Abstract
A genome-wide association study was conducted using microarray technology to identify genes that may be associated with the vulnerability to develop heroin addiction, using DNA from 104 individual former severe heroin addicts (meeting Federal criteria for methadone maintenance) and 101 individual control subjects, all Caucasian. Using separate analyses for autosomal and X chromosomal variants, we found that the strongest associations of allele frequency with heroin addiction were with the autosomal variants rs965972, located in the Unigene cluster Hs.147755 (experiment-wise q = 0.053), and rs1986513 (q = 0.187). The three variants exhibiting the strongest association with heroin addiction by genotype frequency were rs1714984, located in an intron of the gene for the transcription factor myocardin (P= 0.000022), rs965972 (P = 0.000080) and rs1867898 (P = 0.000284). One genotype pattern (AG-TT-GG) was found to be significantly associated with developing heroin addiction (odds ratio (OR) = 6.25) and explained 27% of the population attributable risk for heroin addiction in this cohort. Another genotype pattern (GG-CT-GG) of these variants was found to be significantly associated with protection from developing heroin addiction (OR = 0.13), and lacking this genotype pattern explained 83% of the population attributable risk for developing heroin addiction. Evidence was found for involvement of five genes in heroin addiction, the genes coding for the μ opioid receptor, the metabotropic receptors mGluR6 and mGluR8, nuclear receptor NR4A2 and cryptochrome 1 (photolyase-like). This approach has identified several new genes potentially associated with heroin addiction and has confirmed the role of OPRM1 in this disease.
Keywords: addiction, gene, heroin, microarray, polymorphism
Introduction
Addiction to opiates is a chronic, relapsing brain disease that, if left untreated, can cause major medical, social and economic problems. Approximately one-third of people self-exposed to opiates will become addicted.1 A major contributing factor to the development of addiction is genetic predisposition. Epidemiological studies in men have found that approximately 40–60% of the risk of developing an addiction to heroin is genetically determined2–4 and, in a single study, the genetic component specific to heroin addiction was 38%.3
Genetic variants in and around several genes have been found to be associated with opiate addiction.5–9 These include variants in genes coding for two receptors of the opioid system, the μ10–12 and κ opioid receptors.13 Other variants that have been found to be associated with opiate addiction are located in and around the dopamine D214–16 and D4 receptors,17–20 the serotonin transporter,21,22 the serotonin 1B receptor23 and genes for the enzyme catechol-O-methyltransferase. 24,25
Studies have provided evidence that several chromosomal regions are linked to the vulnerability to develop heroin, alcohol, nicotine or other drug abuse or addiction.26–39 Techniques for the genome-wide association of variants have become available including Affymetrix GeneChip microarrays. Microarrays have been used extensively in the study of cancer to identify variants associated with disease40 and, recently, for linkage analysis of opioid dependence.41 The only other published microarray study to identify variants associated with drug and alcohol addiction have used pooled but not individual samples and the 1494,42 10 000,43 100 000,44,45 and 500 000 single nucleotide polymorphism Affymetrix GeneChips.45
In this study, we have used Affymetrix 10K GeneChips to locate and identify genes that may be associated with vulnerability to develop heroin addiction. Genome-wide scans were performed on individual subjects who were former severe heroin addicts in methadone maintenance treatment and on control subjects, each of whom had been extensively characterized phenotypically.
Materials and methods
Subjects and phenotyping
Two hundred and five subjects, who were consecutive volunteers in genetic studies conducted by the Laboratory of the Biology of Addictive Diseases at The Rockefeller University and who had met the inclusion criteria defined below, were examined in this study. Former severe heroin addicts (n = 104) or controls (n = 101) were recruited from referrals, newspaper advertisements, posted notices and several clinical resources in New York City. All subjects gave a signed informed consent, which was approved by The Rockefeller University Hospital Institutional Review Board, and gave specific consent for genetic studies. The cohort consisted of Caucasian individuals based on the ethnic/cultural background of the subjects, their parents, grandparents and great grandparents.
The Addiction Severity Index46 was administered, and urine analyses were performed for multiple drugs of abuse on all subjects. Case subjects were former severe heroin addicted patients who met Federal guidelines for methadone maintenance treatment (1 year or more of daily multiple injections of heroin or other opiates).47 Control subjects had no current alcohol or illicit drug use (one or more instance of drinking to intoxication or any illicit drug use (except cannabis) in the last 30 days) or a previous history of alcohol or illicit drug use (illicit drug use or drinking to intoxication three or more times per week for 6 months or more) and no use of cannabis (three or more times per week) for more than 4 years. Fifty-one females were in the control group and 35 females in the heroin addiction group.
Microarrays
Following the protocol recommended by the manufacturer (Affymetrix, Santa Clara, CA, USA), 250 ng of genomic DNA in 5 μl water was processed. The reaction mixture was hybridized to Affymetrix 10K 2.0 GeneChips. Microarrays were scanned at 570nm using an Affymetrix GeneChip Scanner 3000. Acquisition of data was performed using the GeneChip Operating System (GCOS) version 1 (Affymetrix). Analyses were performed using the GTYPE genotype Analysis Software version 4 (Affymetrix). The Gene- Chip arrays are fully annotated and the information on the variants is available online from the NetAffx Analysis Center (http://www.affymetrix.com/analysis/index.affx).
Initially, Affymetrix 10K 2.0 GeneChips were used to genotype DNA from 104 former severe heroin addicts and 101 control subjects. Prior to analysis, it was decided that we would use the 100 GeneChips with the highest variant call rates from each group. The microarrays used for the heroin addiction group had an average call rate of 97.23% (range 92.59–99.67%), and the call rates for the control group microarrays were an average of 96.72% (range 87.05–99.44%).
Statistics
To minimize the effects of population stratification, we studied only individuals who were of Caucasian ethnicity. This was done to avoid possible type I and II errors that may occur when cases and controls are of different ethnic proportions. The STRUCTURE 2.2 program was employed to test for population stratification using the 10 ancestry informative markers on the 10K GeneChip.48,49
Analyses were performed on the variants on autosomes separately from the analysis of the variants on the X chromosomes. Prior to the analysis, we decided to exclude autosomal variants according to the following criteria: (1) the variant had a ‘missing rate’ (no genotype call) ≥25% among the 200 subjects, (2) all case and control subjects had the same genotype or (3) the P-value of Hardy–Weinberg Equilibrium test in the control group was less than 1×10−6 (equivalent to 0.01 experiment-wise using a Bonferroni correction for 10 000 tests; χ2 test). Of the 9941 autosomal variants on each Affymetrix 10K, 2.0 GeneChip, 414 were excluded leaving 9527 autosomal variants for analysis.
The variants on the X chromosome were analyzed separately in the female (n = 86) and the male (n = 114) groups. No variants located on the Y chromosome are on the 10K GeneChip. We excluded X chromosomal variants using the same criteria as for the autosomal variants. Of the 263 X chromosomal variants on each 10K GeneChip, 23 were excluded leaving 240 X chromosomal variants for analysis of allele and genotype frequency in the female group.
For the X chromosome variants in the male group, we excluded variants if (1) the variant had a ‘missing rate’ ≥25% among the 114 subjects, or (2) the variant was called as heterozygous in any of the male samples. Of the 263 X chromosomal variants, 48 were excluded leaving 215 X chromosomal variants for analysis of allele frequency in the male group.
Likelihood ratio tests were conducted to test for differences in genotype frequency and allele frequency between the cases and controls. Both analysis of genotype and allele frequency were performed for the X chromosomal variants in the female group, whereas only the analysis of allele frequency was performed for X chromosomal variants in the male group. The QVALUE program (http://faculty.washington.edu/~jstorey/qvalue/) was applied to calculate the minimum false discovery rate (q-value), which corrects for multiple testing of association with addiction in the genotype and allele frequency association analyses.50,51
Using the three variants with the smallest P-values when analyzed by genotype frequency, genotype patterns were determined. Those genotype patterns with frequency ≥5% in all observations, whereas merging all other patterns into a ‘rare’ class, were tested for differences in pattern frequencies using the Fisher’s exact test. To adjust for multiple testing, experiment-wise significance was determined in 100 000 randomizations (permutation test written for the analysis of these data by J.O.).
