Abstract
Background
The oriental river prawn, Macrobrachium nipponense, is an important aquaculture species in China, even in whole of Asia. The androgenic gland produces hormones that play crucial roles in sexual differentiation to maleness. This study is the first de novo M. nipponense transcriptome analysis using cDNA prepared from mRNA isolated from the androgenic gland. Illumina/Solexa was used for sequencing.
Methodology and Principal Finding
The total volume of RNA sample was more than 5 ug. We generated 70,853,361 high quality reads after eliminating adapter sequences and filtering out low-quality reads. A total of 78,408 isosequences were obtained by clustering and assembly of the clean reads, producing 57,619 non-redundant transcripts with an average length of 1244.19 bp. In total 70,702 isosequences were matched to the Nr database, additional analyses were performed by GO (33,203), KEGG (17,868), and COG analyses (13,817), identifying the potential genes and their functions. A total of 47 sex-determination related gene families were identified from the M. nipponense androgenic gland transcriptome based on the functional annotation of non-redundant transcripts and comparisons with the published literature. Furthermore, a total of 40 candidate novel genes were found, that may contribute to sex-determination based on their extremely high expression levels in the androgenic compared to other sex glands,. Further, 437 SSRs and 65,535 high-confidence SNPs were identified in this EST dataset from which 14 EST-SSR markers have been isolated.
Conclusion
Our study provides new sequence information for M. nipponense, which will be the basis for further genetic studies on decapods crustaceans. More importantly, this study dramatically improves understanding of sex-determination mechanisms, and advances sex-determination research in all crustacean species. The huge number of potential SSR and SNP markers isolated from the transcriptome may shed the lights on research in many fields, including the evolution and molecular ecology of Macrobrachium species.
Introduction
The oriental river prawn, Macrobrachium nipponense (Crustacea; Decapoda; Palaemonidae), is an important commercial prawn species, that is widely distributed in freshwater and low-salinity estuarine regions in China and other Asian countries [1]–[7] with an aquaculture production of 205,010 tons annually for aquaculture only [8]. As known, within many other Macrobrachium species, the males grow faster and gain more weight at harvest time than females. Facing stiff market competition, Macrobrachium producers require improvement in fish production and performance traits to obtain more profit. Thus, culture of all-male populations would be necessary for economic purpose. Therefore, the long-term goals of the M. nipponense aquaculture industry include making genetic improvements and gaining a better understanding of sex-differentiation in this species. Some cDNA libraries and transcriptome-level datasets have been generated and serving as a basis for functional genomics approaches aimed at improving the aquaculture performance of this species [9]–[11]. However, little information on sex-determination and sex-differentiation related genes in the androgenic gland of M. nipponense have been reported, therefore, the mechanism of sex-determination remains unclear.
The androgenic gland is found in most crustacean species, it produces hormones that play crucial roles in sexual differentiation to maleness, including the development of the testes and male sexual characteristics [12]. Androgenic gland hormone (AGH) and its relative producers are recognized as important factors when the sex-differentiation and determination mechanisms are concerned, their functions have been widely studied in many crustaceans [13]–[15]. In Macrobrachium rosenbergii, the males undergoes sex reversal to the females after androgenic gland was ablated from males. The all-male population was generated when the “reversal females” were mating with normal male M. rosenbergii [16]–[18]. Considering the fact that ablation or implantation of the androgenic gland at a certain stage of development can result in sex reversal to male or female [12], [19]–[21], the expression pattern of androgenic gland hormone genes in crustaceans have received much attention in recent years [22]–[27]. Previous studies have focused on morphology and anatomy on the M. nipponense androgenic gland [28]. However, no information to date was reported on genetic underpinnings of the androgenic gland in M. nipponense. More novel genes, potentially involved in the sex-determination mechanism, are necessary to be identified from the androgenic gland. Such basic information is essential to better understand the molecular mechanisms involving in sex-determination/differentiation, in turn, it will benefit monosex crustacean aquaculture production.
The transcriptome is the total set of transcripts, mRNA, and non-coding RNA, in one or a population of cells during a specific developmental stage or in response to a particular physiological condition using high-throughput technology [29]. Transcriptome analysis provides the foundation for gene structure and function research and determines when genes are expressed and how they are regulated. The development of next-generation sequencing (NGS) technologies allows the acquisition of more sequence data per run at a substantially lower cost than in long-read technologies [30]. Compared with the microarray methods, NGS, developed by Illumina/Solexa, is able to generate over one billion bases of high-quality sequences per run at less than 1% of the cost of capillary-based methods, and is expected to dominate future analysis of eukaryotic transcriptomes [31]–[34]. Transcriptome analysis is now being widely applied to elucidate genetic factors conferring economically significant traits and/or phenotypes and to manage genetic diversity in cultured crustacean species [9]–[11], [35]–[38]. To date, there are only 81,411 expressed sequence tags (ESTs) from M. nipponense in the public databases, but the classical androgenic gland genes are largely absent from these gene sets [11].
In this study, we generate the first M. nipponense transcriptome using cDNA prepared from mRNA which was isolated from the androgenic gland. The EST sequences generated were assembled and annotated with putative functions where possible, and database mining was performed to identify sex-determination and differentiation related genes. A variety of markers potentially useful for genomic population studies including simple sequence repeats (SSRs) located within coding regions and single nucleotide polymorphisms (SNPs) detected amongst deep coverage sequence region reads are also reported.
Results and Discussion
Transcriptome Analysis
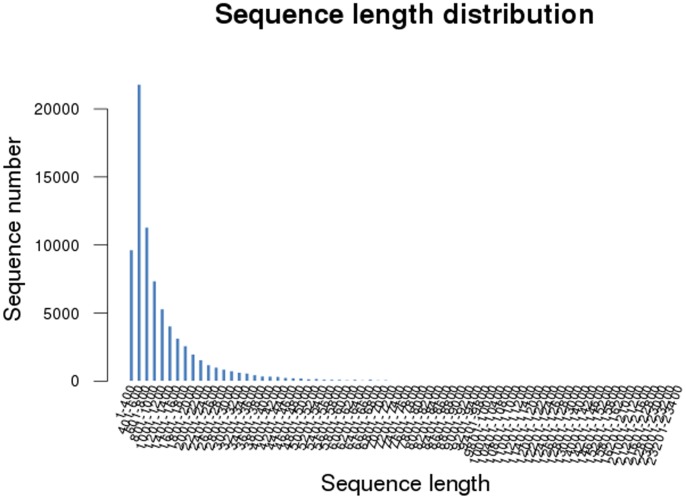
To the best of our knowledge, this is the first comprehensive study of the M. nipponense androgenic gland transcriptome sequenced by Illumina/Solexa. In the present study, the transcriptome profile was used to study the sex-determination and differentiation genes in the androgenic gland using next-generation sequencing technologies. Interestingly, the results revealed that there were genes highly expressed in the androgenic gland, which may be potentially involved in the regulatory mechanism of sex-determination in the oriental river prawn. The reads produced by the Illumina Hiseq2000 were used for clustering and de novo assembly. The reads yielded a total of approximately 357.8 million 200 bp paired-end raw reads with a total size of 7,229,376,990. After eliminating adapter sequences and filtering out the low-quality reads (the number of bases in each read was less than 25 bp) before de novo assembly by the SeqPreq program, Illumina Hiseq2000 sequencing yielded a total of 70,853,361 (Table S1, Table S2) high-quality transcriptome reads with a total size of 6,841,680,044 bp. M. nipponense is a non-model organism and has no reference genome sequence. Therefore, the raw data were assembled de novo using the Trinity program resulting in 78,408 contigs, ranging from 351 to 23,217 bp ( Table 1 ). Most of the contigs (27.8%) were 401–600 bp in length, followed by 601–800 bp (14.42%), and 1–400 bp in length (12.3%) ( Figure 1 ). These 78,408 isosequences yielded a total of 57,619 non-redundant transcripts with an average of 1,244.19 bp because of alternative splicing, thus, two or more isosequences may be matched to one transcript. In prawn, the earliest cDNA libraries were constructed in 2001, based on hemocytes and hepatopancreas from Litopenaeus vannamei and L. setiferus, in order to discovery immune genes and a total of approximately 2045 randomly selected clones were sequenced [39]. Three cDNA libraries (based on the material from the testes, ovaries, and milti-tissues) were constructed in previous studies on M. nipponense by Roche 454 GS FLX sequencing [9]–[11]. However, only limited numbers of genes were obtained from these three cDNA libraries. Compared with those studies, a great number of genes were generated in the current study, taking advantage of Illumina Hiseq2000 NGS which can sequence in higher throughput and provide more candidate genes. Besides, the average length of each isosequence is much longer than the previous studies after clustering and de novo assembly in current study, which can promote further studies on these isosequences, including RT- PCR and western-blot. The 57,619 non-redundant transcripts in this study provide a transcriptome database for future analyses of sex-determination and sex- differentiation related genes in androgenic gland tissue. Therefore, this transcriptome dataset accelerates the understanding toward the sex-determination mechanisms in M. nipponense, and other crustaceans.
Table 1. Summary of Illumina Hiseq2000 assembly and analysis of M.nipponense transcriptomic sequences.
| Number | |
| Total genes | 57619 |
| Total isogenes | 78408 |
| Total residues | 97554839 |
| Average length | 1244.19 |
| Largest isogene | 23217 |
| Smallest isogene | 351 |
Figure 1. Contig length distribution of M. nipponense transcriptomic ESTs.
