded tissue sections of kidney biopsy samples from patients with normal kidneys and patients with diabetic nephropathy (n=5 per group). There was a reduction of mitochondrial cytochrome C oxidase in the diabetic kidneys compared with control kidneys (Figure 4, A and B). Second, mitochondrial DNA in urine exosomes was measured because urine exosomes are largely derived from renal epithelial cells17 and may reflect the intracellular constituents of glomerular and tubular epithelial cells. Consistent with the kidney biopsy findings, we observed a reduction in urine exosomal mitochondrial (mtDNA) in patients with diabetic kidney disease versus controls (Figure 4C). The reduction in mitochondrial protein and mtDNA indicate an overall reduction in mitochondrial biogenesis.
Figure 4.
Mitochondrial biogenesis reduced in diabetic nephropathy. (A) Representative immunostaining of cytochrome C oxidase (complex IV) subunit II staining in normal and diabetic kidney (original magnification ×40). (B) Semi-quantitative analysis (n=5 per group; P<0.05). (C) Copy number of exosome-protected, deoxyribonuclease I–resistant mtDNA in urinary exosomes from patients with diabetic kidney disease (diabetes) versus healthy controls (controls) (controls, 432±147 copies/ng; diabetes, 36±18 copies/ng; P≤0.01; n=16 per group). (D) Gene expression of PGC1α from samples from patients who underwent pretransplant biopsies (controls, n=8), patients with diabetic nephropathy (diab neph, n=14), and patients with minimal-change disease (min change, n=6).
To explain a pathway by which mitochondrial proteins may be reduced in diabetic kidney disease, we examined gene expression of PGC1α, a key regulator of mitochondrial biogenesis.18,19 Quantitative RT-PCR for PGC1α was performed on microdissected cortical tubulointerstitial samples from patients with diabetic kidney disease, minimal-change disease (a nonprogressive proteinuric disease), and pretransplant biopsies as controls (Figure 4D). The PGC1α mRNA expression was reduced in samples with diabetic kidney disease (fold-change, 0.4; P<0.05), whereas the expression was unchanged in minimal-change disease compared with controls.
Discussion
The results of the present study demonstrate that urine metabolomics can be a clinically useful platform to provide a metabolic signature and provide novel biochemical insights in patients with diabetic kidney disease. Of 94 urine metabolites that were examined, 13 metabolites were consistently and significantly reduced in urine from patients with diabetic kidney disease compared with healthy controls. Twelve of the 13 metabolites remained statistically significant compared with values in patients with diabetes and no CKD, and 5 of the 13 remained significant compared with values in patients with CKD from FSGS. The regulation of several of the metabolites altered in diabetic kidney disease may be in part due to reduced renal filtration and possibly via downregulation of the OATs. Using biochemical and systems biology tools, we further demonstrate that diabetic kidney disease is characterized by suppressed mitochondrial function. Independent studies with exosomal analysis and immunohistochemistry validated the hypothesis that mitochondria are reduced in patients with diabetic kidney disease.
To our knowledge, this is the first study to measure the urine metabolome in patients with diabetic kidney disease with a highly quantitative, targeted gas chromatography-mass spectrometry method that has been optimized for urine and used for clinical diagnosis. A recent study from our collaborators20 studied the association of urine metabolites with progression to albuminuria over a mean follow-up of 5.5 years. Using liquid chromatography-mass spectrometry, they identified hippuric acid to be decreased in the group with progressive disease, and S-(3-oxododecanoyl) cysteamine and acylcarnitines were increased. In our study, we found 4-OH hippurate to be marginally reduced in patients with diabetic kidney disease, similar to the finding for the related hippurate in the FinnDiane study;20 both compounds are glycine esters of a single exogenous metabolite or its hydroxylation derivative. In our study, several of the metabolites that were altered in patients with diabetic kidney disease have been described to be regulated by the OAT transporters.13 Oat1 and Oat3 are the rate-limiting step in the renal proximal tubule uptake of numerous drugs, toxins, and metabolites and play a key role in energy metabolism.21,22 Our study is the first to find that OAT1 and OAT3 are reduced in patients with diabetic kidney disease; on the basis of data from the Oat1 knockout mouse,13 it is possible that the OAT1/3 reduction may contribute to reduced urinary levels of many of the metabolites identified.