Variant analysis
Studies have demonstrated that linkage disequilibrium between two variants can extend past 100 000 nucleotides.52,53 If a variant was found within 100 000 nucleotides (100 kb) of an annotated gene, the gene and location of the variant relative to that gene are indicated. Sequences containing the variants were analyzed for predicted transcription factor-binding sites using Transcription Element Search System.54 Predicted regulatory potential was determined using Evolutionary and Sequence Pattern Extraction through Reduced Representation,55 and mammalian conservation was evaluated using the Vertebrate Multiz Alignment & PhastCons Conservation (28 species), both as implemented in the UCSC Genome Browser (March 2006 assembly, http://gemome.ucsc.edu/).
Results
DNA from 104 former severe heroin addicts and 101 control subjects, all of whom were Caucasian, were analyzed individually by microarray analysis using Affymetrix 10K 2.0 GeneChips. Of these subjects, the 100 former severe heroin addicted subjects and the 100 control subjects whose microarrays with the highest variant call rates were chosen for further study. To rule out population stratification in the cases and controls, population substructure was evaluated with the ancestry informative markers on the 10K GeneChip.49 When we compared our cases with controls, we found only one overlapping cluster (K=1 or K= 3). Next, we combined our Caucasian subjects with the Council on Education for Public Health Human Genome Diversity Project Cell Line Panel (CEPH-HGDP) samples used in the study by Lao et al.49 The CEPH-HGDP cohort consists of 1035 individuals derived from 51 populations from America, Central and East Asia, Europe, the Middle East, sub-Saharan Africa and Oceania. When we ran our Caucasian case and control subjects with the CEPH–HGDP samples using three ancestral populations (K = 3), a burn-in period of 100 000 iterations and 1 million Markov chain Monte Carlo replications after burn-in, we obtained the results shown in the histogram in Supplementary Figure S1. The cases and controls used in this study are similar in their mixing of ancestral populations, and appear more homogeneous than the CEPH-HGDP subjects from Europe. Hence, we found no population stratification in the distribution of the ancestral populations in our cases and controls. Next, using these ancestry informative markers, we calculated In, the Informativeness for Assignment.56 For our controls and cases, we calculated In = 0.0180, demonstrating that there is essentially no difference in allele frequencies of these ancestry informative markers between our case and control populations, as In is less than 3% of its maximal possible value.
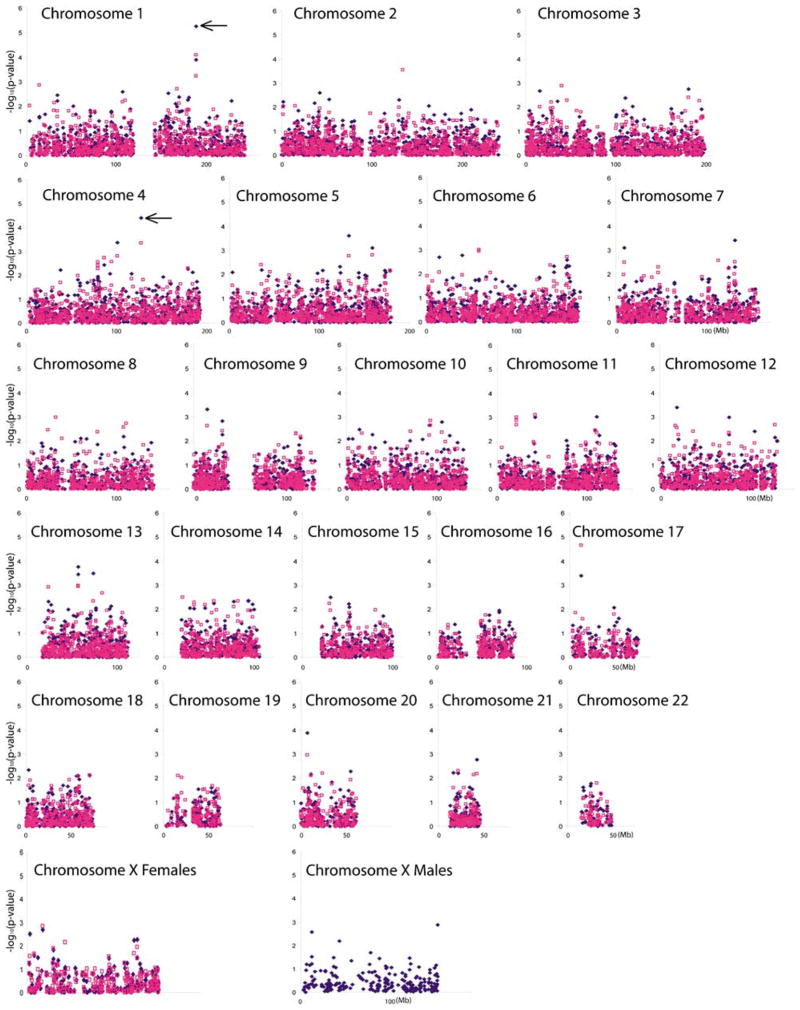
After exclusion (see Materials and methods) of variants due to low call rate, lack of heterozygosity (for example, lack of variation in the genotypes), or lack of Hardy–Weinberg equilibrium, variants were evaluated for association of genotype and allele frequencies with heroin addiction. The negative log10 P-values of the association of the genotype and the allele frequency of each variant with heroin addiction versus the chromosomal location of the variant are shown in Figure 1. The variants located on the X chromosome were evaluated separately from the autosomal variants; also, for the X chromosomal variants, the male and female groups were analyzed independently for association of allele frequency with heroin addiction. The association of the genotype frequency of the X chromosomal variants with heroin addiction was examined in the female group. The genotype frequency association of the X chromosomal variants with heroin addiction in males cannot be analyzed, as males carry only one X chromosome.
Figure 1.
Distribution of the negative log10 P-values of the association of genotype and allele frequency with heroin addiction of each variant across the genome. The negative log10 P-values for the point-wise significance of the genotype frequency (pink squares) and allele frequency (blue diamonds) association analyses with heroin addiction (ordinate) are plotted versus their location on their respective chromosome in the genome in Mb (1 million bases) (abscissa). The two most significantly associated variants, with q-values of less than 0.2 by experiment-wise association of genotype or allele frequency with heroin addiction were rs965972 and rs1986513, which are located on chromosomes 1 and 4, respectively, and are indicated with ←.
Association of allele frequency with heroin addiction
The 25 variants with the smallest point-wise P-values of association of allele frequency with heroin addiction are listed in Table 1 (autosomal variants analyzed in the total cohort and X chromosomal variants analyzed separately in the male and female groups). The 200 variants with the smallest P-values are listed in Supplementary Table S1 (autosomal variants analyzed in the total cohort and X chromosomal variants analyzed separately in the male and female groups). The variants most significantly associated experiment-wise with a q-value of less than 0.2 were rs965972 and rs1986513 that are located on chromosomes 1 and 4, respectively, in a region devoid of nearby genes (not within 100 kb of any known gene). No X chromosomal variants were found to be significantly associated with heroin addiction when examined separately in the male and female groups (Table 1).
Table 1.
