Abstract
Objective
Both protective T cell genotypes and NK cell genotypes have been associated with delayed progression to AIDS and shown to be co-inherited in HIV-1 infected subjects who limit viral replication in absence of antiretroviral therapy (“controllers”). However, a comparative analysis of the genotype and function of the innate and adaptive immune compartments in HIV-1 infected controller subjects has been understudied to date.
Design
Here, we simultaneously tested NK and T cell function in controllers to investigate the mechanism(s) that might account for host-immune control over viral replication.
Methods
We measured CD8 T cell responses against HIV-1 utilizing overlapping 15-mer peptides spanning the HIV-1 Consensus Clade B Gag protein and tested NK cell degranulation and cytokine secretion against tumor target cells following IFN-alpha stimulation.
Results
Among a cohort of 37 controllers, the presence of protective MHC-Class I HLA alleles (such as HLA-B*57) was not correlated with HIV-specific CD8 responses. In contrast, the inheritance of a protective KIR3DL1*h/*y receptor genotype along with the corresponding HLA-Bw4*80I ligand was associated with significantly heightened target cell-induced NK degranulation and cytokine secretion following IFN-alpha stimulation (p=0.0201, n=13). Interestingly, we observed a significant inverse association between the IFN-alpha stimulated NK response to K562 cells and the HIV-specific CD8 T cell response to Gag among elite controllers. (rho=−0.8321, p=0.0010, n=12).
Conclusions
Together, these results suggest that heightened NK responses can be evidenced independently of HIV-specific T cell responses in HIV-1 infected elite controllers.
Keywords: HIV-1, AIDS, T Cells, NK Cells, HLA, KIR, Elite Controllers
Introduction
HIV-1 infected controllers comprise a small fraction (less than 1%) of HIV-1 infected subjects that maintain continuous control over viral replication without the need of anti-retroviral therapy [1, 2]. Controllers have generally been defined as either having undetectable HIV-1 RNA levels using conventional assays (e.g. below 50 copies RNA/mL; “elite controllers”) or having low but detectable levels of viral replication (e.g., 50 to 2000 copies RNA/mL; “viremic controllers”) [3]. While controllers cannot eradicate HIV-1 from the body, they represent the highest echelon of immune control exhibited by infected patients over HIV-1 replication. Consequently, the nature of the protective immune response in controllers is of considerable interest.
Host factors identified in HIV-1 infected controller subjects include cellular proteins that limit viral entry or replication within CD4 target cells [4, 5]. Non-host factors associated with HIV-1 infected controller status include the reduced replicative capacity of viruses from some controller subjects [6–9]. Immunologically, recent data indicates that CD8+ T cell responses directed against the HIV-1 Gag protein are heightened in some controllers [10–12], as are effector molecules important for CTL activity such as T-bet and Perforin [13, 14]. Likewise, protective MHC Class I (MHC-1) alleles important for CD8+ T cell recognition of target cells (such as HLA-B*57) are positively correlated with delayed progression to AIDS [15–18] and enriched in HIV-1 controller cohorts [10, 12]. Together, these studies highlight a role for the adaptive T cell response in maintaining low viral loads among controller subjects. However, immunological heterogeneity has been observed among individuals in HIV-1 controllers cohorts, with greater than half of HIV-1 controllers lacking protective T cell HLA-B alleles or exhibiting low to undetectable HIV-specific CD8 T cell responses [10, 12]. This latter data suggests that alternative mechanisms of immune control, such as innate immune (NK) effector responses, may also contribute to sustained viral control along with traditional adaptive immune responses in some controller subjects.
More recent data has shown a consistent association between certain NK cell Killer Inhibitory Receptor (KIR) alleles and virus control, suggesting that NK cells may also have a functional role in controlling HIV replication [19, 20]. Inheritance of a KIR3DL1 inhibitory receptor genotype that exhibits high cell surface expression (KIR3DL1*h) is associated with delayed progression to AIDS when co-inherited with it’s corresponding HLA-Bw4*80I ligand [21]. Similarly, KIR3DS1, an activating allele of the same locus, has also been associated with delayed progression to AIDS in conjunction with HLA-B alelles [22]. Inheritance of protective KIR3DL1*h/*y receptor genes along with their corresponding HLA-Bw4*80I ligands is believed to lead to high cell surface expression and strong NK licensing [23], while NK cells expressing KIR3DS1 have been shown to produce more IFN-γ [24] and mediate stronger inhibition of HIV-1 replication [25].
In addition to serving a protective HLA allele, HLA-B*57 serves as one of the strongest ligands for the KIR3DL1 family of receptors [19, 26]. The combined genotype of HLA-B*57 with KIR3DL1*h/*y confers more protection from HIV-1 disease progression than either genotype alone suggesting that a synergistic response between the adaptive and innate immune compartment may work in concert to limit viral replication [20, 21]. However, a comparative analysis of the genotype and function of the innate and adaptive immune compartments in HIV-1 infected controller subjects has been understudied to date. We hypothesized that increased NK activity may account for the sustained control over viral replication in a subset of controllers with protective KIR3DL1 alleles but lacking strong HIV-1 specific CD8+ T cell responses. In support of this hypothesis, we observed a significant inverse association between the IFN-alpha stimulated NK response to K562 cells and the HIV-specific CD8 T cell response to Gag among elite controllers. Specifically, the inheritance of protective KIR3DL1*h/*y receptor genotype associated with strong NK responses was significantly correlated with a decrease in HIV-1 Gag-specific CD8 responses among elite controllers. Together, these results indicate that heightened NK responses following stimulation can be evidenced independently of HIV-specific T cell responses in HIV-1 infected controllers that limit viral replication naturally.
Materials and Methods
Subject Criteria and Clinical Assessment
HIV-infected adults were sampled from the University of California, San Francisco-based SCOPE cohort and whole blood was shipped overnight to The Wistar Institute by air courier for analysis. Samples were properly insulated and quality control was maintained to ensure the viability and functional output of innate and adaptive immune cells from all subjects as previously described [27]. Controllers were defined as HIV-seropositive individuals who were not receiving antiretroviral therapy and who had either undetectable HIV-1 RNA levels using conventional assays (<50 copies/mL by Abbott RealTime PCR or <75 copies/mL by bDNA; “elite controllers”) or low but detectable levels of viral replication (e.g., 50 to 2000 copies RNA/mL; “viremic controllers”) for a minimum of 5 years (Table 1). Isolated episodes of viremia above these thresholds were allowed. All participants provided informed consent, and this research was approved by the institutional review board of the University of California San Francisco, the National Institutes of Health and The Wistar Institute.
