Abstract
A cell embedded in a multicellular organism will experience a wide range of mechanical stimuli over the course of its life. Fluid flows or neighboring cells actively exert stresses on the cell, while the cell’s environment presents a set of passive mechanical properties that constrain its physical behavior. Cells respond to these varied mechanical cues through biological responses that regulate activities such as differentiation, morphogenesis, and proliferation, as well as material responses involving compression, stretching, and relaxation. Here, we break down recent studies of mechanotransduction into categories based on the input mechanical stimuli acting upon the cell and the output response of the cell. This framework provides a useful starting point for identifying overlaps in molecular players and sensing modalities, and it highlights how different timescales involved in biological and material responses to mechanical inputs could serve as a means for filtering important mechanical signals from noise.
What kind of mechanotransduction?
Mechanical stimuli can influence a myriad of cellular behaviors, including motility, proliferation, and differentiation. The conversion of mechanical signals into biologically significant outputs has been a subject of study for over 100 years [1], with efforts to understand implications for developmental biology and disease progression accelerating over the last decade. Mechanical inputs including substrate stiffness, compressive strain, and fluid shear stress have been shown to elicit responses that are all termed ‘mechanotransduction.’ But how similar are these? What pathways are shared, and what are distinct? When are the material properties of a cell relevant to its biological behavior and when do they serve as shock absorbers for cells subject to mechanical noise? As more is understood about the molecules linking mechanical stimuli with cell and tissue behavior, a single word will likely become inadequate to communicate the diversity of molecular mechanisms at work. Just as the word ‘chemotransduction’ would poorly capture the diversity of receptor-ligand mediated activity, the word ‘mechanotransduction’ does not communicate the multiple modes through which forces and mechanics can influence cells. In this review, we discuss recent advances in understanding cellular responses to force, displacement, and stiffness, and we organize the results into distinct categories so that similarities and differences can be identified.
Parsing types of inputs and outputs in mechanotransduction
‘Mechanotransduction’ encompasses a diverse set of biological behaviors, and cells exhibit an equally wide array of material behaviors. It has been shown that cells directly detect forces and displacements via cell-cell [2, 3] and cell-ECM contacts [4], as well as extracellular matrix (ECM) stiffness through specific adhesions [5, 6]. Response of cells to these mechanical inputs involves both passive material behavior (e.g. deformation) and specialized biological activity.
At times, cellular responses to different mechanical inputs appear to be similar. For example, focal adhesions mature and strengthen in response to an externally applied force [7] or increased ECM stiffness [8, 9••]. Mesenchymal stem cell fate decisions can be modulated by constrained cell shape [10] or by substrate stiffness [11]. At other times, cellular responses to mechanical inputs can appear inconsistent. Cyclic stress applied to cartilage induces very different gene expression [12] than sustained compression [13], and changing extracellular stiffness induces a third distinct set of differentiation-related behaviors [14•]. While all are important examples of mechanotransduction, these behaviors result from mechanically distinct inputs, making experimental comparisons and mechanistic interpretation difficult.
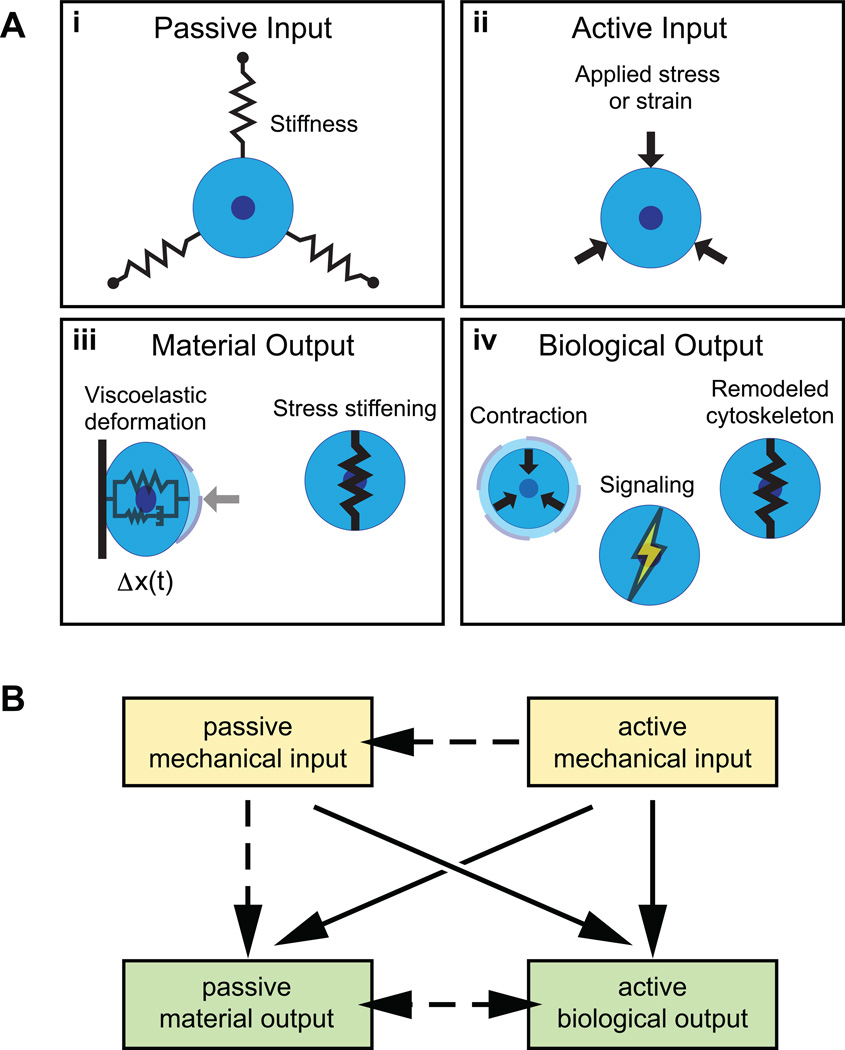
To aid interpretation, we propose a framework for classifying recent mechanotransduction experiments in terms of their inputs and outputs. We define mechanical inputs to be either ‘active’ or ‘passive’ (Figure 1). Passive inputs are physical properties of the environment that, by definition, cannot perturb a cell on their own (Figure 1a-i). Substrate stiffness and viscosity, matrix alignment, and adhesive affinity are all examples of passive inputs. In contrast, active inputs are stimuli that act directly upon the cell and cause some deformation in all or part of the cell (Figure 1a-ii). Externally applied forces and displacements, fluid shear, osmotic pressure, and acoustic waves are examples of active inputs. Importantly, a cell is not required to expend its own energy to sense an active input, although this does not preclude it from doing so.