Top 25 variants sorted by ascending P-value based on analysis of the association of allele frequency with heroin addiction
| Rank order | Variant | Control allele (N, frequency) | Case allele (N, frequency) | P-valuea (q-value) | Cytoband | Variant locationb | Distance (nucleotides) | Gene | Gene description | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| A | B | A | B | ||||||||
| 1 | rs965972 | 1 (0.01) | 199 (1.00) | 19 (0.10) | 181 (0.91) | 5.55 ×10−6 (0.053) | 1q31.2 | ||||
| 2 | rs1986513 | 176 (0.88) | 24 (0.12) | 196 (0.98) | 4 (0.02) | 3.89 ×10−5 (0.187) | 4q28.1 | ||||
| 3 | rs1408830 | 149 (0.76) | 47 (0.24) | 116 (0.58) | 84 (0.42) | 1.28 ×10−4 (0.323) | 1q31.2 | Intronc | HRPT2 | Hyperparathyroidism 2 | |
| Upd | 43 432 | B3GALT | UDP-Gal:betaGlcNAc β 1,3-galactosyltransferase, polypeptide 2 | ||||||||
| 4 | rs720010 | 125 (0.63) | 75 (0.38) | 87 (0.44) | 113 (0.57) | 1.34 ×10−4 (0.323) | 20p12.3 | Intron | BC038533 | IMAGE clone5165367 | |
| 5 | rs2016056 | 137 (0.75) | 45 (0.25) | 102 (0.57) | 78 (0.43) | 1.74 ×10−4 (0.334) | 13q21.2 | ||||
| 6 | rs31347 | 196 (0.98) | 4 (0.02) | 179 (0.90) | 21 (0.11) | 2.46 ×10−4 (0.342) | 5q31.1 | Intron | FSTL4 | Follistatin-like 4 | |
| 7 | rs508596 | 50 (0.30) | 118 (0.70) | 82 (0.49) | 86 (0.51) | 3.33 ×10−4 (0.342) | 13q22.2 | Intron | LMO7 | LIM domain only 7 | |
| 8 | rs950064 | 54 (0.27) | 146 (0.73) | 88 (0.44) | 112 (0.56) | 3.63 ×10−4 (0.342) | 13q21.2 | ||||
| 9 | rs718656 | 157 (0.90) | 17 (0.10) | 139 (0.76) | 43 (0.24) | 3.93 ×10−4 (0.342) | 7p22.3 | Intron | EXOC4 | Exocyst complex component 4 | |
| 10 | rs966162 | 196 (0.98) | 4 (0.02) | 178 (0.90) | 20 (0.10) | 4.07 ×10−4 (0.342) | 12p12.3 | ||||
| 11 | rs1714984 | 33 (0.17) | 167 (0.84) | 63 (0.32) | 137 (0.69) | 4.07 ×10−4 (0.342) | 17p12 | Intron | MYOCD | Myocardin | |
| 12 | rs951299 | 92 (0.46) | 108 (0.54) | 58 (0.29) | 142 (0.71) | 4.26 ×10−4 (0.342) | 4q23 | Up | 18 149 | TSPAN5 | Transmembrane 4 superfamily member 9 |
| 13 | rs3866796 | 104 (0.53) | 94 (0.47) | 139 (0.70) | 61 (0.31) | 4.97 ×10−4 (0.367) | 9p22.3 | Up | 72 503 | SNAPC3 | Small nuclear RNA-activating complex, polypeptide 3, 50 kDa |
| Up | 42 955 | C9orf52 | Chromosome 9 open reading frame 52 | ||||||||
| 14 | rs2421057 | 47 (0.24) | 153 (0.77) | 78 (0.39) | 122 (0.61) | 7.87 ×10−4 (0.515) | 5q33.3 | ||||
| 15 | rs952985 | 192 (0.97) | 6 (0.03) | 177 (0.89) | 23 (0.12) | 8.04 ×10−4 (0.564) | 7p21.3 | ||||
| 16 | rs1944932 | 64 (0.33) | 132 (0.67) | 97 (0.49) | 101 (0.51) | 9.42 ×10−4 (0.564) | 11q23.1 | Intron | BC022056 | IMAGE clone 4694422 | |
| 17 | rs1381784 | 39 (0.20) | 153 (0.80) | 69 (0.35) | 127 (0.65) | 1.00 ×10−3 (0.564) | 11p12 | ||||
| 18 | rs1877115 | 93 (0.47) | 105 (0.53) | 62 (0.31) | 138 (0.69) | 1.06 ×10−3 (0.564) | 12q21.2 | Up | 57 085 | BC034620 | IMAGE clone 4829846 |
| 19 | rs75041e | 9 (0.18) | 40 (0.82) | 1 (0.02) | 62 (0.98) | 1.30 ×10−3 (0.660) | Xq28 | Intron | GABRA3 | γ-aminobutyric acid GABAA receptor subunit α 3 | |
| 20 | rs448104 | 48 (0.25) | 144 (0.75) | 77 (0.40) | 115 (0.60) | 1.53 ×10−3 (0.717) | 9p21.1 | ||||
| 21 | rs728453 | 13 (0.07) | 185 (0.93) | 33 (0.17) | 167 (0.84) | 1.65 ×10−3 (0.7179) | 10q25.1 | Intron | SORCS3 | Sortilin-related VPS10 domain containing receptor 3 | |
| 22 | rs1399925 | 77 (0.39) | 123 (0.62) | 48 (0.24) | 152 (0.76) | 1.70 ×10−3 (0.717) | 21q22.3 | Intron | SLC37A1 | Solute carrier family 37 (glycerol-3-phosphate transporter), member 1 | |
| 23 | rs726108 | 67 (0.34) | 131 (0.66) | 39 (0.20) | 157 (0.80) | 1.72 ×10−3 (0.717) | 6p21.2 | Intron | DNAH8 | Dynein, axonal, heavy polypeptide | |
| 24 | rs956395 | 144 (0.75) | 48 (0.25) | 118 (0.60) | 78 (0.40) | 1.79 ×10−3 (0.717) | 3q26.33 | Intron | PEX5L | PXR2b protein | |
| 25 | rs2213602f | 91 (0.99) | 1 (0.01) | 56 (0.88) | 8 (0.12) | 1.99 ×10−3 (0.749) | Xp22.13 | Downg | 72 976 | CXorf20 | LOC139105 |
P-value (point-wise) is from likelihood ratio χ2 test, d.f. = 1.
Variant location is given when variant is found with 100 000 nucleotides of an annotated gene.
Variant is located in an intron of specified gene.
Distance the variant is located upstream of the transcription start site of the specified gene.
X chromosomal variants found in the male group only.
X chromosomal variant found in female group only.
Distance the variant is located downstream of the polyadenylation site of the specified gene.
Data are from the NetAffx web site; verified and corrected using the USCS Genome Browser.
The autosomal variant rs965972 (located at 1q31.2) had the smallest point-wise P-value (P = 0.00000555) for association of allele frequency with heroin addiction. The q-value for this variant was found to be 0.053. This QVALUE approach, which uses the false discovery rate method, is a more appropriate and more powerful approach than the Bonferroni correction for experiment-wise significance. This variant is in the Unigene cluster Hs.147755, a cluster of three expressed sequence tags cloned from kidney and testis, and is found in a region predicted to have high regulatory potential;55 it is a T→C transition that creates a consensus CREB transcription factor-binding site. The other variant, rs1986513, was also found to be significantly associated with heroin addiction with a point-wise P-value = 0.0000389 and an experiment-wise q-value = 0.187. This variant is located at 4q28.1 in a region of high conservation in mammals, and is an A→T transversion that creates consensus-binding sites for the TATA-binding factor, TFIID and GATA-1, -2 and -3 transcription factors.
Association of genotype frequency with heroin addiction
The variants that were most significantly associated with heroin addiction by genotype frequency were rs1714984, rs965972 and rs1867898, which are located on chromosomes 17p12, 1q31.2 and 2q21.2, respectively (Table 2). The 25 variants with the smallest P-values based on association of heroin addiction with genotype frequency are presented in Table 2 (autosomal variants analyzed in the total cohort and X chromosomal variants analyzed in the female group). Supplementary Table S2 (autosomal variants analyzed in the total cohort and X chromosomal variants analyzed in the female group) lists the 200 variants with the smallest P-values. The variant rs1714984 is located in the second intron of the myocardin gene MYOCD and has the smallest pointwise P-value of 0.0000224. However, after correction for multiple testing, this variant had a q-value of 0.206, which is not significant experiment-wise. The variants with the next smallest P-values were rs965972 (P = 0.0000795; discussed above) and rs1867898 (P = 0.00028), both of which are significant point-wise, but not experiment-wise. The variant rs1867898 is in a region predicted to have high regulatory potential.
Table 2.