In addition, 12 of the 13 urine metabolites that made up the metabolomic signature for diabetic kidney disease were identified to be produced in mitochondria or are largely regulated by mitochondrial function. The reduction of mitochondrial function suggested by the metabolomic signature was confirmed with urine exosomal analysis of mtDNA and immunohistochemistry demonstrating reduced mitochondrial content. The basis for reduced mitochondrial content and function will be difficult to address with clinical samples. A potential mechanism to explain reduced mitochondrial content in the diabetic kidney is reduction of the co-activator PGC1α. In an independent series of samples, PGC1α mRNA levels were reduced in human diabetic kidneys. Because PGC1α is the major co-activator to regulate mitochondrial biogenesis,18 a reduction in PGC1α would be expected to lead to reduced mitochondrial biogenesis. Of note, PGC1α gene expression is reduced in muscle tissue of patients with type 2 diabetes and may be due to epigenetic alterations of the PGC1α promoter.19 In a recent separate study with a mouse model of diabetic kidney disease, PGC1α was found to be reduced in the diabetic kidney in association with reduction of mitochondrial biogenesis. Stimulation of AMPK led to upregulation of PGC1α, stimulation of mitochondrial biogenesis and improvement in renal functional and histologic parameters.23
Because our study had a cross-sectional design, we cannot assess cause and effect. The change in urine metabolites may be partly due to reduction in glomerular filtration itself or may precede and potentially contribute to renal functional decline. Plasma measurement of the metabolome using the same methods as for urine revealed that only 7 of the 13 metabolites were measurable in plasma. Increased plasma levels of 4 of the 7 may be due to reduced renal clearance; however, 2 metabolites were unchanged in the circulation and 1 metabolite was significantly reduced in plasma. As urine levels of several of the same metabolites are regulated by OATs, further evaluation of kidney function in the OAT1/3 knockout mice is warranted. Future longitudinal studies in patients will help to determine whether the urine metabolomic signature provides additive predictive value for kidney functional decline and associated comorbid outcomes. In addition, studies with more patients in the nondiabetic FSGS group will be required to define a distinct metabolomic signature for CKD with FSGS.
In conclusion, urine metabolomics provides a novel, noninvasive method to identify biomarkers and biochemical insights that are associated with kidney function and are highly consistent across patient populations with diabetes and kidney disease. The distinct metabolomic signature indicates that mitochondrial function is reduced in patients with diabetic kidney disease relative to healthy controls, and independent studies confirmed this hypothesis. Ultimately, these findings may identify new therapeutic targets for diabetic kidney disease and serve as novel biomarkers of kidney function.
CONCISE METHODS
Study Populations
Five separate clinical groups were obtained for analysis. A control group of healthy persons (n=23) and a screening group of 24 consecutive patients with type 2 diabetes and the presence of CKD (eGFR ≤ 60 ml/min per 1.73 m2) were enrolled from the San Diego, California, region. The screening group provided 24-hour urine collections during the same time interval as the control group. An independent validation group was composed of 61 patients who had a history of diabetes and reduced eGFR and also had undergone 24-hour urine collections. The patients in the validation group included patients with type 1 (n=12) or type 2 (n=49) diabetes with kidney disease from various geographic locations within the United States (enrolled from the Pirfenidone Study24) and from Finland (as part of the FinnDiane Study).2 Additional cohorts included patients from both the Pirfenidone and FinnDiane studies with type 1 diabetes (n=32) and patients with type 2 diabetes (n=41) who had an eGFR >60 ml/min per 1.73 m2 at the time of 24-hour urine collection. A group with biopsy-proven refractory FSGS (n=12) and with stored aliquots of 24-hour urine collections (to reflect nondiabetic CKD) were also evaluated by urine metabolomics using the same platform. All groups had urine aliquots stored at ≤−70°C for 3 months to 6 years before analysis. The institutional review board of the University of California, San Diego (UCSD); Helsinki University Central Hospital, Finland; and the National Institutes of Health Clinical Research Center, Bethesda, Maryland, approved the study, and all participants provided written informed consent.