Gene Ontology Assignments and COG Analysis
To identify their putative functions, all of the isosequences were compared with the non-redundant protein database and nucleotide sequences in NCBI using Blastp and Blastx at an E-value of <10−5 in the priority order of the Kyoto Encyclopedia of Genes and Genomes (KEGG) database and Cluster of Orthologous Groups (COG) database.
A total of 70,702 non-redundant transcripts, which matched Nr, were annotated, while the other unannotated transcripts represent novel genes whose functions have not yet been identified. These unannotated transcripts may also play a vital role in the metabolism of M. nipponense; however, further research is required. A total of 5,365 out of 78,408 isosequences contain ORFs, with an average protein length of 719.3 bp and a mean nucleotide length of 2,157.9 bp. This implies that these 5,465 isosequences, which might be translated into amino acids, play an essential role in M. nipponense metabolism. Additional function analysis of these isosequences conducted using the COG, GO, and KEGG pathway databases are necessary.
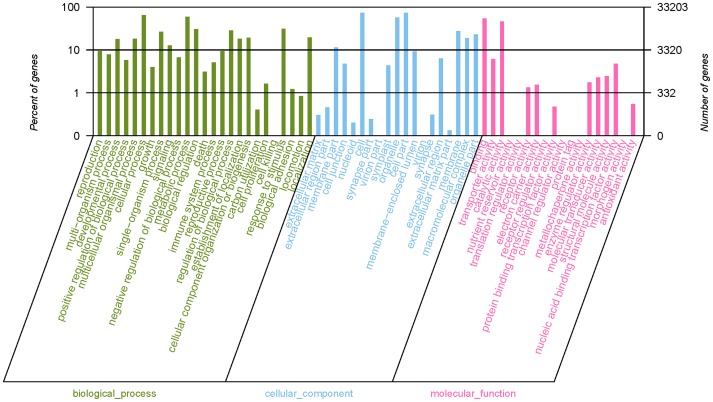
Gene Ontology (GO) divides gene products into three categories (molecular function, cellular component and biological process), aimed to provide a structured and controlled vocabulary for describing gene products. GO terms were assigned to 33,203 M. nipponense contigs based on BLAST matches to proteins with known functions, including 40,399 sequences assigned to the molecular function category, 102,835 to the cellular component category, and 133,527 to the biological process component (Table S3). The matched contigs were comprised of 62 functional groups ( Figure 2 ). Gene Ontology (GO) can provide a structured and controlled vocabulary for describing gene products in three categories: molecular function, cellular component, and biological process [40]. Analyses of the transcriptomes of other crustaceans have identified ESTs possessing similar arrays of potential metabolic functions [9]–[11], [35]–[38]. The total number of GO terms in this study was much larger than that of unique sequences, because many contigs can be assigned to more than one GO term. In the molecular function category, the number of contigs in each GO term ranged from 1 to 24,163. Cell, cell part, and cellular process had most abundant contigs (>20,000). However, there were also 9 functional groups in which the numbers of contigs were less than 10.
Figure 2. Gene ontology classification of non-redundant transcripts.
By alignment to GO terms, 33203 isogenes were mainly divided into three categories with 62 functional groups: biological process (25 functional groups), cellular component (19 functional groups), and molecular function (18 functional groups). The left y-axis indicates the percentage of a specific category of genes existed in the main category, whereas the right y-axis indicates the number of a specific category of genes existed in main category.
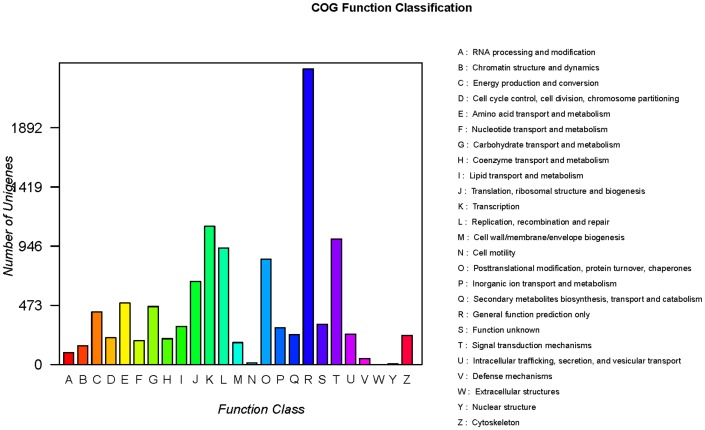
The additional analysis revealed that 13,817 isosequences matched known genes in the COG database. Based on their predicted functions, these unigenes were classified into 25 functional categories (Table S4). Among these 25 functional categories, a cluster for General function prediction only represents the largest group with 4,162 unique sequences, followed by signal transduction mechanisms (2,258), posttranslational modification, protein turnover, chaperones (1,947), and transcription (1,785). Clusters for cell motility, extracellular structures, and nuclear structure represent the smallest groups, in which the numbers of sequences were <30 ( Figure 3 ). Similar to the data in the GO category, the total number of COG sequences was >13,817 because several sequences were involved in more than one functional category.
Figure 3. Cluster of orthologous groups (COG) classification of putative proteins.
A total of 13817 putative proteins were classified functionally into 25 molecular families in the COG database.
The results obtained in this study showed that candidate novel genes with special functions can be easily identified according to the Gene Ontology assignment and COG analysis. Biological functions were mostly predicted to be involved in developmental process, growth and cell proliferation functional groups in “GO assignments” and the general functional prediction only and signal transduction mechanisms functional categories in “COG analysis”, which are more likely to contain the sex-determination and differentiation related genes in M. nipponense. The genes in these functional groups in current study were dramatically more abundant than those in previous studies in M. nipponense [9]–[11], providing more candidate selections for further analysis of sex-determination and differentiation mechanism in M. nipponense.
KEGG Analysis
KEGG database can map the unique sequence into defined metabolic pathway. KEGG analysis was used to identify the potential candidate transcripts in biological pathways in the ladybird. A total of 17,868 isosequences matched the metabolic pathways in the KEGG Pathway database, mapped onto 317 predicted metabolic pathways, and grouped into amino acid metabolism, genetic information processing, cellular processes, and environmental information processing (Table S5). The numbers of unique sequences mapped to various pathways ranged from 1 to 3,500. The main metabolic pathways of unique sequences in the M. nipponense transcriptome are metabolic pathways, biosynthesis of secondary metabolites, microbial metabolism in diverse environments, spliceosome, RNA transport, protein processing in the endoplasmic reticulum, and purine metabolism, in which the numbers of unique sequences were more than 400. Identifying these pathways in the database was acceptable, considering the growth process habits of M. nipponense. KEGG analysis dramatically advances the researches on the relationship between different genes in the transcriptome of androgenic gland in depth. Some metabolic pathways, including spliceosome, and RNA degradation may promote the analysis of sex-determination and differentiation mechanism, because many sex-determination and differentiation related genes were existed in these pathways, such as Transformer-2, and the series of heat-shock protein family. Although not all of the major genes reported in the putative KEGG pathways were found in the current study, this information provides insight into the specific responses and functions involved in the molecular processes of M. nipponense metabolism and sex-determination.
Research on Sex- related Genes
Sex determination is a fundamental and very important biological process involved in the development of sexual characteristics in organisms, thereby leading to sex-specific traits manifested in behavior, physiology, and morphology. Male and female generally have different alleles or even different genes, responsible for the regulation of their sexual morphology [41]. A total of 47 sex-related gene families were identified from the M nipponense androgenic gland transcriptome based on the functional annotation of non-redundant transcripts (Table 2). Most of these functional genes were identified based on comparisons with published data for other species [9]–[11], [42]–[44], but some were identified according to their GO classification.
Table 2. Sex- or reproduction- related ESTs identified in the androgenic gland transcriptome of M. nipponense.
