Abstract
Purpose
Nine previously reported associations between single nucleotide polymorphisms (SNPs) and breast cancer outcomes from the Shanghai Breast Cancer Study (Stage 1) were further evaluated in relation to disease-free survival (DFS) and overall survival (OS) among 5,192 additional breast cancer patients (Stage 2).
Methods
Hazard ratios (HR) and 95% confidence intervals (CI) were calculated by proportional hazards regression in models adjusted for age, disease stage, estrogen and progesterone receptor status, and treatment regimens.
Results
Two SNPs had generally consistent results and significant associations with OS in combined analyses. Compared to women with MMP7 rs11225297 AA genotypes, OS was moderately better for women with AT genotypes (HR: 0.8, 95% CI: 0.7-1.0) and much better for women with TT genotypes (HR: 0.4, 95% CI: 0.2-0.8). Compared to women with MMP8 rs11225395 CC genotypes, OS was slightly better for women with CT genotypes (HR: 0.9, 95% CI: 0.7-1.1) and moderately better for women with TT genotypes (HR: 0.6, 95% CI: 0.4-0.9). Joint analysis showed significant dose-response relationships with increasing numbers of rare alleles for both OS (p<0.001) and DFS (p=0.001)
Conclusions
A functional variant in MMP8 and a SNP in high linkage disequilibrium with a functional variant in MMP7 were significantly associated with breast cancer survival in a large two-stage survival study among Chinese women. This supports the hypothesis that SNPs in MMP genes may influence breast cancer prognosis; additional research on these and other SNPs in genes important in metastasis, angiogenesis, and the regulation of the tumor microenvironment is warranted.
Keywords: breast cancer, survival, genetic variants, replication, matrix metalloproteinases
Introduction
Established prognostic factors for women with breast cancer include age at diagnosis, stage of disease as classified by tumor size, nodal involvement, and the presence of metastases (TNM), tumor hormone receptor status, and Her-2/neu growth factor status. As individual variation in prognosis is only partially explained by these and other tumor or patient characteristics, common inherited genetic variation is proposed to also influence breast cancer survival. Candidate genes and pathways that have been investigated include, but are not limited to, cell proliferation and apoptosis, inflammation and angiogenesis, steroid hormone metabolism and signaling, and drug metabolizing enzymes. Many associations between common single nucleotide polymorphisms (SNPs) and breast cancer outcomes have been previously reported by both our research group (1-8), and others (9-11). However, to date, few breast cancer survival associations have been replicated (12, 13). In order to evaluate if previously reported associations between candidate gene polymorphisms and breast cancer survival could be replicated, we evaluated nine SNPs in eight genes with published results from the Shanghai Breast Cancer Study (SBCS, Stage 1) in an independent set (Stage 2) of breast cancer patients recruited from the second phase of the SBCS and the Shanghai Breast Cancer Survival Study (SBCSS).
Methods
Study population
Subjects were participants of the Shanghai Breast Cancer Study (SBCS) and the Shanghai Breast Cancer Survival Study (SBCSS), two large population based studies of women in urban Shanghai. The SBCS is a two-phase (SBCS-I and SBCS-II) case-control study; study design and data collection procedures have been previously described (14, 15). The SBCSS is a prospective cohort study of breast cancer cases; study design and data collection procedures have been previously described (16). In this analysis, Stage 1 included SBCS-I cases, and Stage 2 included SBCS-II and SBCSS cases. All included participants provided written informed consent, and approval was granted from relevant institutional review boards in both China and the United States.
Study Stage 1 cases were identified through a rapid case ascertainment system supplemented by the Shanghai Cancer Registry. Recruitment occurred between August 1996 and March 1998, and identified a total of 1,602 eligible breast cancer cases. In person interviews with structured questionnaires were completed by 1,459 (91.1%) women. Reasons for nonparticipation included refusal (N=109, 6.8%), death before interview (N=17, 1.1%), and inability to be located (N=17, 1.1%). Clinical characteristics and patient treatment information was abstracted from medical records using a standard protocol. Cancer diagnoses were confirmed by histological examination by two senior pathologists. Patients were followed through July 2005 by active follow-up surveys, as well as death certificate linkage with the Vital Statistics Unit of the Shanghai Center for Disease Control and Prevention. A total of 1,378 (94.4%) patients were directly contacted, or if deceased, contact was made with the next of kin (N=266, 19.3%). Status of the remaining 77 patients was determined by death registry linkage; 47 of these were found be deceased. The remaining 30 patients were assumed to be alive six months prior to the date of the death certificate linkage to allow for the possible delay of record entry. Four subjects had insufficient information for record linkage, and were considered to be lost to follow-up.
