Abstract
Background
Genetic factors influence the risk for posttraumatic stress disorder (PTSD), a potentially chronic and disabling psychiatric disorder that can arise after exposure to trauma. Candidate gene association studies have identified few genetic variants that contribute to PTSD risk.
Methods
We conducted genome-wide association analyses in 1578 European Americans (EAs), including 300 PTSD cases, and 2766 African Americans, including 444 PTSD cases, to find novel common risk alleles for PTSD. We used the Illumina Omni1-Quad microarray, which yielded approximately 870,000 single nucleotide polymorphisms (SNPs) suitable for analysis.
Results
In EAs, we observed that one SNP on chromosome 7p12, rs406001, exceeded genome-wide significance (p = 3.97×10−8). A SNP that maps to the first intron of the Tolloid-Like 1 gene (TLL1) showed the second strongest evidence of association, although no SNPs at this locus reached genome-wide significance. We then tested six SNPs in an independent sample of nearly 2000 EAs and successfully replicated the association findings for two SNPs in the first intron of TLL1, rs6812849 and rs7691872, with p values of 6.3×10−6 and 2.3×10−4, respectively. In the combined sample, rs6812849 had a p value of 3.1 ×10−9. No significant signals were observed in the African American part of the sample. Genome-wide association study analyses restricted to trauma-exposed individuals yielded very similar results.
Conclusions
This study identified TLL1 as a new susceptibility gene for PTSD.
Keywords: American populations, genome-wide association study, posttraumatic stress disorder, TLL1
Posttraumatic stress disorder (PTSD) is a severe anxiety disorder that can develop after a traumatic event that usually involves actual or threatened death, serious injury, or sexual violation. Posttraumatic stress disorder is often a lasting consequence of a traumatic experience that causes intense fear, hopelessness, and horror. Distinguished from the acute response to trauma, the stress reactions of PTSD patients do not resolve quickly. Instead, symptoms can last for long periods of time, over which they may increase in severity. The rate of PTSD is especially high among veterans. One study analyzing retrospective data observed that among American World War II prisoners of war, the rate of PTSD was about 50% 1 year after repatriation and 29% 40 years after repatriation (1). A study of Vietnam combat veterans found a lifetime prevalence of PTSD of 30.9% in men (2). In the general population, the lifetime prevalence of PTSD was about 6.8% (3).
Not everyone that experiences a traumatic event develops PTSD. Risk of PTSD is influenced by characteristics of the trauma and of the individual. For example, experiencing a direct threat of death (4), being female, having a pre-existing psychiatric disorder, and having experienced childhood maltreatment (5,6) have been identified as predisposing to PTSD. In addition, genetic factors play important roles in PTSD risk. Twin studies have consistently shown that genetic components account for at least one third of the variance in risk of PTSD (7,8). Candidate gene association studies have implicated genes in the dopaminergic system (9), the serotonergic system (10), the hypothalamic-pituitary-adrenal axis (11,12), and other neurotransmitter systems (13,14) to be directly associated with PTSD or to interact with childhood maltreatment to regulate PTSD risk. Despite this, candidate gene association studies have failed to identify conclusively a genetic variant that exerts a main effect on risk of PTSD (15). A variant mapped to the serotonin transporter protein gene, SLC6A4, has, however, been shown in several studies to affect risk for PTSD and related symptoms in interaction with the environment (16–18). The candidate gene approach is usually limited to variants mapped to genes for which there are prior hypotheses about their roles in the etiology of PTSD. Given our limited understanding of the pathophysiology of PTSD, the majority of risk alleles cannot be identified by this method. Genome-wide association studies (GWAS) have successfully identified risk loci for more than 100 complex diseases (19), including many psychiatric disorders.
In this study, we performed genome-wide association analyses in 1578 European Americans (EAs) (300 PTSD cases) and 2766 African Americans (AAs) (444 PTSD cases) to find novel common risk alleles for PTSD. We also evaluated the most significant genomic regions in an independent sample of more than 3000 subjects. We replicated two intronic single nucleotide polymorphisms (SNPs) mapped to TLL1 in EAs, one of which exceeded a genome-wide significant threshold for association.
Methods and Materials
Subjects
A total of 9340 subjects (GWAS: n = 5799, replication: n = 3541) were recruited for genetic studies of alcohol, cocaine, and opioid dependence at five US sites using similar methodologies: Yale University School of Medicine (GWAS: n = 2360, replication: n = 1737), the University of Connecticut Health Center (GWAS: n = 2303, replication: n = 1360), the University of Pennsylvania School of Medicine (GWAS: n = 476, replication: n = 228), the Medical University of South Carolina (GWAS: n = 459, replication: n = 192), and McLean Hospital, an affiliate of Harvard Medical School (GWAS: n = 201, replication: n = 24). The institutional review board at each of the participating sites approved the study. Written informed consent was obtained from all participants. Certificates of confidentiality for the studies were issued by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Identification of Lifetime Trauma Exposure and PTSD Diagnosis
Sample collection and diagnostic interviews were performed by trained interviewers using the Semi-Structured Assessment for Drug Dependence and Alcoholism (SSADDA) to derive diagnoses for lifetime psychiatric and substance use disorders based on DSM-IV criteria (20,21).
In the PTSD section of the interview, 12 types of traumatic events were assessed: experienced direct combat in a war; seriously physically attacked or assaulted; physically abused as a child; seriously neglected as a child; raped; sexually molested or assaulted; threatened with a weapon; held captive or kidnapped; witnessed someone being badly injured or killed; involved in a flood, fire, or other natural disaster; involved in a life-threatening accident; suffered a great shock because one of the above events happened to someone close to you; and other. Participants were asked to list up to three traumatic events and describe the trauma in detail. Those reporting traumatic experiences were then interviewed for potential PTSD symptoms. After the data were scored, PTSD diagnoses were generated based on DSM-IV criteria. The interrater and test-retest reliability (κ) of the PTSD diagnosis using the SSADDA were .59 and .76, respectively (21).