| Transcripts | Length (bp) | E-value | Accession number | Hits |
| Insulin-like androgenic gland specificfactor (IAG) | 854–2541 | 0 | ref|NP_563742.1| | 4 |
| DEAD-box ATP-dependent RNA helicase | 397–4215 | 0 | ref|NP_187299.1| | 42 |
| Sex-lethal | 1981–2058 | 0 | ref|XP_003250344.1| | 2 |
| Transformer-2 protein | 452–4421 | 0–1.00E-05 | ref|XP_001513669.2| | 8 |
| Extra sex combs | 1291–1534 | 0 | gb|AAC05332.1| | 2 |
| FTZ-F1 | 382–2037 | 0 | gb|ADK46871.1| | 5 |
| FOXL2 | 3196 | 8.00E-42 | gb|ABP63571.1| | 1 |
| ECM | 437–5934 | 0 | ref|NM_179755.3| | 5 |
| FEM1 | 2066–2741 | 0 | ref|NP_001153369.1| | 4 |
| DHH | 377–2647 | 0–3.00E-28 | ref|NP_193244.2| | 13 |
| START | 430–2598 | 0–2.00E-16 | ref|NM_124797.4| | 8 |
| GATA | 382–2598 | 0–7.00E-14 | ref|NM_001036712.1| | 31 |
| Pumilio | 363–2504 | 0–6.00E-07 | ref|NM_001202701.1| | 9 |
| Argonaute | 380–3427 | 0–2.00E-39 | ref|NM_179453.2| | 13 |
| Chromobox protein | 2954–3086 | 0 | gb|BT060302.1| | 2 |
| Akt | 1520–2755 | 0 | ref|NM_128222.5| | 3 |
| Wnt | 854–2802 | 0–4.00E-40 | gb|EFX66479.1| | 8 |
| Heat Shock Protein | 275–2541 | 0–9.00E-33 | gb|AAM67147.1 | 86 |
| Male reproductive-related protein | 691–6177 | 0–8.00E-42 | gb| EF364539.1| | 12 |
| DNM | 638–2896 | 4.00E-20 | gb|EF208559.1| | 8 |
| Tsunagi | 798 | 0 | gb|EFX81381.1| | 1 |
| Gustavus | 3301 | 0 | gb|GU462157.1| | 1 |
| Cytochrome P450 | 352–2369 | 0–9.00E-14 | ref|NP_189262.1| | 175 |
| Cathepsin A | 2384 | 0 | gb|ADO65982.1| | 1 |
| Cathepsin B | 370–1470 | 0 | gb|EFA01289.1| | 5 |
| Cathepsin D | 731–923 | 0 | sp|Q05744| | 2 |
| Cathepsin L | 1406–1717 | 0 | emb|X99730.1| | 2 |
| PIGS | 443–3186 | 0 | ref|NP_187374.2| | 4 |
| Cyclin | 125–4125 | 0–7.00E-43 | ref|NP_180363.1| | 93 |
| CDC | 456–287 | 0–9.00E-41 | ref|NP_566911.1| | 24 |
| BCS | 364–1566 | 0–1.00E-12 | emb|CAB52469.1| | 7 |
| Cyclophilin | 728–1819 | 0–1.00E-05 | ref|NP_194968.2| | 15 |
| WWP | 3123–4408 | 0 | ref|NP_197706.1| | 2 |
| Dynactin | 796–1677 | 0 | gb|ACD13590.1| | 3 |
| Flotillin | 370–5757 | 0–7.00E-30 | ref|NP_197908.1| | 9 |
| PSMB | 871–1721 | 0 | ref|NP_565156.1| | 15 |
| TCP | 363–2504 | 0–7.00E-33 | ref|NP_172520.1| | 25 |
| Ubiquitin-conjugating enzyme E2 | 365–4010 | 0–6.00E-44 | ref|NP_565440.1| | 67 |
| E3 ubiquitin-protein ligase | 352–10699 | 0–8.00E-29 | ref|NP_192209.2| | 177 |
| Ubiquitin carboxyl-terminal esterase L3 | 1204 | 0 | gb|ACO36738.1| | 1 |
| Ubiquitin carboxyl-terminal hydrolaseisozyme L5 | 1757 | 0 | gb|ACM43511.1| | 1 |
| Ubiquitin carboxyl-terminal hydrolase | 354–3818 | 0–4.00E-39 | ref|NP_563719.1| | 79 |
| Ubiquitin-binding protein | 1349 | 0 | ref|XM_001180530.1| | 1 |
| Ferritin | 172–1116 | 0–3.00E-22 | gb|EU371046.1| | 4 |
| Ferritin heavy-chain subunit | 524–1100 | 0 | ref|NP_990417.1| | 3 |
| Ferritin light-chain subunit | 4589 | 0 | gb|FJ446525.1| | 1 |
| Ferritin peptide | 972 | 2.00E-42 | gb|DQ205422.1| | 1 |
In the M. nipponense androgenic gland transcriptome generated in this study, an important series of transcription factors homologous known as sex-determination related genes in other species, were found through high-throughput technology, these included insulin-like androgenic gland specific factor (IAG), transformer-2 (tra-2), sex lethal (sxl), and so on. IAG function is believed to be similar to that of the isopod AG hormone, which was the first to be structurally elucidated and belongs to the insulin superfamily of proteins, considered as key regulator of male sex-determination [23]. Recently, its homologs were found to be expressed in the AGs of several decapod crustaceans. The gene has been cloned and studied in several crustacean species, including Callinectes sapidus, M. rosenbergii, Macrobrachium nipponense, and Penaeus monodon [22]–[27]. IAG was exclusively and abundantly expressed in the androgenic gland in M. nipponense based on RT-PCR analysis [24]. Further studies will focus on the precise function of this gene in M. nipponense and determine how this gene will affect male sex-determination in this species. Sxl and Tra-2 have been cloned from M. nipponense, based on the construction of a testis cDNA library. During embryogenesis, Sxl and Tra-2 reached their highest levels at the nauplius stage. During the larval stage, Sxl and Tra-2 have similar expression patterns, in which the expression of both genes gradually increased from day 1 post hatching (L1) to day 10 (L10) and decreased to their lowest levels at the end of metamorphosis, suggesting that both Sxl and Tra-2 are involved in M. nipponense sex-determination. A reasonable explanation is that Sxl may act with Tra-2 to play complex and important roles in embryogenesis, metamorphosis, somatic sexual development, and sex-differentiation [45], [46].
FTZ-F1 is a member of the nuclear hormone receptor superfamily [47], [48] and it was originally considered to be involved in the regulation of the transcription of fushi tarazu (ftz) in Drosophila [49]. Two isoforms, α- and β-FTZ-F1, are transcribed from the same gene. Α-FTZ-F1 expressed in the early-stage of embryo, containing ftz expression, whereas β-FTZ-F1 expressed in the late stage embryo when ftz is absent [50]–[51]. Its homologues, an essential factor in sex determination in mammals, have been identified in human, mouse and a number of teleost species [52]–[55]. FEM1 is a signal-transducing regulator in the C. elegans sex-determination pathway, played an essential role in sex determination in C. elegans [56]. The homologues of FEM1, including FEM1A, FEM1B and FEM1C, have been identified in human and house mouse. The expression of a single FEM1 transcript and protein showed no significant difference in both sexes, suggesting its activity was regulated by primarily posttranscriptional and posttranslational [56]. The genes, introduced above, were also reported in previous study as important sex-determination genes [11], indicating these genes are valuable sex-determination genes for further studies.
Chromobox proteins, members of a conserved family, are thought to be located on the W chromosome in chicken [57]. Female heterogamy (ZW) exists in both M. nipponense and P. monodon since this protein was also identified in the ovary cDNA library of these species [9]–[10], [58]; it is involved in the packaging of chromosomal domains into representative heterochromatic states [59]. It has been speculated that the sex of M. rosenbergii is determined by both heterogamous (ZW) females and homogamous (ZZ) males [60]. However, chromobox proteins existed in both the testis and the androgenic gland, implying female heterogamy is the main factor in M. nipponense sex-determination.
Besides, several important sex-differentiation related genes were also identified in the current study. Many studies have shown that ubiquitin may be relevant to heat shock proteins [61]–[63]. For instance, heat shock proteins (HSP) may help ubiquitin and target non-repairable proteins to the proteasome [64]. In the present study, a total of 6 ubiquitin gene families and 8 HSP gene families were found in the transcriptome. The ubiquitin–proteasome affects many biological processes, including cell degradation and protein homeostasis maintenance. A series of ubiquitin-conjugating enzyme E2 (E2) transcripts are essential for oogenesis and spermatogenesis, as its expression levels are significantly different at the various stages of testis and ovary development [65]. Ubc9, which belongs to the family of ubiquitin-conjugating enzyme E2, has been confirmed to have effects on M. nipponense embryogenesis and oogenesis [66]. A series of E3 ubiquitin-protein ligase (E3) transcripts are involved in the regulation of binding the target protein substrate and transferring ubiquitin from the E2 cysteine to a lysine residue on the target protein [67]. Ubiquitin carboxyl-terminal hydrolase L3 (uch-l3) and ubiquitin carboxyl-terminal hydrolase L5 (uch-l5) were also found in this transcriptome, functioning during the meiotic differentiation of spermatocytes into spermatids [68]. Uchl3 and uchl5 are widely expressed throughout the entire body, including the testis/ovary, neuronal cells, and in spermatocytes and spermatids. HSPs are a class of functionally related proteins found in all living organisms, including bacteria, plants, animals, and humans [69]. Their expression increases under stressful conditions, such as exposure to elevated temperature. Heat shock proteins have several different families classified according to their molecular weight [70], [71]. For example, Hsp 90 prevents protein aggregation and increases Hsp expression [72]. Hsp90 is involved in M. nipponense ovarian development, playing a regulatory role in ovary maturation [73]. Heat shock cognate 70 kDa proteins are synthesized in haploid cells during spermatogenesis and are mainly activated at the spermatid stage [74]. The ubiquitin and HSP genes in this study may work in combination, affecting the sex-determination process.
Cathepsins, which are contained in many types of cells in all animals, tear apart other proteins. In crustaceans, cathepsin A has functions in the innate immune system [75]. Cathepsin B was reported to control the developmental processes in insects and other arthropods. Cathepsin D is necessary for the formation of the yolk [76]. Cathepsin L regulates the development of the ovary in many species, including L. vannamei, M. nsis, and Bombyx sp. Cathepsin A, B, D, and L were found in this study, as the same as the M. nipponense testis cDNA library, implying these four types of cathepsins may play an essential role in the male developmental process. In particular, cathepsin A and D may be especially important because these two cathepsins were not found in the M. nipponense ovary cDNA library.