Top 25 variants sorted by ascending p-value based on analysis of the association of genotype frequency with heroin addiction
| Rank order | Variant | Control genotype, N (frequency) | Case genotype, N (frequency) | P-valuea (q-value) | Cytoband | Variant location | Distance (nucleotides)b | Gene | Gene description | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||
| AA | AB | BB | AA | AB | BB | ||||||||
| 1 | rs1714984 | 7 (0.07) | 19 (0.19) | 74 (0.74) | 7 (0.07) | 49 (0.49) | 44 (0.44) | 2.24 ×10−5 (0.2064) | 17p12 | Intronc | MYOCD | Myocardin | |
| 2 | rs965972 | 0 (0.00) | 1 (0.01) | 99 (0.99) | 2 (0.02) | 15 (0.15) | 83 (0.83) | 7.95 ×10−5 (0.3663) | 1q31.2 | ||||
| 3 | rs1867898 | 5 (0.05) | 44 (0.44) | 51 (0.51) | 9 (0.09) | 18 (0.18) | 73 (0.73) | 2.84 ×10−4 (0.6834) | 2q21.2 | ||||
| 4 | rs1986513 | 79 (0.79) | 18 (0.18) | 3 (0.03) | 96 (0.96) | 4 (0.04) | 0 (0.00) | 4.42 ×10−4 (0.6834) | 4q28.1 | ||||
| 5 | rs1408830 | 56 (0.57) | 37 (0.38) | 5 (0.05) | 33 (0.33) | 50 (0.50) | 17 (0.17) | 5.94 ×10−4 (0.6834) | 1q31.2 | Intron | HRPT2 | Hyperpara-thyroidism 2 | |
| Upd | 43 432 | B3GALT | UDP-Gal: βGlcNAc β 1,3-galactosyl- transferase, polypeptide 2 | ||||||||||
| 6 | rs1381784 | 1 (0.01) | 37 (0.39) | 58 (0.60) | 11 (0.11) | 47 (0.48) | 40 (0.41) | 8.05 ×10−4 (0.6834) | 11p12 | ||||
| 7 | rs1459874 | 32 (0.32) | 63 (0.63) | 5 (0.05) | 37 (0.37) | 43 (0.43) | 20 (0.20) | 1.01 ×10−3 (0.6834) | 11p14.3 | ||||
| 8 | rs1358815 | 13 (0.13) | 55 (0.56) | 30 (0.31) | 14 (0.14) | 31 (0.31) | 54 (0.55) | 1.02 ×10−3 (0.6834) | 6p11.2 | Intron | PRIM2A | DNA primase large subunit | |
| 9 | rs954009 | 1 (0.01) | 44 (0.44) | 55 (0.55) | 10 (0.10) | 26 (0.26) | 63 (0.64) | 1.03 ×10−3 (0.6834) | 8p12 | Downe | 32 893 | FUT10 | Fucosyltransferase 10 (α (1,3) fucosyl-transferase) |
| 10 | rs2016056 | 53 (0.58) | 31 (0.34) | 7 (0.08) | 29 (0.32) | 44 (0.49) | 17 (0.19) | 1.07 ×10−3 (0.6834) | 13q21.2 | ||||
| 11 | rs720010 | 42 (0.42) | 41 (0.41) | 17 (0.17) | 20 (0.20) | 47 (0.47) | 33 (0.33) | 1.11 ×10−3 (0.6834) | 20p12.3 | Intron | BC038533 | IMAGE clone 5165367 | |
| 12 | rs1964875 | 8 (0.09) | 53 (0.58) | 30 (0.33) | 10 (0.11) | 30 (0.32) | 54 (0.57) | 1.12 ×10−3 (0.6834) | 6p11.2 | Intron | PRIM2A | Primase, polypeptide 2A, 58 kDa | |
| 13 | rs950064 | 7 (0.07) | 40 (0.40) | 53 (0.53) | 17 (0.17) | 54 (0.54) | 29 (0.29) | 1.16 ×10−3 (0.6834) | 13q21.2 | ||||
| 14 | rs720651 | 14 (0.14) | 55 (0.56) | 30 (0.30) | 36 (0.36) | 39 (0.39) | 24 (0.24) | 1.22 ×10−3 (0.6834) | 13q12.13 | Intron | ATP8A2 | ATPase, amino-phospholipid transporter-like, Class I, type 8A, member 2 | |
| 15 | rs2100690 | 34 (0.34) | 60 (0.60) | 6 (0.06) | 45 (0.45) | 37 (0.37) | 18 (0.18) | 1.28 ×10−3 (0.6834) | 3p22.1 | Intron | MYRIP | Myosin VIIA and Rab interacting protein | |
| 16 | rs727668 | 59 (0.67) | 20 (0.23) | 9 (0.10) | 67 (0.74) | 23 (0.26) | 0 (0.00) | 1.38 ×10−3 (0.6834) | 1p36.21 | ||||
| 17 | rs1903231 | 30 (0.30) | 63 (0.64) | 6 (0.06) | 36 (0.36) | 43 (0.43) | 21 (0.21) | 1.39 ×10−3 (0.6834) | 11p14.3 | ||||
| 18 | rs2213602f | 45 (0.98) | 1 (0.02) | 0 (0.00) | 24 (0.75) | 8 (0.25) | 0 (0.00) | 1.43 ×10−3 (0.6834) | Xp22.13 | ||||
| 19 | rs724585 | 1 (0.01) | 36 (0.38) | 57 (0.61) | 7 (0.07) | 18 (0.18) | 73 (0.74) | 1.45 ×10−3 (0.6834) | 10q32.32 | Down | 22 761 | PPP1R3C | Protein phosphatase 1, regulatory (inhibitor) subunit 3C |
| Down | 90 924 | HECTD2 | HECT domain containing 2 | ||||||||||
| 20 | rs951299 | 22 (0.22) | 48 (0.48) | 30 (0.30) | 7 (0.07) | 44 (0.44) | 49 (0.49) | 1.55 ×10−3 (0.6834) | 4q23 | Up | 18 149 | TSPAN5 | Transmembrane 4 superfamily member 9 |
| 21 | rs2421057 | 6 (0.06) | 35 (0.35) | 59 (0.59) | 12 (0.12) | 54 (0.54) | 34 (0.34) | 1.56 ×10−3 (0.6834) | 5q33.3 | ||||
| 22 | rs31347 | 96 (0.96) | 4 (0.04) | 0 (0.00) | 81 (0.81) | 17 (0.17) | 2 (0.02) | 1.74 ×10−3 (0.6889) | 5q31.1 | Intron | FSTL4 | Follistatin-like 4 | |
| 23 | rs1587014 | 3 (0.03) | 15 (0.15) | 82 (0.82) | 0 (0.00) | 33 (0.33) | 67 (0.67) | 1.85 ×10−3 (0.6889) | 8p23.2 | ||||
| 24 | rs1343762 | 22 (0.25) | 41 (0.47) | 25 (0.28) | 33 (0.35) | 52 (0.56) | 8 (0.09) | 1.86 ×10−3 (0.6889) | 4q21.23 | Intron | BC005018 | IMAGE clone 3638910 | |
| 25 | rs950995 | 6 (0.06) | 36 (0.36) | 58 (0.58) | 0 (0.00) | 25 (0.25) | 75 (0.75) | 1.94 ×10−3 (0.6889) | 6q25.3 | Intron | TFB1M | Transcription factor B1, mitochondrial | |
Abbreviation: SNP, single nucleotide polymorphism.
Data are from the NetAffx web site; verified and corrected using the USCS Genome Browser.
P-value (point-wise) is from likelihood ratio χ2 test, d.f. = 2.
SNP location is given when variant is found with 100 000 nucleotides of an annotated gene.
Variant is located in the intron of specified gene.
Distance the variant is located upstream of the transcription start site of the specified gene.
Distance the variant is located downstream of the polyadenylation site of the specified gene.
X chromosomal variant (one only from the female group).
Genotype patterns associated with heroin addiction
In Table 3, common genotype patterns (that is, patterns with frequencies ≥5%) for the three variants (rs1714984, rs965972 and rs1867898) with the smallest P-values of association of genotype with heroin addiction and all the other 10 patterns merged into a ‘rare’ class were tested for differences in pattern frequencies between cases and controls. The overall significance of these genotype patterns was P = 0.00000000267 from Fisher’s exact test. After correcting for multiple testing (permutation), a significant experiment-wise P-value of 0.026 was found. The genotype pattern AG-TT-GG, which was found in 32% of the heroin addicts, has an odds ratio (OR) of 6.25 for being associated with the vulnerability to develop heroin addiction. In contrast, the genotype pattern GG-CT-GG, which was found in 29% of the controls, was associated with a protective effect from developing heroin addiction with an OR of 0.13 (1/OR = 7.76).