Figure 1.
Schematic representation of active and passive mechanical inputs and active biological and passive material outputs, in the context of stiffness, force, and displacement. (a) Substrate stiffness (i) is a passive mechanical input. Applied stress or strain (ii) is an active mechanical input. Viscoelastic deformation and stress stiffening (iii) are passive material outputs. Changes in contractile behavior, cytoskeleton remodeling, and the activation of signaling pathways are active biological outputs. (b) Diagram highlighting major relationships (solid arrows) between input and output types. In the context of a cell, more complex relationships often occur (dashed arrows).
Similarly, we classify outputs as passive ‘material’ responses or active ‘biological’ responses. Material responses, such as viscoelastic creep or stress stiffening, also happen to nonliving materials in response to a mechanical input (Figure 1a-iii). Biological responses are behavioral outputs including cytoskeletal assembly and dissassembly, signal transduction, and gene expression that typically involve energy consumption and/or the conversion of mechanical signals into biochemical signals (Figure 1a-iv). When considering a cell’s response to a mechanical input, biological and material outputs can feed back on each other to produce complex behaviors. The proposed categorization helps to highlight how, when, and where these outputs are distinct and where feedback could be occurring.
Modes of mechanotransduction
Although biological and material responses occur simultaneously in response to mechanical inputs (Figure 1b), experimental and computational studies tend to focus on one type of response or the other. Here, we highlight examples from recent literature in each of the four categories we have defined.
Active mechanical input, material output
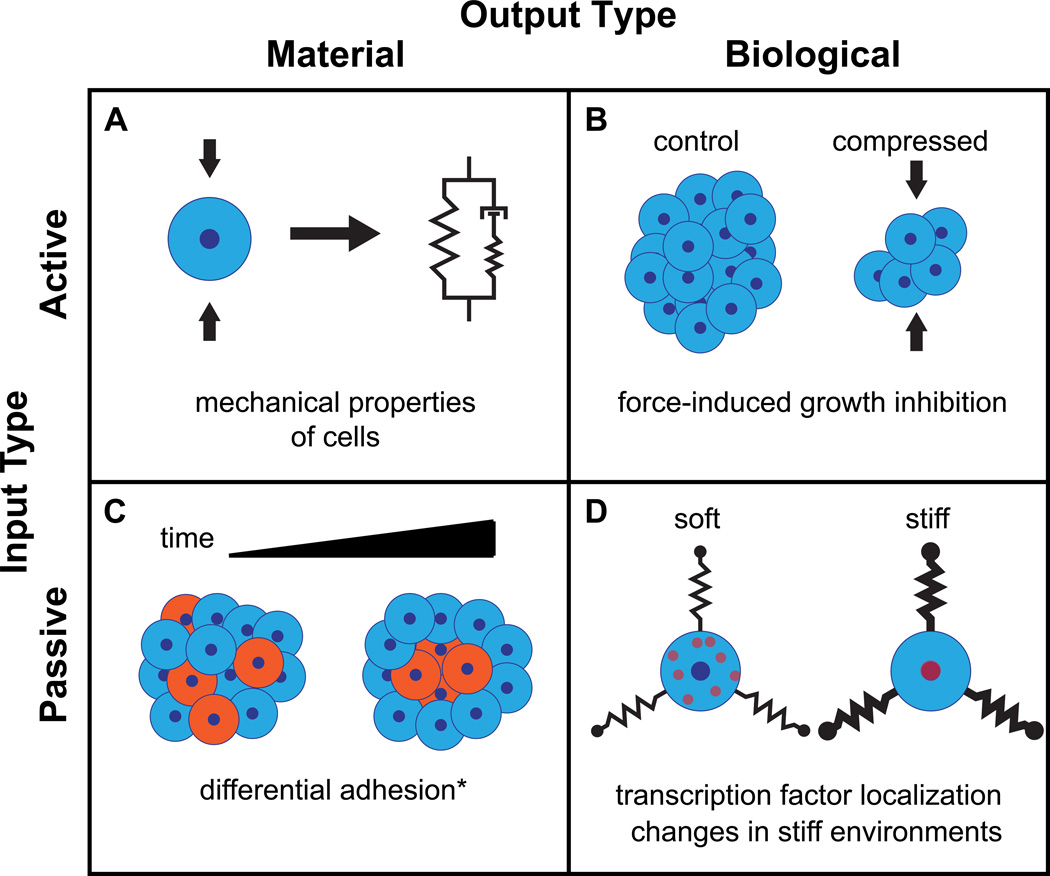
Cells are often described in terms of their mechanical properties based on constitutive equations and models from material science. Elasticity, for example, involves a passive material output (deformation) in response to an active input (force) (Figure 2a). There are many techniques to quantify the deformability of single cells and tissues in response to active inputs, including atomic force microscopy (AFM) [15, 16], shear flow [17•], micropipette aspiration [18, 19], and magnetic twisting cytometry [20]. Based on these measurements, constitutive models have been developed to represent cells as linear viscoelastic [21], power law [22] and poroelastic [23] materials. However, mechanical measurements of cells are made under the assumption that the cell does not mount an active response that changes the measured property on the timescale of the measurement.
