Abstract
Maternal mRNAs are translationally regulated during early development. Zar1 and its closely related homolog, Zar2, are both crucial in early development. Xenopus laevis Zygote arrest 2 (Zar2) binds to the Translational Control Sequence (TCS) in maternal mRNAs and regulates translation. The molecular mechanism of Zar1 has not been described. Here we report similarities and differences between Xenopus Zar1 and Zar2. Analysis of Zar sequences in vertebrates revealed two Zar family members with conserved, characteristic amino acid differences in the C-terminal domain. The presence of only two vertebrate Zar proteins was supported by analyzing Zar1 synteny. We propose that the criteria for naming Zar sequences are based on the characteristic amino acids and the chromosomal context. We also propose reclassification of some Zar sequences. We found that Zar1 is expressed throughout oogenesis and is stable during oocyte maturation. The N-terminal domain of Zar1 repressed translation of a reporter construct in immature oocytes. Both Zar1 and Zar2 bound to the TCS in the Wee1 and Mos 3′ UTRs using a zinc finger in the C-terminal domain. However, Zar1 had much higher affinity for RNA than Zar2. To show the functional significance of the conserved amino acid substitutions, these residues in Zar2 were mutated to those found in Zar1. We show that these residues contributed to the different RNA binding characteristics of Zar1 compared to Zar2. Our study shows that Zar proteins have generally similar molecular functions in the translational regulation of maternal mRNAs, but they may have different roles in early development.
Keywords: Zygote arrest, RNA binding protein, Xenopus oocyte, Translational control
1. Introduction
1.1. Zygote arrest family of proteins
Zygote arrest (Zar) family proteins have been implicated in the early mitotic cleavages of the embryo, the maternal to zygote transition and epidermalization of the embryo [1–3]. Zar-family expression is generally confined to the oocyte and early embryo in all species tested, but in some species Zar proteins are also expressed in the testis [3–8]. There are “Zar1”, “Zar2”, “Zar1-like”, “Zar1-like protein-like” and “similar to Zar1” protein sequences in the databases but there are no established criteria to differentiate between Zar homologs. Therefore, there is a need to re-evaluate Zar sequences to identify the number of family members and to name the sequences accordingly.
Although the importance of Zar family proteins in early development is clear, their molecular mechanism of action has been harder to elucidate. One reason is that their amino acid sequences do not show homology to other proteins. Zar proteins share extensive homology in their C-terminal domains including invariant cysteines that were suggested to comprise a zinc finger, but whether this zinc finger bound to nucleic acid, protein or lipid was unknown [3, 5, 6]. Indeed, Zar proteins had been thought to regulate chromatin structure and gene expression. However, recently we showed that the conserved cysteines in the C-terminal domain in Xenopus Zar2 were required for binding to maternal mRNA sequences [9]. The molecular function of Zar1 has not been described.
1.2. Translational control of maternal mRNAs
Translational control of maternal mRNAs is an evolutionarily conserved strategy to control gene expression from meiosis until activation of the zygotic genome [10]. Maternal mRNAs contain multiple cis-elements in their 3′ untranslated regions that determine when, where and to what extent each mRNA is translated [11–13]. The best characterized cis element is the CPE and its trans-acting factor is CPEB [14, 15]. In general, CPEs and CPEB repress translation in immature oocytes and stimulate translation in maturing oocytes. This stimulation of translation is accompanied by cytoplasmic polyadenylation of the maternal mRNA. Wee1 is a developmentally important protein that is encoded by a maternal mRNA. Wee1 protein is absent in immature oocytes and is synthesized after meiosis I [16, 17]. Wee1 is a negative regulator of Cdk1 and therefore delays entry into M-phase, a function that is thought to elongate the first mitotic division [16, 18] and allow cell movements in gastrulation [19]. The 3′ UTR of Wee1 mRNA contains two cytoplasmic polyadenylation elements (CPEs) and two Translational Control Sequences (TCSs) that regulate translation [17, 20]. The TCSs repress translation in immature oocytes and during maturation they confer cytoplasmic polyadenylation to the mRNA and stimulate translation. The function of the TCS is mediated by its trans-acting factor Zar2. Like the TCS, Zar2 represses translation in immature oocytes and this repression is relieved in maturing oocytes [9]. Mos is another developmentally important protein that is encoded by a maternal mRNA. The Mos protein is absent in immature oocytes and starts to be synthesized shortly after re-entry into the meiotic cell cycle. Appropriate translational control of the Mos mRNA is crucial for timely oocyte maturation and prevention of premature mitotic cell cycles [21–23]. The Mos 3′ UTR contains a CPE, a Musashi binding element (MBE), and also a TCS [9, 12, 24–26]. Zar2 interacts with the Wee1 and Mos TCSs [9]. Because of the similarity between Zar1 and Zar2 we hypothesize that Zar1 also binds to the TCSs in the Wee1 and/or Mos mRNAs and regulates translation. Moreover, because Zar1 and Zar2 do not appear to be redundant, as mice that are null for only Zar1 are infertile [3], we hypothesize that there are some differences in their functions.
The objectives of this study are to determine the molecular function of Zar1 and to characterize similarities and differences between Zar1 and Zar2. We also formally classify Zar sequences.
2. Material and methods
2.1. Cloning of full length zar1 and subsequent plasmids
pXen1 Zar1a: As we found that Xenopus laevis had a Zar2a and a Zar2b[9], we set out to identify Zar1a and Zar1b sequences. BLAST® was used to find X. laevis EST sequences that aligned with X. laevis zar1 (GenBank ID: AY283176) [3]. DY565955, DY545225, DC114665, DC101644, CA987692, BP708289, BJ094813, BJ093826, and AW640468 were essentially identical to sections of Zar1 already in GenBank (AY283176), and were designated Zar1a. ESTs DY564080, DC046119 and EB480588 had a few conserved differences from the Zar1 sequences above and were designated Zar1b. First Choice RLM-RACE Kit (Ambion) was used to isolate full length 5′ ends of Zar1a and Zar1b from immature Xenopus oocytes, according to the manufacturer’s directions. The 5′ RACE primers were: outer zar1a 5′-CTT CAT CTG TCT TGT CCA TCT TCA and inner zar1a 5′-CCT CAC CCT TCT CTT CCA GAT TGA and outer zar1b 5′-CTT GGT CCT TGT CCA TCT TAG and inner zar1b 5′-CCT CAG TCT TCT CTG ACA GAT TTT. These primers showed that there was approximately 140 nt of Zar1a 5′ sequence (GenBank KC476498) that resulted in 12 more N-terminal amino acids than GenBank AY283176. Compared to Zar1a, the Zar1b 5′ sequence was much shorter, so new primers were ordered to a common sequence in both Zar1a and Zar1b: outer zar1 5′-AAC TGG AAC TTT GGC CTT GTC TGA and inner zar1 5′-CTC CTT CTG AGT AAA GTT CTG CTG GGC. The common primers were used with a 5′ RACE cDNA preparation from a different frog, which confirmed the 12 extra 5′ codons in Zar1a and extended the Zar1b 5′ sequence by approximately 60 nucleotides. The new 5′ sequences were submitted to GenBank, Zar1a (GenBank KC476498) and Zar1b (GenBank KC476499). The new Zar1a 5′ sequence was spliced onto AY283176 to make BK008757. The new Zar1b 5′ sequence was spliced with DY564080 and EB480588 to make BK008758. Only Zar1a is shown in this study.
pXen Zar1-MS2: Full length Zar1 was cloned by RT-PCR from total RNA from immature Xenopus oocytes using primers: Zar1 forward 5′-ATG GTA CCC TCG AGG ATG GCT AGC TTC TCA GAG and Zar1 reverse 5′-CCT AGC CCG GGC AAT GAT ATA CTT GAA GCT. PCR products were digested with XhoI and XmaI (underlined) and ligated into pXen C-MS2 [9] cut with XhoI and XmaI. Full length Zar1 was fused 5′ of MS2.