Genotyping and Analysis
DNA was extracted from immortalized cell lines or directly from blood or saliva. For the GWAS analysis, 5799 DNA samples of subjects interviewed using SSADDA were genotyped on the Illumina HumanOmni1-Quad v1.0 microarray (Illumina, San Diego, California) containing 988,306 autosomal SNPs. Genome-wide association study genotyping was conducted at the Center for Inherited Disease Research and the Yale Center for Genome Analysis. All DNA samples were assigned to one of the above two locations and were genotyped using the same type of chip. Single nucleotide polymorphism genotypes were called using GenomeStudio software V2011.1 and genotyping module version 1.8.4 (Illumina).
Principal components analysis was conducted using the Eigensoft package (22,23) to identify population groups of the GWAS subjects. After pruning the GWAS SNPs for linkage disequilibrium (R2) greater than 80%, 145,472 SNPs that were common to the GWAS dataset and HapMap panel were analyzed to characterize the underlying genetic architecture of the samples. The first principal components score distinguished AAs and EAs; these groups were subsequently analyzed separately. Quality control steps were performed using PLINK software (24). We removed individuals with discordant sex information, an overall call rate of less than 98%, and a heterozygosity rate (excluding sex chromosomes) outside three standard deviations of the mean of all individuals.
To exclude any pair of individuals within EAs and AAs who were as closely related as second degree, we calculated estimated genome-wide alleles shared identity-by-descent (IBD) for every pair of individuals using PLINK software and randomly removed one individual from each pair with an IBD value greater than .25 if both individuals had the same lifetime PTSD phenotype; if, within a pair of individuals with IBD larger than .25, one was a PTSD case and the other a PTSD control or PTSD diagnosis unknown, we removed the other subject to maximize the sample size of PTSD cases. A total of 310 EAs and 685 AAs were removed because of relatedness, leaving 1578 EAs and 2766 AAs for analysis. To increase statistical power, we also performed GWAS analysis without removing the related subjects but adjusted the genetic structure using factored spectrally transformed linear mixed models (25). This left 1846 EAs and 3392 AAs for analysis after quality control steps.
To address potential population stratification, we used the PLINK nearest neighbor method and filtered out individuals outside three standard deviations of the first four multidimensional scaling (MDS) factors. The first four MDS factors were also adjusted in the GWAS analyses to control for population substructure, as when the first 10 MDS factors were adjusted, the genomic inflation factor λs were not improved.
For SNP quality control, we excluded SNPs with a call rate <98%, SNPs with minor allele frequencies <1%, and SNPs inconsistent with Hardy-Weinberg equilibrium at a p < 1 ×10−6 in control subjects. We did not exclude such markers from the affected part of the sample, since cases can show deviations from Hardy-Weinberg equilibrium at loci associated with disease (26).
We first performed GWAS analyses on all subjects after quality control with logistic regression models, adjusted by the first four MDS factors, sex, and age. European Americans and AAs were analyzed separately. To evaluate the influence of trauma exposure specifically, we restricted the GWAS analyses to subjects with self-reported trauma experiences. In this secondary analysis, the covariates of MDS factors, sex, and age were the same as the primary analysis.
We estimated the local ancestry around the two TLL1 SNPs (rs6812849 and rs7691872) of all GWAS AA subjects using the HAPMIX program (27). Logistic regression tests of the two TLL1 SNPs and PTSD in AAs were performed after adjustment for age, sex, and local European ancestry scores. The detailed information of estimating local ancestry is provided in Supplemental Methods in Supplement 1.
Based on the GWAS results in EAs, six SNPs (rs406001, rs382903, rs450378, rs6812849, rs1503292, and rs7691872) from two genomic regions (7p12 and 4q32) were scheduled for genotyping using TaqMan assays (Life Technologies, Carlsbad, California) in another 3112 subjects for follow-up and in 5504 of the 5799 GWAS subjects to estimate genotyping error rates. Single nucleotide polymorphisms rs382903 and rs1503292 failed quality control. In the 3112 follow-up subjects, 2752 were genotyped for 96 ancestry informative markers (AIMs), all SNPs, to estimate ancestry proportion by STRUCTURE software (28). Ninety of these AIMs were selected from SNPs included on the Illumina OminQuad genotyping microarray using a Bayesian strategy to maximize allele frequency differences between European, African, Asian, and other ancestry; the remainders were markers of frequent interest in psychiatric genetics. Subjects with African ancestry proportion scores <.5 were classified as EAs; otherwise, they were classified as AAs. We did not have AIMs information for the remaining 360 subjects, and self-reported population group was used to classify them. Of the 2752 subjects for whom AIMs were available, only 1.05% of the self-reported EAs were not grouped as EAs by analysis of the AIMs, indicating that overall, the self-identification of race was highly accurate.
After that, we followed up another four SNPs using a custom Illumina GoldenGate Genotyping Universal-32, 1536-plex micro-array assay (Illumina, San Diego, California). The majority of SNPs included in the custom array were selected for studies of other phenotypes. We followed up the top two most significant SNPs in the GWAS results of AAs and another two SNPs that map to obvious candidate loci and were among the top 10 most significant SNPs in the GWAS results of EAs. A total of 2553 subjects were genotyped and 2441 were retained after quality control. Among them, 505 did not have AIMs information and were grouped using self-reported population information.