Figure 2.
Examples of modes of mechanotransduction behavior, sorted by input and output types. (a) Mechanical property measurements in cells make use of assaying the passive material response to an active mechanical input [15, 16, 17•]. (b) Compression applied to cell aggregates inhibiting proliferation [27] is an example of an active mechanical input influencing an active biological output. (c) Differential adhesion governing cell sorting in aggregates is a passive, diffusion-driven behavior that occurs downstream of the passive material input of adhesive affinity [40]. (*There are no passive material responses that are directly due to stiffness.) (d) Changes in transcription factor localization is an active biological output that can occur in response to the passive mechanical input of substrate stiffness [53••].
Active mechanical input, biological output
Biological outputs in response to active mechanical inputs have a long history of study, particularly in musculoskeletal [1, 24, 25] and vascular [26] tissues. There is growing interest in this type of mechanotransduction in other cell types, particularly with relation to development and cancer. Sustained compressive stress limits the growth of tumor spheroids in culture by activating apoptosis [27] (Figure 2b) and also increases motility of malignant epithelial cells [28]. Active inputs have also been shown to steer growth and development in branching morphogenesis in endothelial cells [29, 30] and in mammary epithelial cells [31•].
Force and displacement cues may be sensed by active biological outputs at the subcellular level. Recently, two reports describe how forces applied to cells can drive actin-associated transcription factor re-localization [32••, 33•], providing a molecular link between the load-bearing actin cytoskeleton and gene regulation. Active inputs can also drive focal adhesion strengthening [34, 35], endocytosis of adhesion molecules [36], disassembly of pre-endocytic caveolin complexes [37••], and activation of regulators of actin assembly and myosin-driven contraction [38, 39].
Passive mechanical input, material output
A passive material output in response to a passive mechanical input includes any behavior that a nonliving material would have as a result of its own intrinsic properties. Cell sorting within an embryo or aggregate of cells based on adhesive affinity, as suggested by the differential adhesion hypothesis [40] is an example of this type of mechanotransduction (Figure 2c). Similarly, sorting based on cortical tension [41] and, at the subcellular scale, receptor clustering during T-cell activation due to size exclusion [42•] indicate that passive mechanical constraints can have profound organizational implications for biological systems.
Passive mechanical input, biological output
Extracellular matrix stiffness, sensed via actomyosin contraction (reviewed [5, 6]), is perhaps the most widely studied passive mechanical input to cells. Stiffness alters force transmission [43, 44•, 45] and modulates the magnitude of contraction force a cell generates, with increasing stiffness correlated with increasing contraction force [46, 47, 48, 49]. Active gel theory [50, 51, 52] combines the constitutive equations of passive material response with active contractile elements to quantitatively describe a material that could respond to stiffness changes.
Recently, the identification of active translocation of a transcription factor in response to substrate stiffness (Figure 2d) has provided the first evidence of a molecular behavior that links external stiffness with lineage-specifying gene expression. The transcriptional co-activator YAP/TAZ localizes to the nucleus in cells cultured on stiff substrates, and this localization behavior is upstream of stiffness-driven differentiation [53••] (Figure 2d).
In addition to transcriptional regulation, stiffness can also change the sensitivity of cells to well-known growth factors. Chondrocyte differentiation via transforming growth factor-β is significantly amplified in cells cultured on substrates with physiologically normal stiffness [14•], and epidermal growth factor signaling is amplified on stiff substrates in breast epithelial cells [54]. Stiffness-driven gene regulation may also be influenced by nuclear shape in a stiffness-dependent fashion [55].
Discussion: Sensing and sorting mechanical signals
Sensing active and passive inputs
This active-passive framework points to a distinction in how cells sense different mechanical inputs. To sense and respond to a passive mechanical input, a cell must interact actively with its surroundings. In the case of stiffness, cells pull against their surroundings via actomyosin contraction, a component of sensing behavior that is upstream of many downstream responses [5, 6]. For example, increasing ECM stiffness inhibits branching morphogenesis in both endothelia and epithelia in an actomyosin contraction dependent manner [56, 57, 58]. These cell-generated contractions in a stiffer environment result in higher forces across the cell [46, 48], often called ‘cell tension.’ The tensional homeostasis model [59, 60, 61] proposes that a cell regulates its tension through contractile machinery. This tension set point is thought to be improperly regulated when a cell is in an environment of the wrong stiffness, driving disease progression.