Table 3.
Genotype patterns of the three autosomal variants with the smallest P-values from Table 2
| Genotype pattern |
Case (na) | Control (n) | Total (n) | ORb | 1/OR | ||
|---|---|---|---|---|---|---|---|
| rs1714984 | rs965972 | rs1867898 | |||||
| AG | TT | GG | 32 | 7 | 39 | 6.25 | 0.16 |
| GG | CT | GG | 5 | 29 | 34 | 0.13 | 7.76 |
| AG | CT | GG | 6 | 11 | 17 | 0.52 | 1.94 |
| GG | TT | GG | 26 | 40 | 66 | 0.53 | 1.90 |
| GG | CC | GG | 7 | 4 | 11 | 1.81 | 0.55 |
| 10 rare patternsc | 24 | 9 | 33 | 3.19 | 0.31 | ||
|
| |||||||
| Total | 100 | 100 | 200 | ||||
Abbreviation: OR, odds ratio.
Point-wise P-value = 2.67×10−9, Fisher’s exact test, experiment-wise P-value = 0.026 (permutation).
Number of cases or controls.
OR of genotype pattern versus all other patterns combined.
Genotype patterns with a frequency < 5% are combined in the rare patterns category. The 10 rare genotype patterns were AA-CT-GG, AA-TT-AA, AA-TT-AG, AA-TT-GG, AG-CC-GG, AG-CT-AG, AG-TT-AA, AG-TT-AG, GG-CT-AG and GG-TT-AG.
As shown in Table 3, when the genotype pattern of AG-TT-GG, the pattern associated with heroin addiction, was compared to all the other patterns combined, the sensitivity of the analysis was 0.32 (95% confidence interval (CI) 0.26, 0.38; that is, the probability that a former severe heroin addict would be predicted as a former severe heroin addict; the true positive rate), the specificity was 0.93 (95% CI 0.89, 0.97; that is, the probability that a non-addict would be predicted as non-addict; the true negative rate), and the random correct prediction rate was 0.50 (95% CI 0.43, 0.57; that is, the probability of making a correct prediction when there is no association between the disease and the risk factor). The associated population attributable risk for this genotype pattern was 0.27 (95% CI 0.16, 0.37), indicating that this pattern explains 27% of the population attributable risk of developing the disease of heroin addiction in this cohort. This genotype pattern in particular is associated with an increased vulnerability to develop heroin addiction.
In a similar approach, when the genotype pattern GG-CT-GG, which was associated with protection from developing heroin addiction, was compared to all other patterns combined, the sensitivity of the analysis was 0.29 (95% CI 0.23, 0.35; that is, the probability that a control subject would be predicted as a control; the true positive rate), the specificity was 0.95 (95% CI 0.92, 0.98; that is, the probability that an addict would be predicted as an addict; the true negative rate). The population attributable risk for lacking this genotype pattern was 0.83 (95% CI 0.57, 0.93), indicating that lacking this pattern explains 83% of the attributable risk of developing heroin addiction in this cohort.
Association of variants in or near candidate genes
Our laboratory compiled a list of 240 genes several years ago, which we had hypothesized may play a role in the vulnerability to develop addiction. We selected these genes because either we (and others) had shown their altered expression in response to acute and chronic exposure to specific drugs of abuse or they encoded proteins or peptides involved in the pathophysiology of addictions (for example, signal transduction molecules). From this list of 240 genes, there were 109 genes with variants on the array located within 100 kb of those genes; theses genes are listed in Supplementary Table S3. An independent list of 252 genes had been compiled by the Gershon laboratory, which contains genes that are hypothesized to be involved in affective disorders.57 One hundred and fifty-three genes from the Gershon laboratory list were not in our list (Supplementary Table S3) and these are listed in Supplementary Table S4. Of these, 74 genes had at least one variant represented on the GeneChip within 100 kb. In total, there were 393 genes examined and there were variants within 100 kb of 183 of these genes. None of the variants listed in Supplementary Tables S3 or S4 had experiment-wise significance by either association of allele or genotype frequency with heroin addiction.
The four variants with the lowest P-values from the association of allele frequency with heroin addiction and the four variants with the lowest P-values from the association of genotype frequency with heroin addiction from Supplementary Tables S3 and S4 were combined into a single list comprised of five variants that represent the union of the two lists (Table 4). The variant rs1074287 is 11 634 nucleotides upstream of the μ opioid receptor gene OPRM1 (allelic P-value = 0.0055, genotypic P-value = 0.031) and rs953741 is 75 kb downstream of the metabotropic receptor subunit mGluR6 gene GRM6 (allelic P-value = 0.0071, genotypic P-value = 0.0068). The variant rs1034576 is 15 kb downstream of the metabotropic receptor subunit mGluR8 gene GRM8 (allelic P-value = 0.0052, genotypic P-value = 0.0058), rs1405735 is located 45 kb upstream of the nuclear receptor subfamily 4, group A, member 2 gene NR4A2 (allelic P-value = 0.031, genotypic P-value = 0.024) and rs1861591 is in an intron of the cryptochrome 1 (photolyase-like) gene CRY1 (allelic P-value = 0.0040, genotypic P-value = 0.013).
Table 4.
Five genes containing the variants with the four smallest P-values by association of allele frequency and the four smallest P-values by the association of genotype frequency with heroin addiction from the hypothesis-based gene lists of the Kreek and Gershon-gene lists (Supplementary Tables S3 and S4)
| Symbol | Gene | Gene locationa | Variant | SNP location | Distance from closest annotated gene (nucleotides) | Allele | Case (n) | Control (n) | P-value by allele frequency (rankb) | P-value by genotype frequency (rankc) |
|---|---|---|---|---|---|---|---|---|---|---|
| GRM8 | mGluR8 | 7q31.33 | rs1034576 | Downd | 15 742 | A G |
50 150 |
28 172 |
0.0052 (2) | 0.0058 (1) |
| CRY1 | Cryptochrome 1 (Photolyase-like) | 12q23.3 | rs1861591 | Introne | A G |
62 118 |
88 90 |
0.0040 (1) | 0.0125 (3) | |
| GRM6 | mGluR6 | 5q35.3 | rs953741 | Down | 75 486 | A G |
163 37 |
140 60 |
0.0071 (4) | 0.0068 (2) |
| OPRM1 | μ opioid receptor | 6q25.2 | rs1074287 | Upf | 11 634 | C T |
52 114 |
35 153 |
0.0055 (3) | 0.0309 (8) |
| NR4A2 | Nuclear receptor subfamily 4, group A, member 2 | 2q24.1 | rs1405735 | Up | 44 632 | C G |
44 154 |
63 135 |
0.0312 (11) | 0.0236 (4) |
Abbreviation: OR, odds ratio.
The four genes from Supplementary Tables S3 and S4 with the lowest P-values from the association of allele frequency with heroin addiction and the four genes with the lowest P-values from the association of genotype frequency with heroin addiction were combined into a single list of six genes, which represents the union of the two lists of four genes.
Gene location from UCSC Genome Browser, March 2006 assembly.
Rank sorted by smallest P-value (point-wise) of the association of allele frequency with heroin addiction.
Rank sorted by smallest P-value (point-wise) of the association of genotype frequency with heroin addiction.
Distance the variant is located downstream of the polyadenylation site of the specified gene.
Variant is located in the intron of specified gene.
Distance the variant is located upstream of the transcription start site of the specified gene.
Discussion
Using a genome-wide association approach with a case/control design of individual subjects and of single nucleotide polymorphisms, this study has identified the two variants rs965972 and rs1986513 that are associated with heroin addiction, as well as specific autosomal genotype patterns that explained risk for and protection from developing heroin addiction in this cohort. In addition, this study has confirmed an association of the μ opioid receptor gene OPRM1 with heroin addiction.