Table 1.
Cohort Characteristics
| Elite Controllers (n=14) | Viremic Controllers (n=23) | |
|---|---|---|
| VL copies/ml | Median 40 copies/ml (IQR 0) | Median 556 copies/ml (IQR 770) |
| CD4 Count | Median 701 cells/μl (IQR 718) | Median 572 cells/μl (IQR 405) |
| Years Since HIV-1 Diagnosis | Median 21 years (IQR 9 years) | Median 21 years (IQR 16) |
Allele Genotyping
Participating elite and viremic controller subjects recruited from the University of California, San Francisco-based SCOPE cohort were selected based upon previous genotyping for protective HLA and KIR3DL1*h/*y genotypes conducted at the Immunogenetics Laboratory in The National Cancer Institute. Briefly, 2×106 PBMC from HIV-1 Infected controllers were frozen in DNAzol (Molecular Research Center, Cincinnati, OH) and genotyped as previously described [28]. The HLA class I loci were typed by the sequence based typing method as recommended by the 13th International Histo-compatibility Workshop (http://www.ihwg.org/tmanual/TMcontents.htm). We defined the following HLA-B alleles (HLA-B*13, HLA-B*2705, HLA-B*57, HLA-B*5801, HLA-B*8100) as protective as shown in Supplementary Table 1 based upon previous studies highlighting their association with lower viral loads and increased frequency in HIV-1 controller cohorts [10, 12].
The protective NK KIR3DL1 receptor genotype was defined as described previously [21, 29] and referred to as KIR3DL1*h/*y (Supplementary Table 1). Corresponding ligands for the KIR3DL1 receptor were identified among HLA-B or HLA-A alleles with Isoleucine at amino acid position 80 as previously described [22, 30]. Only controllers possessing a KIR3DL1*h/*y genotype in conjunction with their corresponding HLA-Bw4*80I ligands were defined as possessing protective KIR3DL1 genotype and were compared to controllers lacking either the KIR3DL1*h/*y genotype or the requisite HLA-Bw4*80I ligands. Due to their low frequency and potentially confounding effects, subjects inheriting the KIR3DS1 activating receptor were excluded from the analysis.
Flow Cytometry
All cell surface antibodies and isotype controls were pre-conjugated and used at the recommended dilution of 0.25 μg antibody per million cells in PBSA (PBS with 0.09% sodium azide). PBMCs were stained with antibodies to phenotypic and functional markers for 15 minutes at RT°C in the dark and washed twice. Cells were then fixed and permeabilized with the Cytofix/Cytoperm kit (BD Biosciences, San Jose, CA) as described by the manufacturer. Intra-cellular staining was carried out in 1X Perm/Wash Buffer (BD) for 15 minutes at RT°C in the dark with 0.25 μg of anti-IFN-gamma FITC antibody (BD) per million cells. A minimum of one hundred thousand events were collected on a BD LSR-II Flow Cytometer and samples were subsequently analyzed with FlowJo software (Tree Star Incorporated, Ashland OR). Prior to analysis, all samples were gated by forward (height and area) and side scatter to exclude doublets and dead cells.
NK Target Cell Degranulation and Intracellular Cytokine Staining Assay
0.5×106 PBMC were co-cultured alone (no target control) or with K562 cells at a 10:1 effector/target ratio in the presence of 10 μl anti-CD107a monoclonal antibody, 0.133 μl Golgi-stop (BD) and 5 μg/mL Brefeldin A (BD) for 4 hours in a 200 μl volume. PBMC were washed, stained with antibodies to NK Cell phenotypic markers (CD56/CD3) and intra-cellular staining for IFN-gamma was carried out as described above. NK cells were gated by CD56+/CD3− expression and the percentage of NK cells staining positive for CD107a and/or IFN-gamma following incubation with K562 cells was determined after subtraction of background levels of staining in the absence of target cells.
T Cell Peptide Stimulation and Intracellular Cytokine Staining Assay
0.5×106 PBMC were co-cultured with a 1 μg/mL mixture of overlapping 15-mer peptides spanning the HIV-1 Consensus Clade B Gag protein (AIDS Research and Reference Reagent Repository, NIH) in the presence of 2.5 μl CD28/CD49d co-stimulation (BD) and 5 μg/mL Brefeldin A for 18 hours in a 200 μl volume. Alternatively, PBMC were stimulated with a 1 μg/mL mixture of a CEF peptide pool comprising 23 peptides consisting of sequences derived from the human Cytomegalovirus, Epstein-Barr and Influenza Viruses (AIDS Research and Reference Reagent Repository, NIH). Unstimulated and 5 μg/mL SEB (Staphylococcal Enterotoxin B, Sigma Aldrich) stimulated PBMC were used as negative and positive controls, respectively. PBMC were washed, stained with antibodies to T Cell phenotypic markers (CD8/CD3) and intra-cellular staining for IFN-gamma was carried out as described above. CD8 T cells were gated by CD8+/CD3+ staining and the percentage of cells staining positive for CD107a and/or IFN-gamma was determined after subtraction of background levels of staining in unstimulated control cells.
Statistical Analysis
All graphic presentations and statistical analyses were performed with Prism software (GraphPad Software, La Jolla, CA). Statistical analysis of two groups was carried out using a Mann-Whitney unpaired, non-parametric T test and graphic comparisons are displayed as median with interquartile range. Correlations between two variables were carried out using a non-parametric Spearman Correlation of untransformed data with a 95% confidence interval. In all cases, p values were two-sided with alpha <0.05.