Active inputs could bypass the internal force generation step. Applied forces can align, stretch, and unfold sensory molecules independent of cell-generated forces. These molecules can deform in a way that translates the input directly into biochemical signals, such as the opening of an ion channel (reviewed in [62]) or the stretching of a protein that reveals cryptic binding sites [63]. Active and passive inputs can be sensed by activating similar signaling pathways, such as Src activation through integrins by force [4, 64] or stiffness [65]. Given such an instance of different inputs with seemingly the same biological outputs, some conservation in the mechanisms of sensing active and passive inputs is likely.
Significant overlap appears to exist between responses to an active force input and to force generated actively by the cell. Local active force inputs alter focal adhesion structure and stimulate increases in size and strength of adhesions [34, 35, 66, 67]. Additionally, active actomyosin-driven force generation is required for focal adhesion maturation [8, 68, 69•], much as it is for stiffness sensing. The similarity in responses at the level of focal adhesions to both active and passive mechanical inputs has drawn much attention to the assembly and disassembly of focal adhesions as key structures in stiffness sensing in adherent, and often motile, cells [9••, 70••]. To date, however, experimental evidence does not yet explain how force and stiffness generate propagating and intersecting signals.
Enacting responses on differential timescales
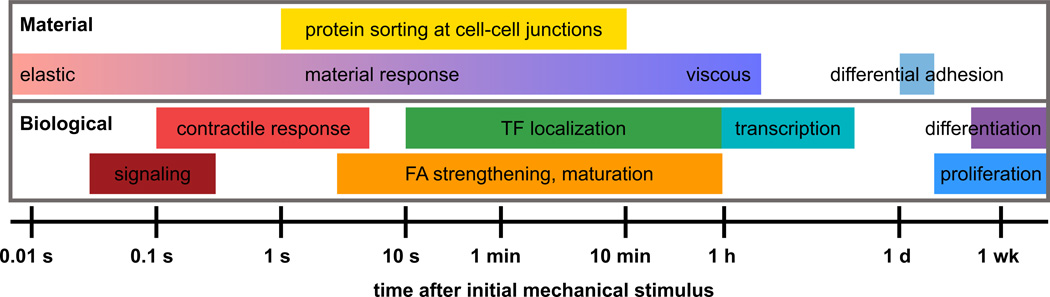
The categorization of output behaviors into active biological responses and passive material responses highlights the role of response timescale in separating cellular functions (Figure 3). Passive material responses to active mechanical inputs dominate on short timescales, during which the cell must maintain sufficient structural integrity to perform its functions within its native tissue environment. This first response to forces must occur quickly, meaning the cell does not have time to actively rearrange its cytoskeleton or express new proteins. Elastic deformation is the primary component of this behavior on microsecond timescales before viscous drag and contractile responses allow the cell to safely deform and resist forces.
Figure 3. Mechanical response behaviors span a wide range of biologically relevant timescales.
Passive material responses to mechanical stimuli can span many timescales, as seen in material response behavior, protein sorting [42•], and differential adhesion [40]. Active biological responses to mechanical stimuli occur on disparate timescales, as seen in the activation of signaling [4, 64]; changes in contraction behavior [47, 49]; focal adhesion strengthening, phosphorylation, and growth [8, 35, 66]; transcriptional regulation [32••, 33•, 53••], and differentiation and proliferation [11, 27].
Early evidence of this behavior comes from single cell experiments. Through the use of feedback control, the stiffness a cell experiences while contracting between two parallel, rigid surfaces can be changed on sub-second timescales [47, 49]. Intriguingly, a contracting cell adjusts its force and velocity almost instantaneously in response to a step change in stiffness [43], a behavior that is identical to a swelling hydrogel [44•]. In these cases, contractility itself does not appear to be regulated by stiffness, rather serving as a generator of force or displacement.
On longer timescales, cells have more freedom to change their functional states in response to new mechanical inputs. Active responses contribute to the response on longer timescales where the cell has enough time to adapt to its mechanical environment. A simple example is muscle tissue. During physical activity, muscle cells must maintain tissue integrity while still exerting forces on their surrounding environment. On short timescales, muscles have an intrinsic stiffness that resists deformation, but also can isometrically contract to stiffen if needed [71]. Activity triggers long-term gene expression [72], allowing cells to adapt by growing bigger and changing their contractile ability in anticipation of future activity.
The active biological behaviors that maintain cell and tissue structure in response to mechanical perturbations may not necessarily be involved in the interpretation of mechanical signals that direct long timescale biological behaviors. Reinforcing adhesion structures is advantageous to maintaining tissue layer integrity, but transient deformations – such as a stubbed toe – are unlikely to be of biological importance and probably do not alter development or cause cancer.
Temporal separation of active biological outputs is potentially a useful strategy by which a cell can filter mechanical inputs. At each step in the propagation of a signal generated by a mechanical input, the signal can decay via processes including diffusion, phosphatase activity, focal adhesion disassembly, and protein degradation. Additionally, differentiated cells may exhibit different active biological outputs due to suppression of specific gene expression in the differentiated state. Further experiments will be necessary to determine how some mechanical inputs result in meaningful biological responses while others are ignored.
Conclusions: Integrating Active and Passive
Studies from the rapidly expanding field of mechanotransduction have illustrated that cells can behave as both passive materials and active systems in response to mechanical cues. At the level of an organism, it is becoming clear that mechanical inputs can serve as one solution to the problem of coordinating behavior among tens, hundreds, or thousands of cells within a tissue. How a cell integrates material and biological responses to give rise to multicellular function and behavior remains an open question. Studies combining careful manipulation of mechanical inputs with in vivo and in vitro culture models will help us understand how cells speak to each other mechanically.