Study Stage 2 included SBCS-II and SBCSS participants, which due to overlapping recruitment times and practices had 1,486 patients enrolled in both studies. Briefly, SBCS-II recruitment occurred between April 2002 and February 2005, and identified a total of 2,386 eligible breast cancer cases. Eligibility criteria between SBCS-I and SBCS-II were identical, except that age limits were expanded from 25 to 65 years in SBCS-I, to 20 to 70 years in SBCS-II. Of eligible SBCS-II cases, 1,988 (83.3%) completed questionnaires that included detailed information on demographic, reproductive, and other factors. Reasons for nonparticipation in SBCS-II included refusal (N=327, 13.7%) and the inability to be located (N=71, 3.0%). The SBCSS recruited women aged 20 to 75 years who were diagnosed with primary breast cancer between March 2002 and April 2006, identified from the population-based Shanghai Cancer Registry. Of 6,299 cases identified, 5,042 (80.0%) provided written informed consent and completed in-person interviews with structured questionnaires covering demographic, reproductive, medical and lifestyle characteristics approximately 6 months after cancer diagnoses. Reasons for nonparticipation included refusal (N=757, 12.0%), absence during enrollment (N=258, 4.1%), inability to be contacted (N=83, 1.3%), or health or communication issues (N=159, 2.5%). Cancer diagnoses were confirmed by a combination of medical record review and central review of pathological slides. Stage 2 participants were followed through June 2009 by active follow-up surveys, as well as annual death certificate linkage with the Vital Statistics Unit of the Shanghai Center for Disease Control and Prevention, last conducted in October 2008.
DNA extraction, SNP selection, and genotyping
Blood or buccal cell samples were donated and available for 1,193 (81.8%) cases from SBCS-I, and 5,381 (97.0%) cases from SBCS-II and SBCSS. Genomic DNA was extracted from buffy coats using commercial DNA purification kits according to manufacturer instructions. Polymorphisms analyzed in the current study were selected from published SBCS-I results of SNPs found to have significant or marginal associations with breast cancer survival. Nine SNPs in 8 genes were included in this analysis (Table 1). Stage 1 genotyping was conducted by a variety of methods, including PCR-RFLP (CCND1 rs9344, COMT rs4680, and TGFB1 rs1800470) (1, 4, 7), TaqMan allelic discrimination assays (MMP8 rs11225395, MMP12 rs652438, PAI1/SERPINE1 rs1799889, and VEGFA rs2010963) (2, 3, 5, 6) and Affymetrix Targeted genotyping (MMP7 rs11568818 and rs11225297) (8). Stage 2 genotyping was also conducted by a variety of methods including Taqman allelic discrimination assays (PAI1/SERPINE1 rs1799889 and TGFB1 rs1800470), Sequenom iPLEX MassARRAY assays (CCND1 rs9344, MMP7 rs11568818 and rs11225297, MMP12 rs652438, and VEGFA rs2010963), and the Affymetrix Genome-Wide Human SNP Array 6.0 (COMT rs4680, MMP7 rs11225297, and MMP8 rs11225395). Stringent quality control was employed for all methods included. Genotyping in Stage 1 had an average quality control rate of 98.3 (range: 95.5-100%); genotyping in Stage 2 had an average quality control rate of 98.7% (range: 96.5-100%).
Table 1.
Breast Cancer Patients Genotyped and Evaluated for Replication of SNP-Survival Associations, the Shanghai Breast Cancer Study and the Shanghai Breast Cancer Survival Study
| Characteristic * | Stage 1 ** | Stage 2 ** |
|---|---|---|
| Patients, N | 1,115 | 5,192 |
| Disease-Free Survival Time, years | 5.9 (2.2) | 3.8 (1.6) |
| Overall Survival Time, years | 6.5 (1.8) | 4.3 (1.3) |
| Age at Diagnosis, years | 47.8 (7.9) | 53.2 (9.9) |
| TNM Stage of Disease | ||
| 0-I | 280 (26.9) | 1,840 (38.5) |
| II | 640 (61.4) | 2,463 (51.5) |
| III-IV | 123 (11.8) | 479 (10.0) |
| Estrogen Receptor Status | ||
| Positive | 495 (63.6) | 3,248 (64.4) |
| Negative | 283 (36.4) | 1,793 (35.6) |
| Progesterone Receptor Status | ||
| Positive | 498 (64.8) | 2,955 (58.8) |
| Negative | 271 (35.2) | 2,069 (41.2) |
| Surgery | ||
| Yes | 1,109 (100) | 5,149 (99.8) |
| No | 0 (0) | 13 (0.3) |
| Chemotherapy | ||
| Yes | 1,046 (94.8) | 4,696 (91.0) |
| No | 58 (5.3) | 466 (9.0) |
| Radiotherapy | ||
| Yes | 428 (44.7) | 1,592 (30.8) |
| No | 530 (55.3) | 3,570 (69.2) |
| Tamoxifen | ||
| Yes | 717 (77.6) | 2,567 (52.1) |
| No | 207 (22.4) | 2,357 (47.9) |
Mean (standard error) or N (%) for each variable
Column percents may not sum to 100 due to rounding error
Statistical analysis
Hardy-Weinberg equilibrium (HWE) was evaluated by comparing observed and expected genotype frequencies in both separate and combined study stages. Associations between SNPs and patient or clinical characteristics were evaluated with the χ2 test. Survival time was defined as beginning at the time of cancer diagnosis, and ending at either relapse or breast cancer death for disease-free survival (DFS) or any death for overall survival (OS), or else censored at the date of last contact. Hazard ratios and their corresponding 95% confidence intervals (HR and 95%CI, respectively) were determined by Cox proportional hazards regression. Estimates of effect were estimated in both age-adjusted models, and in models that included additional adjustment for clinical predictors, including TNM, steroid hormone receptor status (ER and PR), and treatment, including chemotherapy, radiotherapy, and tamoxifen. In analyses of the two study stages combined, additional adjustment for study stage was included. Five-year survival rates were determined by Kaplan-Meier functions; differences in survival curves were assessed by the log-rank test. In the current analysis, Stage 1 results may differ from those previously published due to additionally genotyped participants, exclusion of previously included participants, additional follow-up time for participants included, and the use of major allele homozygotes as the reference group. All statistical tests were two-tailed. P-values less than 0.05 were considered statistically significant; all analyses were conducted with SAS v 9.1 (SAS Institute, Cary, NC).