Cyclins are necessary for the progression of cells through the cell cycle by activating the cyclin-dependent kinase enzymes [77]. In this study, a dozen transcripts, related to the cyclin family subtype were found. These may play essential roles in the reproductive and developmental processes, they included cyclin A, B, D, and cyclin-dependent kinases (CDKs). Cyclin A and D are involved in many mechanisms, including egg maturation, meiosis, and the normal cell cycle [78], [79]. Cyclin B is involved in mitosis; it rises abruptly through the cell cycle until mitosis and then decreases significantly because of the degradation of cyclin B [80]. CDKs, present in all eukaryotes, are also involved in regulating transcription, mRNA processing, and the differentiation of nerve cells and their regulatory function in the cell cycle and are considered to be evolutionarily conserved [81].
Novel Genes from Androgenic Gland
In this study, we compared the expression levels of genes in the M. nipponense androgenic gland with genes in the transcriptomes of other sex glands, including the vasa deferentia, ovaries and testes. These transcriptomes were constructed by our lab and the literatures being written currently. We divided the androgenic gland transcriptome genes into two main parts. One comprises the genes specifically expressed in the androgenic gland (21,246 genes) (Table S6), and the other comprises the genes generally expressed in sex glands (35,400 genes) (Table S7). A total of 40 candidate novel genes were found in the androgenic gland. Expression levels of these genes were significantly higher in the androgenic gland compared to the other sex glands, suggesting that they potentially play important roles in the sex-determination mechanism of M. nipponense. In the group of genes specifically expressed in the androgenic gland, 24 were considered to be the candidate genes because their expression levels were >1,000 ( Table 3 ). In the generally expressed gene group, the IAG expression pattern was used as reference because it plays an essential role in the androgenic gland. A total of 16 genes were found to have similar expression patterns to IAG ( Table 4 ). Interestingly, of the 40 candidate genes, 10 were uncharacterized proteins, the functions still remain unclear, but they may play essential roles in the androgenic gland because they are highly expressed there. However, this has yet to be confirmed.
Table 3. High expression levels of genes in androgenic gland specially expressed gene group.
| AG | |
| Slow-tonic S2 tropomyosin | 35859.99 |
| Slow-tonic S2 tropomyosin | 35322.07 |
| Slow tropomyosin isoform | 6497.87 |
| Beta-glucosidase 23 | 5577 |
| Slow tropomyosin isoform | 4756.74 |
| Transketolase | 4295 |
| Nitric oxide synthase | 3159.14 |
| Uncharacterized protein | 2652.28 |
| Conserved hypothetical protein | 2213.42 |
| Calmin-like protein | 1896.84 |
| Similar to ankyrin 2,3/unc44 | 1866.91 |
| Glycogen debranching enzyme-like | 1855.11 |
| Dihydropteroate synthase | 1838 |
| Glutamine synthetase cytosolic isozyme 1–2 | 1655.61 |
| Aquaporin PIP2-1 | 1384.64 |
| Endoplasmin-like protein | 1352 |
| Hypothetical protein CaO19.7238 | 1231.24 |
| Glycine cleavage system H protein | 1171 |
| Tubulin binding cofactor C domain-containing protein | 1163 |
| Uncharacterized protein | 1141 |
| Uncharacterized protein | 1041 |
| Uncharacterized protein | 1023.71 |
| ATP synthase subunit gamma | 1015 |
| 40S ribosomal protein S6-1 | 1012 |
Note: AG means androgenic gland. Genes in this table only expressed in androgenic gland and were not detected in vasa deferentia, ovary and testis. Nmuber means the gene expression level in androgenic gland.
Table 4. Genes with similar expression pattern with IAG in generally expressed gene group.
| AG | VD | O | T | |
| Troponin I | 64234.01 | 1301.32 | 9 | 97.62 |
| Uncharacterized protein | 60606.33 | 3145.35 | 4.07 | 123.7 |
| Uncharacterized protein | 50406 | 3535.28 | 11 | 60.13 |
| Uncharacterized protein | 35327.41 | 2353.87 | 3.93 | 72.92 |
| Troponin T-like isoform 4 | 34579.06 | 202.28 | 33.54 | 35.08 |
| SERCA | 21300.36 | 261.57 | 9.35 | 63.52 |
| IAG | 20198.69 | 4912.33 | 6 | 5 |
| Lit v 3 allergen myosin light chain | 20123 | 946.23 | 1 | 29 |
| Actin 1 | 19808.65 | 138.52 | 3 | 3 |
| Chlorophyll a–b binding protein 4 | 16184 | 32 | 14 | 15 |
| Troponin C2 | 15186.76 | 1136.6 | 1 | 36.2 |
| Beta-adaptin-like protein C | 14389 | 24 | 22 | 18 |
| Chlorophyll a–b bindingprotein CP26 | 14270 | 10 | 11 | 7 |
| Sarcoplasmic calcium-binding protein | 13419 | 37 | 7 | 10 |
| Uncharacterized protein | 13217 | 597 | 24 | 22 |
| Muscle LIM protein isoform 1 | 11554.41 | 1946.92 | 6.67 | 18 |
Note: AG means androgenic gland. VD indicated vasa deferentia. O indicates ovary. T means testis. Nmuber means the gene expression level in each tissue.
Tropomyosin (4 genes) and troponin (3 genes) may have important effects on the androgenic gland ( Table 3 , Table 4 ). Tropomyosin is highly expressed in the androgenic gland, implying it plays a vital role there. Tropomyosins control the function of actin filaments in both muscle and non-muscle cells. They are often divided into muscle and non-muscle tropomyosin isoforms. In the muscle sarcomere, muscle tropomyosin isoforms regulate interactions between actin and myosin, playing a pivotal role in regulated muscle contraction. Non-muscle tropomyosin isoforms function in all cells, controlling and regulating the cell’s cytoskeleton and other key cellular functions [82]. Troponin is a complex of three regulatory proteins (troponin C, troponin I, and troponin T), which are integral to contraction in skeletal and cardiac muscles [83]. Troponin is attached to the protein tropomyosin and lies within the groove between actin filaments in the muscle tissue [83]. The high expression levels of both tropomyosin and troponin families imply that genes from these families act together affecting the androgenic gland. Furthermore, many genes from these 40 candidate novel genes were involved in glucose metabolism, including beta-glucosidase 23, glycogen debranching enzyme-like, and glutamine synthetase cytosolic isozyme 1–2. Research on these potential candidate novel genes and analysis of the relationship between them and genes expressed in other tissues, including identifying the genes up- and downstream of sex-determination related genes, may significantly advance all-male aquaculture having positive economic effects.
Identification of Molecular Markers
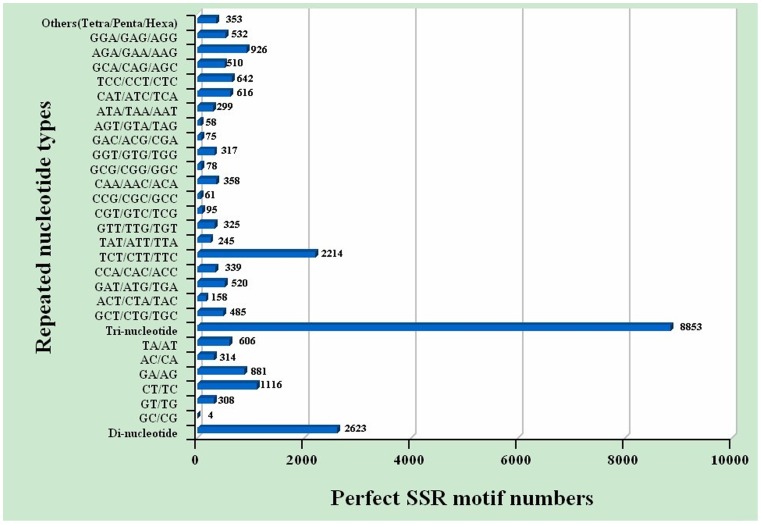
In this study, a large number of SSR and SNP markers were obtained (Table S8, Table S9). A total of 12,437 SSRs were obtained in the transcriptomic dataset, including 71.18% tri-nucleotide, 25.96% di-nucleotide, and 2.85% tetra/penta/hexa-nucleotide repeats ( Figure 4 ). Among the tri-nucleotide repeat motifs, (TCT/CTT/TTC)n with a total of 2,214 SSRs and a frequency of 17.80% was the most common type, dramatically more than the other types of tri-nucleotide repeat motifs ( Figure 4 ). There was a bias towards tri-nucleotide repeat motifs composed of C and T. (CT/TC)n, (GA/AG)n, and (TA/AT)n were the three dominant di-nucleotide types. Compared with the three previously constructed M. nipponense cDNA libraries, the volume in our study was a lot higher. In addition, 968 out of 12,437 EST-SSRs from the androgenic gland transcriptome were screened out. The frequency of these EST-SSRs was 11.4%. 673 SSRs were dinucleotide repeats, accounting for 69.5% of all SSR sequences, followed by 239 trinucleotide repeat SSRs and 26 tetranucleotide repeat SSRs. The SSRs with 5 or more nucleotides accounted for 0.03%. One hundred thirty-five pairs of primers were synthesized randomly, and 72 pairs of these exhibited clear bands. Finally, 14 markers were polymorphic in the test population of 32 individuals. The repeat motifs are listed in Table 5 and the repetitions ranged from 5 to 26. The average allele number was 7 per locus, ranging from 4 to 13. The observed heterozygosity ranged from 0.4125 to 0.8938 and expected heterozygosity ranged from 0.6786 to 0.9332. The PIC value ranged from 0.613 to 0.899. Four loci (E-WXM9, E-WXM10, E-WXM11, and E-WXM62) showed significant departure from HWE in the test population (P<0.05) ( Table 5 ). These 14 EST-SSR markers may to some extent improve further studies, including those on the construction of linkage groups. We intend to isolate more SSR markers from this transcriptome in the future.