Results
Protective HLA and KIR Genotypes in Elite and Viremic Controllers
To investigate the role of protective HLA and KIR genotypes in influencing innate and adaptive immune function, we utilized a well-characterized cohort of HIV-1 infected controllers. This cohort was comprised of 14 elite controllers and 23 viremic controllers that exhibited a mean duration of infection of over 20 years without the need for anti-retroviral therapy (Table 1). As documented previously for HIV-1 controller cohorts [10, 12], we observed a pronounced enrichment of protective HLA-B alleles associated with delayed progression to AIDS among both viremic and elite controllers in our study (data not shown). Similarly, we confirmed that both viremic and elite controllers exhibited a high frequency of protective KIR3DL1*h/*y receptor genotypes associated with delayed progression to AIDS [21] (data not shown). To investigate the impact of protective HLA and KIR3DL1*h/*y genotypes on predicting innate and adaptive immune function among elite and viremic controllers, we recruited subjects based upon the presence or absence of protective T cell HLA-B alleles and protective NK KIR3DL1*h/*y genotypes as described in the Materials and Methods and shown in Supplementary Table 1.
Influence of KIR3DL1*h/*y receptor genotype on NK activity in Controllers
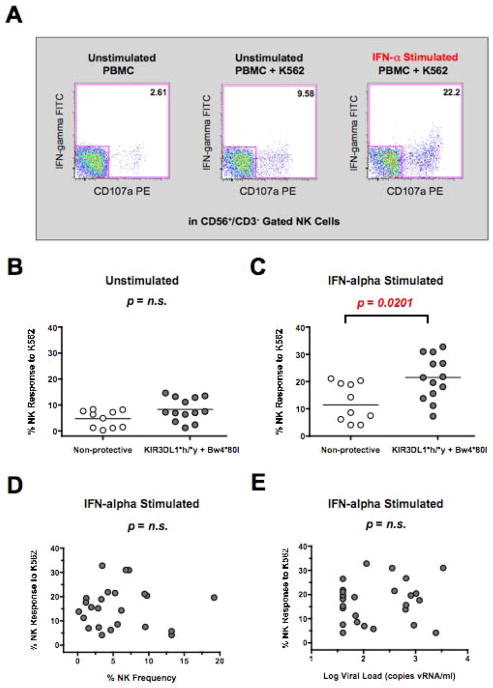
First, we measured if target cell-induced NK functional activity was correlated with the inheritance of protective KIR3DL1*h/*y receptor genotypes and their corresponding HLA-Bw4*80I ligands. As we documented previously [31], we observed that NK cell CD107a degranulation and IFN-gamma cytokine production in response to K562 tumor cells was dramatically increased in the presence of IFN-alpha stimulation (Figure 1A). Following IFN-alpha stimulation of PBMC, NK function against K562 target cells was significantly augmented (p=0.0201) in controllers possessing a protective KIR3DL1*h/*y receptor genotype along with their corresponding HLA-Bw4*80I ligand (Figure 1C). The functional capacity of IFN-alpha stimulated NK cells from subjects inheriting the KIR3DL1*h/*y genotype in conjunction with their corresponding HLA-Bw4*80I ligands was significantly increased compared to controllers lacking either the protective KIR3DL1*h/*y genotype or the requisite HLA-Bw4*80I ligand (Supplementary Figure 1). In unstimulated PBMC, the enhanced functional output of NK cells from controllers possessing a protective KIR3DL1*h/*y receptor genotype along with their corresponding HLA-Bw4*80I ligand was also observed but the effect was not statistically significant (Figure 1B). These findings suggest that target cell-induced NK functional activity following IFN-alpha stimulation was correlated with the inheritance of protective KIR3DL1*h/*y receptor genotypes and their corresponding HLA-Bw4*80I ligands.
Figure 1. Influence of Protective KIR3DL1*h/*y Genotype on NK Cell Function.
(A) CD107a and IFN-gamma staining on CD56+/CD3- NK cells gated from unstimulated or IFN-alpha stimulated PBMC incubated in the presence or absence of K562 target cells. NK cells were gated by CD56+/CD3− expression and the percentage of NK cells staining positive for CD107a and/or IFN-gamma following incubation with K562 cells was determined after subtraction of background levels of staining in the absence of target cells (Panel 1). (B–C) Composite graph of constitutive (B) or IFN-alpha stimulated (C) K562-specific NK response from controllers possessing or lacking a protective KIR3DL1*h/*y genotype along with their corresponding HLA-Bw4*80I ligands. (D–E) Spearman correlation of IFN-alpha induced K562-specific NK function as described in panel B (y-axis) and NK frequency (D) or the log viral load (E) (x-axis). Comparisons between two groups were performed using an unpaired, non-parametric Mann-Whitney T-test and correlations between two variables were carried out using a non-parametric Spearman test. In all cases, p-value were two-tailed with a 95% confidence interval (n.s., not significant).
NK cell frequency was similar between controllers possessing or lacking the KIR3DL1*h/*y receptor genotype along with their corresponding HLA-Bw4*80I ligand (data not shown), and NK frequency within the PBMC was not correlated with NK functional output (Figure 1D). While high viral loads have been shown to inversely impact NK function [32–34], we did not observe that NK function was negatively impacted by low levels of viral replication (Figure 1E). Similarly, the NK response to K562 cells among elite controllers and viremic controllers possessing the protective KIR3DL1*h/*y receptor genotypes and their corresponding HLA-Bw4*80I ligands was comparable (data not shown). This data indicates that the heightened NK response among both elite and viremic controllers inheriting protective NK receptor genotypes is maintained in HIV-1 infected subjects exhibiting either total or partial control over viral replication in contrast to the loss of function expected in non-controllers with higher levels of viral replication.
Influence of Protective HLA-B alleles on HIV-1 Gag-specific T cell Responses among Controllers
To characterize the role of protective HLA-B alleles in predicting T cell function in controllers, we measured the capacity of CD8+/CD3+ gated T cells from fresh PBMC to produce interferon-gamma or degranulate in response to HIV-1 peptide pools. We focused on HIV-1 Gag-specific T cells responses due to their enhanced functional activity [35] and pronounced effect on reducing viral load among HIV-1 infected subjects [36]. As shown in Figure 2B, there was a wide heterogeneity in Gag-specific CD8+ T cell responses among controllers with some subjects exhibiting low or undetectable Gag-specific responses and others exhibiting very high levels. However, Gag-specific CD8+ T cell responses were not significantly different between controllers possessing or lacking protective HLA-B alleles (Figure 2B). SEB-specific and CEF-specific (CMV, EBV and Flu) CD8+ T cell responses also did not differ between controllers possessing or lacking protective HLA alleles (data not shown). There was an increased trend in Gag-specific CD8+ T cell responses in viremic controllers versus elite controllers (data not shown), potentially due to the effect of a low-level of viral replication on stimulating Gag-specific CD8+ recall responses. However, no significant correlation between viral load and the frequency of Gag-specific responses was observed (Figure 2C).