Results
Nine SNPs in eight genes (Table 1) previously reported to be associated with breast cancer survival among SBCS-I participants (Stage 1) were selected for evaluation in Stage 2 among SBCS-II and SBCSS participants. Minor allele frequencies of the SNPs ranged from 8.6% (MMP7 rs11568818, Stage 1) to 48.4% (TGFB1 rs1800470, Stage 2). SNP genotype frequencies were found to be in HWE except for MMP12 rs652438 in Stage 1 (p=0.003), and TGFB1 rs1800470 in Stage 2 (p=0.003). Among corresponding Stage 1 controls, genotypes for MMP12 rs652438 did not deviate from HWE; no corresponding Stage 2 controls were genotyped for TGFB rs1800470, so HWE could not be assessed.
A total of 6,307 Chinese breast cancer patients with genotyping data and follow-up data were evaluated in the current study (Table 2); not all participants were genotyped for all variants. Stage 1 included 1,115 SBCS-I cases, and Stage 2 included 5,192 SBCS-II and SBCSS breast cancer cases. As Stage 2 participants were more recently recruited, Stage 1 participants had longer follow-up time than Stage 2 participants; mean follow-up times for Stage 1 and Stage 2 participants were 6.5 and 4.3 years, respectively. All together, these 6,307 women accumulated more than 29,665 person-years at risk; a total of 724 deaths occurred. Breast cancer cases in the two study stages originated from the same source population, and so were generally comparable. Differences between Stage 1 and Stage 2 patients likely arose from changes in age eligibility criteria, calendar year of diagnosis, and changes in treatment regimens for breast cancer. Stage 2 patients were older (p-value<0.001), more likely to have earlier disease stages (p-value<0.001), and less likely to be progesterone receptor positive (p-value=0.002), or treated with chemotherapy (p-value<0.001), radiotherapy (p-value<0.001), or tamoxifen (p-value<0.001) than Stage 1 patients. None of these differences remained significant after adjustment for calendar year of diagnosis (data not shown).
Table 2.
SNPs Evaluated for Replicated Survival Associations, the Shanghai Breast Cancer Study and the Shanghai Breast Cancer Survival Study
| Gene | SNP | Alternate Denotation (if applicable) | Gene Location 1 | Alleles 2 | Minor Allele Frequency 3
|
SBCS-I Report | |
|---|---|---|---|---|---|---|---|
| Stage 1 | Stage 2 | ||||||
| CCND1 | rs9344 | rs603965, A870G | exon 4 (S) | A/G | 43.9 | 44.9 | Shu et al, 2005 |
| COMT | rs4680 | Met158Val | exon 4 (N) | G/A | 26.4 | 26.9 | Long et al, 2007 |
| MMP7 | rs11568818 | promoter | A/G | 8.9 | 8.6 | Beeghly-Fadiel et al, 2009 | |
| MMP7 | rs11225297 | 3′ FR | A/T | 18.2 | 20.8 | Beeghly-Fadiel et al, 2009 | |
| MMP8 | rs11225395 | promoter | C/T | 38.2 | 36.7 | Decock et al, 2007 | |
| MMP12 | rs652438 | A1082G, Asn357Ser | exon 8 (M) | A/G | 10.7 | 11.2 | Shin et al, 2005 |
| PAI1/SERPINE1 | rs1799889 | 4G/5G | promoter | 4G/5G | 45.0 | 45.3 | Zhang et al, 2006 |
| TGFB1 | rs1800470 | T+29C (merged with rs1982073) | exon 1 (M) | C/T | 48.3 | 48.4 | Shu et al, 2004 |
| VEGFA | rs2010963 | G405C | 5′ UTR | G/C | 40.6 | 42.6 | Lu et al, 2005 |
Coding SNPs were either synonymous (S), nonsynonymous (N), or missense (M)
Major/minor alleles as determined by the genotype distributions among SBCS and SBCSS cases