Urine and Plasma Metabolomics
Aliquots of the frozen 24-hour urine collection were thawed and analyzed for creatinine content before being processed for analysis at the UCSD Biochemical Genetics and Metabolomics Laboratory. Urine and plasma samples underwent oximation of ketoacids with pentafluorobenzylhydroxylamine, lyophilization, isolation of organic acids by liquid partition chromatography on silica (42% 2-methyl-2-butanol in chloroform), evaporation of the eluate, and silylation of the dry residue with Trisil-N,O-bis (trimethylsilyl) trifluoroacetamide.25 Urine aliquots corresponding to 1 μmol of creatinine or 2 μl of plasma were applied by injection onto a 30 m × 0.32 mm column (Agilent DB-5) in a gas chromatogram (Agilent 5890) and eluted with a 4°C/min gradient of 70–300°C; analytes were detected by electron impact mass spectrometry (Agilent 5973 mass selective detector). Each compound was identified by spectrum and confirmed ratio of a qualifying ion and quantifying ion. The quantifying ion’s integrated current was used to estimate concentration based on standard curves for targeted metabolites or based on a ratio to 4-nitrophenol or the oximated derivative of 2-ketocaproic acid. As per our procedure, approved by the College of American Pathologists, we maintain 4- to 6-point calibration curves on 83 adducts of 76 compounds; for other compounds (e.g., when authentic standards are not available), concentrations are estimated relative to the quantity of the appropriate internal standard (4-nitrophenol or 2-oxocaproate). The results are reported in µmol organic acid per mmol creatinine for urine and μmol/L for plasma. Similar analyses were performed in the urine of OAT1 and OAT3 knockout mice as previously described.13
Biochemical and Protein-Metabolite Network
The 13 metabolites that were significantly different for diabetic kidney disease were searched for using the global map of human metabolic pathway in the KEGG database (http://www.genome.jp/kegg/pathway/map/map01100.html). Of the metabolites we searched, only 7 could be mapped to the pathway. For these, we were able to identify their associated enzymes. For the rest, we manually listed the enzymes based on consensus knowledge from the UCSD Biochemical Genetics Laboratory’s internal database. We then downloaded human protein-protein interactions from the BioGRID database (http://thebiogrid.org/) and searched for interactions involving the known enzymes. The interaction network was constructed and visualized through Cytoscape (http://www.cytoscape.org/).
Kidney Sections
Unstained slides of kidney samples from biopsy specimens diagnosed as diabetic nephropathy (n=5) or normal (n=5) kidneys were obtained from Dr. Agnes Fogo of Vanderbilt University. These samples were exempt from the requirement of informed consent, according to the institutional review board's approved use of organs and tissues from deceased donors for research. Unstained sections were processed for immunostaining using standard protocols. Sections were incubated first with mouse anti–cytochrome C oxidase, subunit II (Abcam) primary antibody, and subsequently with biotin-conjugated α-mouse secondary antibody (Santa Cruz Biotechnology) and horseradish peroxidase-streptavidin (BD Biosciences). Labeling was visualized with chromogen diaminobenzidine (Vector Labs, SK-4100), and sections were counterstained with hematoxylin. Sections were digitally scanned at 20× magnification using the Aperio Scanscope at Sanford Burnham Medical Research Institute (La Jolla, CA). Staining was assessed using a semi-quantitative scoring method by an observer masked to the identity of the sections. Significance was determined by a t test.
Urine Exosome Analysis
Aliquots of 24-hour urine collections were thawed, and exosomes were purified and concentrated 100-fold by volume exclusion. Exosome protein was measured by Pierce BCA assay and total extra- and intra-exosomal double-stranded DNA was quantified by PicoGreen fluorescence. Extraexosomal DNA was hydrolyzed by treatment with DNAse I. Intraexosomal mtDNA was quantified by real-time quantitative PCR using two primer pairs: one directed to the ND4 region of mtDNA in the major arc and the other pair directed to the 16S region in the minor arc. Copy numbers of mtDNA are reported in copies per µg of exosomal DNA.