As shown in Table 1, the variant most significantly associated by allele frequency with heroin addiction was rs965972. This variant is located at chromosome 1q31.2, in the Unigene cluster Hs.147755 in a cluster of three expressed sequence tags cloned from kidney and testis, is in a region of predicted high regulatory potential, and alters a consensus CREB-binding site. The next most significant variant was rs1986513, which was found to be in a region of high mammalian conservation. It is interesting that both of these variants are found in intergenic regions. These regions of the genome, found between genes, have been shown to contain sequences that code for noncoding RNAs, such as micro RNAs and Piwi-interacting RNAs (a recently discovered class of small RNAs expressed mainly during spermatogenesis),58 which are required for many cellular functions, including gene regulation and replication. The rs1986513 variant also creates several new consensus transcription factor-binding sites, including TFIID.
It is evident that complex diseases are caused by the interaction of multiple loci. Empirical evidence has demonstrated that interactions of loci (even when they are not on the same chromosome) can contribute substantially to complex traits. A goal of genome-wide association studies is to identify genotype patterns that contribute to a specific phenotype, as genes may interact with each other to produce disease and analysis of genotype patterns may reflect such joint gene effects better than looking at one gene at a time.59–61 Information revealed by genotype patterns is more accurate than that from haplotypes, which are generally derived from statistical programs and not measured directly.62
Using the three autosomal variants with the highest significance by association of genotype frequency with heroin addiction, we employed a strategy in which variants from different genes were analyzed together to identify common genotype patterns of unlinked alleles that are associated with heroin addiction. Using this method, we found one specific genotype pattern to be associated with heroin addiction, and a different genotype pattern with protection from heroin addiction. Further analysis revealed that the specific genotype pattern associated with heroin addiction had a population attributable risk of 27%. The other genotype pattern that was found to be significantly associated with the absence of heroin addiction had a population attributable risk of 83% when lacking this pattern. The three variants used to generate these genotype patterns were rs1714984, rs965972 and rs1867898. The variant rs1714984 resides in the second intron of the myocardin gene MYOCD. Myocardin functions as a muscle-specific transcriptional coactivator that activates specific smooth and cardiac muscle genes in conjunction with the transcription factor, serum response factor.63 The second variant rs965972 was discussed above, while the third variant rs1867898 is found in a region of high predicted regulatory potential.
We also found a point-wise significant association of a variant upstream of the μ opioid receptor gene OPRM1 with heroin addiction. First, this variant was found as the variant with the 62nd most significant association of allele frequency with heroin addiction out of the 9982 total variants (Supplementary Table S1). Second, OPRM1 had the fourth most significant P-value for the association of allele frequency of the 348 genes in the combined lists from our laboratory and that of the Gershon57 laboratory (Table 4). The confirmation of an association of the μ opioid receptor gene OPRM1 with heroin addiction demonstrates the utility of our genome-wide scan procedure with extensively characterized individual subjects in a case/control design.
Five genes (Table 4) from our list of genes that may be involved in drug addiction and other neuropsychiatric disorders (Supplementary Table S3) and from the list of Gershon57 that may be involved in affective disorder vulnerability (Supplementary Table S4) were identified as having a strong association with heroin addiction. These are the μ opioid receptor gene OPRM1; the metabotropic receptor genes GRM6 and GRM8, which encode the metabotropic receptor subunits mGluR6 and mGluR8, respectively; the NR4A2 gene, which encodes a nuclear receptor that is a member of the steroid-thyroid hormone-retinoid receptor superfamily; and CRY1, which encodes cryptochrome 1 (photolyase-like). Although the variants in or near these genes did not reach experiment-wise significance they are interesting to consider for future studies in other or larger cohorts, as they were selected on the basis of hypotheses as to their possible role in addiction and affective disorders. To our knowledge, no variants in GRM6, GMR8, NR4A2 or CRY1 have been shown to be associated with any addictive disease.
Gelernter et al.35 reported a linkage study on opioid dependence in a cohort of 393 families who were recruited at four study sites by having two siblings with cocaine dependence or at two study sites by having two siblings with opioid dependence. In two of five defined clusters, they found a LOD score of 3.46 for marker D17S787 at 17q22 for ‘non-opioid users’ (defined as no opioid abuse or dependence but where 80% had cocaine dependence and 42% had alcohol dependence (SSADDA criteria)). Nearby, at chromosome 17q25.1, a LOD score of 3.06 for marker D17S785 was found for ‘heavy-opioid users’ (defined as having opioid dependence using opioid ‘heavily’ for 3 years or more, of whom 85% also had cocaine dependence, and 47% had alcohol dependence). We found one variant rs1828096 at chromosome 17q22, which was ranked 106th by allele frequency (P = 0.00869; Supplementary Table S1) and 171st by genotype frequency (P = 0.0160; Supplementary Table S2). This variant is located 969 kb from D17S787 and 22 million nucleotides from D17S785.
In another linkage study, Tsuang and co-workers found two chromosomal regions that were associated with point-wise significance with heroin dependence in Han Chinese.39 One of these regions was at chromosome 4q31.21 (Z-score = 2.19, uncorrected P = 0.014). The marker with the highest nonparametric linkage score in this chromosomal region was D4S1644, located 16 million nucleotides from rs1986513. In our study, this variant had the second most significant association of allele frequency (point-wise P = 0.000039) with heroin addiction, but was not within 100 kb of any known gene. The second identified marker in the Tsuang study was D17S1880 (located at chromosome 17q11.2; Z-score = 2.36, uncorrected P = 0.009).
Recently, Lachman et al.41 reported a linkage study of opioid dependence in a cohort of 591 subjects comprising 296 ‘subject-described’ families. The variants we found to be significantly associated with heroin addiction are not on the same chromosome as the two linkage peaks reported by Lachman and co-workers.
The Uhl group found 38 ‘nominally reproducibly positive’ variants associated with nonspecific substance abuse based on a mathematical transformation of allele frequency in a study of European- and African-American pooled cohorts using the Affymetrix 10K GeneChip.43 None of these 38 variants were among our top 200 variants with the smallest P-values, as determined by association of allele or genotype frequency with heroin addiction (Supplementary Tables S1 and S2).
In another recent study by the Uhl group using a pooled sample approach and the Affymetrix 100K GeneChip, 51 ‘clustered positive’ chromosomal regions were identified that contained at least three variants associated with alcohol dependence based on allele frequency differences in a European- American cohort.44 Many of these regions contained genes involved in cell adhesion, cell signaling, development, gene regulation or Mendelian disorders. One region that was identified on chromosome 8 near the ‘disintegrin and metalloproteinase domain 5’ gene ADAM5 contained the variant rs724322. We also identified this variant in our list of top 200 variants with the smallest P-values as determined by association of genotype frequency with addiction (P = 0.0078; Supplementary Table S2). This variant lies 26 kb downstream from the C8orf4 gene (also known as TC1), which encodes a positive regulator of the Wnt/ beta-catenin signaling pathway.64 In addition, this variant is located 8687 nucleotides upstream of the Unigene alignment Hs.190261, a cluster of expressed sequence tags from testis and mixed tissues.
The study reported here identified two variants that are significantly associated with vulnerability to develop heroin addiction. We also identified a specific genotype pattern that explained in our cohort 27% of the population attributable risk for developing heroin addiction, whereas another protective genotype pattern was identified, and lacking this genotype pattern explained 83% of the population attributable risk for developing heroin addiction. Furthermore, a number of genes were found to be associated with heroin addiction, including those coding for the μ opioid receptor, the metabotropic receptors mGluR6 and mGluR8, the nuclear receptor NR4A2 and cryptochrome 1. If future studies confirm the role of these genes in heroin addiction, then one or several of these genes may be the potential targets for therapeutic interventions.
Supplementary Material
Acknowledgments
We thank Dorothy Melia, RN, Kathy Bell, RN, Elizabeth Ducat, NP, Lisa Borg, MD, Pauline McHugh, MD, James Schluger, MD, and Heather Hofflich, DO for recruiting, screening and assessment of study subjects and Connie Zhao, PhD for the processing of the microarrays. We thank Oscar Lao, PhD, Erasmus University Medical Centre Rotterdam, The Netherlands, for assistance with the analysis of population structure and for providing the genotypes of the CEPH-HGDP subjects. We also thank the late K Steven LaForge, PhD, for his role in the planning of these genetic studies. This work was supported in part by NIH-NIDA P60-05130 (MJK), NIH-NIDA K05-00049 (MJK), NIH-RR UL1RR024143 (BC), NIH-MH R01-44292 (JO) and NSFC grant 30730057 from the Chinese Government (JO).