Minor allele frequency among SBCS and SBCSS cases
Associations with DFS and OS were evaluated in Stage 1, Stage 2, and in combined analyses (Table 3). Two SNPs were found to have generally consistent results between the two study stages, and significant dose-response associations with OS in analyses of both study stages combined. Compared to women with MMP7 rs11225297 AA genotypes, women with AT genotypes had 20% better OS (HR: 0.8, 95% CI: 0.7-1.0), and women with TT genotypes had 60% better OS (HR: 0.4, 95% CI: 0.2-0.8). Similarly, compared to women with MMP8 rs11225395 CC genotypes, women with CT genotypes had 10% better OS (HR: 0.9, CI: 0.7-1.1), and women with TT genotypes had 40% better OS (HR: 0.6, 95% CI: 0.4-0.9). The remaining SNPs were not significantly associated with either DFS or OS in combined analyses of Stage 1 and Stage 2 participants using additive models. A recessive model of TGFB rs1800470 (C/T) (also known as T+29C, which merged with rs1982073) did show a significant association with OS (HR: 0.8, 95% CI: 0.6-1.0), although significance was attenuated after adjustment for known prognostic factors (p=0.077). All results were unaltered when either in situ (N=202) or stage 4 (N=39) cases were excluded from analysis, or when models included only adjustment for age (data not shown).
Table 3.
Two Stage Survival Analysis, the Shanghai Breast Cancer Study and the Shanghai Breast Cancer Survival Study
| Gene and SNP * Study Stage | Overall Survival HR (95% CI) **
|
Disease Free Survival HR (95% CI) **
|
||||
|---|---|---|---|---|---|---|
| AB | BB | p-value | AB | BB | p-value | |
| CCND1 rs9344 (A/G) | ||||||
| Study Stage 1 | 1.4 (1.0-1.9) | 1.6 (1.1-2.3) | 0.015 | 1.3 (1.0-1.7) | 1.3 (0.9-1.9) | 0.080 |
| Study Stage 2 | 1.0 (0.8-1.2) | 1.0 (0.7-1.3) | 0.820 | 0.9 (0.8-1.1) | 0.9 (0.7-1.1) | 0.200 |
| Combined | 1.1 (0.9-1.2) | 1.1 (0.9-1.4) | 0.325 | 1.0 (0.9-1.2) | 1.0 (0.8-1.2) | 0.744 |
| COMT rs4680 (G/A) | ||||||
| Study Stage 1 | 1.0 (0.8-1.4) | 1.3 (0.8-2.3) | 0.393 | 1.0 (0.8-1.3) | 1.7 (1.1-2.7) | 0.084 |
| Study Stage 2 | 1.5 (1.0-2.2) | 1.3 (0.6-2.9) | 0.123 | 1.2 (0.8-1.8) | 0.9 (0.4-2.1) | 0.560 |
| Combined | 1.1 (0.9-1.5) | 1.3 (0.8-2.0) | 0.151 | 1.1 (0.9-1.3) | 1.5 (1.0-2.1) | 0.094 |
| MMP7 rs11568818 (A/G) | ||||||
| Study Stage 1 | 1.1 (0.8-1.5) | 6.7 (2.4-18.7) | 0.137 | 1.1 (0.8-1.5) | 5.0 (1.8-13.9) | 0.146 |
| Study Stage 2 | 1.0 (0.7-1.4) | 0.5 (0.1-3.7) | 0.821 | 1.1 (0.8-1.4) | 0.4 (0.1-2.9) | 0.978 |
| Combined | 1.0 (0.8-1.3) | 2.0 (0.8-4.9) | 0.478 | 1.1 (0.9-1.3) | 1.6 (0.7-3.8) | 0.380 |
| MMP7 rs11225297 (A/T) | ||||||
| Study Stage 1 | 0.7 (0.5-0.9) | 0.3 (0.1-0.9) | 0.002 | 0.8 (0.7-1.1) | 0.4 (0.2-0.8) | 0.016 |
| Study Stage 2 | 0.9 (0.7-1.2) | 0.6 (0.2-1.3) | 0.243 | 0.9 (0.7-1.1) | 1.2 (0.8-2.0) | 0.927 |
| Combined | 0.8 (0.7-1.0) | 0.4 (0.2-0.8) | 0.003 | 0.9 (0.8-1.1) | 0.8 (0.5-1.2) | 0.095 |
| MMP8 rs11225395 (C/T) | ||||||
| Study Stage 1 | 0.8 (0.6-1.1) | 0.5 (0.3-0.8) | 0.007 | 0.8 (0.6-1.0) | 0.8 (0.5-1.2) | 0.094 |
| Study Stage 2 | 0.9 (0.6-1.3) | 0.8 (0.4-1.5) | 0.375 | 1.1 (0.7-1.6) | 1.1 (0.6-2.1) | 0.594 |
| Combined | 0.9 (0.7-1.1) | 0.6 (0.4-0.9) | 0.021 | 0.9 (0.7-1.1) | 0.9 (0.7-1.2) | 0.353 |
| MMP12 rs652438 (A/G) | ||||||