Figure 4. Distribution of simple sequence repeat (SSR) nucleotide classes among different nucleotide types found in the transcriptome of M. nipponense.
Table 5. Characterization of 14 polymorphic EST-SSR makers in M. nipponense.
| Locus | Primer sequence(5-3) | Size(bp) | Ta(°C) | Na | Ho | H E | PIC |
| E-WXM3 | F:AGTTGCTGTGCCACCTGC | 213–278 | 58 | 5 | 0.7135 | 0.8235 | 0.802 |
| R:AAGCCACCACTGCCCTGT | |||||||
| E-WXM7 | F: GCAGTTTACATATCGGAACA | 295–336 | 54 | 8 | 0.6625 | 0.8388 | 0.821 |
| R: CGATCTGGTGGAGTTGAT | |||||||
| E-WXM9 | F:AACATTAAACCGTCTGAA | 338–402 | 54 | 5 | 0.5000 | 0.8297 | 0.899+ |
| R:ACCCTATGCGTCCTAACT | |||||||
| E-WXM10 | F: ACCCATCCAATCAAACAC | 241–254 | 54 | 4 | 0.5312 | 0.7178 | 0.791+ |
| R:TGAGCATCAGCAGCATTA | |||||||
| E-WXM11 | F:GTCCGAGCCTCCTTCTTC | 234–248 | 56 | 4 | 0.4125 | 0.6786 | 0.613+ |
| R:TCCACCTCCTTTGCCACT | |||||||
| E-WXM14 | F:CCCTCGTGAGATGATGTG | 348–416 | 55 | 7 | 0.6875 | 0.8333 | 0.799 |
| R:CAGGACTGAGTGGCAAAA | |||||||
| E-WXM16 | F:GCAGTGAATTATTGTGCTCCTA | 303–335 | 56 | 7 | 0.6700 | 0.8145 | 0.776 |
| R:TCCTGTGGCTCTGCTTTG | |||||||
| E-WXM24 | F:AAGGTTCGTTCATGCGTTAG | 289–358 | 56 | 6 | 0.7923 | 0.8520 | 0.725 |
| R:CGGATATTATTTCTGTTGGGTT | |||||||
| E-WXM29 | F: GATTTATCTCAGGTGGGT | 182–194 | 54 | 4 | 0.7500 | 0.8436 | 0.874 |
| R: ACTAAGGAAACAGGCATT | |||||||
| E-WXM33 | F: AACATTCACTGGCTCTTCG | 188–236 | 54 | 13 | 0.8938 | 0.9216 | 0.890 |
| R: CCACTACTGTTTCTATCCACC | |||||||
| E-WXM62 | F:GCTTGTAGAAACCCGTAG | 134–189 | 52 | 13 | 0.7188 | 0.9132 | 0.672+ |
| R:CTCTGACCTGCTTAGAAAA | |||||||
| E-WXM89 | F:GTTACCCAACCAGGCATT | 282–333 | 56 | 10 | 0.7688 | 0.8249 | 0.682 |
| R:GCATTTTCAGACGCACATAA | |||||||
| E-WXM93 | F:GCCAAGAAGCCGAAGACT | 235–298 | 56 | 8 | 0.7138 | 0.8652 | 0.798 |
| R:TTTTGACAGCAAGGGGAT | |||||||
| E-WXM147 | F:ATTGTCGTAGGCTCACGT | 243–280 | 50 | 9 | 0.8688 | 0.9332 | 0.657 |
| R:AAAATTGGTCTTGCTCCC |
Note: Ta, annealing temperature; Na number of alleles; H O observed heterozygosity; H E expected heterozygosity; PIC, polymorphic information content.
indicates significant deviation from HWE (P<0.05).
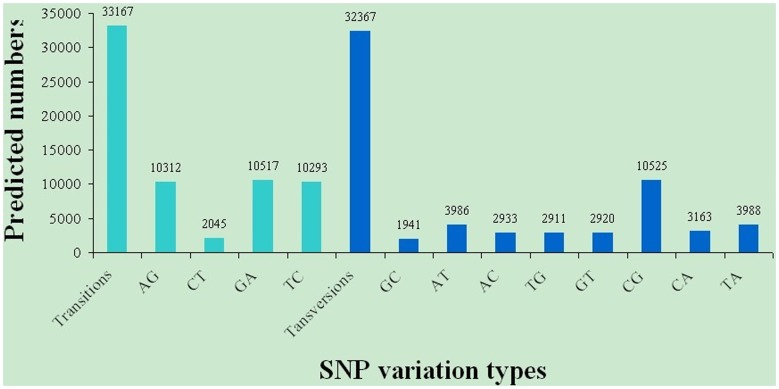
SNPs were identified from alignments of multiple sequences used for contig assembly. By excluding those that had a mutation frequency of bases less than 1%, a total of 65,535 SNPs were obtained, of these 33,167 were putative transitions (Ts) and 32,367 were putative transversions (Tv), giving a mean Ts: Tv ratio of 1.02∶1 across the M. nipponense androgenic gland transcriptome ( Figure 5 ). The AG/GA, CT/TC, and CG/GC SNPs were the most common. In contrast, CA/AC, AT/TA, and TG/GT types were the fewest SNP types because of the differences in the base structure and the number of hydrogen bonds between different bases. Compared with the three existing M. nipponense cDNA libraries, the volume of SNPs in our study was also a lot higher. The transcriptomes, sequenced by Roche 454 GS FLX, generally have missing SNPs [11], [37], mainly because of the experimental methodology. However, there are no missing SNPs in this study, suggesting Illumina/Solexa sequencing is an ideal method for future transcriptome construction.
Figure 5. Distribution of putative single nucleotide polymorphisms (SNP) in the transcriptome of M. nipponense.
SSRs, or microsatellites, are polymorphic loci present in genomic DNA, consisting of repeated core sequences of 2–6 base pairs in length [84]. SNPs (single-nucleotide polymorphisms) are the most common type of variation in the genome. SNPs provide the best genome coverage for analyzing the performance and production of traits. Genome with high-density SNP coverage is a powerful tool for whole genome association studies because it allows for the detection of linkage disequilibrium [85]. Thus, based on the advantages of SSRs and SNPs, the development of such markers for this species was desirable. It is envisaged that the potential markers identified here within the ESTs will provide an invaluable resource for studying the evolution and molecular ecology of M. nipponense, for genome mapping, and quantitative trait loci (QTL) analysis. However, many of the putative M. nipponense SNPs identified could simply represent allelic variants and future studies are planned to validate which ones are real.
Conclusion
This is the first report on the transcriptome of the M. nipponense androgenic gland by de novo assembly using the Illumina Hiseq2000. The 57,619 non-redundant transcripts identified and assembled will facilitate gene discovery in M. nipponense. A total of 47 sex-determination and sex-differentiation process related gene families were identified. Many candidate novel genes, potentially involved in the sex-determination mechanism, were identified in the androgenic gland for the first time and are worthy of further investigation. In addition, a large number of SNPs and SSRs were predicted and can be used for subsequent marker development, genetic linkage, and QTL analysis. Such findings generated by pyrosequencing in M. nipponense provide a new resource for future investigations in this economically important species, especially in understanding the sex-determination and differentiation mechanism of M. nipponense.
Materials and Methods
Ethics Statement
The prawns were obtained from the Tai Lake in Wuxi, China. We got the permission from the Tai Lake Fishery Management Council. M. nipponense is not an endangered or protected species in China, which can be used for experimental materials. All the experimental animal programs involved in this study were approved by committee of Freshwater Fisheries Research Institute, and followed the experimental basic principles. Androgenic gland from each prawn was sheared under MS222 anesthesia, and all efforts were made to minimize suffering.
Prawn and Tissue Preparation
A total of 100 healthy adult male M. nipponense with a wet weight ranging from 4.9 to 6.2 g (average = 5.5 g), and a total length ranging from 6.1 cm to 7.2 cm (average = 6.8 cm), were obtained from Tai Lake in Wuxi, China (120°13′44″ E, 31°28′22″ N). These specimens were transferred to a 500 L tank and maintained in aerated freshwater at room temperature (26°C) for 72 h prior to tissue collection. The androgenic gland from 100 individuals was collected and immediately frozen in liquid nitrogen until used for RNA extraction for transcriptome sequencing, in order to prevent total RNA degradation.