Figure 2. Influence of Protective HLA Genotype on HIV-1 Specific CD8 T Cell Function.
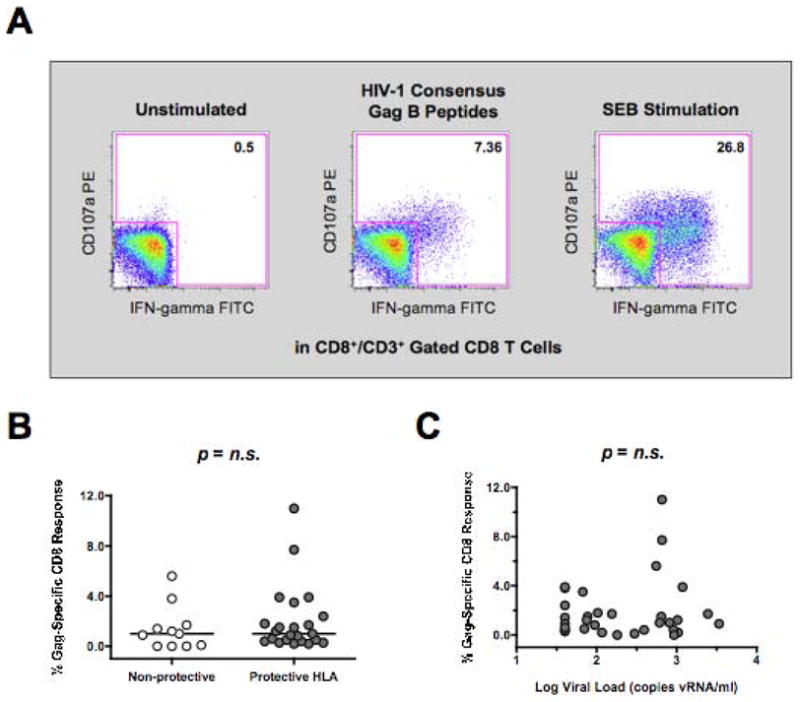
(A) CD107a or IFN-gamma staining on CD8+/CD3+ gated T cells from PBMCs incubated alone, with overlapping 15-mer peptides to HIV-1 Gag, or with the super-antigen SEB for 18 hours. (B) Composite graph of HIV-specific Gag induced CD107a or IFN-gamma staining on CD8+/CD3+ gated T cells from HIV-1 controller subjects possessing or lacking protective HLA alleles as described in Methods Section. (C) Spearman correlation of HIV-1 Gag-specific CD8+ T cell responses (y-axis) and the log viral load (x-axis). Comparisons between two groups were performed using an unpaired, non-parametric Mann-Whitney T-test and correlations between two variables were carried out using a non-parametric Spearman test. In all cases, p-value were two-tailed with a 95% confidence interval (n.s., not significant).
Inverse Relationship Between T Cell and NK activity Among Elite Controllers
Due to the observed heterogeneity in T Cell responses among controllers (Figure 2B), we hypothesized that increased NK activity may account for the sustained control over viral replication in a subset of controllers lacking strong HIV-1 specific CD8+ T cell responses. Due to the potential confounding effect of viral replication on HIV-specific T cell responses in viremic controllers, we focused exclusively on elite controllers. As shown in Figure 3A, we observed that elite controllers possessing low HIV-1 specific CD8+ T cell responses (x-axis) did indeed possess heightened IFN-alpha induced NK responses against K562 target cells (y-axis). The inverse association between HIV-1 specific CD8+ T cell responses and NK activity was significant (rho= −0.8321, p=0.001, n=12), and was not observed when NK activity was plotted against CEF-specific CD8+ T cell responses against the combined epitopes of CMV, EBV and Flu (Figure 3B). Finally, we tested if the observed inverse association between HIV-1 specific CD8+ T cell responses and NK activity was due to inheritance patterns in protective KIR3DL1*h/*y genotypes. As shown in Figure 3C, elite controllers that possessed protective KIR3DL1*h/*y alleles along with their corresponding HLA-Bw4*80I alleles had significantly lower (p=0.0087) CD8 responses to HIV-1 Gag than controllers lacking protective KIR3DL1*h/*y alleles. Importantly, pre-stimulation with IFN-alpha does not alter the absolute number of CD8+ cells nor the frequency of HIV-specific CD8+ T cells (Supplemental Figure 2), consistent with the innate immune responses being the primary effector response activated by IFN-alpha. Together, these results suggest that inheritance patterns of protective KIR3DL1*h/*y genotypes may influence the magnitude of both anti-viral NK cell and CD8 T cell responses among elite controllers.
Figure 3. Comparison of CD8 T Cell and NK Cell Function in Elite Controllers.