Although we excluded related individuals in our primary GWAS analyses, a small portion (5.9%) of the replication samples were relatives. Therefore, generalized estimating equations (GEE) were applied to fit the logistic regression model in the replication samples to account for the correlated data from individuals in the same family. The GEE logistic regression estimates the same model as the standard logistic regression. Unlike standard logistic regression, GEE logistic regression allows for dependence (such as related individuals) within clusters (29). The logistic regression models were adjusted by age and sex.
Results
After quality control of the GWAS data and removal of the related subjects, 1633 EAs and 2847 AAs were retained. We further removed 55 EAs and 81 AAs with unknown lifetime PTSD diagnosis. Final GWAS analyses included 1578 EAs (300 PTSD cases) and 2766 AAs (444 PTSD cases). Demographic information of the subjects is listed in Table 1.
Table 1.
Subject Demographic Information
| Cases
|
Control Subjects
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Subjects (n) | % Female | Age (SD) | % SD | Subjects (n) | % Female | Age (SD) | % SD | |
| GWAS Subjects | ||||||||
| AA | 444 | 54.3 | 41.5 (8.7) | 96.8 | 2322 | 43.1 | 41.2 (9.3) | 79.5 |
| EA | 300 | 60.3 | 37.7 (9.8) | 100 | 1278 | 35.4 | 38.4 (11.3) | 93 |
| Replication Subjects | ||||||||
| AA | 89 | 51.7 | 41.0 (9.5) | 88.8 | 655 | 46.6 | 41.1 (11.2) | 52.5 |
| EA | 207 | 58.5 | 38.4 (10.8) | 90.8 | 1692 | 44.9 | 39.6 (13.9) | 42 |
AA, African Americans; Age (SD), Age (standard deviation); EA, European Americans; GWAS, genome-wide association study; % SD, % of substance dependence; subjects who met criteria for alcohol dependence, cocaine dependence, and/or opioid dependence based on DSM-IV.
In EAs and AAs, 768,146 and 870,103 SNPs, respectively, passed quality control steps. Based on Bonferroni correction for multiple comparisons, the threshold of genome-wide significance was 6.51 ×10−8 for EAs and 5.75×10−8 for AAs. The genomic inflation factor λs were 1.010 and 1.003 for EAs and AAs, respectively. The quartile-quartile plots are shown in Figures 1A and B. In EAs (Table 2), one SNP, rs406001 on chromosome 7p12, exceeded the genome-wide significant threshold for association with p = 3.97×10−8 (Figure 2A). Another two SNPs in the same region, rs382903 and rs450378, were the second and fourth most significant SNPs. The three SNPs are located approximately 630 kilobase downstream of the nearest gene Cordon-Bleu (COBL). The region showing the second strongest evidence of association was chromosome 4q32 (Figure 2B), which is in the first intron of Tolloid-Like 1 gene (TLL1). The top SNP rs6812849 reached a p value of 2.99×10−7. Another two SNPs in the same region, rs7691872 and rs1503292, were the sixth and seventh most significant SNPs. The SNPs ranked as the eighth and ninth most significant were located in the introns of NDRG1 and GABBR2, respectively, with p values of 3.1 × 10−6 and 5.4 × 10−6. Compared with EAs, GWAS results for AAs were less remarkable; no SNPs had p < 1 ×10−6 (Table 3).
Figure 1.
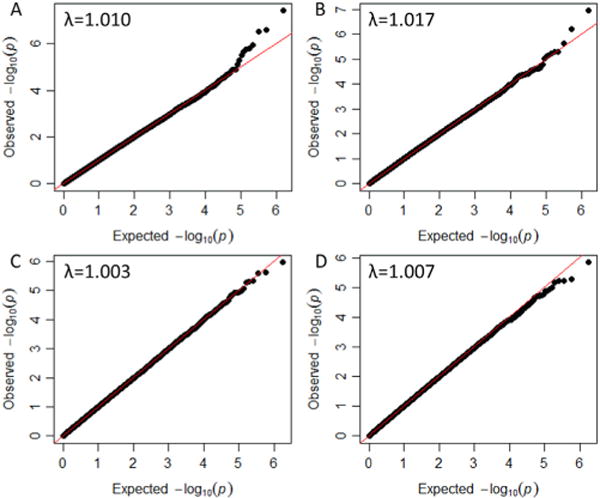
Quintile-Quintile plots for whole cohort of European Americans (A), European Americans with trauma experiences (B), whole cohort of African Americans (C), and African Americans with trauma experiences (D). The genomic inflation factor (λ) is the ratio of the observed median p value to its expected value.
Table 2.