Material responses of cells are, perhaps, underappreciated and overlooked contributors to biological behaviors. A cell that can passively move down an energy landscape in a useful fashion in response to an applied force or displacement via passive reorganization gains a significant resource expenditure advantage over a cell that cannot. We anticipate that much of the energy transmitted to a cell via active mechanical inputs is simply dissipated, providing cells with a filter to prevent every mechanical input a cell is exposed from stimulating a long timescale, genetic response.
How a cell distinguishes useful mechanical signals from unimportant mechanical noise is an open question. Considering mechanical inputs as active or passive provides a useful framework to ask clear questions about a cell’s sensing and response processes. The timescales of observed biological responses to mechanical stimuli suggest that most long timescale response behaviors require either a constant input signal, such as environmental stiffness, or the recording and storage of a transient input signal. This latter case is particularly puzzling given that many transient mechanical inputs are ignored. Studies with clearly defined inputs and outputs will continue to be needed to describe detailed mechanisms of how a cell measures, records, and responds to mechanical cues in its environment.
Acknowledgements
We are grateful to Dr. Alba Diz Muñoz and Win Pin Ng for helpful comments on this manuscript. This work was supported by grants from NSF and NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Recommended Reading
Papers of particular interest, within the period of the review, have been highlighted as:
• Of special interest.
•• Of outstanding interest.
- 1.Wolff J. Das gesetz der transformation der knochen. Berlin: Quarto. 1892 [Google Scholar]
- 2.le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, de Rooij J. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II--dependent manner. The Journal of cell biology. 2010;189:1107–1115. doi: 10.1083/jcb.201001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leckband DE, le Duc Q, Wang N, de Rooij J. Mechanotransduction at cadherin-mediated adhesions. Curr Opin Cell Biol. 2011;23:523–530. doi: 10.1016/j.ceb.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–1045. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 5.Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell. 2010;19:194–206. doi: 10.1016/j.devcel.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoffman BD, Grashoff C, Schwartz MA. Dynamic molecular processes mediate cellular mechanotransduction. Nature. 2011;475:316–323. doi: 10.1038/nature10316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Riveline D, Zamir E, Balaban NQ, Schwarz US, Ishizaki T, Narumiya S, Kam Z, Geiger B, Bershadsky AD. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J Cell Biol. 2001;153:1175–1186. doi: 10.1083/jcb.153.6.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pasapera AM, Schneider IC, Rericha E, Schlaepfer DD, Waterman CM. Myosin II activity regulates vinculin recruitment to focal adhesions through FAK-mediated paxillin phosphorylation. J Cell Biol. 2010;188:877–890. doi: 10.1083/jcb.200906012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kawano T, Kidoaki S. Elasticity boundary conditions required for cell mechanotaxis on microelastically-patterned gels. Biomaterials. 2011;32:2725–2733. doi: 10.1016/j.biomaterials.2011.01.009. •• Using photolithography to generate substrates with well-controlled boundaries between regions of hydrogels with different stiffness, the authors show that fibroblasts sense a substrate stiffness for durotaxis with a threshold difference of 30–40 kPa over a distance of ~50 µm, about the length of a cell. This study establishes quantitative limits on the gradient of stiffness a cell can sense.
- 10.McBeath R, Pirone DM, Nelson CM, Bhadriraju K, Chen CS. Cell shape, cytoskeletal tension, and RhoA regulate stem cell lineage commitment. Dev Cell. 2004;6:483–495. doi: 10.1016/s1534-5807(04)00075-9. [DOI] [PubMed] [Google Scholar]
- 11.Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 12.Wong M, Siegrist M, Cao X. Cyclic compression of articular cartilage explants is associated with progressive consolidation and altered expression pattern of extracellular matrix proteins. Matrix Biology. 1999;18:391–399. doi: 10.1016/s0945-053x(99)00029-3. [DOI] [PubMed] [Google Scholar]
- 13.Murata M, Bonassar LJ, Wright M, Mankin HJ, Towle CA. A role for the interleukin-1 receptor in the pathway linking static mechanical compression to decreased proteoglycan synthesis in surface articular cartilage. Archives of Biochemistry and Biophysics. 2003;413:229–235. doi: 10.1016/s0003-9861(03)00129-2. [DOI] [PubMed] [Google Scholar]
- 14. Allen JL, Cooke ME, Alliston T. ECM stiffness primes the TGFβ pathway to promote chondrocyte differentiation. Molecular Biology of the Cell. 2012;23:3731–3742. doi: 10.1091/mbc.E12-03-0172. • TGF-β1 signaling is stiffness sensitive in chondrocytes. When cultured in environments matching native cartilage stiffness, cells increased Sox9, Col2α1, and Aggregcan expression.
- 15.Harris AR, Charras GT. Experimental validation of atomic force microscopy-based cell elasticity measurements. Nanotechnology. 2011;22:345102. doi: 10.1088/0957-4484/22/34/345102. [DOI] [PubMed] [Google Scholar]
- 16.Lopez JI, Kang I, You W, McDonald DM, Weaver VM. In situ force mapping of mammary gland transformation. Integr Biol (Camb) 2011;3:910–921. doi: 10.1039/c1ib00043h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gossett DR, Tse HTK, Lee SA, Ying Y, Lindgren AG, Yang OO, Rao J, Clark AT, Di Carlo D. Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc Natl Acad Sci U S A. 2012;109:7630–7635. doi: 10.1073/pnas.1200107109. • This study describes a technique that achieves rapid, high-throughput measurement of cell stiffness by imaging suspended cells as they deform under fluid flow.