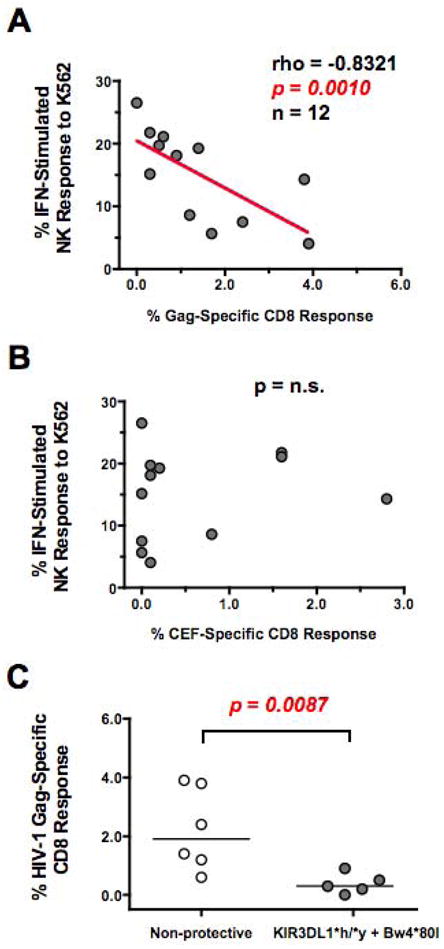
(AB) Spearman correlation of (A) HIV-1 Gag-specific or (B) CEF (CMV, EBV and Flu)-specific CD8+ T cell response (x-axis) and IFN-alpha stimulated, K562-induced NK response (y-axis) from elite controllers. (C) Composite graph of HIV-1 Gag-specific CD8+ T cell response from elite controller subjects possessing or lacking a protective KIR3DL1*h/*y genotype along with their corresponding HLA-Bw4*80I ligands. Comparisons between two groups were performed using an unpaired, non-parametric Mann-Whitney T-test and correlations between two variables were carried out using a non-parametric Spearman test. In all cases, p-value were two-tailed with a 95% confidence interval (n.s., not significant).
Discussion
Here, we simultaneously tested the functional capacity of both NK cells and T cells in HIV-1 infected controller subjects to investigate correlates of immune control over viral replication. We hypothesized that increased NK activity may account for the sustained control over viral replication in a subset of controllers lacking strong HIV-1 specific CD8+ T cell responses. In support of this hypothesis, we observed a significant inverse association between the IFN-alpha stimulated NK response to K562 cells and the HIV-specific CD8 T cell response to Gag among elite controllers. (Figure 3A). The inheritance of protective KIR3DL1*h/*y receptor genotype previously associated with strong NK responses was also associated with a statistically significant decrease in HIV-1 Gag-specific CD8 responses among elite controllers (Figure 3C). These findings suggest that NK responses and T cell responses can be observed independently in HIV-1 infected controllers, and that inheritance of protective NK cell KIR3DL1*h/*y genotypes may influence the magnitude of both anti-viral NK cell and CD8+ T cell responses. Interestingly, a negative regulatory role of NK cells on anti-viral CD8+ T cells was recently described by Waggoner at al. [37], and this is the first observation of a similar relationship between NK function and CD8 activity in HIV-1 infected Elite Controllers.
Importantly, we documented that both NK cells and CD8+ T cells from controllers exhibit strong effector function when tested in vitro despite detectable levels of viremia in most controller subjects (Figures 1 and 2). High levels of viremia have been shown to negatively impact NK function [32–34], however, we did not observe that NK activity was inversely affected by the lower viral loads exhibited by controllers from our cohort (Figure 1E). NK cells from controllers also remained responsive to IFN-alpha stimulation which has been shown to be necessary for NK-mediated lysis against several virally infected target cell types including herpesvirus infected fibroblasts [38–42] and HIV-1 infected autologous CD4+ primary T cells [31]. Accordingly, we observed that NK cell CD107a degranulation and IFN-gamma cytokine production in response to K562 tumor cells was dramatically increased in the presence of Interferon-alpha stimulation, particularly among controllers possessing a protective KIR3DL1*h/*y genotype along with their corresponding HLA-Bw4*80I ligand (Figure 1C). The enhanced functional output of NK cells from controllers possessing a protective KIR3DL1*h/*y receptor genotype along with their corresponding HLA-Bw4*80I ligand was also observed in unstimulated PBMC but the effect was not statistically significant (Figure 1B) as has been described previously [43]. These findings may suggest that an enhanced ability to respond to IFN-alpha stimulation from PDC may be associated with the NK stimulatory effect observed in subjects inheriting the KIR3DL1*h/*y genotype. Further studies will be needed to conclude if both constitutive levels of NK lysis and IFN-alpha stimulated NK lysis are simultaneously increased in subjects inheriting the KIR3DL1*h/*y genotype. Our analysis of PDC frequency and TLR9-stimulated PDC secretion of INF-alpha in subjects from our cohort is consistent with recent data [44] showing that PDC responses are maintained among controllers (data not shown).
Genetically, we observed that Gag-specific CD8+ T cell responses were not significantly different between controllers possessing or lacking protective HLA-B alleles (Figure 2B). Future work will be needed to address if protective HLA alleles have an effect on the magnitude of HIV-1 specific Gag responses as we did not measure the effect of protective HLA alleles on the breadth or polyfunctionality of Gag-specific CD8+ T cell responses. In contrast, we confirmed that inheritance of protective KIR3DL1*h/*y genotype along with their corresponding HLA-Bw4*80I ligand among controllers is associated with significantly increased NK function following IFN-alpha stimulation (Figure 1C) when compared to controllers lacking either the protective KIR3DL1*h/*y genotype or the requisite HLA-Bw4*80I ligands (Supplementary Figure 1). The protective HLA-B*57 allele, in addition to presenting peptides that are better able to limit viral replication [45], also serves as a primary ligand for the NK KIR3DL1 receptor. As compared to other HLA-Bw4*80I ligands, HLA-B*57 has been observed to independently augment NK function [19]. Here, we could not assess the independent effect of HLA-B*57 compared to other HLA-Bw4*80I ligands on NK function due to the pronounced enrichment of controllers inheriting both the KIR3DL1*h/*y genotype with HLA-B*57 (data not shown). The strong co-inheritance pattern of protective KIR3DL1*h/*y genotype with HLA-B*57 in our cohort of controllers suggests that a synergistic response between the adaptive and innate immune compartment may work in concert to limit viral replication. While our studies have relied on the universally recognized K562 target cell system to measure NK cytotoxic function, it will be of interest to confirm if autologous CD4+ T cells from elite controllers super-infected with HIV-1 are lysed by interferon-alpha stimulated NK cells irrespective of low HIV-specific CD8+ T cell responses as indicated by our results with K562 cells.
In support of a role for NK pressure in limiting HIV-1 replication, recent data from several groups has identified specific HIV-1 viral escape mutations from CTL responses that can also modulate NK recognition of virally infected cells [46, 47]. Future studies may need to focus on a combination of potential innate and adaptive immune factors to fully elucidate what dominant variables allow for continued elite controller status over time in this rare subset of HIV-1 infected subjects. As elite controllers have been seen as a model for achieving a functional cure in HIV-1 infected subjects [48], these findings underscore the premise that activation of either innate or adaptive mechanisms of host-mediated control may be associated with control of viral replication in the absence of anti-retroviral therapy.