RNA Isolation for RNA-seq
The androgenic gland tissues from the 100 individuals were pooled to provide sufficient RNA for transcriptome sequencing. The androgenic glands were extracted under an Olympus SZX16 microscope. Total RNA was extracted by using the UNlQ-10 Column Trizol Total RNA Isolation Kit (Sangon) following the manufacturer’s protocol. The OD260/280 and OD260/230 should range from 1.8 to 2.0 and >2.0, respectively, to ensure the purity of the RNA sample. To guarantee the transcriptome quality, the total volume of the RNA sample was >5 µg. RNA integrity was confirmed using a 2100 Bioanalyzer (Agilent Technologies, Inc.) with a minimum RNA integrity number (RIN) value of 7.0. The samples for transcriptome analysis were prepared using a Truseq™ RNA Sample Prep Kit (Illumina) according to the manufacturer’s recommendations. Briefly, mRNA was isolated from >5 µg of total RNA using oligo (dT) magnetic beads. mRNA was cut into short fragments by adding fragmentation buffer. First-strand cDNA was synthesized using random hexamer-primers, taking these short fragments as templates. RNaseH, buffer, dNTPs, and DNA polymerase I was used to synthesize second-strand cDNA. Short fragments were purified with Takara’s PCR extraction kit (Takara Bio, Inc.). Sequencing adapters were ligated to short fragments and resolved by agarose gel electrophoresis. Proper fragments were selected and purified and subsequently PCR amplified to create the final cDNA library template.
Analysis of the Transcriptome Results
The transcriptome was sequenced using the Illumina HiSeq™ 2000. Four fluorescently labeled nucleotides and a specialized polymerase were used to determine the clusters base by base in parallel. The size of the library was approximately 200 bp and both ends of the library were sequenced. The 200 bp raw paired-end reads were generated on the Illumina sequencing platform. Image deconvolution and quality value calculations were performed using Illumina GA pipeline v1.6. The raw reads were cleaned by removing adaptor sequences, empty reads, and low quality sequences (reads with unknown sequences ‘N’ or less than 25 bp). The clean reads were assembled into non-redundant transcripts using the Trinity, which has been developed specifically for the de novo assembly of transcriptomes using short reads. To obtain non-redundant transcripts, we removed short sequences (100 bp in length) and partially overlapping sequences. The resulting sequences were used for BLAST searches and annotation against the Nr protein, the Swissprot, the COG, and the KEGG databases using an E-value cut-off of 10−5. Functional annotation by GO terms (www.geneontology.org) was analyzed by the Blast2go software. The COG and KEGG pathway annotations were performed using Blastall software against the COG and KEGG databases.
Expression Analysis
The abundance of each tag was normalized to one transcript per million, in order to allow comparison between various sex glands, including the androgenic gland, vasa deferentia, ovaries and testes. The raw reads were cleaned by removing low quality sequences including ambiguous nucleotides and adaptor sequences. The calculation of unigene expression was conducted by using RSEM software [86]–[87].RSEM is an accurate and user-friendly software tool for quantifying transcript abundances from RNA-Seq data. It is particularly useful for quantification with de novo transcriptome assemblies because it does not rely on the existence of a reference genome [86].
Molecular Marker Detection
All ESTs from the M. nipponense androgenic gland transcriptome with a total number of 17,060 were converted into FASTA format, and screened for the presence of SSRs using SSRHunter software (http://www.bio-soft.net). The primers were designed using PRIMER 5.0 software (http://www.bbioo.com/download/58-166-1.html). Thirty-two wild oriental river prawn individuals were collected as a test population from Taihu Lake in China. Genomic DNA was extracted from the muscle of these individuals using traditional proteinase-K digestion and phenol–chloroform extraction protocols [88]. PCR amplification was carried out in a 25 µl reaction volume, containing 1×PCR buffer (Tiangen, Beijing, China), 30–50 ng genomic DNA, 0.25 µM for each primer, 150 µmol/l dNTPs, 1.5 mM MgCl2, and 0.5 U Taq DNA polymerase (Tiangen, Beijing, China). The PCR reaction conditions were as follows: the DNA was first denatured at 94°C for 3 min; followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 50–58°C for 30 s, and elongation at 72°C for 40 s; with a final elongation at 72°C for 10 min. PCR products were size-fractionated on 6% polyacrylamide gels by silver staining. The pBR322 DNA/BsuRI (Hae III) marker (Fermentas, Shenzhen, China) was used to determine the allele sizes. POPGEN32 software was used to analyze allele frequencies, Ho, and He for each locus and statistical significance of HWE [89]. PIC was estimated using the formula of Botstein et al. [90].
Data deposition
The Illumina Hiseq 2000 reads of M. nipponense were submitted to www.ffrc.cn/gene/list.asp.
Supporting Information
Summary of BLASTx results for contigs of M. nipponense .
(XLS)
Summary of BLASTx results for contigs with ORF of M. nipponense .
(XLS)
Categories of Gene Ontology of M. nipponense unique sequences.
(XLS)
Categories of COG of M. nipponense unique sequences.
(XLS)
KEGG summary of M. nipponense unique sequences.
(XLS)
Summary of genes specifically expressed in androgenic gland.
(XLS)
Summary of genes generally expressed in sexual gland.
(XLS)
Summary of putative SSRs from M. nipponense transcriptome.
(XLS)
Summary of putative SNPs from M. nipponense transcriptome.
(XLS)
Acknowledgments
We thank for the help from Liwen editing company for the revised English. We also thank for the help from lab member, Fajun Li (Wuxi Fishery College, Nanjing Agricultural University) to obtain the androgenic gland from M. nipponense. We acknowledge Fan Yang and Binxun Liu (Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd.) for their kind help in sequencing and bioinformatics analysis.
Funding Statement
The project was supported by the National Natural Science Foundation of China (Grant No.31272654) (http://www.nsfc.gov.cn/Portal0/InfoModule_544/29249.htm), the National Science & Technology Supporting Program of the 12th Five-year Plan of China (Grant No. 2012BAD26B04) (http://www.most.gov.cn/kjbgz/201208/t20120821_96332.htm) and the Science & Technology Supporting Program of Jiangsu Province (Grant No. BE2012334)(http://www.jiangsu.gov.cn/xxgk/bmhsxwj/bmwj/201206/t20120626_740122.html).The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Ma KY, Feng JB, Lin JY, Li JL (2011) The complete mitochondrial genome of Macrobrachium nipponense . Gene 487: 160–165. [DOI] [PubMed] [Google Scholar]
- 2. Feng JB, Li JL, Cheng X (2008) Research progress on germplasm resource exploitation and protection of Macrobrachium nipponense . J Shanghai Fish Univ 17: 371–376. [Google Scholar]
- 3. Yu H, Miyake S (1972) Five species of the genus Macrobrachium (Crustacea, Decapoda, Palaemonidae) from Taiwan. Ohmu 3: 45–55. [Google Scholar]
- 4. Cai Y, Shokita S (2006) Report on a collection of freshwater shrimps (Crustacea: Decapoda: Caridea) from the Philippines, with descriptions of four new species. Raffles B Zool 54: 245–270. [Google Scholar]
- 5. De Grave S, Ghane A (2006) The establishment of the oriental river prawn, Macrobrachium nipponense (de Haan, 1849) in Anzali Lagoon, Iran. Aquat Invasions 1: 204–208. [Google Scholar]
- 6. Mirabdullaev IM, Niyazov DS (2005) Alien decapods (Crustacea) in Uzbekistan. Abstracts of the II International Symposium Invasion of alien species in Holarctic, Borok, Russia. 2: 113–114. [Google Scholar]
- 7. Salman SD, Page TJ, Naser MD, Yasser AG (2006) The invasion of Macrobrachium nipponense (De Haan, 1849) (Caridea: Palaemonidae) into the southern Iraqi marshes. Aquat Invasions 1: 109–115. [Google Scholar]
- 8.Bureau of Fishery, Ministry of Agriculture, P.R.C. (2009) Fisheries economic statistics. In: China Fishery Yearbook. Beijing China Agricultural Press: 236.