- 18.Evans EA. Minimum energy analysis of membrane deformation applied to pipet aspiration and surface adhesion of red blood cells. Biophys J. 1980;30:265–284. doi: 10.1016/S0006-3495(80)85093-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houk AR, Jilkine A, Mejean CO, Boltyanskiy R, Dufresne ER, Angenent SB, Altschuler SJ, Wu LF, Weiner OD. Membrane tension maintains cell polarity by confining signals to the leading edge during neutrophil migration. Cell. 2012;148:175–188. doi: 10.1016/j.cell.2011.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang N, Butler JP, Ingber DE. Mechanotransduction across the cell surface and through the cytoskeleton. Science. 1993;260:21. doi: 10.1126/science.7684161. [DOI] [PubMed] [Google Scholar]
- 21.Leipzig ND, Athanasiou KA. Unconfined creep compression of chondrocytes. Journal of biomechanics. 2005;38:77–85. doi: 10.1016/j.jbiomech.2004.03.013. [DOI] [PubMed] [Google Scholar]
- 22.Desprat N, Richert A, Simeon J, Asnacios A. Creep function of a single living cell. Biophysical journal. 2005;88:2224–2233. doi: 10.1529/biophysj.104.050278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mitchison T, Charras G, Mahadevan L. Implications of a poroelastic cytoplasm for the dynamics of animal cell shape. 2008;19:215–223. doi: 10.1016/j.semcdb.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mellon S, Tanner K. Bone and its adaptation to mechanical loading: a review. International Materials Reviews. 2012;57:235–255. [Google Scholar]
- 25.Grodzinsky AJ, Levenston ME, Jin M, Frank EH. Cartilage tissue remodeling in response to mechanical forces. Annual review of biomedical engineering. 2000;2:691–713. doi: 10.1146/annurev.bioeng.2.1.691. [DOI] [PubMed] [Google Scholar]
- 26.Shyy JY, Chien S. Role of integrins in endothelial mechanosensing of shear stress. Circ Res. 2002;91:769–775. doi: 10.1161/01.res.0000038487.19924.18. [DOI] [PubMed] [Google Scholar]
- 27.Cheng G, Tse J, Jain RK, Munn LL. Micro-environmental mechanical stress controls tumor spheroid size and morphology by suppressing proliferation and inducing apoptosis in cancer cells. PLoS One. 2009;4:e4632. doi: 10.1371/journal.pone.0004632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, Munn LL. Mechanical compression drives cancer cells toward invasive phenotype. Proc Natl Acad Sci U S A. 2012;3:911–916. doi: 10.1073/pnas.1118910109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsumoto T, Yung YC, Fischbach C, Kong HJ, Nakaoka R, Mooney DJ. Mechanical strain regulates endothelial cell patterning in vitro. Tissue Eng. 2007;13:207–217. doi: 10.1089/ten.2006.0058. [DOI] [PubMed] [Google Scholar]
- 30.Song JW, Munn LL. Fluid forces control endothelial sprouting. Proc Natl Acad Sci U S A. 2011;108:15342–15347. doi: 10.1073/pnas.1105316108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo C, Ouyang M, Yu J, Maslov J, Price A, Shen C. Long-range mechanical force enables self-assembly of epithelial tubular patterns. Proceedings of the National Academy of Sciences. 2012;109:5576–5582. doi: 10.1073/pnas.1114781109. • Mammary epithelial cells can mechanically signal to each other along collagen fibers on large size scales (up to 600 µm) via traction forces to guide migration during tubule formation.
- 32. McGee KM, Vartiainen MK, Khaw PT, Treisman R, Bailly M. Nuclear transport of the serum response factor coactivator MRTF-A is downregulated at tensional homeostasis. EMBO Rep. 2011;12:963–970. doi: 10.1038/embor.2011.141. •• The transcriptional coactivator MRTF-A associates with G-actin in the cytoplasm and translocates to the nucleus in response to serum stimulation in fibroblasts cultured on floating collagen gels but not on attached collagen gels. In attached collagen gels, MRTF-A translocates to the nucleus within a few seconds in response to mechanical perturbation with a microneedle.
- 33. Venkatesan Iyer K, Pulford S, Mogilner A, Shivashankar GV. Mechanical activation of cells induces chromatin remodeling preceding MKL nuclear transport. Biophys J. 2012;103:1416–1428. doi: 10.1016/j.bpj.2012.08.041. • The actin associated transcription co-factor MKL undergoes cytoplasm-to-nucleus translocation with tens of seconds to a few minutes after application of shear force via magnetic beads adhered to the dorsal surface of HeLa cells. Additionally, chromatin rearranges on faster timescales in response to the same type of mechanical perturbation.
- 34.Galbraith CG, Yamada KM, Sheetz MP. The relationship between force and focal complex development. J Cell Biol. 2002;159:695–705. doi: 10.1083/jcb.200204153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matthews BD, Overby DR, Mannix R, Ingber DE. Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J Cell Sci. 2006;119:508–518. doi: 10.1242/jcs.02760. [DOI] [PubMed] [Google Scholar]
- 36.Kiyoshima D, Kawakami K, Hayakawa K, Tatsumi H, Sokabe M. Force- and Ca2+-dependent internalization of integrins in cultured endothelial cells. J Cell Sci. 2011;124:3859–3870. doi: 10.1242/jcs.088559. [DOI] [PubMed] [Google Scholar]
- 37. Sinha B, Köster D, Ruez R, Gonnord P, Bastiani M, Abankwa D, Stan RV, Butler-Browne G, Vedie B, Johannes L, Morone N, Parton RG, Raposo G, Sens P, Lamaze C, Nassoy P. Cells respond to mechanical stress by rapid disassembly of caveolae. Cell. 2011;144:402–413. doi: 10.1016/j.cell.2010.12.031. •• Caveolin complexes on the membrane of lung endothelial cells and HeLa cells rapidly disassemble in response to osmotic stress in an ATP- and actin-independent manner. These complexes serve as preassembled membrane reservoirs to buffer membrane tension against rapid mechanical shocks.