Intrarenal Gene Expression Analysis
Human renal biopsy specimens were collected in an international multicenter study, the European Renal cDNA Bank–Kröner-Fresenius biopsy bank (ERCB-KFB; see the acknowledgments for participating centers).26 Biopsy specimens were obtained from patients after informed consent and with approval of the local ethics committees. In a hybridization experiment, Affymetrix HG-U133A microarrays were initially hybridized with cDNA from cortical tubulo-interstitial specimen.27 For PGC-1α confirmatory real-time RT-PCR analyses were performed on microdissected specimen from clinically indicated biopsies from additional patients with diabetic kidney disease (n=14) or minimal-change disease (n=6) or from pretransplant kidney biopsies from living donors as controls (n=8).
Statistical Analyses
Distributions of all metabolites were checked, and because of the skewed distributions, natural log transformation was applied to all metabolites, with 1 added where appropriate. To initially compare the screening group with the control group, analysis of covariance was used, adjusting for age, race, and sex. All 94 metabolites were compared in this initial analysis, and a false discovery rate method was used to determine a significance cut point.28 A total of 17 metabolites met the false discovery rate threshold. These 17 were carried forward for a validation analysis using a Bonferroni-adjusted significance level of P=0.0029 (0.05/17). Because metabolites were natural log transformed, for ease of interpretation, results are presented as a percentage (95% confidence interval) compared with the healthy control sample for each group. Percentages were obtained using the transformation (eβ − 1) × 100. Unadjusted geometric means (95% confidence intervals) of the metabolites were also calculated. Additional analysis of covariance models, adjusted for the above-listed confounders and clinical factors (i.e., mean arterial pressure, body mass index, hemoglobin A1c), were used to compare patients with diabetes mellitus and CKD to those with diabetes mellitus without CKD and FSGS (CKD-DM) groups.
To determine whether a parsimonious profile could distinguish between the 5 groups (i.e., control, screening, validation, and type 1 and type 2 diabetes without CKD), we performed a principal components analysis with varimax rotation of the factors. Scree plots were used to determine an adequate number of components, and results of PC1 versus PC2 were plotted for all groups.
To examine a dose-response relationship between eGFR and urine albumin-to-creatinine ratio levels in the DM+CKD group, with the validated metabolites, we applied linear regression models adjusted for age, race, and sex, with separate terms for eGFR or the log2 of urine albumin-to-creatinine ratio terms for each group. The screening and validation groups were combined for this analysis.
SAS software, version 9.2 (SAS Institute, Inc., Cary, NC), and the free software package R (http://www.r-project.org/) were used for all statistical analyses.
Disclosures
K.S. is founder of Clinical Metabolomics (ClinMet) Inc.
Supplementary Material
Acknowledgments
We thank Dr. Agnes Fogo of Vanderbilt University for providing slides from human kidney biopsies and all participating centers of the European Renal cDNA Bank - Kröner-Fresenius biopsy bank and their patients for their cooperation (participating centers listed on http://www.research-projects.uzh.ch/p9291.htm).
K.S. was supported by grants from the Juvenile Diabetes Research Foundation (VA Merit Award 5101BX000277) and National Institute of Diabetes and Digestive and Kidney Diseases (1DP3DK094352). B.K. and A.V.M. were supported by a National Institutes of Health T32 Training Grant. The Finnish Diabetic Nephropathy Study was supported by grants from the Folkhälsan Research Foundation, the Wilhelm and Else Stockmann Foundation, and the Liv och Hälsa Foundation. P.G. and C.F. were supported by the Folkhausen Research Foundation. R.K.N. and L.W. were supported in part by the UCSD Christini Fund, the Wright Family Foundation, and the Lennox Foundation. C.D.C. was supported by grants from the Else Kröner-Fresenius Foundation (A62/04) and the NCCR Kidney. T.I. was funded by the National Resource for Network Biology (P41 GM103504) and the San Diego Center for Systems Biology (P50 GM085764).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2013020126/-/DCSupplemental.