Supplementary Material
Composite graph of IFN-alpha stimulated, K562-specific NK response from controllers possessing or lacking a protective KIR3DL1*h/*y genotype along with their corresponding HLA-Bw4*80I ligands. CD107a and IFN-gamma staining on CD56+/CD3- NK cells gated from IFN-alpha stimulated PBMC incubated in the presence or absence of K562 target cells. NK cells were gated by CD56+/CD3− expression and the percentage of NK cells staining positive for CD107a and/or IFN-gamma following incubation with K562 cells was determined after subtraction of background levels of staining in the absence of target cells.
(A) Composite graphs showing the lack of effect of IFN-alpha pre-stimulation on the percentage of total CD8+ T cells and HIV-specific CD8+ T cells in response to Autologous HIV-1 Infected CD4 Primary target Cells. (B–C) Unstimulated (B) or IFN-alpha stimulated (C) PBMC were incubated alone, with uninfected or with HIV-1 SPL-3 infected CD4 cells at a 10:1 effector to target cell ratio for 4 hours in the presence of anti-CD107a PE antibody and Brefeldin A. PBMC were then stained for CD8 phenotypic surface markers and permeabilized for intra-cellular cytokine staining. The percentage of CD8 cells staining positive for CD107a and/or IFN-gamma following incubation with target cells was determined after subtraction of background levels of staining in the absence of target cells (no target control).
Protective HLA-B genotypes (shown in blue) and protective KIR3DL1*h/*y Receptor Genotypes (shown in green) are highlighted as described in Materials and Methods section. N/D is not-determined and these subjects were excluded from the analysis of NK function based upon genotype.
Acknowledgments
This study was supported by grants from the National Institutes of Health (R21 AI078870, NIDA R01 DA028775, R01 AI073219, RO1 AI065279, Core grant P30 CA10815), the Philadelphia Foundation, and funds from the Pennsylvania Commonwealth Universal Research Enhancement Program. The UCSF-based SCOPE cohort of controllers was supported in part by the Centers for AIDS Research at UCSF (PO AI27763), CFAR Network of Integrated Systems (R24 AI067039), the UCSF CTSI (UL1 RR024131), NIAID (RO1 AI087145, K24AI069994, AI 76174), amfAR, and the Ragon Institute. This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: Complete Set of HIV-1 Consensus B Gag (15 amino acids in length) peptides, CEF peptide pool, 23 peptides (8–12 amino acids in length), consisting of sequences derived from the human Cytomegalovirus, Epstein-Barr virus and Influenza Virus.
Footnotes
Authorship
Costin Tomescu (Completed all of the in vitro assays, co-wrote the manuscript, completed statistical analysis), Fuh-Mei Duh (Completed all of the HLA genotyping), Rebecca Hoh (Coordinated subject recruitment, analyzed subject behavioral data), Anne Viianni and Kara Harvill (Coordinated subject blood draws and organized subject behavioral data), Maureen P. Martin (Completed all of the KIR genotyping, edited the manuscript), Mary Carrington (Coordinated KIR, HLA and CCR5 genotyping, edited the manuscript), Steven G. Deeks (Coordinated HIV-1 Infected Controller cohort, co-coordinated the study design, edited manuscript), Luis J. Montaner (Coordinated the study, analyzed all of the functional data, co-wrote the manuscript). The authors have no conflicts of interest.
References
- 1.Grabar S, Selinger-Leneman H, Abgrall S, Pialoux G, Weiss L, Costagliola D. Prevalence and comparative characteristics of long-term nonprogressors and HIV controller patients in the French Hospital Database on HIV. AIDS. 2009;23:1163–1169. doi: 10.1097/QAD.0b013e32832b44c8. [DOI] [PubMed] [Google Scholar]
- 2.Hubert JB, Burgard M, Dussaix E, Tamalet C, Deveau C, Le Chenadec J, et al. Natural history of serum HIV-1 RNA levels in 330 patients with a known date of infection. The SEROCO Study Group. Aids. 2000;14:123–131. doi: 10.1097/00002030-200001280-00007. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG, Walker BD. Human Immunodeficiency Virus Controllers: Mechanisms of Durable Virus Control in the Absence of Antiretroviral Therapy. Immunity. 2007;27 doi: 10.1016/j.immuni.2007.08.010. in press. [DOI] [PubMed] [Google Scholar]
- 4.Buzon MJ, Seiss K, Weiss R, Brass AL, Rosenberg ES, Pereyra F, et al. Inhibition of HIV-1 integration in ex vivo-infected CD4 T cells from elite controllers. J Virol. 2011;85:9646–9650. doi: 10.1128/JVI.05327-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen H, Li C, Huang J, Cung T, Seiss K, Beamon J, et al. CD4+ T cells from elite controllers resist HIV-1 infection by selective upregulation of p21. J Clin Invest. 2011;121:1549–1560. doi: 10.1172/JCI44539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brumme ZL, Li C, Miura T, Sela J, Rosato PC, Brumme CJ, et al. Reduced replication capacity of NL4-3 recombinant viruses encoding reverse transcriptase-integrase sequences from HIV-1 elite controllers. J Acquir Immune Defic Syndr. 2011;56:100–108. doi: 10.1097/QAI.0b013e3181fe9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miura T, Brockman MA, Brumme ZL, Brumme CJ, Pereyra F, Trocha A, et al. HLA-associated alterations in replication capacity of chimeric NL4-3 viruses carrying gag-protease from elite controllers of human immunodeficiency virus type 1. J Virol. 2009;83:140–149. doi: 10.1128/JVI.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura T, Brumme CJ, Brockman MA, Brumme ZL, Pereyra F, Block BL, et al. HLA-associated viral mutations are common in human immunodeficiency virus type 1 elite controllers. J Virol. 2009;83:3407–3412. doi: 10.1128/JVI.02459-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miura T, Brumme ZL, Brockman MA, Rosato P, Sela J, Brumme CJ, et al. Impaired replication capacity of acute/early viruses in persons who become HIV controllers. J Virol. 