pXen N-Zar1-MS2: 1–159 aa were kept and the C-terminal 160–307 aa were deleted from pXen Zar1-MS2. NcoI sites (underlined) were introduced by PCR to remove the C-terminal domain: N-terminal Zar1 reverse primer 5′-CGA TCC ATG GCT CAC CCT TCT CTT CCA G and MS2 forward primer 5′-ATG CCC ATG GCC CGG GAT GGC TTC TAA CTT TAC. The PCR product was cut with NcoI and self-ligated.
pXen-C-Zar1: pXen1 [27] was a kind gift from Dr. Angus MacNicol, University of Arkansas for Medical Sciences, AR, USA. Amino acids 165–307 were PCR amplified from cDNA from immature Xenopus oocytes using primers: forward, 5′-GAT CCT CGA GGG ATG GGC TCT GAA GGA GGG AGG and reverse, 5′-GAT CCT CGA GTC AAA TGA TAT ACT TGA A. The PCR product was cut with XhoI (underlined) and ligated into XhoI-cut pXen1. Cysteine to alanine mutations were made in pXen-C-Zar1 and were performed with QuikChange (Agilent Technologies) mutagenesis methods with the following codon changes: C215A, TGT→GCT; C242A, TGC→GCC; C259A, TGT→GCT; and C287A, TGT→GCT.
pGEX 6P-3 C-Zar1 (amino acids 165–307): Using pXen-C-Zar1 as the template, C-Zar1 was amplified with forward primer 5′-CCA GGG ATC CGG CTC TGAA GGA GGG AGG C and reverse primer 5′-GTC GAG CGG CCG CTC AAA TGA TAT ACT TGA AGC TAA AAG and ligated into pGEX 6P-3 vector (GE Lifesciences) using BamHI and NotI (underlined).
pGEX 6P-3 C-Zar2mut12: To mutate Zar2 amino acids in pGEX 6P-3 C-Zar2 [9] to conserved Zar1 amino acids, the following codon changes were used: F213Y, TTC→TAC; K219N, AAA→AAC; T220I, ACC→ATC; I230V, ATT→TAT; S231Q, TCT→CAA; L241F, CTT→TTT; A256D, GCC →CAC; Q258T, CAA→ACA; I276V, ATA→GTA; L278P, CTG→CCG; E285D, GAA→GAC; Y301F, TAC→TTC. Mutations were introduced by QuikChange.
pXen N-MS2, pXen N-Zar2-MS2, pXen rluc (Renilla luciferase), pXen fluc (firefly luciferase), and pXen fluc-2x-SL (pXen fluc with stem-loops) have been described before [9].
All plasmids were sequenced to verify integrity using the University of Colorado Cancer Center DNA Sequencing and Analysis Core. For in vitro transcription, all plasmids were linearized with PstI unless otherwise noted. 5′ capped RNA was synthesized in vitro with SP6 mMessage mMachine transcription kit (Ambion). RNA quality was assessed using gel electrophoresis. Nucleotide and amino acid sequence alignments were performed with MacVector 11.1.2.
2.2. Multiple sequence alignment
Zar1 and Zar2 protein sequences were obtained from the public genome databases NCBI (http://www.ncbi.nlm.nih.gov/) and ENSEMBL (http://www.ensembl.org/index.html). Initial sequences were queried through word searches (i.e. Zygote Arrest, Zar1, Zar1-like, Zar2) in NCBI databases. Zar1 and Zar2 protein and nucleotide sequences identified from this preliminary search were used to query ENSEMBL, NCBI, and BLAST browsers for more sequences resembling zygote arrest proteins. Originally, 218 sequences were obtained; however, 53 sequences were duplicated within and between GenBank and ENSEMBL resulting in the elimination of one of the two sequences from the data pool. A multiple sequence alignment was performed in MacVector 11.1.2 using CLUSTALW, a progressive approach in which successive alignments were performed with different algorithms. An initial alignment was performed on 165 non-redundant sequences using the PAM substitution matrix [28]. Penalties were assigned as follows: open gap, 10; extend gap, 0.05; delay divergent, 40%; and gap separation distance, 8. Sequences were manually evaluated for quality and integrity of conserved regions. Sequences were removed from the alignment if they did not meet the following criteria: 250–500 amino acids, 12 specific conserved cysteine residues, and a WESAY motif in the C-terminal domain. Prior to removing sequences based on these criteria, attempts to patch the sequences were made by searching EST and genomic databases for nucleotide sequences to evaluate them for point mutations, frame-shifts, or annotation errors. The 92 remaining sequences, representing 40 different species, were manually evaluated for residues that distinguish Zar1 from Zar2. A final alignment was performed with only one sequence from each species. The representative sequence for each organism was chosen manually based on similarity scores and the number of times the sequence has been referenced in literature; however, sequences obtained from the RefSeq database were typically prioritized as representative of the species.
2.3. Synteny
Zar2 sequences identified from Fig. 1 were queried in the NCBI Gene database. The genomic context was evaluated for both chromosome placement and neighboring genes. Where genomes were not annotated thoroughly, BLAST was used to identify neighboring genes.
Fig. 1.

Amino acid alignment of Zar proteins in different vertebrate species showing that there are two Zar family members. Invariant cysteines are highlighted in yellow. Distinguishing residues are highlighted in green (Zar1) or red (Zar2). The position of the peptide used to raise antibodies (33–49) is shown in horizontal blue shading and labeled above the sequences. The positions of the truncations for the N-terminal domain (aa 1–159) and the C-terminal domain (165–307) of Zar1 are shown by blue boxes. For comparison, the position of the truncation in Zar2 [9] is also shown with a blue box. Note that Zar1 and Zar2 do not have homology where the truncations were made. Armadillo, ENSDNOP00000014704, ENSDNOP00000007014; bushbaby, ENSOGAP00000020507; chimpanzee, XP_001151745; chicken, NP_001165014; cod, ENSGMOP00000008302; cow, NP_001069671, NP_001120912; dog, XP_534509; dolphin, ENSTTRP00000009993; elephant, XP_003415917; gibbon, ENSNLEP00000021119, ENSNLEP00000001223; gorilla, ENSGGOP00000007480, ENSGGOP00000015415; green anole, P-Z1L_XP_003225355, XP_003219269; human, NP_783318, NP_001130043; hyrax, ENSPCAP00000005605; marmoset, XP_002745908, XP_002749004; medaka, ENSORLP00000000308; microbat, ENSMLUP00000009433; monkey, XP_001103446; mouse, NP_777366, NP_001153165; opossum, XP_001372028, XP_001377086; orangutan, XP_002814775, XP_002824199; panda, ENSAMEP00000003206, XP_002918270; pig, NP_001123428, ENSSSCP00000009969; pika, ENSOPRP00000014016; pufferfish, CAG00418, NP_001027939; rat, NP_852050, XP_002724811; rabbit, XP_002720629; shrew, ENSSARP00000002461; sickleback, ENSGACP00000018707; tarsier, ENSTSYP00000000438; tilapia, XP_003439989, XP_003458321; wallaby, ENSMEUP00000011769; Xenopus laevis, NP_001083958 (with extra 12 amino acids identified in this study), JQ776638; Xenopus tropicalis, NP_001016947; zebrafish, NP_919362.
2.4. Custom Zar1 antibody preparation
Anti-N-Zar1 antibodies were raised in rabbit against peptides in the N-terminus (amino acids 33–49) or C-terminus (amino acids 267–286) of Zar1 and purified by peptide column (Proteintech). These peptides were in equivalent positions as the peptides we used to raise Zar2 antibodies [9]. Both rabbits that were immunized with a peptide from the N-terminal domain made antibodies suitable for western blot, but not for immunoprecipitation. In contrast, neither rabbit immunized with a peptide from the C-terminal domain made antibodies specific to Zar1. To test antibody specificity, 1.5 μg/ml of the immunizing peptide was incubated for 1 h at room temperature with the primary antibody before adding to the transfer membrane.
2.5. Rabbit reticulocyte lysate protein expression
pXen-C-Zar1 and cysteine mutants were expressed in rabbit reticulocyte lysates using TnT SP6 coupled reticulocyte lysate system (Promega), according to the manufacturer’s instructions. To confirm C-Zar1 protein expression, western blots with a GST antibody were performed.
2.6. Kd calculations
Specific complex and free probe were imaged and quantified using the Licor Odyssey software. Percentage of bound probe was calculated as specific complex / (specific complex + free probe) and plotted against concentration of protein added to EMSA binding reaction using KaleidaGraph (Synergy software).