| Study Stage 1 | 1.5 (1.1-2.2) | 0.5 (0.1-2.1) | 0.184 | 1.4 (1.0-1.9) | 0.6 (0.2-1.8) | 0.311 |
| Study Stage 2 | 0.8 (0.6-1.2) | 0.4 (0.1-1.8) | 0.141 | 0.9 (0.7-1.2) | 0.9 (0.4-2.2) | 0.490 |
| Combined | 1.1 (0.8-1.3) | 0.5 (0.2-1.3) | 0.759 | 1.1 (0.9-1.3) | 0.8 (0.4-1.6) | 0.752 |
| PAI1 rs1799889 (4G/5G) | ||||||
| Study Stage 1 | 0.8 (0.6-1.0) | 0.7 (0.5-1.0) | 0.037 | 0.8 (0.6-1.0) | 0.6 (0.4-0.9) | 0.006 |
| Study Stage 2 | 1.2 (0.9-1.6) | 1.2 (0.9-1.8) | 0.219 | 1.1 (0.8-1.4) | 1.1 (0.8-1.5) | 0.434 |
| Combined | 1.0 (0.8-1.2) | 1.0 (0.8-1.3) | 0.804 | 1.0 (0.8-1.1) | 0.9 (0.7-1.1) | 0.232 |
| TGFB1 rs1800470 (C/T) | ||||||
| Study Stage 1 | 1.1 (0.8-1.6) | 1.0 (0.7-1.5) | 0.944 | 1.0 (0.7-1.2) | 0.9 (0.6-1.2) | 0.427 |
| Study Stage 2 | 1.2 (0.9-1.5) | 0.8 (0.6-1.2) | 0.326 | 1.2 (0.9-1.5) | 1.0 (0.7-1.4) | 0.841 |
| Combined | 1.2 (0.9-1.4) | 0.9 (0.7-1.2) | 0.523 | 1.1 (0.9-1.3) | 1.0 (0.8-1.2) | 0.774 |
| VEGFA rs2010963 (G/C) | ||||||
| Study Stage 1 | 0.7 (0.5-1.0) | 0.7 (0.5-1.0) | 0.033 | 0.8 (0.6-1.0) | 0.8 (0.6-1.1) | 0.102 |
| Study Stage 2 | 1.3 (1.0-1.7) | 1.0 (0.7-1.5) | 0.602 | 1.1 (0.9-1.4) | 1.0 (0.7-1.3) | 0.904 |
| Combined | 1.0 (0.8-1.2) | 0.9 (0.7-1.1) | 0.338 | 1.0 (0.8-1.1) | 0.9 (0.7-1.1) | 0.320 |
Gene name, SNP rs number, and alleles (major / minor alleles among genotyped SBCS/SBCSS participants)
Proportional hazards regression; adjusted for age at diagnosis, disease stage, ER, PR, and treatment (chemotherapy, radiotherapy, and tamoxifen); combined analysis additionally adjusted for study stage; AA major allele homozygotes (reference group); AB heterozygotes; BB minor allele homozygotes; p-value for additive trend
To evaluate the combined effect of MMP7 rs11225297 and MMP8 rs11225395 on breast cancer survival, joint analysis was conducted. Significant dose-response relationships from Cox proportional hazards regression were evident for both OS (p<0.001) and DFS (p=0.001) (Table 4). Similarly, Kaplan-Meier survival functions were significantly different for women with increasing numbers of minor alleles for both OS (p<0.008) and DFS (p<0.029) (Figure 1). For women with zero, one or two, and three or four minor alleles, five-year OS rates were 78.4%, 83.9%, and 90.0%, and five-year DFS rates were 71.6%, 77.6%, and 80.1%, respectively.
Table 4.
Joint Effect of MMP7 rs11225297 and MMP8 rs11225395 on Breast Cancer Survival, the Shanghai Breast Cancer Study and the Shanghai Breast Cancer Survival Study
| MMP7 rs11225297 & MMP8 rs11225395 | Overall Survival
|
Disease Free Survival
|
||||
|---|---|---|---|---|---|---|
| Cases / Events | HR (95% CI) * | 5 year % ** | Cases / Events | HR (95% CI) * | 5 year % ** | |
| Minor Alleles, N | ||||||
| 0 | 249 / 68 | 1.0 (reference) | 78.4 | 246 / 82 | 1.0 (reference) | 71.6 |
| 1-2 | 590 / 122 | 0.9 (0.7-1.1) | 83.9 | 573 / 150 | 0.8 (0.7-1.1) | 77.6 |
| 3-4 | 77 / 9 | 0.4 (0.2-0.7) | 90.0 | 72 / 15 | 0.5 (0.3-0.9) | 80.1 |
| p-value | <0.001 | 0.008 | 0.001 | 0.029 | ||
Cox Proportional Hazards regression, adjusted for age at diagnosis, disease stage, ER, PR, and treatment; p-value for trend
Five year survival rate determined from Kaplan-Meier analysis; p-value from Log-Rank test
Figure 1. Kaplan-Meier Functions for Joint Effect of MMP7 rs11225297 and MMP8 rs11225395 on Breast Cancer Survival.