- 38.Guilluy C, Swaminathan V, Garcia-Mata R, O'Brien ET, Superfine R, Burridge K. The Rho GEFs LARG and GEF-H1 regulate the mechanical response to force on integrins. Nat Cell Biol. 2011;13:722–727. doi: 10.1038/ncb2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higashida C, Kiuchi T, Akiba Y, Mizuno H, Maruoka M, Narumiya S, Mizuno K, Watanabe N. F- and G-actin homeostasis regulates mechanosensitive actin nucleation by formins. Nat Cell Biol. 2013;15:395–405. doi: 10.1038/ncb2693. [DOI] [PubMed] [Google Scholar]
- 40.Phillips HM, Steinberg MS. Equilibrium measurements of embryonic chick cell adhesiveness, I. Shape equilibrium in centrifugal fields. Proceedings of the National Academy of Sciences. 1969;64:121–127. doi: 10.1073/pnas.64.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krieg M, Arboleda-Estudillo Y, Puech P, K๏er J, Graner F, Müller DJ, Heisenberg C. Tensile forces govern germ-layer organization in zebrafish. Nat Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 42. James JR, Vale RD. Biophysical mechanism of T-cell receptor triggering in a reconstituted system. Nature. 2012;487:64–69. doi: 10.1038/nature11220. • T-cell receptor clustering and activation were reconsistuted in a recombinant mammalian cell system. Segregation of the T-cell receptor from its cognate phosphatase CD45 is sufficient to achieve activation. The authors propose that CD45 is excluded from regions of cell-cell contact due to its size.
- 43.Mitrossilis D, Fouchard J, Pereira D, Postic F, Richert A, Saint-Jean M, Asnacios A. Realtime single-cell response to stiffness. Proc Natl Acad Sci U S A. 2010;107:16518–16523. doi: 10.1073/pnas.1007940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Crow A, Webster KD, Hohlfeld E, Ng WP, Geissler P, Fletcher DA. Contractile equilibration of single cells to step changes in extracellular stiffness. Biophys J. 2012;102:443–451. doi: 10.1016/j.bpj.2011.11.4020. • Single fibroblasts respond to a step change in extracellular stiffness with changes to their contraction velocity and force on a timescale of seconds. This behavior is similar to that of an expanding hydrogel, and can be described with linear viscoelastic elements and a constant velocity actuator.
- 45.Polackwich RJ, Koch D, Arevalo R, Miermont AM, Jee KJ, Lazar J, Urbach J, Mueller SC, McAllister RG. A novel 3D fibril force assay implicates Src in tumor cell force generation in collagen networks. PLOS ONE. 2013;8:e58138. doi: 10.1371/journal.pone.0058138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saez A, Buguin A, Silberzan P, Ladoux B. Is the mechanical activity of epithelial cells controlled by deformations or forces? Biophys J. 2005;89:L52–L54. doi: 10.1529/biophysj.105.071217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mitrossilis D, Fouchard J, Guiroy A, Desprat N, Rodriguez N, Fabry B, Asnacios A. Single-cell response to stiffness exhibits muscle-like behavior. Proc Natl Acad Sci U S A. 2009;106:18243–18248. doi: 10.1073/pnas.0903994106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lam WA, Chaudhuri O, Crow A, Webster KD, Li T, Kita A, Huang J, Fletcher DA. Mechanics and contraction dynamics of single platelets and implications for clot stiffening. Nat Mater. 2011;10:61–66. doi: 10.1038/nmat2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webster KD, Crow A, Fletcher DA. An AFM-based stiffness clamp for dynamic control of rigidity. PLoS One. 2011;6:e17807. doi: 10.1371/journal.pone.0017807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kruse K, Joanny JF, Jülicher F, Prost J, Sekimoto K. Generic theory of active polar gels: a paradigm for cytoskeletal dynamics. Eur Phys J E Soft Matter. 2005;16:5–16. doi: 10.1140/epje/e2005-00002-5. [DOI] [PubMed] [Google Scholar]
- 51.Juelicher F, Kruse K, Prost J, Joanny J. Active behavior of the cytoskeleton. Physics Reports. 2007;449:3–28. [Google Scholar]
- 52.Marcq P, Yoshinaga N, Prost J. Rigidity sensing explained by active matter theory. Biophys J. 2011;101:L33–L35. doi: 10.1016/j.bpj.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Dupont S, Morsut L, Aragona M, Enzo E, Giulitti S, Cordenonsi M, Zanconato F, Le Digabel J, Forcato M, Bicciato S, Elvassore N, Piccolo S. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. •• The transcriptional co-activator YAP/TAZ is required for stiffness-dependent lineage specification in mesenchymal stem cells. Additionally, YAP/TAZ localizes to the nucleus or the cytoplasm in cells cultured on stiff or soft substrates, respectively. This study establishes a molecular link between stiffness-sensing and differentiation in mesenchymal stem cells.