References
- 1.Rosolowsky ET, Skupien J, Smiles AM, Niewczas M, Roshan B, Stanton R, Eckfeldt JH, Warram JH, Krolewski AS: Risk for ESRD in type 1 diabetes remains high despite renoprotection. J Am Soc Nephrol 22: 545–553, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Groop PH, Thomas MC, Moran JL, Wadèn J, Thorn LM, Mäkinen VP, Rosengård-Bärlund M, Saraheimo M, Hietala K, Heikkilä O, Forsblom C, FinnDiane Study Group : The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes. Diabetes 58: 1651–1658, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Susztak K, Böttinger E, Novetsky A, Liang D, Zhu Y, Ciccone E, Wu D, Dunn S, McCue P, Sharma K: Molecular profiling of diabetic mouse kidney reveals novel genes linked to glomerular disease. Diabetes 53: 784–794, 2004 [DOI] [PubMed] [Google Scholar]
- 4.Ewens KG, George RA, Sharma K, Ziyadeh FN, Spielman RS: Assessment of 115 candidate genes for diabetic nephropathy by transmission/disequilibrium test. Diabetes 54: 3305–3318, 2005 [DOI] [PubMed] [Google Scholar]
- 5.Sharma K, Lee S, Han S, Lee S, Francos B, McCue P, Wassell R, Shaw MA, RamachandraRao SP: Two-dimensional fluorescence difference gel electrophoresis analysis of the urine proteome in human diabetic nephropathy. Proteomics 5: 2648–2655, 2005 [DOI] [PubMed] [Google Scholar]
- 6.Newgard CB, An J, Bain JR, Muehlbauer MJ, Stevens RD, Lien LF, Haqq AM, Shah SH, Arlotto M, Slentz CA, Rochon J, Gallup D, Ilkayeva O, Wenner BR, Yancy WS, Jr, Eisenson H, Musante G, Surwit RS, Millington DS, Butler MD, Svetkey LP: A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 9: 311–326, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang TJ, Larson MG, Vasan RS, Cheng S, Rhee EP, McCabe E, Lewis GD, Fox CS, Jacques PF, Fernandez C, O’Donnell CJ, Carr SA, Mootha VK, Florez JC, Souza A, Melander O, Clish CB, Gerszten RE: Metabolite profiles and the risk of developing diabetes. Nat Med 17: 448–453, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweetman L, Nyhan WL: Detailed comparison of the urinary excretion of purines in a patient with the Lesch-Nyhan syndrome and a control subject. Biochem Med 4: 121–134, 1971 [DOI] [PubMed] [Google Scholar]
- 9.Nyhan WL, James JA, Teberg AJ, Sweetman L, Nelson LG: A new disorder of purine metabolism with behavioral manifestations. J Pediatr 74: 20–27, 1969 [DOI] [PubMed] [Google Scholar]
- 10.Aramaki S, Lehotay D, Nyhan WL, MacLeod PM, Sweetman L: Methylcitrate in maternal urine during a pregnancy with a fetus affected with propionic acidaemia. J Inherit Metab Dis 12: 86–88, 1989 [DOI] [PubMed] [Google Scholar]
- 11.Natarajan L, Pu M, Messer K: Exact statistical tests for the intersection of independent lists of genes. Ann Appl Stat 6: 521–541, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nigam SK, Bush KT, Bhatnagar V: Drug and toxicant handling by the OAT organic anion transporters in the kidney and other tissues. Nat Clin Pract Nephrol 3: 443–448, 2007 [DOI] [PubMed] [Google Scholar]
- 13.Eraly SA, Vallon V, Vaughn DA, Gangoiti JA, Richter K, Nagle M, Monte JC, Rieg T, Truong DM, Long JM, Barshop BA, Kaler G, Nigam SK: Decreased renal organic anion secretion and plasma accumulation of endogenous organic anions in OAT1 knock-out mice. J Biol Chem 281: 5072–5083, 2006 [DOI] [PubMed] [Google Scholar]