2.7. Electrophoretic mobility shift assay (EMSA), luciferase assay, statistical analysis, oocyte isolation, culture and microinjection, western blot, analysis of RNA destabilization, and bacterial protein expression and purification
All procedures were performed as described previously [9]. Adult female X. laevis were housed and sacrificed according to internationally recognized guidelines and with the approval of the University of Colorado Denver Institutional Animal Care and Use Committee. Antibodies used for the supershift were anti-GST (Santa Cruz) and anti-Tubulin (Sigma). GST-C-Zar1, GST-C-Zar2 and GST-C-Zar2mut12 proteins were purified from Escherichia coli BL21 (DE3) (Novagen). The Mos RNA probe for EMSA was the last 50 nt of the Mos 3′ UTR including five adenylyl residues. The Mos probe was synthesized by IDT and contained a 5′ Cy5. The unrelated competitor RNA for the EMSA was the β-globin 5′ UTR from pXen [27]. The plasmid was linearized with BamHI and transcribed with SP6 mMessage mMachine to make a 154 nt RNA. Western blots were quantified with the Licor Odyssey.
3. Results
3.1. There are two Zar family proteins based on conserved amino acid changes
First, the full length Zar1 cDNA sequence was identified and isolated from Stage VI X. laevis (Xl) oocytes using 5′ RACE (rapid amplification of cDNA ends) with two different 5′ primer sets from the cDNA of two different frogs. We identified a sequence (GenBank ID: KC476498) that when translated was identical to Xl Zar1 (GenBank ID: AY283176), but with an extra 12 amino acids at the very N-terminus (MASFSEEAMDRY), which we called Zar1a (GenBank ID: BK008757). With these extra N-terminal amino acids, Xl Zar1a is 307 amino acids, the same length as Zar2b [9]. We also found a very closely related sequence by 5′ RACE that we called Zar1b (GenBank ID: KC476499) that also had an open reading frame that included these extra 12 amino acids (Supplementary Fig. S1A). The Xenopus tropicalis (Xt) Zar1 sequence (GenBank ID: NP_001016947) has 4 of these 12 extra amino acids (Fig. S1A). However, the Xt Zar1 sequence is derived from several ESTs that may not contain the very 5′ end. When Xt genomic sequence is analyzed, nucleotides with high homology to the 5′ Xl Zar1 are found. Like Xl Zar1a and Zar1b, the Xt Zar1 sequence has three potential starting methionines (Fig. S1B). The most 5′ potential starting codon is associated with the strongest Kozak sequence. When these nucleotides are translated, the Xt Zar1 N-terminal 12 amino acids are identical to Xl Zar1b (Fig. S1C). Two upstream in-frame stop codons are also conserved between Xl and Xt Zar1 suggesting that all the 5′ open reading frame has been found. All experiments using the N-terminal domain of Zar1a in this study include the extra 12 amino acids.
To analyze Zar sequences from many different species, BLAST® was used to identify amino acid sequences with homology to Xl Zar2b (GenBank ID: JQ776638) and the sequences were aligned using MacVector (Fig. 1). Sequences were retrieved only from vertebrate species ranging from fish to mammals. Like other studies, we did not find any obvious homology with Drosophila or Caenorhabditis elegans sequences. While the twelve most N-terminal amino acids of Zar1 are conserved between Xl and Xt, they are not conserved across all vertebrates. However, the newly extended N-terminus of Xl Zar1 is a more comparable length to that in other species. Two blocks of homology were observed in the Zar proteins, one block in the N-terminal domain and the other block consisting of most of the C-terminal domain. The twelve conserved cysteines noted in previous studies were observed to be invariant across all species (Fig. 1, yellow). Sequences retrieved had the following names: Zar1, Zar2, “Zar1-like”, “Zar1-like protein-like” and “similar to Zar1”. However, these sequences fell into just two groups based on conserved amino acid differences in the C-terminal domain. Amino acids identified that were Zar1-specific were highlighted in green and amino acids that were Zar2-specific were highlighted in red. In three positions there was total conservation of the differences: Y213, V230 and F241. Note that amino acid positions refer to X. laevis Zar1a, but positions in Zar2b are the same. In six positions there were up to three mismatches: N219, Q231, V276, P278, D285 and F301. There were up to 6 mismatches in two positions: D256, and M258. In one position (I220) there were up to 11 mismatches, but amino acid similarity was completely conserved between a hydrophobic amino acid (Zar1) and a polar amino acid (Zar2). These twelve amino acids were evaluated to be characteristic of distinguishing between Zar1 and Zar2 from amphibians to mammals. Fish were not as easily categorized and just six amino acid differences could distinguish between Zar1 and Zar2: Y213, V230, Q231, F241, V276 and F301. Because the C-terminal domain is the RNA-binding domain [9], these conserved amino acid changes could be predicted to influence RNA-binding characteristics.
There was also a block of homology in the N-terminal domain of Zar proteins. There were Zar family member-specific differences in three positions in this region, but the conservation was not to the same extent as in the C-terminal domain. There were no obvious protein motifs identified in this conserved N-terminal region. Because the N-terminal domain is the translational regulation domain of Zar2 [9], and because Zar1 and Zar2 are homologous in this region, it could be predicted that Zar1 and Zar2 regulate translation in a similar manner.
For experiments later in the study, Zar1 and Zar2 have been truncated to an N-terminal domain (1–159) and a C-terminal domain (165–307). Fig. 1 (blue boxes) and S1A show where the boundaries of these truncations lie. Essentially the truncation is made about half way along the poorly conserved middle section of the Zar proteins. The truncation site in Zar1 was chosen based on the position of the truncation in Zar2 [9]. This position in Zar2 was chosen because it was the site of the original cloned C-terminal fragment of Zar2 that interacted with RNA as found in a yeast three hybrid screen.
3.2. Chromosomal context classifies Zar1 versus Zar2
To support the finding that there are only two Zar family members in vertebrates, the chromosomal positions of Zar genes were analyzed. Fig. 2A shows that Zar1 genes, as defined by the conserved amino acid differences described above, are found tail-to-tail with Fryl (Furry-like), and in tandem with Slc10a4 and Slain2 in most vertebrates, from fish to mammals. Only mouse Zar1 did not fit the pattern. Mouse has a Zar1 pseudogene in synteny with Fryl, Slc10a4 and Slain2 but this pseudogene is not expressed; the expressed Zar1 is just proximal to the pseudogene on chromosome 5 [4]. Synteny for Zar2 has already been described for some species, where it is found tail-to-tail with Fry (Furry) in all species tested, and head-to-head with Brca2 in many species except in fish [6,29]. We extend these observations by showing Zar2 gene context in additional species (Fig. 2B). We found Zar2 tail-to-tail with Fry in all additional species and head-to-head with Brca2 in all additional species except in fish. In some organisms Zar1 or Zar2 protein sequences have not yet been identified, although they are expected to be present. In the chimpanzee, no Zar2 (or Zar1-like) sequence has been identified by predictive software, despite the genome being completely sequenced. As synteny analysis has been used to identify genes in animals and yeast [30–32], we used this method with the goal of identifying chimpanzee Zar2. In primates (human, gorilla, orangutan), Zar1 is found on chromosome 4 and Zar2 (Zar1-like) is found on chromosome 13. Human Zar2 sequence was queried against the chimpanzee genome using tBLASTn and hits were evaluated for identity matches and gene context. As expected, one of the hits was the previously annotated Zar1 on chromosome 4. However, a match with greater identity, including the twelve characteristic amino acids of Zar2, was found on chromosome 13 (NW_003457787.1), between Fry (Gene ID: 452524 at 10,794 bp) and Brca2 (Gene ID: 452526 at 11,728 bp). Thus, the chimpanzee Zar2 gene was identified by utilizing the conserved amino acid differences and synteny.
Fig. 2.