Top Ten Most Significant SNPs in the GWAS Discovery Results for European Americans
| CHR | SNP | Nearest Gene | Distance (bp) | p Value in EA GWAS | p Value in Replication EA Samples | p value in Combined EA Samples |
|---|---|---|---|---|---|---|
| Whole Cohort of European Americans | ||||||
| 7 | rs406001 | COBL | 621900 | 3.97E-08 | .95 | 2.77E-04 |
| 7 | rs382903 | COBL | 648684 | 2.70E-07 | NT | NT |
| 4 | rs6812849 | TLL1 | intron | 2.99E-07 | 6.28E-06 | 3.13E-09 |
| 7 | rs450378 | COBL | 624219 | 1.19E-06 | .25 | 2.03E-04 |
| 20 | rs4491792 | OTOR | 8016 | 1.57E-06 | NT | NT |
| 4 | rs1503292 | TLL1 | intron | 1.71E-06 | NT | NT |
| 4 | rs7691872 | TLL1 | intron | 2.22E-06 | 2.30E-04 | 1.22E-07 |
| 8 | rs2272651 | NDRG1 | intron | 3.10E-06 | .76 | 5.36E-05 |
| 9 | rs2779551 | GABBR2 | intron | 5.36E-06 | .28 | .019 |
| 10 | rs16907840 | MTRNR2L5 | 188808 | 7.59E-06 | NT | NT |
| Trauma Exposed European Americans | ||||||
| 7 | rs406001 | COBL | 621900 | 1.14E-07 | .41 | 3.41E-03 |
| 7 | rs382903 | COBL | 648684 | 6.10E-07 | NT | NT |
| 4 | rs6812849 | TLL1 | intron | 2.37E-06 | 1.59E-03 | 1.22E-06 |
| 7 | rs450378 | COBL | 624219 | 4.89E-06 | .41 | 5.59E-04 |
| 1 | rs4430311 | AKT3 | 9107 | 5.11E-06 | NT | NT |
| 20 | rs4491792 | OTOR | 8016 | 5.96E-06 | NT | NT |
| 1 | rs2236805 | PAX7 | iTRPS1 intron | 6.72E-06 | NT | NT |
| 4 | rs1425392 | TECRL | 117034 | 7.37E-06 | NT | NT |
| 13 | rs9538394 | DIAPH3 | 311296 | 9.12E-06 | NT | NT |
| 10 | rs16907840 | MTRNR2L5 | 188808 | 1.58E-05 | NT | NT |
bp, base pair; CHR, chromosome; EA, European Americans; GWAS, genome-wide association study; NT, not tested; SNP, single nucleotide polymorphism.
Figure 2.
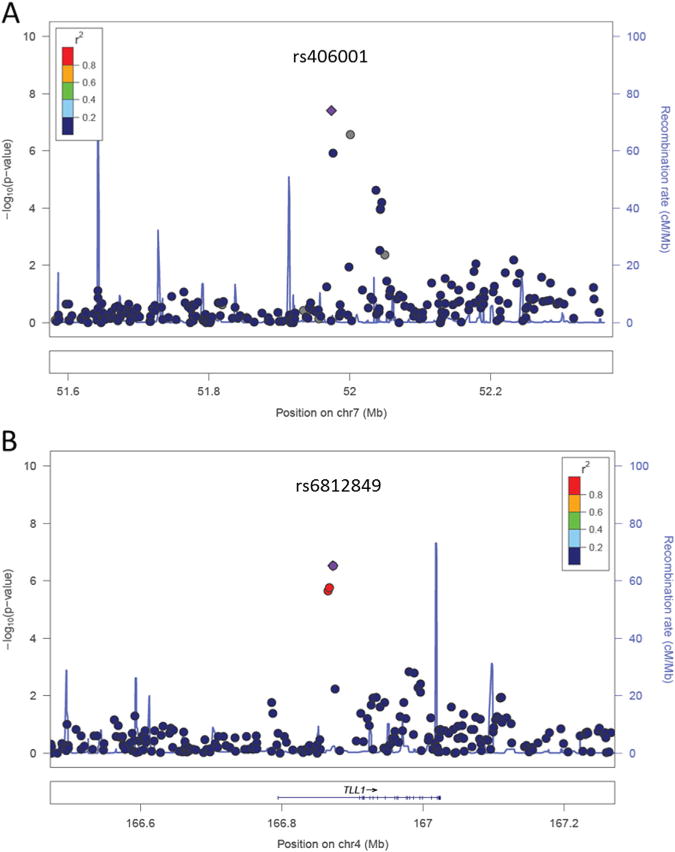
Regional Manhattan plots of the top two strongest association regions in European Americans: chromosome 7p12 (A) and chromosome 4q32 (B). The single nucleotide polymorphisms are color coded based on the linkage disequilibrium with the most significant single nucleotide polymorphism in each region, rs406001 and rs6812849, respectively.
Table 3.
Top Ten Most Significant SNPs in the GWAS Results of the African Americans
| CHR | SNP | Nearest Gene | Distance (bp) | p Value in AA GWAS | p Value in Replication AA Samples | p Value in Combined AA Samples |
|---|---|---|---|---|---|---|
| Whole Cohort of African Americans | ||||||
| 8 | rs7014900 | TRPS1 | 842417 | 1.10E-06 | .45 | 1.31E-04 |
| 2 | rs13006863 | SLC4A5 | intron | 2.45E-06 | .015 | 2.25E-03 |
| 14 | rs7158872 | RPS29 | 617197 | 2.54E-06 | NT | NT |
| 11 | rs2305272 | NTM | intron | 4.70E-06 | NT | NT |
| 2 | 2-21091512 | APOB | intron | 5.07E-06 | NT | NT |
| 8 | rs16904179 | FAM49B | intron | 5.42E-06 | NT | NT |
| 11 | rs6589574 | SIK3 | intron | 8.73E-06 | NT | NT |
| 3 | rs4955793 | ZMAT3 | intron | 1.06E-05 | NT | NT |
| 7 | rs6969903 | AHR | 92683 | 1.11E-05 | NT | NT |
| 17 | rs7207499 | PRKCA | intron | 1.16E-05 | NT | NT |
| Trauma Exposed African Americans | ||||||
| 8 | rs7014900 | TRPS1 | 842417 | 1.46E-06 | .14 | 3.90E-04 |
| 11 | rs2305272 | NTM | intron | 5.22E-06 | NT | NT |
| 14 | rs7158872 | RPS29 | 617197 | 6.02E-06 | NT | NT |
| 2 | rs13006863 | SLC4A5 | intron | 6.03E-06 | .022 | 2.14E-03 |
| 6 | rs2572096 | FBXL4 | 55222 | 7.07E-06 | NT | NT |
| 8 | rs1500899 | C8orf37 | 253528 | 9.94E-06 | NT | NT |
| 2 | 2-21091512 | APOB | intron | 1.18E-05 | NT | NT |
| 21 | rs11909487 | USP25 | 40117 | 1.30E-05 | NT | NT |
| 10 | rs7076096 | GRIN1 | intron | 1.36E-05 | NT | NT |
| 16 | rs9923585 | ABCC1 | intron | 1.74E-05 | NT | NT |
AA, African Americans; bp, base pair; CHR, chromosome; GWAS, genome-wide association study; NT, not tested; SNP, single nucleotide polymorphism.