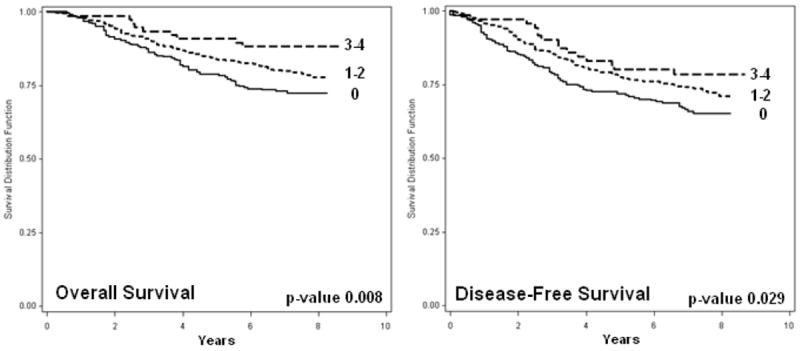
Overall and Disease-Free Kaplan-Meier Survival Functions by the number of minor alleles for two MMP Polymorphisms (MMP7 rs11225297 and MMP8 rs11225395); p-values from the Log-Rank test.
Discussion
Nine SNPs in eight genes previously reported to be associated with breast cancer survival among participants of the SBCS-I (Stage 1) were selected and evaluated for replication among SBCS-II and SBCSS participants (Stage 2). Results for five SNPs (CCND1 rs9344, MMP7 rs11568818, MMP12 rs652438, PAI1/SERPINE1 rs1799889, and VEGFA rs2010963) were neither consistent between the two study stages, nor significant in combined analyses. One SNP (TGFB1 rs1800470) had results that were generally consistent between the two study stages, and significant in an age-adjusted recessive model for OS, although significance was attenuated after adjustment for additional prognostic factors. Current results for this SNP differ from previously published results (1) as updated SBCS-I data was included and women with two copies of the major allele were used as the reference group. Another SNP (COMT rs4680) also had generally consistent results between the two study stages for OS, however the primary Stage 1 finding was for a recessive association for DFS (7), which was not replicated in Stage 2. Two SNPs had generally consistent associations with overall survival among participants in the two study stages, and significant associations in combined analyses. MMP8 rs11225395 was associated with a greater than 15% reduction in the risk of death per rare allele (HR: 0.82; 95% CI: 0.69-0.97), while MMP7 rs11225297 was associated with a greater than 20% reduction in the risk of death per rare allele (HR: 0.78; 95% CI: 0.66-0.92).
Evidence supporting these associations includes biologic plausibility for a role in cancer progression for MMPs, as well as a demonstrated functional relevance for both polymorphisms. The matrix metalloproteinases are family of zinc-dependent endopeptidases responsible for the degradation of all protein components of the extracellular matrix (ECM). In addition to a well-known role in cancer metastasis, the MMPs have also been implicated in cell proliferation, cell survival, cell migration, and angiogenesis (17-19). MMP7 is a minimal domain metalloproteinase with broad substrate specificity; it is normally expressed by both the ductal and glandular epithelium of the breast (20, 21). Tumors were found to express higher MMP7 levels than normal breast tissues, and high expression was associated with poor clinical outcome (22). We previously reported that two MMP7 variants were associated with breast cancer survival among SBCS-I participants (8); however, only one of these had a consistent association with OS in the current analysis. MMP7 rs11225297 is in high LD (D′=0.96) with rs12184413, which was associated with breast cancer risk among SBCS-I and II participants and demonstrated to influence nuclear protein binding by in vitro experiments (14). MMP8 is expressed primarily by neutrophils and cleaves interstitial collagens. A paradoxical role for MMP8 in metastasis has been described; serum MMP8 levels were significantly higher in breast cancer patients with moderate lymph node involvement than in controls and lymph node negative patients, but were also significantly higher than patients with extensive lymph node metastasis (23). MMP8 rs11225395 was first shown to be associated with lymph node metastasis among breast cancer patients in the Leuven Breast Cancer Study, and then found to be associated with breast cancer survival among SBCS-I participants (6). Allelic differences in promoter activity and nuclear protein binding were demonstrated for MMP8 rs11225395 (6). Results of the current analysis support the hypothesis that individual variation in the region downstream of the MMP7 gene and in the promoter of MMP8 may influence breast cancer prognosis.