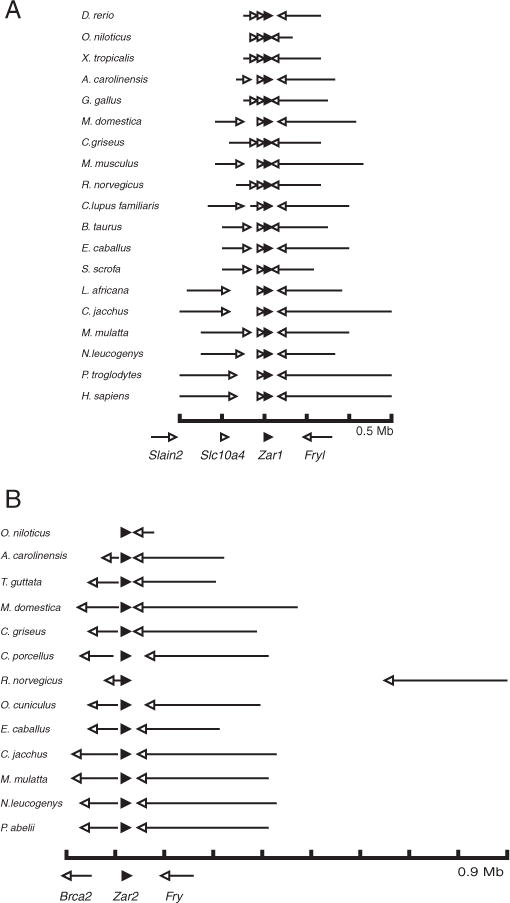
Comparison of gene context for Zar1 and Zar2. The scale shown at the bottom represents nucleotides, with each tick mark being 0.1 Mb. The direction of each gene is represented by an arrow. (A) Zar1 gene context. Zar1 gene (black arrowhead) is shown in context with Furry-like (Fryl), Slc10a4 and Slain2 (open arrowheads). (B) Zar2 gene context. Zar2 gene (black arrowhead) is shown in context with Brca2 and Furry (Fry) (open arrowheads).
3.3. Zar protein family classification
There have been Zar1, Zar1-like, Zar1-like protein-like and Zar2 sequences identified in different species. However, no criteria have been formally established that distinguish between these various classes of Zar proteins. Using the RefSeq database from NCBI (a collection of curated, non-redundant genomic DNA, transcript and protein sequences), Zar sequences were sorted into two groups based on conserved amino acid differences identified in Fig. 1 and synteny described in Fig. 2 and [6, 29]. Currently, Zar family naming is not in agreement with the distribution of these characteristic amino acids or synteny. Table 1 shows that at least seven Zar1 sequences have been misclassified as Zar1-like (Zar2) sequences. At least nine Zar2 sequences have been misclassified as Zar1-like protein-like and at least one Zar2 sequence has not yet been classified and is still referred to as hypothetical protein. One Zar2 sequence (Silurana, XP_002934069) has been misclassified as a Zar1 sequence, yet shares the 12 conserved C-terminal amino acids and syntenic context of Zar2 sequences. Note that in chicken and tilapia both Zar1 and Zar2 sequences are both called Zar1-like, but are distinct family members based onproposed naming criteria. Asthe “-like” notation is a general marker of similarity, we prefer that Zar2 is used for genes that are similar to Zar1 but contain the conserved amino acid differences as shown in Fig. 1.
Table 1.
Categorization of vertebrate Zar sequences into two groups (Zar1 and Zar2) based on sequence alignments in Fig. 1. The headings of the columns, Zar1 and Zar2 (underlined), refer to the proposed new naming of these sequences based on conserved family member-specific amino acid differences and syntenic context (where available). The names of the sequences within the columns refer to the current names of Zar sequences in the RefSeq collection of NCBI. Only one Zar1 sequence or Zar2 sequence from each species was used. Seven “Zar1-like” sequences should be classified as Zar1 rather than Zar2, and one Zar2 is misclassified as Zar1. Moreover, “Zar1-like protein-like” sequences aligned with Zar2. RefSeq names that have been misclassified are shown in bold. We propose this new naming of “Zar1” and “Zar2” for Zar-family proteins.
| Species | Common name |
Zar1
|
Zar2
|
||
|---|---|---|---|---|---|
| RefSeq name | Accession # | RefSeq name | Accession # | ||
| Xenopus laevis | African frog | Zar1 | NP_001083958 | Zar2 | NP_001153159 |
| Papio anubis | Baboon | Zar1 | XP_003898710 | Zar1-like | XP_003913788 |
| Otolemur garnettii | Bushbaby | Zar1 | XP_003794640 | Zar1-like | XP 003797650 |
| Felis catus | Cat | Zar1-like protein-like | XP_004001387 | ||
| Pan troglodytes | Chimpanzee | Zar1 | XP_001151745 | ||
| Pan paniscus | Chimp (pygmy) | Zar1 | XP 003816080 | Zar1-like | XP_003826913 |
| Gallus gallus | Chicken | Zar1-like | XP_003641256 | Zar1-like | NP_001165014 |
| Bos taurus | Cow | Zar1 | NP_001069671 | Zar1-like | NP 001120912 |
| Canis lupus familiaris | Dog | Zar1 | XP 003431884 | Zar1-like protein like | XP_534509 |
| Loxodonta africana | Elephant | Zar1-like | XP_003415917 | ||
| Nomascus leucogenys | Gibbon | Zar1 | XP_003258518 | ||
| Gorilla gorilla | Gorilla | Zar1 | XP 004038703 | Zar1-like | XP 004054397 |
| Anolis carolinensis | Green anole | Zar1-like | XP 003225355 | Zar1-like protein-like | XP 003219269 |
| Cricetulus griseus | Hamster | Zar1-like | XP 003512255 | Zar1-like protein like | XP 003504792 |
| Equus caballus | Horse | Zar1-like | XP_003364734 | Zar1-like protein-like | XP_003363240 |
| Homo sapiens | Human | Zar1 | NP_783318 | Zar1-like (Zar2) | NP_001130043 |
| Callithrix jacchus | Marmoset | Zar1 | XP_002745908 | Zar1-like | XP 002749004 |
| Macaca mulatta | Monkey | Zar1 | XP_001103446 | Zar1-like protein-like | XP_002800766 |
| Saimiri boliviensis | Monkey (squirrel) | Zar1 | XP_003933726 | Zar1-like | XP_003920405 |
| Mus musculus | Mouse | Zar1 | NP 777366 | Zar1-like | NP 001153165 |
| Monodelphis domestica | Opossum | Zar1-like | XP_001372028 | Zar1-like protein-like | XP_001377086 |
| Pongo abelii | Orangutan | Zar1 | XP_002814775 | Zar1-like | XP 002824199 |
| Ailuropoda melanoleuca | Panda | Zar1-like protein like | XP_002918270 | ||
| Sus scrofa | Pig | Zar1 | NP_001123428 | Zar1-like | XP_001929265 |
| Takifugu rubripes | Pufferfish | Zar1 | NP_001027939 | Zar1-like | XP 003976951 |
| Oryctolagus cuniculus | Rabbit | Hypothetical Protein | XP_002720629 | ||
| Tetraodon nigroviridis | Pufferfish | Zar1 | NP_001027939 | ||
| Rattus norvegicus | Rat | Zar1 | NP_852050 | Zar1-like protein-like | XP_002724811 |
| Ovis aries | Sheep | Zar1 | XP_004010112 | Zar1-like | XP_004012186 |
| Sarcophilus harrisii | Tasmanian devil | Zar1-like | XP_003761163 | ||
| Oreochromis niloticus | Tilapia | Zar1-like | XP_003439989 | Zar1-like | XP_003458321 |
| Oncorhynchus mykiss | Trout | Zar1 | NP_001118153 | ||
| Xenopus (Silurana) tropicalis | Western clawed frog | Zar1 | NP_001016947 | Zar1 | XP_002934069 |
| Danio rerio | Zebrafish | Zar1 | NP_919362 | Zar1-like | NP_001186296 |
3.4. Expression of Zar1 during Xenopus oogenesis and oocyte maturation
Next, we wanted to compare the expression of Zar1 during oogenesis and oocyte maturation so we developed custom antibodies to equivalent peptides that we used for Zar2 [9]. The N-terminal peptide (Figs. 1 and S1A) is distinct to that of Zar2 and it produced antibodies that performed well for immunoblot (Fig. 3A) but not for immunoprecipitation (data not shown). To test antibody recognition we expressed N-Zar1-MS2 in immature oocytes. The Zar1 antibody recognized a band at the same size as a band recognized by the anti-MS2 antibody (Fig. 3A, left panel). To test specificity, the antibody was incubated with the immunizing peptide before being used in the western blot. This pretreatment prevented recognition of N-Zar1-MS2 by the Zar1 antibody. A band at the expected size of endogenous Zar1 (39 kDa) was also recognized by the Zar1 antibody. Therefore, we concluded that the Zar1 antibody recognizes endogenous Zar1. A Zar1 antibody raised in a duplicate rabbit also had the same recognition pattern (data not shown). To test if the Zar1 antibody recognized Zar2, the N-terminal domains of Zar1 and Zar2 fused to MS2 coat protein were expressed in immature oocytes. Equivalent amounts of Zar1 and Zar2 fusion proteins were expressed as seen when blotted with MS2 antibodies (Fig. 3A, right panel). When the lysates were blotted with anti-Zar1 or anti-Zar2 antibodies, only the relevant fusion protein was detected, demonstrating that the Zar1 and Zar2 antibodies were specific to their individual homologs.