- 14.Ohtsuki S, Kikkawa T, Mori S, Hori S, Takanaga H, Otagiri M, Terasaki T: Mouse reduced in osteosclerosis transporter functions as an organic anion transporter 3 and is localized at abluminal membrane of blood-brain barrier. J Pharmacol Exp Ther 309: 1273–1281, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Wikoff WR, Nagle MA, Kouznetsova VL, Tsigelny IF, Nigam SK: Untargeted metabolomics identifies enterobiome metabolites and putative uremic toxins as substrates of organic anion transporter 1 (Oat1). J Proteome Res 10: 2842–2851, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaler G, Truong DM, Khandelwal A, Nagle M, Eraly SA, Swaan PW, Nigam SK: Structural variation governs substrate specificity for organic anion transporter (OAT) homologs. Potential remote sensing by OAT family members. J Biol Chem 282: 23841–23853, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou H, Cheruvanky A, Hu X, Matsumoto T, Hiramatsu N, Cho ME, Berger A, Leelahavanichkul A, Doi K, Chawla LS, Illei GG, Kopp JB, Balow JE, Austin HA, 3rd, Yuen PS, Star RA: Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int 74: 613–621, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spiegelman BM: Transcriptional control of mitochondrial energy metabolism through the PGC1 coactivators. Novartis Found Symp 287: 60–63, discussion 63–69, 2007 [PubMed] [Google Scholar]
- 19.Barrès R, Osler ME, Yan J, Rune A, Fritz T, Caidahl K, Krook A, Zierath JR: Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab 10: 189–198, 2009 [DOI] [PubMed] [Google Scholar]
- 20.van der Kloet FM, Tempels FW, Ismail N, van der Heijden R, Kasper PT, Rojas-Cherto M, van Doorn R, Spijksma G, Koek M, van der Greef J, Mäkinen VP, Forsblom C, Holthöfer H, Groop PH, Reijmers TH, Hankemeier T: Discovery of early-stage biomarkers for diabetic kidney disease using ms-based metabolomics (FinnDiane study). Metabolomics 8: 109–119, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ahn SY, Nigam SK: Toward a systems level understanding of organic anion and other multispecific drug transporters: A remote sensing and signaling hypothesis. Mol Pharmacol 76: 481–490, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu W, Dnyanmote AV, Nigam SK: Remote communication through Slc and Abc drug transporter pathways: an update on the remote sensing and signaling hypothesis. Mol Pharmacol 79: 795–805, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dugan L, You Y-H, Ali S, Diamond-Stanic M, Miyamoto S, DeCleves AE, Andreyev A, Quach T, Ly S, Shekhtman G, Nguyen W, Chepetan A, Le TP, Wang L, Xu M, Paik KP, Fogo A, Viollet B, Murphy A, Brosius F, Naviaux RK, Sharma K: Reduced superoxide and mitochondrial function in diabetes is AMPK dependent. J Clin Invest, in press, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sharma K, Ix JH, Mathew AV, Cho M, Pflueger A, Dunn SR, Francos B, Sharma S, Falkner B, McGowan TA, Donohue M, Ramachandrarao S, Xu R, Fervenza FC, Kopp JB: Pirfenidone for diabetic nephropathy. J Am Soc Nephrol 22: 1144–1151, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmann G, Aramaki S, Blum-Hoffmann E, Nyhan WL, Sweetman L: Quantitative analysis for organic acids in biological samples: Batch isolation followed by gas chromatographic-mass spectrometric analysis. Clin Chem 35: 587–595, 1989 [PubMed] [Google Scholar]
- 26.Cohen CD, Frach K, Schlöndorff D, Kretzler M: Quantitative gene expression analysis in renal biopsies: A novel protocol for a high-throughput multicenter application. Kidney Int 61: 133–140, 2002 [DOI] [PubMed] [Google Scholar]
- 27.Schmid H, Boucherot A, Yasuda Y, Henger A, Brunner B, Eichinger F, Nitsche A, Kiss E, Bleich M, Gröne HJ, Nelson PJ, Schlöndorff D, Cohen CD, Kretzler M, European Renal cDNA Bank (ERCB) Consortium : Modular activation of nuclear factor-kappaB transcriptional programs in human diabetic nephropathy. Diabetes 55: 2993–3003, 2006 [DOI] [PubMed] [Google Scholar]
- 28.Benjamini Y, Hochberg Y: Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Stat Soc, Series B (Methodological) 57: 289–300, 1995 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.