Evidence that imposes caution in the interpretation of the current findings includes the inconsistency between associations for DFS and OS. However, it is plausible that a factor of interest may not be associated with both DFS and OS; DFS is a measure of breast cancer progression, while OS includes all causes of death. Additional evidence that imposes caution on these findings is the lack of independent significance among participants in the second study stage. However, the power of a joint analysis is a staged study design has been shown to be superior to that of a replication-based analysis (24). Further, when results from Stage 1 and Stage 2 were evaluated for significant differences in findings, only CCND1 rs9344 and DFS, and PAI1 rs1799889 and OS were statistically different (data not shown). Differences between associations in Stage 1 and Stage 2 may also be due to the difference in follow-up times between these participants, as well as differences in patient characteristics or clinical parameters. However, no SNP genotypes were significantly related to disease stage or tumor characteristics, except for TGFB rs1800470 which was associated with PR only among Stage 2 participants (data not shown). Further, statistical adjustment for study stage or calendar year of diagnosis did not materially alter our results.
In summary, nine common SNPs previously reported to be associated with breast cancer survival were evaluated in a second study population. Two polymorphisms, MMP7 rs11225297 and MMP8 rs11225395, were found to have results that were generally consistent between the two study populations and significant associations with overall survival in combined analyses. Both SNPs are either functional or in linkage disequilibrium with functional variants, and are in genes related to metastasis and the regulation of the tumor microenvironment. However, as associations among Stage 2 participants were not independently significant, a cautious interpretation of these findings is required. Additional evaluation of functionally relevant polymorphisms in MMP7 and MMP8 with breast cancer survival is warranted.
Acknowledgments
The authors wish to thank Dr. Fan Jin for her contributions to data collection and study implementation, Dr. Wanqing Wen for assistance with Kaplan-Meier analyses, and Ms. Bethanie Rammer Hull and Ms. Jacqueline Stern for manuscript editing and submission. We also gratefully acknowledge the participants and research staff of the Shanghai Breast Cancer Study and Shanghai Breast Cancer Survival Study.
Funding Statement: This research was supported by grants from the National Institute of Health, National Cancer Institute (R01 CA064277 and R01 CA124558, PI: W. Zheng and R01 CA118229, PI: XO Shu), and the Department of Defense Breast Cancer Research Program (DAMD 17-02-1-0607, PI: XO Shu). Dr. Beeghly-Fadiel is supported in part by a grant from the National Institutes of Health, National Institute of Child Health and Human Development (5K12 HD043483-09; PI: N. Brown).
Footnotes
Disclosure Statement: The authors have no conflict of interests to declare.
References
- 1.Shu XO, Gao YT, Cai Q, Pierce L, Cai H, Ruan ZX, et al. Genetic polymorphisms in the TGF-beta 1 gene and breast cancer survival: a report from the Shanghai Breast Cancer Study. Cancer Res. 2004;64:836–9. doi: 10.1158/0008-5472.can-03-3492. [DOI] [PubMed] [Google Scholar]
- 2.Lu H, Shu XO, Cui Y, Kataoka N, Wen W, Cai Q, et al. Association of Genetic Polymorphisms in the VEGF Gene with Breast Cancer Survival. Cancer Research. 2005;65:5015–9. doi: 10.1158/0008-5472.CAN-04-2786. [DOI] [PubMed] [Google Scholar]
- 3.Shin A, Cai Q, Shu XO, Gao YT, Zheng W. Genetic polymorphisms in the matrix metalloproteinase 12 gene (MMP12) and breast cancer risk and survival: the Shanghai Breast Cancer Study. Breast Cancer Research. 2005;7:R506–R512. doi: 10.1186/bcr1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shu XO, Moore DB, Cai Q, Cheng J, Wen W, Pierce L, et al. Association of Cyclin D1 Genotype with Breast Cancer Risk and Survival. Cancer Epidemiology Biomarkers & Prevention. 2005;14:91–7. [PubMed] [Google Scholar]
- 5.Zhang X, Shu XO, Cai Q, Ruan Z, Gao YT, Zheng W. Functional Plasminogen Activator Inhibitor-1 Gene Variants and Breast Cancer Survival. Clinical Cancer Research. 2006;12:6037–42. doi: 10.1158/1078-0432.CCR-05-2851. [DOI] [PubMed] [Google Scholar]