Fig. 3.
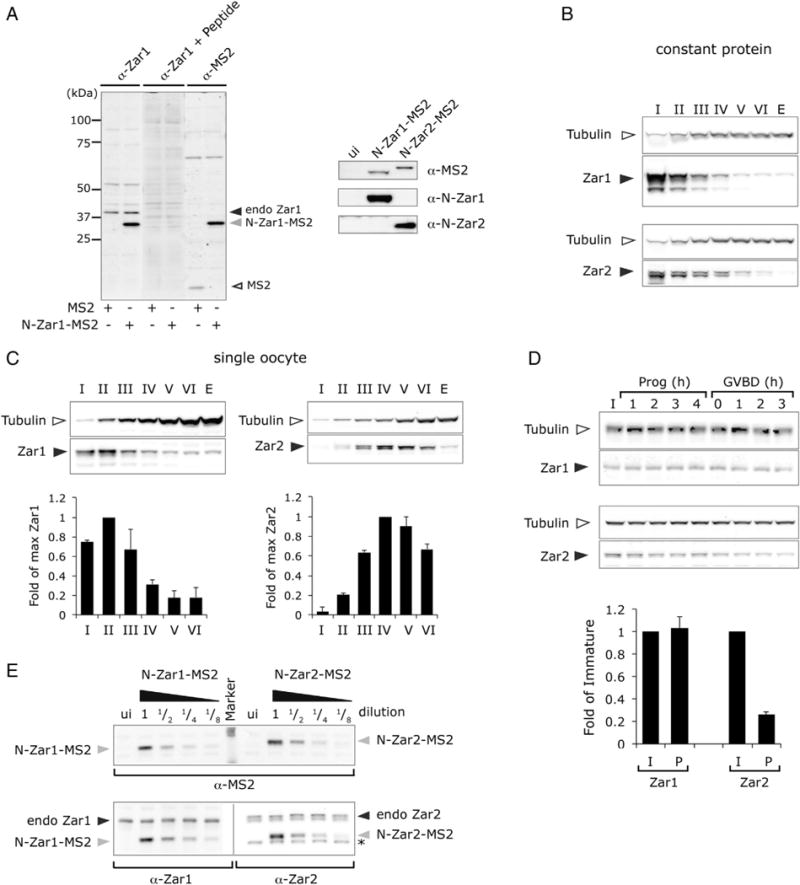
Comparison of Zar1 and Zar2 protein expression during oogenesis and oocyte maturation. (A) Left panel, western blot characterizing the Zar1 antibody used in this study. Lysates from MS2- or N-Zar1-MS2-expressing immature oocytes were analyzed by the Zar1 antibody or MS2 antibody as indicated. The immunizing peptide (+ peptide) was used to block specific antibody sites. Antibodies recognize N-Zar1-MS2 and endogenous Zar1. Right panel, western blots showing that the antibodies are specific to the different Zar family members. Immature oocyte lysates expressing the indicated fusion proteins or uninjected (ui) were analyzed by the indicated antibodies. (B, C, D) Endogenous Zar1 and Zar2 expression during oogenesis and maturation. Western blots on lysates from the same frog using Zar1 N-terminal antibody or Zar2 antibodies (solid arrowhead), and β-Tubulin antibodies (open arrowhead) as a loading control. (B) Constant total protein (3.75 μg) was loaded. (C). Pools of oocytes were lysed and protein equivalent to a single oocyte from Stages I to VI was loaded. Upper panels, representative western blots. Lower panel, quantification of western blots from 2 to 4 frogs. Results shown are means ± SD and are expressed relative to maximum expression, which was arbitrarily set to 1. (D) Upper panels, representative western blots. Pools of oocytes were lysed at different times (h) after progesterone (prog) stimulation and protein equivalent to 0.5 oocyte was loaded. GVBD, germinal vesicle breakdown, a marker of meiosis I. Lower panel, immature (I) and GVBD 3 h (P) time points were quantified from blots from 3 to 4 independent frogs and the amount of Zar protein was normalized to Tubulin. Mean ± SD was expressed relative to the amount of Zar in immature oocytes. (E) There are equivalent levels of endogenous Zar1 and Zar2 proteins in immature oocytes. Immature oocytes were injected with mRNA encoding N-Zar1-MS2 or N-Zar2-MS2 and incubated overnight. Pools of oocytes were lysed and MS2 tagged protein lysates were diluted with uninjected oocyte lysate (1/2 etc). Upper panel, equivalent amounts of exogenous N-Zar1-MS2 and N-Zar2-MS2 (gray arrowhead) were confirmed by MS2 western blot. Lower panel, endogenous Zar1 and Zar2 protein levels (solid arrowhead) relative to exogenous levels were analyzed by western blot using anti-Zar1 and anti-Zar2 antibodies. Uninjected (ui) oocyte lysates were used for comparison. Asterisk (*) indicates a non-specific band that runs just below the N-Zar2-MS2 band.
Next we used the anti-Zar1 antibody to determine Zar1 protein expression during Xenopus oogenesis and oocyte maturation. We compared this to Zar2 expression using antibodies raised to the C-terminal peptide of Zar2, which specifically recognize Zar2 and not Zar1 [9]. Both Zar1 and Zar2 were expressed throughout oogenesis. When constant levels of total protein are analyzed the highest concentrations relative to Tubulin of both Zar1 and Zar2 are in Stage I oocytes (Fig. 3B). However, when protein levels per oocyte were compared, Zar1 and Zar2 expression profiles were different. Zar1 levels reached a maximum in Stages I–III and then declined through Stages IV to VI, whereas Zar2 levels reached a maximum at Stages IV–V and then declined in Stage VI (Fig. 3C). During oocyte maturation, Zar1 levels remained constant, whereas Zar2 levels decreased (Fig. 3D). These data show that Zar1 and Zar2 expression profiles are similar, but there are some differences, which may indicate that Zar1 and Zar2 play different roles in early development. Interestingly, the Zar1 antibody recognized only one band in Stage VI oocytes and during oocyte maturation, but recognized several bands in oocytes that were in earlier stages of oogenesis. These additional bands may represent degradation products, post-translational modification, alternative splicing or non-specific cross-reacting bands. Further studies are necessary to distinguish between these possibilities.
Because Zar1 and Zar2 are both expressed during oogenesis, we wanted to compare their relative expression levels. MS2 fusion proteins were used to calibrate the amount of endogenous protein in an immature Stage VI oocyte, by comparing the levels of the endogenous Zar1 or Zar2 protein to the exogenous MS2-tagged Zar1 or Zar2 protein, and comparing the MS2-tagged proteins to each other. RNA encoding either MS2-N-Zar1 or MS2-N-Zar2 was injected into immature oocytes, which were incubated overnight to express the protein. To determine the relative amount of MS2 fusion proteins that were expressed, MS2-expressing lysates were titrated by serially diluting with uninjected lysate (to keep the amount of endogenous Zar protein constant) and the amount of MS2 fusion protein was measured by western blot with anti-MS2 antibodies. Fig. 3E (upper panel), shows that approximately equivalent amounts of MS2-N-Zar1 and MS2-N-Zar2 were loaded. Lysates were then analyzed with antibodies against the N-terminal peptide of Zar1 or Zar2 (Fig. 3E, lower panel). The levels of endogenous Zar1 or Zar2 (upper band) were compared to the MS2 fusion protein (lower band). Both endogenous Zar1 and Zar2 levels were approximately equivalent to a 1/2 dilution of their respective MS2-fusion protein. Because the MS2-N-Zar1 and MS2-N-Zar2 proteins were expressed at equivalent levels, we conclude that endogenous Zar1 and Zar2 proteins are present in immature Stage VI oocytes in approximately equal amounts.