2010;84:7581–7591. doi: 10.1128/JVI.00286-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Emu B, Sinclair E, Hatano H, Ferre A, Shacklett B, Martin JN, et al. HLA Class I-Restricted T Cell Responses May Contribute to the Control of HIV Infection, but Such Responses are Not Always Necessary for Long-term Virus Control. J Virol. 2008 doi: 10.1128/JVI.02176-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mothe B, Llano A, Ibarrondo J, Zamarreno J, Schiaulini M, Miranda C, et al. CTL responses of high functional avidity and broad variant cross-reactivity are associated with HIV control. PLoS One. 2012;7:e29717. doi: 10.1371/journal.pone.0029717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pereyra F, Addo MM, Kaufmann DE, Liu Y, Miura T, Rathod A, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 13.Hersperger AR, Martin JN, Shin LY, Sheth PM, Kovacs CM, Cosma GL, et al. Increased HIV-specific CD8+ T-cell cytotoxic potential in HIV elite controllers is associated with T-bet expression. Blood. 2012;117:3799–3808. doi: 10.1182/blood-2010-12-322727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hersperger AR, Pereyra F, Nason M, Demers K, Sheth P, Shin LY, et al. Perforin expression directly ex vivo by HIV-specific CD8 T-cells is a correlate of HIV elite control. PLoS Pathog. 2010;6:e1000917. doi: 10.1371/journal.ppat.1000917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costello C, Tang J, Rivers C, Karita E, Meizen-Derr J, Allen S, Kaslow RA. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. Aids. 1999;13:1990–1991. doi: 10.1097/00002030-199910010-00031. [DOI] [PubMed] [Google Scholar]
- 16.McNeil AJ, Yap PL, Gore SM, Brettle RP, McColl M, Wyld R, et al. Association of HLA types A1-B8-DR3 and B27 with rapid and slow progression of HIV disease. Qjm. 1996;89:177–185. doi: 10.1093/qjmed/89.3.177. [DOI] [PubMed] [Google Scholar]
- 17.den Uyl D, van der Horst-Bruinsma IE, van Agtmael M. Progression of HIV to AIDS: a protective role for HLA-B27? AIDS Rev. 2004;6:89–96. [PubMed] [Google Scholar]
- 18.Migueles SA, Laborico AC, Imamichi H, Shupert WL, Royce C, McLaughlin M, et al. The differential ability of HLA B*5701+ long-term nonprogressors and progressors to restrict human immunodeficiency virus replication is not caused by loss of recognition of autologous viral gag sequences. J Virol. 2003;77:6889–6898. doi: 10.1128/JVI.77.12.6889-6898.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kamya P, Boulet S, Tsoukas CM, Routy JP, Thomas R, Cote P, et al. Receptor-Ligand Requirements for Increased NK Cell Polyfunctional Potential in Slow Progressors Infected with HIV-1 Coexpressing KIR3DL1*h/*y and HLA-B*57. J Virol. 2011;85:5949–5960. doi: 10.1128/JVI.02652-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lopez-Vazquez A, Mina-Blanco A, Martinez-Borra J, Njobvu PD, Suarez-Alvarez B, Blanco-Gelaz MA, et al. Interaction between KIR3DL1 and HLA-B*57 supertype alleles influences the progression of HIV-1 infection in a Zambian population. Hum Immunol. 2005;66:285–289. doi: 10.1016/j.humimm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 21.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin MP, Gao X, Lee JH, Nelson GW, Detels R, Goedert JJ, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 23.Yawata M, Yawata N, Draghi M, Little AM, Partheniou F, Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J Exp Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long BR, Ndhlovu LC, Oksenberg JR, Lanier LL, Hecht FM, Nixon DF, Barbour JD. KIR3DS1 Conferral of Enhanced Natural Killer Cell Function in Early HIV-1 Infection. J Virol. 2008 doi: 10.1128/JVI.02449-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alter G, Martin MP, Teigen N, Carr WH, Suscovich TJ, Schneidewind A, et al. Differential natural killer cell-mediated inhibition of HIV-1 replication based on distinct KIR/HLA subtypes. J Exp Med. 2007;204:3027–3036. doi: 10.1084/jem.20070695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boulet S, Kleyman M, Kim JY, Kamya P, Sharafi S, Simic N, et al. A combined genotype of KIR3DL1 high expressing alleles and HLA-B*57 is associated with a reduced risk of HIV infection. AIDS. 2008;22:1487–1491. doi: 10.1097/QAD.0b013e3282ffde7e. [DOI] [PubMed] [Google Scholar]
- 27.Naranbhai V, Bartman P, Ndlovu D, Ramkalawon P, Ndung’u T, Wilson D, et al. Impact of blood processing variations on natural killer cell frequency, activation, chemokine receptor expression and function. J Immunol Methods. 2011;366:28–35. doi: 10.1016/j.jim.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin MP, Carrington M. KIR locus polymorphisms: genotyping and disease association analysis. Methods Mol Biol. 2008;415:49–64. doi: 10.1007/978-1-59745-570-1_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas R, Yamada E, Alter G, Martin MP, Bashirova AA, Norman PJ, et al. Novel KIR3DL1 alleles and their expression levels on NK cells: convergent evolution of KIR3DL1 phenotype variation? J Immunol. 2008;180:6743–6750. doi: 10.4049/jimmunol.180.10.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stern M, Ruggeri L, Capanni M, Mancusi A, Velardi A. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood. 2008;112:708–710. doi: 10.1182/blood-2008-02-137521. [DOI] [PubMed] [Google Scholar]
- 31.Tomescu C, Chehimi J, Maino VC, Montaner LJ. NK Cell Lysis of HIV-1-Infected Autologous CD4 Primary T Cells: Requirement for IFN-Mediated NK Activation by Plasmacytoid Dendritic Cells. J Immunol. 2007;179:2097–2104. doi: 10.4049/jimmunol.179.4.2097. [DOI] [PubMed] [Google Scholar]
- 32.De Maria A, Fogli M, Costa P, Murdaca G, Puppo F, Mavilio D, et al. The impaired NK cell cytolytic function in viremic HIV-1 infection is associated with a reduced surface expression of natural cytotoxicity receptors (NKp46, NKp30 and NKp44) Eur J Immunol. 2003;33:2410–2418. doi: 10.1002/eji.200324141. [DOI] [PubMed] [Google Scholar]