- 9. Qiao H, Fu HT, Jin SB, Wu Y, Jiang SF, et al. (2012) Constructing and random sequencing analysis of normalized cDNA library of testis tissue from oriental river prawn (Macrobrachium nipponense). Comp Biochem Phys D 7: 268–276. [DOI] [PubMed] [Google Scholar]
- 10. Wu P, Qi D, Chen LQ, Zhang H, Zhang XW, et al. (2009) Gene discovery from an ovary cDNA library of oriental river prawn Macrobrachium nipponense by ESTs annotation. Comp Biochem Phys D 4: 111–120. [DOI] [PubMed] [Google Scholar]
- 11. Ma K, Qiu G, Feng J, Li J (2012) Transcriptome Analysis of the Oriental River Prawn, Macrobrachium nipponense Using 454 Pyrosequencing for Discovery of Genes and Markers. PLoS ONE 7: e39727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sagi A, Cohen D, Milner Y (1990) Effect of androgenic gland ablation on morphotypic differentiation and sexual characteristics of male freshwater prawns, Macrobrachium rosenbergii . Gen Comp Endocr 77: 15–22. [DOI] [PubMed] [Google Scholar]
- 13. Li SH, Li FH, Sun Z, Xiang JH (2012) Two spliced variants of insulin-like androgenic gland hormone gene in the Chinese shrimp, Fenneropenaeus chinensis . General and Comparative Endocrinology 2: 246–255. [DOI] [PubMed] [Google Scholar]
- 14. Atsuro O, Yuriko H, Makoto N, Tsuyoshi O, Rinkei K, et al. (2002) Preparation of an active recombinant peptide of crustacean androgenic gland hormone. Peptides 3: 567–572. [DOI] [PubMed] [Google Scholar]
- 15. Morakot S, Charoonroj C, Michael JS, Nantawan S, Napamanee K, et al. (2010) Bilateral eyestalk ablation of the blue swimmer crab, Portunus pelagicus, produces hypertrophy of the androgenic gland and an increase of cells producing insulin-like androgenic gland hormone. Tissue and Cell 5: 293–300. [DOI] [PubMed] [Google Scholar]
- 16. Sagi A, Cohen D, Milne Y (1990) Effect of androgenic gland ablation on morpphotypic diffentiation and sexual characteristics of male freshwater prawns Macrobrachium rosenbetgii . Gen Comp Endorinol 77: 15–22. [DOI] [PubMed] [Google Scholar]
- 17. Sagi A, Cohen D (1990) Growth, maturation and progeny of sex-reversed Macrobrachium rosenbetgii males . World Aquaculture 21(4): 87–90. [Google Scholar]
- 18. Sagi A, Ra’anan Z, Cohen D, Wax Y (1986) Production of Macrobrachium rosenbetgii in momosex population: yield characteristes under intensive monoculture conditions in cages. Aquaculture 51: 265–275. [Google Scholar]
- 19. Nagamine C, Knight AW, Maggenti A, Paxman G (1980) Effects of androgenic gland ablation on male primary and secondary sexual characteristics in the Malaysian prawn, Macrobrachium rosenbergii (de Man) (Decapoda, Palaemonidae), with first evidence of induced feminization in a nonhermaphroditic decapod. Gen Comp Endocr 41: 423–441. [DOI] [PubMed] [Google Scholar]
- 20. Manor R, Aflalo ED, Segall C, Weil S, Azulay D, et al. (2004) Androgenic gland implantation promotes growth and inhibits vitellogenesis in Cherax quadricarinatus females held in individual compartments. Invertebr Reprod Dev 45: 151–159. [Google Scholar]
- 21. Malecha SR, Nevin PA, Ha P, Barck LE, Lamadrid-Rose Y, et al. (1992) Sexratios and sex-determination in progeny from crosses of surgically sex-reversed freshwater prawns, Macrobrachium rosenbergii . Aquaculture 105: 201–218. [Google Scholar]
- 22. Sook J, Chung RM, Sagi A (2011) Cloning of an insulin-like androgenic gland factor (IAG) from the blue crab, Callinectes sapidus: Implications for eyestalk regulation of IAG expression. General and Comparative Endocrinology 1: 4–10. [DOI] [PubMed] [Google Scholar]
- 23. Tomer V, Sagi A (2012) The insulin-like androgenic gland hormone in crustaceans: From a single gene silencing to a wide array of sexual manipulation-based biotechnologies. Biotechnology Advances 6: 1543–1550. [DOI] [PubMed] [Google Scholar]
- 24. Ma KY, Lin JY, Guo SZ, Chen Y, Li JL, et al. (2013) Molecular characterization and expression analysis of an insulin-like gene from the androgenic gland of the oriental river prawn, Macrobrachium nipponense . General and Comparative Endocrinology 185: 90–96. [DOI] [PubMed] [Google Scholar]
- 25. Mareddy VR, Rosen O, Thaggard HB, Manor R, Kuballa AV, et al. (2011) Isolation and characterization of the complete cDNA sequence encoding a putative insulin-like peptide from the androgenic gland of Penaeus monodon . Aqua 3–4: 364–370. [Google Scholar]
- 26. Tomer V, Rivka M, Eliahu DA, Simy W, Shaul R, et al. (2009) Temporal Silencing of an Androgenic Gland-Specific Insulin-Like Gene Affecting Phenotypical Gender Differences and Spermatogenesis. Endocrinology 150(3): 1278–1286. [DOI] [PubMed] [Google Scholar]
- 27. Tomer V, Rivka M, Eliahu DA, Simy W, Isam K, et al. (2011) Expression of an Androgenic-Gland-Specific Insulin-Like Peptide during the Course of Prawn Sexual and Morphotypic Differentiation. ISRN Endocrinology 2011: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu P, Yang LR, Chong JR, Wang YL, Chen K (2002) Research on androgenic gland of oriental river prawn, Macrobrachium nipponense . Water conservancy related fisheries 123: 21–23. [Google Scholar]
- 29. Mardis E (2008) The impact of next-generation sequencing technology on genetics. Trends in Genetics 24: 133–141. [DOI] [PubMed] [Google Scholar]
- 30. Huang W, Marth G (2008) A genome assembly viewer for next-generation sequencing technologies. Genome Res 18: 1538–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cloonan N, Grimmond SM (2008) Transcriptome content and dynamics at singlenucleotide Resolution. Genome Biol 9: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Morozova O, Marra MA (2008) Applications of next-generation sequencing technologies in functional genomics. Genomics 92: 255–264. [DOI] [PubMed] [Google Scholar]
- 33. Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ozsolak F, Milos PM (2011) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12: 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Clark MS, Thorne MAS, Toullec J-Y, Meng Y, Guan LL, et al. (2011) Antarctic Krill 454 Pyrosequencing Reveals Chaperone and Stress Transcriptome. PLoS ONE 6(1): e15919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kawahara-Miki R, Wada K, Azuma N, Chiba S (2011) Expression Profiling without Genome Sequence Information in a Non-Model Species, Pandalid Shrimp (Pandalus latirostris), by Next-Generation Sequencing. PLoS ONE 6(10): e26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Jung H, Lyons RE, Dinh H, Hurwood DA, McWilliam S, et al. (2011) Transcriptomics of a Giant Freshwater Prawn (Macrobrachium rosenbergii): De Novo Assembly, Annotation and Marker Discovery. PLoS ONE 6(12): e27938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pereiro P, Balseiro P, Romero A, Dios S, Forn-Cuni G, et al. (2012) High-Throughput Sequence Analysis of Turbot (Scophthalmus maximus) Transcriptome Using 454-Pyrosequencing for the Discovery of Antiviral Immune Genes. PLoS ONE 7(5): e35369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Grossa PS, Bartletta TC, Browdyb CL, Chapmanb RW, Warra GW (2001) Immune gene discovery by expressed sequence tag analysis of hemocytes and hepatopancreas in the Pacific White Shrimp, Litopenaeus vannamei, and the Atlantic White Shrimp, L. setiferus . Developmental & Comparative Immunology 25 (7): 565–577. [DOI] [PubMed] [Google Scholar]
- 40. 40. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al (2000) Gene Ontology: tool for the unification of biology. Nat Genet 25: 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hake L, O’Connor C (2008) Genetic mechanisms of sex determination. Nature Education 1(1).