3.5. Zar1 represses translation in immature oocytes
The N-terminal domain of Zar2 represses translation in immature oocytes [9] and there is an area of amino acid conservation in the N-terminal domains of both Zar1 and Zar2 (Fig. 1). To test if Zar1 also repressed translation, a tethered assay was used, which tethers a protein of interest to a reporter RNA through viral MS2 stem-loop RNA and coat protein interactions [9, 33]. Similar to N-Zar2-MS2 [9], the MS2 coat protein replaced the C-terminal RNA-binding domain of Zar1 (Fig. 4A). Oocytes were injected with RNA encoding MS2 fusion proteins and incubated overnight. Oocytes were then injected with a mixture of RNA reporters encoding firefly luciferase with MS2 stem-loops (fluc-2x-SL) in the 3′ UTR, and Renilla luciferase (rluc) (Fig. 4A) as a loading control, and incubated for 10 h. The amount of firefly luciferase activity was assessed and expressed a ratio relative to the amount of Renilla luciferase activity. MS2 alone did not affect accumulation of the firefly luciferase (Fig. 4B). However, both N-Zar1-MS2 and N-Zar2-MS2 repressed translation of firefly luciferase in immature oocytes in a dose-responsive fashion (Fig. 4B). More Zar1 RNA had to be injected than Zar2 RNA to see equivalent levels of MS2 fusion protein accumulation (Fig. 4D). When N-Zar2-MS2 and N-Zar1-MS2 protein levels were equivalent (10 ng and 100 ng of injected mRNA respectively), similar repression of reporter translation was observed, about 30% repression. Note that higher levels of N-Zar2-MS2 can repress to a greater extent (50%). Repression required that Zar1 be tethered to the firefly reporter as no repression was observed when stem-loops were omitted (fluc) (Fig. 4C), even though equivalent amounts of N-Zar1-MS2 protein were expressed (Fig. 4E). Repression was not due to degradation of the firefly RNA reporter as equivalent amounts of fluc-2x-SL reporter cDNA were detected by PCR in the presence of the maximum dose (100 ng) of N-Zar1-MS2 (Fig. 4F). Similar results were obtained when full length Zar1 or Zar2 was fused to MS2 (data not shown). Together these data show that Zar1 represses translation in immature oocytes.
Fig. 4.
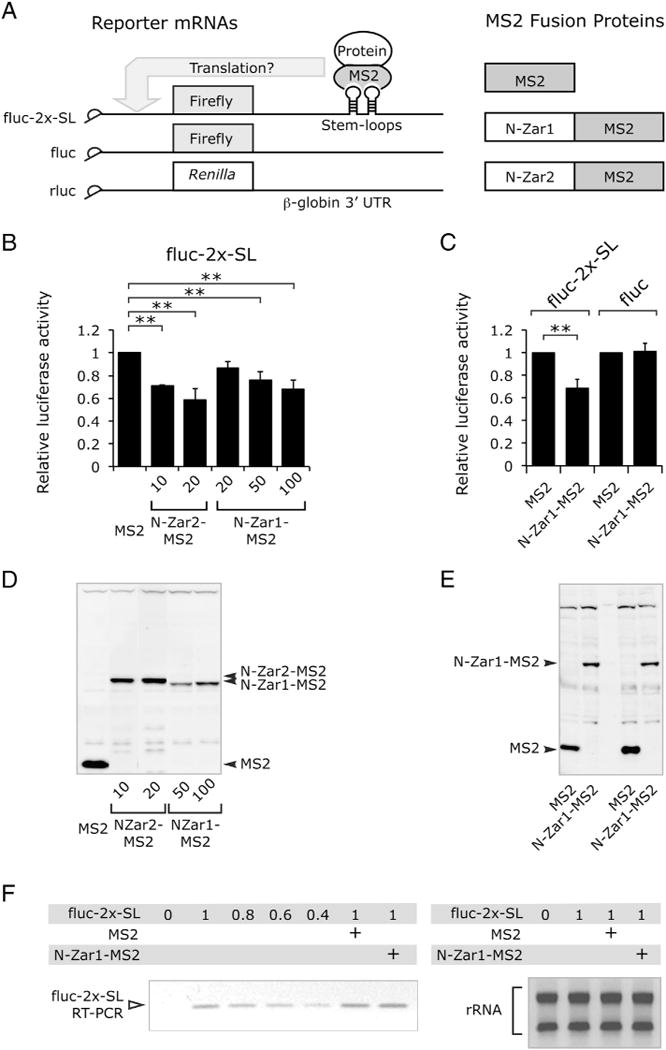
The N-terminal domain of Zar1 represses translation in immature oocytes. (A) Cartoon of the constructs used in this study. (B) Bar chart showing N-Zar1-MS2 represses translation. Oocytes were injected with RNA encoding MS2 fusion proteins and incubated overnight. RNA encoding MS2 was injected at 1 ng, other RNAs were injected as indicated. Oocytes were then injected with a mixture of fluc-2x-SL and rluc (Renilla luciferase, loading control) reporter constructs and harvested after 8–10 h. Bars show mean relative firefly luciferase activity normalized to MS2 alone [9]. Error bars represent SD and differences in mean were considered significant with p < 0.01 (**) as analyzed by one way ANOVA. Both N-Zar2-MS2 and N-Zar1-MS2 repress translation in immature oocytes (n = 3–5). (C) Bar chart showing N-Zar1-MS2 does not repress translation when it is not tethered. Oocytes were injected with RNA encoding MS2 fusion proteins and incubated for 24 h. N-Zar1-MS2 RNA was injected at 100 ng. Oocytes were then injected with a mixture of fluc or fluc-2x-SL, and rluc reporter constructs, and harvested 24 h later (n = 3). (D and E) Western blots with MS2 antibodies showing expression of MS2 fusion proteins at the end of the experiments. (F) Translation repression is not due to degradation of the RNA reporter. Left panel, semi-quantitative PCR showing that there is the same amount of fluc-2x-SL RNA in the presence or absence of 100 ng N-Zar1-MS2 in immature oocytes. The PCR was in its quantitative range as demonstrated by less product formation when 0.6× of the cDNA was added to the PCR reaction. Right panel, ethidium bromide stained gel of total RNA after extraction from oocytes showing equal recovery of rRNA prior to cDNA synthesis.
3.6. Zar1 binds to TCSs in the 3′ UTR of Wee1 mRNAs with a zinc finger
Zar1 has a highly homologous C-terminal domain to Zar2, and Zar2 binds to the TCS in the Wee1 3′ UTR [9]. Therefore, we tested whether Zar1 also binds to the Wee1 3′ UTR (Fig. 5A) by electrophoretic mobility shift assay (EMSA).The conserved C-terminal domain of Zar1 containing all the conserved cysteines (Fig. 1) was fused to GST, expressed in E. coli and purified over glutathione sepharose. The purified protein was mixed with Cy5-labeled Wee1 UTR probe and the binding assessed by retardation of the probe. Fig. 5B shows that a specific complex is formed when the binding reaction contains Zar1. This specific complex contained GST-C-Zar1, as it was supershifted by anti-GST antibodies but not by an irrelevant antibody (anti-Tubulin). Similar to the full length Zar2 [9], the full length Zar1 does not bind RNA in vitro (data not shown). To test if Zar1 was binding specifically to the Wee1 3′ UTR, competition studies were performed (Fig. 5C). Binding of Zar1 to the labeled probe was not competed by a 50-fold molar excess of unrelated and unlabeled RNA transcribed from pXen plasmid [27] that specified the 5′ UTR of β-globin (βg). To test if Zar1 bound specifically to the TCS in the Wee1 3′ UTR, unlabeled Wee1 RNA was used to compete for binding. Unlabeled wild type Wee1 RNA did compete for binding, whereas an RNA with both TCSs disrupted (mt1&2) did not fully compete for binding.