Next, we restricted the GWAS analyses to PTSD control subjects with self-reported traumatic experiences. A total of 468 EAs and 933 AAs who did not report any traumatic events were excluded, leaving 1110 EAs and 1833 AAs for analyses. Distribution of different types of trauma exposure in EAs and in AAs is shown in Figure S1 in Supplement 1. In both populations, witnessed someone being badly injured or killed was the most reported trauma experience, followed by seriously physically attacked or assaulted. In EAs, 32% reported one traumatic experience, 23% reported two traumatic experiences, and 45% reported three or more traumatic experiences. The distribution in AAs was very similar (Figure S2 in Supplement 1). In EAs and AAs, 768,784 SNPs and 871,502 SNPs, respectively, passed quality control steps. The threshold of genome-wide significance was 6.50×10−8 for EAs and 5.74×10−8 for AAs. The genomic inflation factor λs were 1.017 and 1.007 for EAs and AAs, respectively. The quartile-quartile plots are shown in Figure 1C, D. Despite restricting the control subjects to trauma exposed subjects, which removed nearly one third of the sample, the GWAS results for EAs were very similar to those derived from the whole cohort (Figure S3 in Supplement 1). Single nucleotide polymorphism rs406001 on chromosome 7p12 remained the most significant SNP, followed by rs382903 and rs450378 located in the same region of chromosome 7p12 with p values ranked as the second and fourth in the results. The first intron of TLL1 remained the second most significant region. However, probably due to the reduced sample size, no SNPs reached genome-wide significance in the subsample. Genome-wide association study results in AAs also yielded no significant association regions.
We repeated the GWAS analyses again without removing the related subjects. Instead, we adjusted the genetic structure using factored spectrally transformed linear mixed models (25). This left 1846 EAs (308 cases) and 3392 AAs (456 cases) for analysis after quality control steps. Apparently, due to the larger sample size, GWAS results for EAs were more significant than before (Table S1 in Supplement 1). Single nucleotide polymorphism rs406001 on chromosome 7p12 remained the most significant SNP with p = 1.74×10−8. Single nucleotide polymorphisms rs382903 and rs450378 in the same region were the third and thirteenth most significant SNPs, and rs6812849 in the first intron of TLL1 was the second most significant SNP with p = 1.21 × 10−7. Another two SNPs in the first intron of TLL1, rs1503292 and rs7691872, were the fourth and fifth most significant SNPs, respectively. After restricting the analysis to trauma-exposed subjects, the three SNPs on 7p12 were the first, fifth, and thirteenth most significant SNPs; the three SNPs in the first intron of TLL1 ranked as the sixth, seventh, and eighth most significant SNPs. Genome-wide association study results for AAs were also very similar (Table S2 in Supplement 1). No SNPs reached genome-wide significance for AAs.
Based on the GWAS results from EAs, we TaqMan genotyped four SNPs in another 3112 subjects recruited using the same methodology for follow-up. These four SNPs were also genotyped in 5504 of the 5799 GWAS subjects for estimating genotyping error rates. Two of the four SNPs, rs406001 and rs450378, were located on chromosome 7p12. The linkage disequilibrium (R2) between them was .54. The other two SNPs, rs6812849 and rs7691872, were located in the first intron of TLL1. They were in high linkage disequilibrium (R2 = .88). By comparing the genotyping results from TaqMan and the GWAS array, the measured discordance rates for rs406001, rs450378, rs6812849, and rs7691872 were .20%, .65%, .71%, and .29%, respectively.
We excluded 85 of the 3112 replication samples due to an unknown PTSD diagnosis. In the remaining 3027 subjects, 1899 were EAs, including 207 PTSD cases. Of the four SNPs genotyped in the replication sample, the two located at TLL1, rs6812849 and rs7691872, were significantly associated with PTSD with p values of 6.3×10−6 and 2.3×10−4, respectively. Upon combining the GWAS and replication samples, rs6812849 reached genome-wide significance with p = 3.1×10−9, and rs7691872 reached p = 1.2×10−7. When the analyses were restricted to subjects with trauma exposure, the p values for rs6812849 and rs7691872 were 1.6×10−3 and 2.5×10−3, respectively, in the replication sample and 1.2×10−6 and 4.3×10−6, respectively, in the combined sample. The association between PTSD and the two SNPs on chromosome 7p12 was not replicated (Table 2).
In addition to chromosome 7p12 and chromosome 4q32, we followed up another four SNPs using a GoldenGate custom array (Illumina, San Diego, California) in 2553 replication subjects. After quality control, 1658 EAs (134 PTSD cases) and 744 AAs (89 PTSD cases) with known PTSD diagnosis were included in the replication analyses. The first SNP, rs2272651, was located in the seventh intron of NDRG1 and the second SNP, rs2779551, was located in the fourth intron of GABBR2. These two SNPs were the eighth and ninth most significant in the GWAS results for EAs and were selected because they map to obvious candidate loci. Neither of these SNPs was significantly associated with PTSD in the 1658 EA subject replication sample (Table 2). We also tested two SNPs with the most significant p values in the GWAS results of AAs: rs7014900, which is about 842 kilobase away from the nearest gene TRPS1, and rs13006863, which is located in the 14th intron of SLC4A5. Likewise, neither of them was replicated in the 744 AAs (Table 3).