- 54.Kim J, Asthagiri AR. Matrix stiffening sensitizes epithelial cells to EGF and enables the loss of contact inhibition of proliferation. J Cell Sci. 2011;124:1280–1287. doi: 10.1242/jcs.078394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lovett DB, Shekhar N, Nickerson JA, Roux KJ, Lele TP. Modulation of nuclear shape by substrate rigidity. Cellular and Molecular Bioengineering. 2013;1 doi: 10.1007/s12195-013-0270-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miyajima H, Matsumoto T, Sakai T, Yamaguchi S, An SH, Abe M, Wakisaka S, Lee KY, Egusa H, Imazato S. Hydrogel-based biomimetic environment for in vitro modulation of branching morphogenesis. Biomaterials. 2011;32:6754–6763. doi: 10.1016/j.biomaterials.2011.05.072. [DOI] [PubMed] [Google Scholar]
- 57.Fischer RS, Gardel M, Ma X, Adelstein RS, Waterman CM. Local cortical tension by myosin II guides 3D endothelial cell branching. Curr Biol. 2009;19:260–265. doi: 10.1016/j.cub.2008.12.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Myers KA, Applegate KT, Danuser G, Fischer RS, Waterman CM. Distinct ECM mechanosensing pathways regulate microtubule dynamics to control endothelial cell branching morphogenesis. J Cell Biol. 2011;192:321–334. doi: 10.1083/jcb.201006009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Banes AJ, Tsuzaki M, Yamamoto J, Brigman B, Fischer T, Brown T, Miller L. Mechanoreception at the cellular level: the detection, interpretation, and diversity of responses to mechanical signals. Biochemistry and Cell Biology. 1995;73:349–365. doi: 10.1139/o95-043. [DOI] [PubMed] [Google Scholar]
- 60.Brown RA, Prajapati R, McGrouther DA, Yannas IV, Eastwood M. Tensional homeostasis in dermal fibroblasts: mechanical responses to mechanical loading in threedimensional substrates. J Cell Physiol. 1998;175:323–332. doi: 10.1002/(SICI)1097-4652(199806)175:3<323::AID-JCP10>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 61.Paszek MJ, Zahir N, Johnson KR, Lakins JN, Rozenberg GI, Gefen A, Reinhart-King CA, Margulies SS, Dembo M, Boettiger D, Hammer DA, Weaver VM. Tensional homeostasis and the malignant phenotype. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 62.Haswell ES, Phillips R, Rees DC. Mechanosensitive channels: what can they do and how do they do it? Structure. 2011;19:1356–1369. doi: 10.1016/j.str.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP. Stretching single talin rod molecules activates vinculin binding. Science. 2009;323:638–641. doi: 10.1126/science.1162912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Na S, Collin O, Chowdhury F, Tay B, Ouyang M, Wang Y, Wang N. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci U S A. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yeh Y, Hur SS, Chang J, Wang K, Chiu J, Li Y, Chien S. Matrix stiffness regulates endothelial cell proliferation through Septin 9. PloS one. 2012;7:e46889. doi: 10.1371/journal.pone.0046889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lele TP, Pendse J, Kumar S, Salanga M, Karavitis J, Ingber DE. Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J Cell Physiol. 2006;207:187–194. doi: 10.1002/jcp.20550. [DOI] [PubMed] [Google Scholar]
- 67.Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EHK. Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J Cell Sci. 2009;122:1665–1679. doi: 10.1242/jcs.042986. [DOI] [PubMed] [Google Scholar]
- 68.Aratyn-Schaus Y, Gardel ML. Transient frictional slip between integrin and the ECM in focal adhesions under myosin II tension. Curr Biol. 2010;20:1145–1153. doi: 10.1016/j.cub.2010.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Roca-Cusachs P, Del Rio A, Puklin-Faucher E, Gauthier NC, Biais N, Sheetz MP. Integrindependent force transmission to the extracellular matrix by α-actinin triggers adhesion maturation. Proc Natl Acad Sci U S A. 2013;110:E1361–E1370. doi: 10.1073/pnas.1220723110. • The actin crosslinking protein α-actinin has a previously undescribed role in the transmittance of actomyosin-generated force to the ECM via mature focal adhesions through a direct interaction with β3 integrins.
- 70. Plotnikov SV, Pasapera AM, Sabass B, Waterman CM. Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell. 2012;151:1513–1527. doi: 10.1016/j.cell.2012.11.034. •• Individual FAs within fibroblasts exhibit either fluctuating traction forces or stable traction forces. These force fluctuations within individual FAs are required for durotaxis but not for FA maturation, chemotaxis, or haptotaxis, demonstrating that rigidity is sensed via constant, active tugging against the ECM.
- 71.Lambertz D, Souza TOL, Canon F, Xavier LCC, Ferraz KM. Influence of overweight on the active and the passive fraction of the plantar flexors series elastic component in prepubertal children. Journal of Applied Physiology. 2013;114:73–80. doi: 10.1152/japplphysiol.00241.2012. [DOI] [PubMed] [Google Scholar]
- 72.Jensen JH, Conley LN, Hedegaard J, Nielsen M, Young JF, Oksbjerg N, Hornshøj H, Bendixen C, Thomsen B. Gene expression profiling of porcine skeletal muscle in the early recovery phase following acute physical activity. Experimental Physiology. 2012;97:833–848. doi: 10.1113/expphysiol.2011.063727. [DOI] [PubMed] [Google Scholar]