Fig. 5.
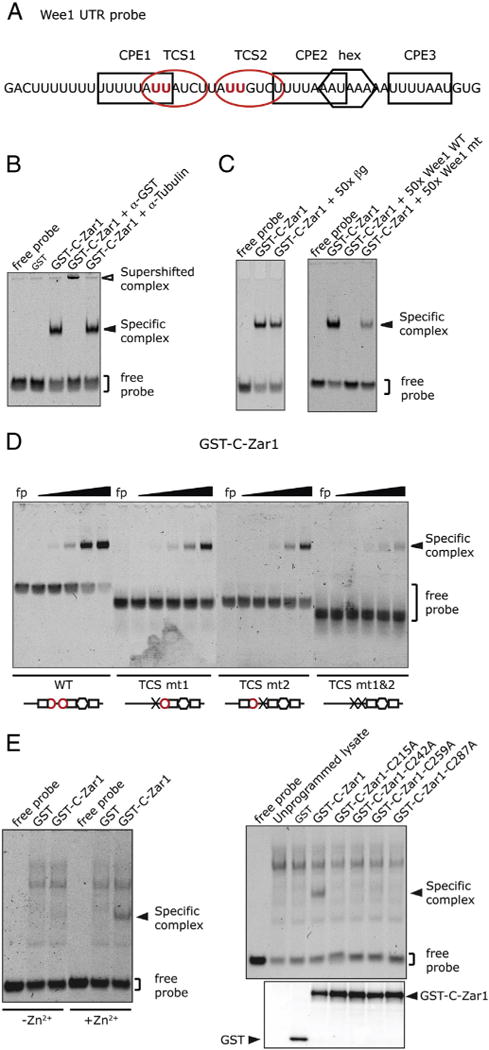
Zar1 binds to TCSs in the Wee1 3′ UTR with a zinc finger. (A) Cartoon of the Wee1 3′ UTR RNA probe used for this study. The CPEs are shown with black boxes, the TCSs are shown with red ovals, and the polyadenylation hexanucleotide is shown with a hexagon. The sites of mutation in the TCSs are marked in bold red. (B) EMSA showing that bacterially expressed GST-C-Zar1 forms specific complexes with Wee1 3′ UTR that can be supershifted. The RNA probe was incubated with GST, or GST-C-Zar1 proteins. Antibodies against β-Tubulin or GST were added where indicated. (C) EMSAs showing that specific complex formation can be competed with unlabeled RNA containing TCSs. The Wee1 RNA probe was incubated with GST-C-Zar1 protein, and a 50-fold molar excess of an unrelated RNA (βg) (left panel), or wild type (WT) or TCS-disrupted (mt) Wee1 3′ UTR (right panel) was used to compete for labeled probe binding. (D) EMSA showing that Zar1 directly binds to the TCSs in the Wee1 UTR. Wee1 probes with mutations (x) in TCS 1 (TCS mt1), TCS 2 (TCS mt2) or both TCSs (TCS mt1&2) were used. Probes were incubated with decreasing amounts of GST-C-Zar1 from 20 ng in a 4-fold dilution series. Below each gel is a diagram of the probe that was used showing which TCSs (red circles) or CPEs (black squares) were present. Mutating both TCSs markedly reduced binding of GST-C-Zar1. (E) Zar1 binds RNA with a zinc finger. Left panel, EMSA showing reduced specific complex formation in reduced Zn2+ conditions. The Wee1 probe was incubated with GST-C-Zar1-expressing reticulocyte lysate. Binding reactions were performed in buffer with (+Zn2+) or without (−Zn2+) zinc chloride. Right panel, upper, EMSA showing no specific complex formation with C-Zar1 cysteine mutations. A series of cysteine to alanine mutations was made in the C-terminal domain of Zar1 within the predicted zinc finger domain (as shown in Fig. 1). Mutant proteins were expressed in reticulocyte lysates and mixed with the Wee1 RNA probe. Right panel, lower, GST-western blot of the protein preparation showing that equivalent amounts of mutant proteins were used in the assay.
Next we tested if Zar1 bound directly to the TCS, by using mutant RNA probes (Fig. 5D). Mutating each TCS individually (mt1 or mt2) reduced binding of Zar1 to the RNA slightly, but mutating both TCSs together (mt1&2) markedly reduced Zar1 binding. Because the specific complex was seen with either of the two mutant probes (mt1 and mt2), this shows that both TCSs were able to bind Zar1. There did not appear to be two independent Zar1 binding events at both TCSs at the same time on the wild type probe, because the specific complex is the same size as on the mutant probes. There is residual binding of Zar1 on the double mutant probe (Fig. 5D), and the unlabeled double mutant RNA retained some ability to compete for labeled probe binding (Fig. 5C) suggesting that the mutation used may not have completely abolished Zar1-binding activity. The C-terminal domain of Zar2 contains a zinc finger that is important for RNA binding [9]. Because of the sequence similarities between the C-terminal domains of Zar1 and Zar2, including the invariant cysteines, we tested if Zar1 also contains a zinc finger that is important for RNA binding. For these experiments, GST-C-Zar1 was expressed in rabbit reticulocyte lysates, hence additional non-specific cross-reacting bands appearing in the gel. Reticulocyte lysates expressing GST-C-Zar1 were mixed with the Wee1 RNA probe in binding buffer containing Zn2+ or in buffer in which Zn2+ had been omitted. Zar1 bound RNA in the presence of Zn2+ but not in the absence of Zn2+ (Fig. 5E, left panel). Similar to Zar2, conserved cysteines in the C-terminal domain of Zar1 were important for RNA binding, as mutating them inhibited binding even though the mutant proteins were expressed to the same level (Fig. 5E, right panel). Together, these data show that Zar1 binds to Wee1 RNA using a zinc finger.
3.7. Zar1 and Zar2 have different RNA binding characteristics
Only 2.5–3 ng of Zar1 fusion protein was used in the EMSAs in Fig. 5, compared to 800 ng of Zar2 protein used in the previous Zar2 study [9], suggesting that Zar1 has a higher affinity for the Wee1 3′ UTR than Zar2. To examine this we titrated the amount of Zar1 and Zar2 proteins required for probe binding. Much less Zar1 protein was required to shift the Wee1 probe than Zar2 protein (Fig. 6A). Over several protein preparations the Kd for Zar1 ranged from 3.5 nM to 40 nM and the Kd for Zar2 ranged from 60 nM to 440 nM.
Fig. 6.