Neither of the TLL1 SNPs, rs6812849 and rs7691872, was significantly associated with PTSD in AAs. Because AA is an admixed population with ~20% European ancestry (30), we estimated the local ancestry around the two TLL1 SNPs of the GWAS AA subjects using HAPMIX (27). Local estimated European ancestry of each subject was adjusted in the logistic regression model. Nevertheless, no significant association was observed (rs6812849, p = .68; rs7691872, p = .48). Among the 2766 AAs, 2266 had local African ancestry ≥50%. We restricted our analysis to these subjects, and no significant association was observed (rs6812849, p = .95; rs7691872, p = .90).
Discussion
In this study, we performed GWAS analyses of PTSD in 1578 EAs (300 PTSD cases) and 2766 AAs (444 PTSD cases). In the primary analyses in EAs, we observed genome-wide significant evidence of association to that trait for one SNP located on chromosome 7p12, rs406001. Two other SNPs in the same region, rs382903 and rs450378, were the second and fourth most significant SNPs. The region showing the second strongest evidence of association was the first intron of TLL1 where three SNPs, rs6812849, rs7691872 and rs1503292, ranked in the top 10 results. When GWAS analyses were restricted to subjects with trauma exposure, despite the exclusion of nearly one third of the subjects, the results were very similar. Chromosome 7p12 and the first intron of TLL1 remained the top two most significant genomic regions. Based on the GWAS results, we genotyped six SNPs in an independent sample of nearly 2000 EAs using TaqMan genotyping or GoldenGate custom array. Two SNPs located in the first intron of TLL1 were significantly associated with PTSD with p values 6.3×10−6 and 2.3×10−4. Combining the GWAS and replication sample, rs6812849 reached genome-wide significance with a p value of 3.1×10−9 and rs7691872 had a p value of 1.2×10−7. These two SNPs in TLL1 were also replicated in subjects with trauma exposure, but the association did not reach genome-wide significance in the combined samples. The two SNPs on 7p12 were not replicated. No genome-wide significant SNPs were observed in AAs.
Mammalian Tolloid-like 1 (mTll1) is one of the four Tolloid proteins that are pleiotropic, astacin-like metalloproteases. This protein is expressed at high levels in the granular layer of the cerebellum of adult mice (30) and in the hippocampus of both juvenile and adult mice (31). The hippocampus is a direct target of stress hormones (32). Cerebellum plays important roles in emotion processing and fear conditioning through connecting with the hypothalamic-pituitary-adrenal axis (33). Studies have found that the development of cerebellum is more dependent on environmental factors than most other brain regions (34). It has been consistently shown that children and adolescents with a history of maltreatment have decreased cerebellar volume (35,36). Although TLL1 has not been previously associated with PTSD, functional studies suggest that glucocorticoids, which regulate stress response in mammals, could decrease mTll1 expression in vitro (31). An elevated number of hippocampal neurons expressing mTll1 were found in mice with increased neurogenesis, which is related to the stress response (31,37). Research on reptilian orthology of TLL1 isolated from turtle showed that the gene cleaves the precursor pro-brain-derived neurotrophic factor (proBDNF) into mature BDNF (38). The mature form of BDNF is involved in learning and memory and is very important in normal neural development.
Previous studies have indicated that genetic factors influence the risk of exposure to trauma, perhaps through personality differences (8). Although exposure to trauma (or perceived exposure to trauma) is essential to PTSD, in the primary GWAS analyses, we did not restrict the control subjects to trauma-exposed subjects. The significant genetic markers found by this method thus contribute to the risk of PTSD after trauma exposure and/or trauma exposure, which lead to PTSD. After restricting the control subjects to those who were trauma exposed, the significant genetic markers would appear to contribute to PTSD risk only. Although the p values were less significant, which was probably due to a smaller sample size, the two most significant genomic regions did not change when this restriction was imposed on the control group. Thus, the two regions identified are most likely to contribute to PTSD, rather than trauma exposure.
Our GWAS analyses of PTSD in AAs did not provide any strong evidence of association. When we compared the results of AAs and EAs, we found that no SNPs ranked among the top 100 most significant overlapped between the two population groups. Further, none of the top 10 most significant SNPs in the GWAS results for EAs were associated with PTSD in AAs at p < .05, and vice versa. We tested the association between the two TLL1 SNPs (rs6812849 and rs7691872) and PTSD in AAs after adjustment for local ancestry but no association was observed, and no association was observed in AAs with local African ancestry larger than 50%. Possible explanations for this observation include population differences in epistasis and in gene expression and regulation.
Although the heritability of PTSD is estimated to be in the range of 30% to 40% by twin studies (7,8), no genetic variants have been conclusively identified to contribute to PTSD risk by candidate gene association studies (15). A study showed that an SNP in gene ADCYAP1R1 was associated with PTSD in women (39). However, it was not replicated in two independent samples (40). Recently, Logue et al. (41) performed a PTSD GWAS study and observed that the RORA gene was a significant risk locus for PTSD. However, our study failed to replicate the results (Table S3 in Supplement 1). In the current GWAS, only one genomic region (TLL1) was successfully replicated and reached genome-wide significance. It is important to ascertain whether this region can be replicated in other independent samples. Due to the relatively small sample size, we likely missed true risk variants. It is also possible that common variants do not make a large contribution to the risk of PTSD.
The findings from this study should be considered in the context of a number of limitations. First, the sample was recruited primarily to study substance dependence traits. There is high comorbidity between PTSD and substance dependence, and although we have replicated findings from other groups relevant to PTSD in this sample (11,12,18,41), it is possible that the findings are not applicable to PTSD in other patient populations. In addition, the size of the GWAS and replication samples was small, and thus the statistical power was low. Finally, our replicated results are specific to EA populations. Previous work regarding FKBP5 (11,12) showed gene×environment effects were applicable only to AA populations. We may, therefore, conclude that the genetic underpinnings of PTSD are at least partially different in these two populations.