- 42. Zeng S, Gong ZY (2002) Expressed sequence tag analysis of expression profiles of zebrafish testis and ovary. Gene 1–2: 45–53. [DOI] [PubMed] [Google Scholar]
- 43. Valentina C, Anna Giulia C, Federica R, Giovanni B, Genciana T, et al. (2008) Genes expressed in Blue Fin Tuna (Thunnus thynnus) liver and gonads. Gene 1: 207–213. [DOI] [PubMed] [Google Scholar]
- 44. Li H, Papadopoulos V, Vidic B, Dym M, Culty M (1997) Regulation of rat testis gonocyte proliferation by platelet-derived growth factor and estradiol: identification of signaling mechanisms involved. Endocrinology 138: 1289–1298. [DOI] [PubMed] [Google Scholar]
- 45.Zhang YP, Qiao H, Zhang WY, Sun SM, Jiang SF, et al. (2013) Molecular cloning and expression analysis of two sex-lethal homolog genes during development in the oriental river prawn, Macrobrachium nipponense. Genetic Molecular Research, IN PRESS. [DOI] [PubMed]
- 46. Zhang YP, Fu HT, Qiao H, Jin SB, Jiang SF, et al. (2013) Molecular cloning and expression analysis of transformer-2 gene during development in Macrobrachium nipponense (de Haan). Journal of the world Aquaculture Society 44 (3): 338–349. [Google Scholar]
- 47. Lavorgna G, Ueda H, Clos J, Wu C (1991) FTZ-F1, a steroid hormone receptor-like protein implicated in the activation of fushi tarazu . Science 252: 848–851. [DOI] [PubMed] [Google Scholar]
- 48. Ueda H, Sun GC, Murata T, Hirose S (1992) A novel-DNA binding motif abuts the zinc finger domain of insect nuclear hormone receptor FTZ-F1 and mouse embryonal long terminal repeat-binding protein. Mol Cell Biol 12: 5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ueda H, Hirose S (1990) Identificati on and purification of a Bombyx mori homologue of FTZ-F1. Nucl Acids Res 18: 7229–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lavorgna G, Karim FD, Thummel CS, Wu C (1993) Potential role for a FTZ-F1 steroid receptor superfamily member in the control of Drosophila metamorphosis . Proc Natl.Acad Sci USA 90: 3004–3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ueda H, Sonoda S, Brown JL, Scott MP, Wu C (1990) A sequence-specific DNA-binding protein that activates fushi tarazu segmentation gene expression. Genes Dev 4: 624–635. [DOI] [PubMed] [Google Scholar]
- 52. Oba K, Yanase T, Nomura M, Morohashi K, Takayanagi R, et al. (1996) Structural characterization of human Ad4bp (SF-1) gene. Biochem Bioph Res Co 226: 261–267. [DOI] [PubMed] [Google Scholar]
- 53. Lala DS, Rice DA, Parker KL (1992) Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol Endocrinol. 6: 1249–1258. [DOI] [PubMed] [Google Scholar]
- 54. Von Hofsten J, Jones I, Karlsson J, Olsson P-E (2001) Developmental expression patterns of FTZ-F1 homologues in zebrafish (Danio rerio). Gen Comp Endocr 121: 146–155. [DOI] [PubMed] [Google Scholar]
- 55. Zhang WM, Zhang YY, Zhang LH, Zhao HH, Li X, et al. (2007) The mRNA expression of P450 aromatase, gonadotropin beta-subunits and FTZ-F1 in the orange-spotted grouper (Epinephelus coioides) during 17 alpha-methyltestosterone- induced precocious sex change. Mol Reprod Dev 74: 665–673. [DOI] [PubMed] [Google Scholar]
- 56. Gaudet J, Vanderlst I, Spence AM (1996) Post-transcriptional regulation of sex determination in Caenorhabditis elegans: widespread expression of the sex-determining gene fem-1 in both sexes. Mol Biol Cell 7: 1107–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yamaguchi K, Hidema S, Mizuno S (1998) Chicken chromobox proteins: cDNA cloning of CHCB1,-2,-3 and their relation to W-heterochromatin. Exp Cell Res 242: 303–314. [DOI] [PubMed] [Google Scholar]
- 58. Rachanimuk P, Rungnapa L, Kanchana S, Sirawut K, Bavornlak K, et al. (2007) Expressed Sequence Tag Analysis for Identification and Characterization of Sex-Related Genes in the Giant Tiger Shrimp Penaeus monodon . Journal of Biochemistry and Molecular Biology 40 (4): 501–510. [DOI] [PubMed] [Google Scholar]
- 59. Jones DO, Mattei MG, Horsley D, Cowell IG, Singh PB (2001) The gene and pseudogenes of Cbx3/mHP1gamma. DNA Seq 12: 147–160. [DOI] [PubMed] [Google Scholar]
- 60. Melecha SR, Nevin PA, Ha P, Barck LE, Lamadrid-Rose Y, et al. (1992) Sex-ratio and sex-determination in progeny from crosses of surgically sex reversed freshwater prawns, Macrobrachium rosenbergii . Aquaculture 105: 201–218. [Google Scholar]
- 61. Ursula B, Milton JS (1985) Ubiquitin Is a Heat Shock Protein in Chicken Embryo Fibroblasts. Molecular and cellular biology 5(5): 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Noel C, Scott R, Martin R (1987) Microinjection of Ubiquitin: Changes in Protein Degradation in HeLa Cells Subjected to Heat-Shock. The Journal of Cell Biology 104: 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kunihiro U, Christiane RL, William W, Eveline S, Olaf G, et al. (2006) Convergence of Heat Shock Protein 90 with Ubiquitin in Filamentous α-Synuclein Inclusions of α-Synucleinopathies. Am J Pathol 168(3): 947–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schwartz AL, Ciechanover A (1999) The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med 50: 57–74. [DOI] [PubMed] [Google Scholar]
- 65. Shen BL, Zhang ZP, Wang YL, Wang GD, Chen Y, et al. (2008) Differential expression of ubiquitin-conjugating enzyme E2r in the developing ovary and testis of penaeid shrimp Marsupenaeus japonicus . Mol Biol Rep 36: 1149–1157. [DOI] [PubMed] [Google Scholar]
- 66. Zhang FY, Chen LQ, Wu p, Zhao WH, Li EC, et al. (2010) cDNA cloning and expression of Ubc9 in the developing embryo and ovary of oriental river prawn, Macrobrachium Nipponense . CBPB 155 (3): 288–293. [DOI] [PubMed] [Google Scholar]
- 67. Ardley HC, Robinson PA (2005) “E3 ubiquitin ligases”. ESSAYS IN BIOCHEMISTRY 41: 15–30. [DOI] [PubMed] [Google Scholar]
- 68. Kwon J, Wang YL, Setsuie R, Sekiguchi S, Sakurai M, et al. (2004) Developmental regulation of ubiquitin c-terminal hydrolase isozyme expression during spermatogenesis in mice. Biol Reprod 71: 515–521. [DOI] [PubMed] [Google Scholar]
- 69. De Maio A (January 1999) “Heat shock proteins: facts, thoughts, and dreams”. Shock (Augusta, Ga.) 11 (1): 1–12. [DOI] [PubMed] [Google Scholar]
- 70. Wu C (1995) “Heat shock transcription factors: structure and regulation”. Annual review of cell and developmental biology 11: 441–69. [DOI] [PubMed] [Google Scholar]
- 71. Raboy B, Sharon G, Parag HA, Shochat Y, Kulka RG (1991) “Effect of stress on protein degradation: role of the ubiquitin system”. Acta biologica Hungarica 42 (1–3): 3–20. [PubMed] [Google Scholar]
- 72. Picard D (2002) Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci 59: 1640–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zhao WH, Chen LQ, Qin JG, Wu P, Zhang FY, et al. (2011) MnHSP90 cDNA characterization and its expression during the ovary development in oriental river prawn, Macrobrachium nipponense . Molecular Biology Reports 38 (2): 1399–1406. [DOI] [PubMed] [Google Scholar]
- 74. Matsumoto M, Kurata S, Fujimoto H, Hoshi M (1993) Haploid specific activations of protamine 1 and hsc70t genes in mouse spermatogenesis. Biochim Biophys Acta1174: 274–278. [DOI] [PubMed] [Google Scholar]
- 75. Li WW, He L, Jin XK, Jiang H, Chen LL, et al. (2011) Molecular cloning‚ characterization and expression analysis of cathepsin A gene in Chinese mitten crab Eriocheir sinensis . Peptides 32(3): 518–525. [DOI] [PubMed] [Google Scholar]
- 76. Retzek H, Steyrer E, Sanders EJ, Nimpf J, Schneider WJ (1992) Molecular cloning and functional characterization of chicken cathepsin D, a key enzyme for yolk formation. DNA Cell Biol 11: 661–672. [DOI] [PubMed] [Google Scholar]
- 77. Galderisi U, Jori FP, Giordano A (2003) “Cell cycle regulation and neural differentiation”. Oncogene 22 (33): 5208–19. [DOI] [PubMed] [Google Scholar]
- 78. Casas E, Betancourt M, Bonilla E, Duculomb Y, Zayas H, et al. (1999) Changes in cyclin B localisation during pig oocyte in vitro maturation. Zygote 7: 21–26. [DOI] [PubMed] [Google Scholar]
- 79. Day PJ, Cleasby A, Tickle IJ, O’Reilly M, Coyle JE, et al. (March 2009) “Crystal structure of human CDK4 in complex with a D-type cyclin”. Proc Natl Acad Sci USA 106 (11): 4166–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Ito M (2000) “Factors controlling cyclin B expression”. Plant Mol Biol 43(5–6): 677–90. [DOI] [PubMed] [Google Scholar]
- 81. Lee M, Nurse P (1987) “Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2.”. Nature 327: 31–35. [DOI] [PubMed] [Google Scholar]
- 82. Gunning PW, Schevzov G, Kee AJ, Hardeman EC (2005) “Tropomyosin isoforms:divining rods for actin cytoskeleton function”. Trends Cell Biol 15 (6): 333–341. [DOI] [PubMed] [Google Scholar]
- 83. Takeda S, Yamashita A, Maeda K, Maeda Y (2003) “Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form”. Nature 424 (6944): 35–41. [DOI] [PubMed] [Google Scholar]
- 84. Queller DC, Strassman JE, Hughes CR (1993) “Microsatellites and Kinship”. Trends in Ecology and Evolution 8 (8): 285–288. [DOI] [PubMed] [Google Scholar]
- 85. Wang DG, Fan JB, Siao CJ, Berno A, Young P, et al. (1998) Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280: 1077–1082. [DOI] [PubMed] [Google Scholar]
- 86. Li Dewey (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome.BMC Bioinformatics. 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN (2010) RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics 26(4): 493–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sambrook J, Russell DW (2011) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, New York.
- 89. Yeh FC, Boyle TJB (1997) Population genetic analysis of co-dominant and dominant markers and quantitative traits. Belg J Bot 129: 157.12. [Google Scholar]
- 90. Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32: 314–331. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of BLASTx results for contigs of M. nipponense .
(XLS)
Summary of BLASTx results for contigs with ORF of M. nipponense .
(XLS)
Categories of Gene Ontology of M. nipponense unique sequences.
(XLS)
Categories of COG of M. nipponense unique sequences.
(XLS)
KEGG summary of M. nipponense unique sequences.
(XLS)
Summary of genes specifically expressed in androgenic gland.
(XLS)
Summary of genes generally expressed in sexual gland.
(XLS)
Summary of putative SSRs from M. nipponense transcriptome.
(XLS)
Summary of putative SNPs from M. nipponense transcriptome.
(XLS)