References
- 1.Kreek MJ. Gene diversity in the endorphin system: SNPs, chips, and possible implications. In: Yudell M, DaSalle R, editors. The Genomic Revolution: Unveiling the Unity of Life. Joseph Henry Press; Washington, DC: 2002. pp. 97–108. [Google Scholar]
- 2.Kendler KS, Jacobson KC, Prescott CA, Neale MC. Specificity of genetic and environmental risk factors for use and abuse/ dependence of cannabis, cocaine, hallucinogens, sedatives, stimulants, and opiates in male twins. Am J Psychiatry. 2003;160:687–695. doi: 10.1176/appi.ajp.160.4.687. [DOI] [PubMed] [Google Scholar]
- 3.Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 4.Tsuang MT, Lyons MJ, Eisen SA, Goldberg J, True W, Lin N, et al. Genetic influences on DSM-III-R drug abuse and dependence: a study of 3372 twin pairs. Am J Med Genet. 1996;67:473–477. doi: 10.1002/(SICI)1096-8628(19960920)67:5<473::AID-AJMG6>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 5.LaForge KS, Yuferov V, Kreek MJ. Opioid receptor and peptide gene polymorphisms: potential implications for addictions. Eur J Pharmacol. 2000;410:249–268. doi: 10.1016/s0014-2999(00)00819-0. [DOI] [PubMed] [Google Scholar]
- 6.Mayer P, Hollt V. Allelic and somatic variations in the endogenous opioid system of humans. Pharmacol Ther. 2001;91:167–177. doi: 10.1016/s0163-7258(01)00154-1. [DOI] [PubMed] [Google Scholar]
- 7.Kreek MJ, Nielsen DA, LaForge KS. Genes associated with addiction: alcoholism, opiate, and cocaine addiction. Neuromolecular Med. 2004;5:85–108. doi: 10.1385/NMM:5:1:085. [DOI] [PubMed] [Google Scholar]
- 8.Kreek MJ, Bart G, Lilly C, LaForge KS, Nielsen DA. Pharmacogenetics and human molecular genetics of opiate and cocaine addictions and their treatments. Pharmacol Rev. 2005;57:1–26. doi: 10.1124/pr.57.1.1. [DOI] [PubMed] [Google Scholar]
- 9.Kreek MJ, Nielsen DA, Butelman ER, LaForge KS. Genetic influences on impulsivity, risk taking, stress responsivity and vulnerability to drug abuse and addiction. Nat Neurosci. 2005;8:1450–1457. doi: 10.1038/nn1583. [DOI] [PubMed] [Google Scholar]
- 10.Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, et al. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Mol Psychiatry. 2004;9:547–549. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci USA. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szeto CY, Tang NL, Lee DT, Stadlin A. Association between mu opioid receptor gene polymorphisms and Chinese heroin addicts. Neuroreport. 2001;12:1103–1106. doi: 10.1097/00001756-200105080-00011. [DOI] [PubMed] [Google Scholar]
- 13.Yuferov V, Fussell D, LaForge KS, Nielsen DA, Gordon D, Ho A, et al. Redefinition of the human kappa opioid receptor gene (OPRK1) structure and association of haplotypes with opiate addiction. Pharmacogenetics. 2004;14:793–804. doi: 10.1097/00008571-200412000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li T, Liu X, Zhao J, Hu X, Ball DM, Loh el W, et al. Allelic association analysis of the dopamine D2, D3, 5-HT2A, and GABA(A)gamma2 receptors and serotonin transporter genes with heroin abuse in Chinese subjects. Am J Med Genet. 2002;114:329–335. doi: 10.1002/ajmg.10200. [DOI] [PubMed] [Google Scholar]
- 15.Lawford BR, Young RM, Noble EP, Sargent J, Rowell J, Shadforth S, et al. The D(2) dopamine receptor A(1) allele and opioid dependence: association with heroin use and response to methadone treatment. Am J Med Genet. 2000;96:592–598. doi: 10.1002/1096-8628(20001009)96:5<592::aid-ajmg3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 16.Xu K, Lichtermann D, Lipsky RH, Franke P, Liu X, Hu Y, et al. Association of specific haplotypes of D2 dopamine receptor gene with vulnerability to heroin dependence in 2 distinct populations. Arch Gen Psychiatry. 2004;61:597–606. doi: 10.1001/archpsyc.61.6.597. [DOI] [PubMed] [Google Scholar]
- 17.Kotler M, Cohen H, Segman R, Gritsenko I, Nemanov L, Lerer B, et al. Excess dopamine D4 receptor (D4DR) exon III seven repeat allele in opioid-dependent subjects. Mol Psychiatry. 1997;2:251–254. doi: 10.1038/sj.mp.4000248. [DOI] [PubMed] [Google Scholar]
- 18.Li T, Zhu ZH, Liu X, Hu X, Zhao J, Sham PC, et al. Association analysis of polymorphisms in the DRD4 gene and heroin abuse in Chinese subjects. Am J Med Genet. 2000;96:616–621. doi: 10.1002/1096-8628(20001009)96:5<616::aid-ajmg6>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 19.Szilagyi A, Boor K, Szekely A, Gaszner P, Kalasz H, Sasvari-Szekely M, et al. Combined effect of promoter polymorphisms in the dopamine D4 receptor and the serotonin transporter genes in heroin dependence. Neuropsychopharmacol Hung. 2005;7:28–33. [PubMed] [Google Scholar]
- 20.Li T, Xu K, Deng H, Cai G, Liu J, Liu X, et al. Association analysis of the dopamine D4 gene exon III VNTR and heroin abuse in Chinese subjects. Mol Psychiatry. 1997;2:413–416. doi: 10.1038/sj.mp.4000310. [DOI] [PubMed] [Google Scholar]
- 21.Gerra G, Garofano L, Santoro G, Bosari S, Pellegrini C, Zaimovic A, et al. Association between low-activity serotonin transporter genotype and heroin dependence: behavioral and personality correlates. Am J Med Genet B Neuropsychiatr Genet. 2004;126B:37–42. doi: 10.1002/ajmg.b.20111. [DOI] [PubMed] [Google Scholar]
- 22.Tan EC, Yeo BK, Ho BK, Tay AH, Tan CH. Evidence for an association between heroin dependence and a VNTR polymorphism at the serotonin transporter locus. Mol Psychiatry. 1999;4:215–217. doi: 10.1038/sj.mp.4000541. [DOI] [PubMed] [Google Scholar]
- 23.Proudnikov D, LaForge KS, Hofflich H, Levenstein M, Gordon D, Barral S, et al. Association analysis of polymorphisms in serotonin 1B receptor gene and heroin addiction: a comparison of molecular and statistically estimated haplotypes. Pharmacogenet Genomics. 2006;16:25–36. doi: 10.1097/01.fpc.0000182782.87932.d6. [DOI] [PubMed] [Google Scholar]
- 24.Cao L, Li T, Xu K, Liu X. Association study of heroin-dependence and −287 A/G polymorphism of catechol-O-methyltransferase gene. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2002;19:499–501. [PubMed] [Google Scholar]
- 25.Horowitz R, Kotler M, Shufman E, Aharoni S, Kremer I, Cohen H, et al. Confirmation of an excess of the high enzyme activity COMT val allele in heroin addicts in a family-based haplotype relative risk study. Am J Med Genet. 2000;96:599–603. [PubMed] [Google Scholar]
- 26.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, et al. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 27.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- 28.Straub RE, Sullivan PF, Ma Y, Myakishev MV, Harris-Kerr C, Wormley B, et al. Susceptibility genes for nicotine dependence: a genome scan and followup in an independent sample suggest that regions on chromosomes 2, 4, 10, 16, 17 and 18 merit further study. Mol Psychiatry. 1999;4:129–144. doi: 10.1038/sj.mp.4000518. [DOI] [PubMed] [Google Scholar]
- 29.Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- 30.Stallings MC, Corley RP, Hewitt JK, Krauter KS, Lessem JM, Mikulich SK, et al. A genome-wide search for quantitative trait loci influencing substance dependence vulnerability in adolescence. Drug Alcohol Depend. 2003;70:295–307. doi: 10.1016/s0376-8716(03)00031-0. [DOI] [PubMed] [Google Scholar]