Supplementary Material
Acknowledgments
This study was supported by National Institutes of Health Grants RC2 DA028909, R01 DA12690, R01 DA12849, R01 DA18432, R01 AA11330, and R01 AA017535 and the Veterans Affairs National Center for Posttraumatic Stress Disorder Research.
We appreciate the recruitment and assessment efforts at Yale University School of Medicine and the APT Foundation by James Poling, Ph.D.; at McLean Hospital by Roger Weiss, M.D.; at the Medical University of South Carolina by Kathleen Brady, M.D., Ph.D., and Raymond Anton, M.D.; and at the University of Pennsylvania by David Oslin, M.D. Genotyping services for a part of our genome-wide association study were provided by the Center for Inherited Disease Research and Yale University (Center for Genome Analysis). The Center for Inherited Disease Research is fully funded through a federal contract from the National Institutes of Health to the Johns Hopkins University (contract number N01-HG-65403). We are grateful to Ann Marie Lacobelle, Michelle Cucinelli, Christa Robinson, and Greg Dalton-Kay for their excellent technical assistance and to the Semi-Structured Assessment for Drug Dependence and Alcoholism interviewers, led by Yari Nuñez and Michelle Slivinsky.
Footnotes
Dr. Kranzler has served as a consultant or advisory board member for Alkermes, Lilly, Lundbeck, Pfizer, and Roche. He is also a member of the American Society of Clinical Psychopharmacology’s Alcohol Clinical Trials Initiative, which is supported by Lilly, Lundbeck, Abbott, and Pfizer. The other authors report no biomedical financial interests or potential conflicts of interest.
Supplementary material cited in this article is available online at http://dx.doi.org/10.1016/j.biopsych.2013.04.013.
References
- 1.Speed N, Engdahl B, Schwartz J, Eberly R. Posttraumatic stress disorder as a consequence of the POW experience. J Nerv Ment Dis. 1989;177:147–153. doi: 10.1097/00005053-198903000-00004. [DOI] [PubMed] [Google Scholar]
- 2.Kulka RA, Schlenger WE, Fairbank JA. Trauma and the Vietnam War: Report of Findings from the National Vietnam Veterans Readjustment Study. New York: Brunner/Mazel; 1990. [Google Scholar]
- 3.Kessler RC, Berglund P, Demler O, Jin R, Merikangas KR, Walters EE. Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Arch Gen Psychiatry. 2005;62:593–602. doi: 10.1001/archpsyc.62.6.593. [DOI] [PubMed] [Google Scholar]
- 4.Kessler RC, Sonnega A, Bromet E, Hughes M, Nelson CB. Posttraumatic stress disorder in the National Comorbidity Survey. Arch Gen Psychiatry. 1995;52:1048–1060. doi: 10.1001/archpsyc.1995.03950240066012. [DOI] [PubMed] [Google Scholar]
- 5.Bromet E, Sonnega A, Kessler RC. Risk factors for DSM-III-R posttraumatic stress disorder: Findings from the National Comorbidity Survey. Am J Epidemiol. 1998;147:353–361. doi: 10.1093/oxfordjournals.aje.a009457. [DOI] [PubMed] [Google Scholar]
- 6.Bramsen I, Dirkzwager AJ, van der Ploeg HM. Predeployment personality traits and exposure to trauma as predictors of posttraumatic stress symptoms: A prospective study of former peacekeepers. Am J Psychiatry. 2000;157:1115–1119. doi: 10.1176/appi.ajp.157.7.1115. [DOI] [PubMed] [Google Scholar]
- 7.True WR, Rice J, Eisen SA, Heath AC, Goldberg J, Lyons MJ, Nowak J. A twin study of genetic and environmental contributions to liability for posttraumatic stress symptoms. Arch Gen Psychiatry. 1993;50:257–264. doi: 10.1001/archpsyc.1993.01820160019002. [DOI] [PubMed] [Google Scholar]
- 8.Stein MB, Jang KL, Taylor S, Vernon PA, Livesley WJ. Genetic and environmental influences on trauma exposure and posttraumatic stress disorder symptoms: A twin study. Am J Psychiatry. 2002;159:1675–1681. doi: 10.1176/appi.ajp.159.10.1675. [DOI] [PubMed] [Google Scholar]
- 9.Segman RH, Cooper-Kazaz R, Macciardi F, Goltser T, Halfon Y, Dobroborski T, Shalev AY. Association between the dopamine transporter gene and posttraumatic stress disorder. Mol Psychiatry. 2002;7:903–907. doi: 10.1038/sj.mp.4001085. [DOI] [PubMed] [Google Scholar]
- 10.Lee HJ, Lee MS, Kang RH, Kim H, Kim SD, Kee BS, et al. Influence of the serotonin transporter promoter gene polymorphism on susceptibility to posttraumatic stress disorder. Depress Anxiety. 2005;21:135–139. doi: 10.1002/da.20064. [DOI] [PubMed] [Google Scholar]
- 11.Binder EB, Bradley RG, Liu W, Epstein MP, Deveau TC, Mercer KB, et al. Association of FKBP5 polymorphisms and childhood abuse with risk of posttraumatic stress disorder symptoms in adults. JAMA. 2008;299:1291–1305. doi: 10.1001/jama.299.11.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie P, Kranzler HR, Poling J, Stein MB, Anton RF, Farrer LA, Gelernter J. Interaction of FKBP5 with childhood adversity on risk for post-traumatic stress disorder. Neuropsychopharmacology. 2010;35:1684–1692. doi: 10.1038/npp.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolassa IT, Kolassa S, Ertl V, Papassotiropoulos A, De Quervain DJ. The risk of posttraumatic stress disorder after trauma depends on traumatic load and the catechol-o-methyltransferase Val(158)Met polymorphism. Biol Psychiatry. 2010;67:304–308. doi: 10.1016/j.biopsych.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 14.Amstadter AB, Koenen KC, Ruggiero KJ, Acierno R, Galea S, Kilpatrick DG, Gelernter J. Variant in RGS2 moderates posttraumatic stress symptoms following potentially traumatic event exposure. J Anxiety Disord. 