- 33.Mavilio D, Benjamin J, Daucher M, Lombardo G, Kottilil S, Planta MA, et al. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc Natl Acad Sci U S A. 2003;100:15011–15016. doi: 10.1073/pnas.2336091100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sirianni MC, Mezzaroma I, Aiuti F, Moretta A. Analysis of the cytolytic activity mediated by natural killer cells from acquired immunodeficiency syndrome patients in response to phytohemagglutinin or anti-CD16 monoclonal antibody. Eur J Immunol. 1994;24:1874–1878. doi: 10.1002/eji.1830240824. [DOI] [PubMed] [Google Scholar]
- 35.Julg B, Williams KL, Reddy S, Bishop K, Qi Y, Carrington M, et al. Enhanced anti-HIV functional activity associated with Gag-specific CD8 T-cell responses. J Virol. 2010;84:5540–5549. doi: 10.1128/JVI.02031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kiepiela P, Ngumbela K, Thobakgale C, Ramduth D, Honeyborne I, Moodley E, et al. CD8+ T-cell responses to different HIV proteins have discordant associations with viral load. Nat Med. 2007;13:46–53. doi: 10.1038/nm1520. [DOI] [PubMed] [Google Scholar]
- 37.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2011:10624. doi: 10.1038/nature10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bandyopadhyay S, Perussia B, Trinchieri G, Miller DS, Starr SE. Requirement for HLA-DR+ accessory cells in natural killing of cytomegalovirus-infected fibroblasts. Journal of Experimental Medicine. 1986;164:180–195. doi: 10.1084/jem.164.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feldman M, Howell D, Fitzgerald-Bocarsly P. Interferon-alpha-dependent and -independent participation of accessory cells in natural killer cell-mediated lysis of HSV-1-infected fibroblasts. J Leukoc Biol. 1992;52:473–482. doi: 10.1002/jlb.52.5.473. [DOI] [PubMed] [Google Scholar]
- 40.Fitzgerald-Bocarsly P, Feldman M, Curl S, Schnell J, Denny T. Positively selected Leu-11a (CD16+) cells require the presence of accessory cells or factors for the lysis of herpes simplex virus-infected fibroblasts but not herpes simplex virus-infected Raji. J Immunol. 1989;143:1318–1326. [PubMed] [Google Scholar]
- 41.Oh SH, Bandyopadhyay S, Miller DS, Starr SE. Cooperation between CD16(Leu-11b)+ NK cells and HLA-DR+ cells in natural killing of herpesvirus-infected fibroblasts. J Immunol. 1987;139:2799–2802. [PubMed] [Google Scholar]
- 42.Perussia B, Fanning V, Trinchieri G. A leukocyte subset bearing HLA-DR antigens is responsible for in vitro alpha interferon production in response to viruses. Nat Immun Cell Growth Regul. 1985;4:120–137. [PubMed] [Google Scholar]
- 43.Boulet S, Song R, Kamya P, Bruneau J, Shoukry NH, Tsoukas CM, Bernard NF. HIV protective KIR3DL1 and HLA-B genotypes influence NK cell function following stimulation with HLA-devoid cells. J Immunol. 2010;184:2057–2064. doi: 10.4049/jimmunol.0902621. [DOI] [PubMed] [Google Scholar]
- 44.Machmach K, Leal M, Gras C, Viciana P, Genebat M, Franco E, et al. Plasmacytoid Dendritic Cells Reduce HIV Production in Elite Controllers. J Virol. 2012 doi: 10.1128/JVI.07114-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinez-Picado J, Prado JG, Fry EE, Pfafferott K, Leslie A, Chetty S, et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol. 2006;80:3617–3623. doi: 10.1128/JVI.80.7.3617-3623.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fadda L, O’Connor GM, Kumar S, Piechocka-Trocha A, Gardiner CM, Carrington M, et al. Common HIV-1 peptide variants mediate differential binding of KIR3DL1 to HLA-Bw4 molecules. J Virol. 2011;85:5970–5974. doi: 10.1128/JVI.00412-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alter G, Heckerman D, Schneidewind A, Fadda L, Kadie CM, Carlson JM, et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature. 2011;476:96–100. doi: 10.1038/nature10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Autran B, Descours B, Avettand-Fenoel V, Rouzioux C. Elite controllers as a model of functional cure. Curr Opin HIV AIDS. 2011;6:181–187. doi: 10.1097/COH.0b013e328345a328. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Composite graph of IFN-alpha stimulated, K562-specific NK response from controllers possessing or lacking a protective KIR3DL1*h/*y genotype along with their corresponding HLA-Bw4*80I ligands. CD107a and IFN-gamma staining on CD56+/CD3- NK cells gated from IFN-alpha stimulated PBMC incubated in the presence or absence of K562 target cells. NK cells were gated by CD56+/CD3− expression and the percentage of NK cells staining positive for CD107a and/or IFN-gamma following incubation with K562 cells was determined after subtraction of background levels of staining in the absence of target cells.
(A) Composite graphs showing the lack of effect of IFN-alpha pre-stimulation on the percentage of total CD8+ T cells and HIV-specific CD8+ T cells in response to Autologous HIV-1 Infected CD4 Primary target Cells. (B–C) Unstimulated (B) or IFN-alpha stimulated (C) PBMC were incubated alone, with uninfected or with HIV-1 SPL-3 infected CD4 cells at a 10:1 effector to target cell ratio for 4 hours in the presence of anti-CD107a PE antibody and Brefeldin A. PBMC were then stained for CD8 phenotypic surface markers and permeabilized for intra-cellular cytokine staining. The percentage of CD8 cells staining positive for CD107a and/or IFN-gamma following incubation with target cells was determined after subtraction of background levels of staining in the absence of target cells (no target control).
Protective HLA-B genotypes (shown in blue) and protective KIR3DL1*h/*y Receptor Genotypes (shown in green) are highlighted as described in Materials and Methods section. N/D is not-determined and these subjects were excluded from the analysis of NK function based upon genotype.