- 6.Decock J, Long JR, Laxton RC, Shu XO, Hodgkinson C, Hendrickx W, et al. Association of Matrix Metalloproteinase-8 Gene Variation with Breast Cancer Prognosis. Cancer Research. 2007;67:10214–21. doi: 10.1158/0008-5472.CAN-07-1683. [DOI] [PubMed] [Google Scholar]
- 7.Long JR, Cai Q, Shu XO, Cai H, Gao YT, Zheng W. Genetic polymorphisms in estrogen-metabolizing genes and breast cancer survival. Pharmacogenet Genomics. 2007;17:331–8. doi: 10.1097/FPC.0b013e32801a3bfe. [DOI] [PubMed] [Google Scholar]
- 8.Beeghly-Fadiel A, Shu XO, Long J, Li C, Cai Q, Cai H, et al. Genetic polymorphisms in the MMP-7 gene and breast cancer survival. Int J Cancer. 2009;124:208–14. doi: 10.1002/ijc.23859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambrosone CB, Ahn J, Singh KK, Rezaishiraz H, Furberg H, Sweeney C, et al. Polymorphisms in Genes Related to Oxidative Stress (MPO, MnSOD, CAT) and Survival After Treatment for Breast Cancer. Cancer Research. 2005;65:1105–11. [PubMed] [Google Scholar]
- 10.Udler MS, Azzato EM, Healey CS, Ahmed S, Pooley KA, Greenberg D, et al. Common germline polymorphisms in COMT, CYP19A1, ESR1, PGR, SULT1E1 and STS and survival after a diagnosis of breast cancer. Int J Cancer. 2009;125:2687–96. doi: 10.1002/ijc.24678. [DOI] [PubMed] [Google Scholar]
- 11.Knechtel G, Hofmann G, Gerger A, Renner W, Langsenlehner T, Szkandera J, et al. Analysis of common germline polymorphisms as prognostic factors in patients with lymph node-positive breast cancer. Journal of Cancer Research and Clinical Oncology. 2010;136:1813–9. doi: 10.1007/s00432-010-0839-2. [DOI] [PubMed] [Google Scholar]
- 12.Azzato E, Driver K, Lesueur F, Shah M, Greenberg D, Easton D, et al. Effects of common germline genetic variation in cell cycle control genes on breast cancer survival: results from a population-based cohort. Breast Cancer Research. 2008;10:R47. doi: 10.1186/bcr2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abraham JE, Harrington P, Driver KE, Tyrer J, Easton DF, Dunning AM, et al. Common Polymorphisms in the Prostaglandin Pathway Genes and Their Association with Breast Cancer Susceptibility and Survival. Clinical Cancer Research. 2009;15:2181–91. doi: 10.1158/1078-0432.CCR-08-0716. [DOI] [PubMed] [Google Scholar]
- 14.Beeghly-Fadiel A, Long JR, Gao YT, Li C, Qu S, Cai Q, et al. Common MMP-7 Polymorphisms and Breast Cancer Susceptibility: A Multistage Study of Association and Functionality. Cancer Research. 2008;68:6453–9. doi: 10.1158/0008-5472.CAN-08-0636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng W, Long J, Gao YT, Li C, Zheng Y, Xiang YB, et al. Genome-wide association study identifies a new breast cancer susceptibility locus at 6q25.1. Nat Genet. 2009;41:324–8. doi: 10.1038/ng.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu XO, Zheng Y, Cai H, Gu K, Chen Z, Zheng W, et al. Soy Food Intake and Breast Cancer Survival. JAMA: The Journal of the American Medical Association. 2009;302:2437–43. doi: 10.1001/jama.2009.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–74. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 18.Hojilla CV, Mohammed FF, Khokha R. Matrix metalloproteinases and their tissue inhibitors direct cell fate during cancer development. Br J Cancer. 2003;89:1817–21. doi: 10.1038/sj.bjc.6601327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martin M, Matrisian L. The other side of MMPs: Protective roles in tumor progression. Cancer and Metastasis Reviews. 2007;26:717–24. doi: 10.1007/s10555-007-9089-4. [DOI] [PubMed] [Google Scholar]
- 20.Saarialho-Kere UK, Crouch EC, Parks WC. Matrix metalloproteinase matrilysin is constitutively expressed in adult human exocrine epithelium. J Invest Dermatol. 1995;105:190–6. doi: 10.1111/1523-1747.ep12317104. [DOI] [PubMed] [Google Scholar]
- 21.Wilson CL, Matrisian LM. Matrilysin: an epithelial matrix metalloproteinase with potentially novel functions. Int J Biochem Cell Biol. 1996;28:123–36. doi: 10.1016/1357-2725(95)00121-2. [DOI] [PubMed] [Google Scholar]
- 22.Jiang WG, Davies G, Martin TA, Parr C, Watkins G, Mason MD, et al. Targeting Matrilysin and Its Impact on Tumor Growth In vivo: The Potential Implications in Breast Cancer Therapy. Clinical Cancer Research. 2005;11:6012–9. doi: 10.1158/1078-0432.CCR-05-0275. [DOI] [PubMed] [Google Scholar]
- 23.Decock J, Hendrickx W, Vanleeuw U, Van Belle V, Van Huffel S, Christiaens MR, et al. Plasma MMP1 and MMP8 expression in breast cancer: Protective role of MMP8 against lymph node metastasis. BMC Cancer. 2008;8:77. doi: 10.1186/1471-2407-8-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–13. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]