2009;23:369–373. doi: 10.1016/j.janxdis.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cornelis MC, Nugent NR, Amstadter AB, Koenen KC. Genetics of post-traumatic stress disorder: Review and recommendations for genome-wide association studies. Curr Psychiatry Rep. 2010;12:313–326. doi: 10.1007/s11920-010-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie P, Kranzler HR, Poling J, Stein MB, Anton RF, Brady K, et al. Interactive effect of stressful life events and the serotonin transporter 5-HTTLPR genotype on posttraumatic stress disorder diagnosis in 2 independent populations. Arch Gen Psychiatry. 2009;66:1201–1209. doi: 10.1001/archgenpsychiatry.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie P, Kranzler HR, Farrer L, Gelernter J. Serotonin transporter 5-HTTLPR genotype moderates the effects of childhood adversity on posttraumatic stress disorder risk: A replication study. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:644–652. doi: 10.1002/ajmg.b.32068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilpatrick DG, Koenen KC, Ruggiero KJ, Acierno R, Galea S, Resnick HS, et al. The serotonin transporter genotype and social support and moderation of posttraumatic stress disorder and depression in hurricane-exposed adults. Am J Psychiatry. 2007;164:1693–1699. doi: 10.1176/appi.ajp.2007.06122007. [DOI] [PubMed] [Google Scholar]
- 19.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierucci-Lagha A, Gelernter J, Chan G, Arias A, Cubells JF, Farrer L, Kranzler HR. Reliability of DSM-IV diagnostic criteria using the semi-structured assessment for drug dependence and alcoholism (SSADDA) Drug Alcohol Depend. 2007;91:85–90. doi: 10.1016/j.drugalcdep.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pierucci-Lagha A, Gelernter J, Feinn R, Cubells JF, Pearson D, Pollastri A, et al. Diagnostic reliability of the Semi-structured Assessment for Drug Dependence and Alcoholism (SSADDA) Drug Alcohol Depend. 2005;80:303–312. doi: 10.1016/j.drugalcdep.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 22.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 23.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8:833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- 26.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price AL, Tandon A, Patterson N, Barnes KC, Rafaels N, Ruczinski I, et al. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 2009;5:e1000519. doi: 10.1371/journal.pgen.1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. Am J Hum Genet. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeger SL, Liang KY. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
- 30.Takahara K, Lyons GE, Greenspan DS. Bone morphogenetic protein-1 and a mammalian tolloid homologue (mTld) are encoded by alternatively spliced transcripts which are differentially expressed in some tissues. J Biol Chem. 1994;269:32572–32578. [PubMed] [Google Scholar]
- 31.Tamura G, Olson D, Miron J, Clark TG. Tolloid-like 1 is negatively regulated by stress and glucocorticoids. Brain Res Mol Brain Res. 2005;142:81–90. doi: 10.1016/j.molbrainres.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 32.McEwen BS. Stress and hippocampal plasticity. Annu Rev Neurosci. 1999;22:105–122. doi: 10.1146/annurev.neuro.22.1.105. [DOI] [PubMed] [Google Scholar]
- 33.Schutter DJ, van Honk J. The cerebellum on the rise in human emotion. Cerebellum. 2005;4:290–294. doi: 10.1080/14734220500348584. [DOI] [PubMed] [Google Scholar]
- 34.Giedd JN, Schmitt JE, Neale MC. Structural brain magnetic resonance imaging of pediatric twins. Hum Brain Mapp. 2007;28:474–481. doi: 10.1002/hbm.20403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bauer PM, Hanson JL, Pierson RK, Davidson RJ, Pollak SD. Cerebellar volume and cognitive functioning in children who experienced early deprivation. Biol Psychiatry. 2009;66:1100–1106. doi: 10.1016/j.biopsych.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carrion VG, Weems CF, Watson C, Eliez S, Menon V, Reiss AL. Converging evidence for abnormalities of the prefrontal cortex and evaluation of midsagittal structures in pediatric posttraumatic stress disorder: An MRI study. Psychiatry Res. 2009;172:226–234. doi: 10.1016/j.pscychresns.2008.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hopkins DR, Keles S, Greenspan DS. The bone morphogenetic protein 1/Tolloid-like metalloproteinases. Matrix Biol. 2007;26:508–523. doi: 10.1016/j.matbio.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keifer J, Sabirzhanov BE, Zheng Z, Li W, Clark TG. Cleavage of proBDNF to BDNF by a tolloid-like metalloproteinase is required for acquisition of in vitro eyeblink classical conditioning. J Neurosci. 2009;29:14956–14964. doi: 10.1523/JNEUROSCI.3649-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ressler KJ, Mercer KB, Bradley B, Jovanovic T, Mahan A, Kerley K, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chang SC, Xie P, Anton RF, De Vivo I, Farrer LA, Kranzler HR, et al. No association between ADCYAP1R1 and post-traumatic stress disorder in two independent samples. Mol Psychiatry. 2012;17:239–241. doi: 10.1038/mp.2011.118. [DOI] [PubMed] [Google Scholar]
- 41.Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, et al. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus [published online ahead of print August 7] Mol Psychiatry. 2012 doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