- 31.Ehlers CL, Wilhelmsen KC. Genomic scan for alcohol craving in Mission Indians. Psychiatr Genet. 2005;15:71–75. doi: 10.1097/00041444-200503000-00012. [DOI] [PubMed] [Google Scholar]
- 32.Bierut LJ, Rice JP, Goate A, Hinrichs AL, Saccone NL, Foroud T, et al. A genomic scan for habitual smoking in families of alcoholics: common and specific genetic factors in substance dependence. Am J Med Genet. 2004;124:19–27. doi: 10.1002/ajmg.a.20329. [DOI] [PubMed] [Google Scholar]
- 33.Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, et al. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- 34.Gelernter J, Liu X, Hesselbrock V, Page GP, Goddard A, Zhang H. Results of a genomewide linkage scan: support for chromosomes 9 and 11 loci increasing risk for cigarette smoking. Am J Med Genet B Neuropsychiatr Genet. 2004;128B:94–101. doi: 10.1002/ajmg.b.30019. [DOI] [PubMed] [Google Scholar]
- 35.Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ehlers CL, Wilhelmsen KC. Genomic screen for substance dependence and body mass index in southwest California Indians. Genes Brain Behav. 2006;6:184–191. doi: 10.1111/j.1601-183X.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- 37.Ehlers CL, Gilder DA, Wall TL, Phillips E, Feiler H, Wilhelmsen KC. Genomic screen for loci associated with alcohol dependence in Mission Indians. Am J Med Genet B Neuropsychiatr Genet. 2004;129B:110–115. doi: 10.1002/ajmg.b.30057. [DOI] [PubMed] [Google Scholar]
- 38.Guerrini I, Cook CC, Kest W, Devitgh A, McQuillin A, Curtis D, et al. Genetic linkage analysis supports the presence of two susceptibility loci for alcoholism and heavy drinking on chromosome 1p22.1-11.2 and 1q21.3-24. 2. BMC Genet. 2005;6:11. doi: 10.1186/1471-2156-6-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glatt SJ, Su JA, Zhu SC, Zhang R, Zhang B, Li J, et al. Genome-wide linkage analysis of heroin dependence in Han Chinese: results from wave one of a multi-stage study. Am J Med Genet B Neuropsychiatr Genet. 2006;141:648–652. doi: 10.1002/ajmg.b.30361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu N, Wang C, Hu Y, Yang HH, Giffen C, Tang ZZ, et al. Genome-wide association study in esophageal cancer using GeneChip mapping 10K array. Cancer Res. 2005;65:2542–2546. doi: 10.1158/0008-5472.CAN-04-3247. [DOI] [PubMed] [Google Scholar]
- 41.Lachman HM, Fann CS, Bartzis M, Evgrafov OV, Rosenthal RN, Nunes EV, et al. Genomewide suggestive linkage of opioid dependence to chromosome 14q. Hum Mol Genet. 2007;16:1327–1334. doi: 10.1093/hmg/ddm081. [DOI] [PubMed] [Google Scholar]
- 42.Uhl GR, Liu QR, Walther D, Hess J, Naiman D. Polysubstance abuse-vulnerability genes: genome scans for association, using 1004 subjects and 1494 single-nucleotide polymorphisms. Am J Hum Genet. 2001;69:1290–1300. doi: 10.1086/324467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu QR, Drgon T, Walther D, Johnson C, Poleskaya O, Hess J, et al. Pooled association genome scanning: validation and use to identify addiction vulnerability loci in two samples. Proc Natl Acad Sci USA. 2005;102:11864–11869. doi: 10.1073/pnas.0500329102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Johnson C, Drgon T, Liu QR, Walther D, Edenberg H, Rice J, et al. Pooled association genome scanning for alcohol dependence using 104,268 SNPs: validation and use to identify alcoholism vulnerability loci in unrelated individuals from the collaborative study on the genetics of alcoholism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:844–853. doi: 10.1002/ajmg.b.30346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu QR, Drgon T, Johnson C, Walther D, Hess J, Uhl GR. Addiction molecular genetics: 639,401 SNP whole genome association identifies many ‘cell adhesion’ genes. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:918–925. doi: 10.1002/ajmg.b.30436. [DOI] [PubMed] [Google Scholar]
- 46.McLellan AT, Luborsky L, Woody GE, O’Brien CP. An improved diagnostic evaluation instrument for substance abuse patients. The addiction severity index. J Nerv Ment Dis. 1980;168:26–33. doi: 10.1097/00005053-198001000-00006. [DOI] [PubMed] [Google Scholar]
- 47.Rettig RA, Yarmolinsky A, editors. Federal Regulation of Methadone Treatment. National Academy Press; Washington, DC: 1995. [PubMed] [Google Scholar]
- 48.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lao O, van Duijn K, Kersbergen P, de Knijff P, Kayser M. Proportioning whole-genome single-nucleotide-polymorphism diversity for the identification of geographic population structure and genetic ancestry. Am J Hum Genet. 2006;78:680–690. doi: 10.1086/501531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storey JD, Tibshirani R. Statistical significance for genome-wide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Storey JD, Taylor JE, Siegmund D. Strong control, conservative point estimation, and simultaneous conservative consistency of false discovery rates: A unified approach. J R Statist Soc B. 2004;66:187–205. [Google Scholar]
- 52.Collins A, Lonjou C, Morton NE. Genetic epidemiology of single-nucleotide polymorphisms. Proc Natl Acad Sci USA. 1999;96:15173–15177. doi: 10.1073/pnas.96.26.15173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]
- 54.Schug J, Overton GC. Computational Biology and Informatics Laboratory, School of Medicine, University of Pennsylvania, 1977. Technical Report CBIL-TR-1997-1001-v0.0. http://www.cbil.upenn.edu/tess.
- 55.King DC, Taylor J, Elnitski L, Chiaromonte F, Miller W, Hardison RC. Evaluation of regulatory potential and conservation scores for detecting cis-regulatory modules in aligned mammalian genome sequences. Genome Res. 2005;15:1051–1060. doi: 10.1101/gr.3642605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, et al. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- 57.Hattori E, Liu C, Zhu H, Gershon ES. Genetic tests of biologic systems in affective disorders. Mol Psychiatry. 2005;10:719–740. doi: 10.1038/sj.mp.4001695. [DOI] [PubMed] [Google Scholar]
- 58.ENCODE Project Consortium. Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sklan EH, Lowenthal A, Korner M, Ritov Y, Landers DM, Rankinen T, et al. Acetylcholinesterase/paraoxonase genotype and expression predict anxiety scores in Health, Risk Factors, Exercise Training, and Genetics study. Proc Natl Acad Sci USA. 2004;101:5512–5517. doi: 10.1073/pnas.0307659101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tosic M, Ott J, Barral S, Bovet P, Deppen P, Gheorghita F, et al. Schizophrenia and oxidative stress: glutamate cysteine ligase modifier as a susceptibility gene. Am J Hum Genet. 2006;79:586–592. doi: 10.1086/507566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Moore JH, Ritchie MD. STUDENTJAMA. The challenges of whole-genome approaches to common diseases. JAMA. 2004;291:1642–1643. doi: 10.1001/jama.291.13.1642. [DOI] [PubMed] [Google Scholar]
- 62.Hoh J, Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nat Rev Genet. 2003;4:701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- 63.Pipes GCT, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 64.Kim B, Koo H, Yang SP, Bang S, Jung Y, Kim YK, et al. TC1(C8orf4) correlates with Wnt/{beta}-catenin target genes and aggressive biological behavior in gastric cancer. Clin Cancer Res. 2006;12:3541–3548. doi: 10.1158/1078-0432.CCR-05-2440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.