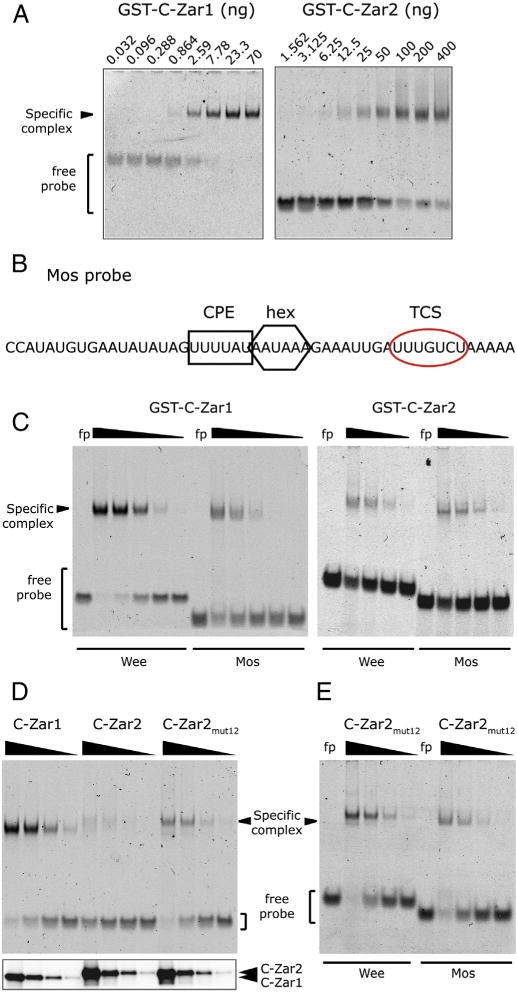
Comparison of RNA binding between Zar1 and Zar2. (A) Probe retardation in response to increasing amounts of bacterially expressed and purified Zar proteins. Left panel, GST-C-Zar1 was added to the binding reactions from 70 ng to 32 pg in a 3-fold dilution series. Right panel, GST-C-Zar2 was added to the binding reactions from 400 ng to 1.56 ng in a 2-fold dilution series. (B) Sequence of the Mos 3′ UTR probe used for this study. The CPE is marked with a black square, the polyadenylation hexanucleotide (hex) with a black hexagon, and the TCS with a red oval. (C) Differential binding of Zar1 and Zar2 to Wee and Mos 3′ UTRs. Left panel, Wee or Mos probes were incubated with decreasing amounts of GST-C-Zar1 from 100 ng in a 5-fold dilution series. Right panel, Wee or Mos probes were incubated with decreasing amounts of GST-C-Zar2 from 3 μg in a 5-fold dilution series. Zar1 binds more strongly to the Wee1 probe than to the Mos probe, while Zar2 binds to both probes with similar affinity. (D and E) Conserved amino acid differences between C-terminal domains of Zar1 and Zar2 contribute to RNA binding characteristics. (D) Upper panel, EMSA showing relative affinity of Zar proteins to the Wee1 UTR. 12 amino acids in GST-C-Zar2 were mutated to those in Zar1 as shown in Fig. 1 (C-Zar2mut12). Decreasing amounts of bacterially expressed GST-C-Zar1, GST-C-Zar2, or GST-C-Zar2mut12 from 625 ng in a 5-fold dilution series were incubated with Wee1 UTR. A representative gel of three different experiments is shown. Lower panel, western blot of the protein preparation, showing equivalent amounts of mutant proteins were used in the assay. (E) Differential binding of C-Zar2mut12 to Wee and Mos 3′ UTRs. Wee or Mos probes were incubated with decreasing amounts of GST-C-Zar2mut12 from 1250 ng in a 5-fold dilution series. Zar2mut12 binds a little more strongly to the Wee1 probe than to the Mos probe.
Putative TCSs have been identified in Wee1, PCM-1 and Mos 3′ UTRs. The TCSs are slightly different: AUUAUCU (Wee1 TCS1), AUUGUCU (Wee1 TCS2) and UUUGUCU (Mos and PCM-1 TCS) [20], prompting us to explore if Zar1 had different sequence specificities to that of Zar2. First, we looked at the data with the TCS mutant RNA probes (Fig. 5D). Both the TCS mt1 and TCS mt2 probes had similar affinity for Zar1. Our previous results showed that Zar2 also bound to TCS mt1 and TCS mt2 with similar affinity [9]. We have shown that Zar2 interacts with the Mos TCS by yeast three hybrid analyses [9]. Next we tested binding of both Zar1 and Zar2 to the Mos 3′ UTR by EMSA. The Mos TCS is the seven most 3′ nucleotides in the UTR (Fig. 6B). When we used an RNA probe that was the last 50 nt of the Mos 3′ UTR, no specific complex was formed with GST-C-Zar2 (data not shown). 3′ UTRs are normally followed by 10–75 adenylyl residues, even in maternal mRNAs that have shortened poly(A) tails. Mos mRNA specifically has poly(A) tail of 40 adenylyls in immature oocytes [34]. Accordingly, we used a probe that was the last 45 nt of the Mos 3′ UTR followed by five adenylyl residues (Fig. 6B), and as a result, both Zar1 and Zar2 formed a specific complex with this Mos probe (Fig. 6C). We verified the specificity of Zar1 binding to the Mos TCS by supershift and competition studies (data not shown). Zar1 bound to the Wee1 3′ UTR slightly better than the Mos 3′ UTR (Fig. 6C). In contrast Zar2 bound to the Wee1 and Mos 3′ UTRs with similar affinity. This indicates that Zar1 and Zar2 may have different RNA sequence specificities.
The distinguishing characteristics between Zar1 and Zar2 are the twelve conserved amino acid substitutions in the C-terminal RNA-binding domain. Therefore we hypothesized that these twelve amino acids are responsible for the difference in RNA binding characteristics. We mutated the residues in Zar2 to those found in Zar1 (Zar2mut12), with the aim of increasing the affinity of Zar2 for RNA. The Zar2 mutations were: F213Y, K219N, T220I, I230V, S231Q, L241F, A256D, Q258T, I276V, L278P, E285D, and Y301F. Note that in the highly conserved 105 C-terminal amino acids (203–307), there were 27 mismatches between Zar1 and Zar2 [9], but only the twelve most conserved differences were targeted for mutation. GST-Zar fusion proteins were expressed in E. coli and purified on glutathione sepharose. All proteins were expressed and purified at the same time for consistency. Purified proteins were mixed with the Wee1 3′ UTR RNA probe and the binding assessed by EMSA. As expected, GST-C-Zar1 had higher affinity for RNA than GST-C-Zar2 (Fig. 6D). When the twelve amino acids in Zar2 were mutated to those found in Zar1 (GST-C-Zar2mut12) the affinity increased, seen by increased complex formation with the same amount of protein (Fig. 6D). These data show that the conserved amino acid changes contribute to the higher RNA binding affinity of Zar1.
Fig. 6C shows that Zar1 binds to the Wee1 3′ UTR more strongly than it binds to the Mos 3′ UTR, whereas Zar2 binds Wee1 and Mos to the same extent. Therefore, the role of the twelve amino acids in sequence specificity was tested. Fig. 6E shows that Zar2mut12 binds to the Wee1 3′ UTR a little more strongly than to the Mos 3′ UTR. This binding profile is more like Zar1 than Zar2. These data show that not only do the twelve conserved amino changes that distinguish Zar1 from Zar2 contribute to affinity, but they also contribute to sequence specificity.
4. Discussion
Zygote arrest proteins are important in early development. Recently, we showed that X. laevis Zar2 binds to maternal mRNAs and regulates translation. Here, we show that the closely related Zar1 also binds to maternal mRNAs and regulates translation. This is the first report of a molecular function for Zar1. We also show that Zar1 and Zar2 may be functionally distinct. Whereas Zar2 is degraded during oocyte maturation, Zar1 is stable throughout oocyte maturation. Zar1 also has different RNA binding characteristics than Zar2. We show that conserved amino acid changes in the C-terminal domain contribute to the difference in RNA binding and these amino acids can be used to distinguish between Zar1 and Zar2 family members.
4.1. Zar1 represses translation in immature oocytes
Similar to Zar2, Zar1 repressed translation in immature oocytes, as shown using the tethered assay and a luciferase reporter (Section 3.5). The amount of repression conferred by Zar1 was of the same magnitude as that of Zar2 when the same amount of protein was expressed, suggesting that the N-terminal domains of the Zar proteins are functionally similar. Indeed, there is a region of conservation in the N-terminal domains of Zar1 and Zar2. The multiple alignment of Zar sequences (Fig. 1) showed few conserved family member-specific amino acid differences in that domain, suggesting that the function is conserved between Zar1 and Zar2.
Both Zar1 and Zar2 bind to the TCS in the Mos 3′ UTR (Section 3.7). The regulated translation of the Mos mRNA has long been known to be important for oocyte maturation [21], however it is still not known how the Mos mRNA is repressed in immature oocytes. The CPE combinatorial code does not account for Mos repression: the Mos mRNA does not have multiple CPEs close to the polyadenylation hexanucleotide nor a pumilio-binding site [13]. The TCS in Mos mRNA has not been formally characterized, but based on its function in the Wee1 3′ UTR, the TCS could contribute to Mos repression via Zar family proteins. Further studies are required to test this hypothesis.
4.2. Zar1 binds the TCS in maternal mRNAs
As Zar2 is a trans-acting factor of the TCS based on its ability to bind to the TCS and repress translation in immature oocytes, then by these same criteria, Zar1 is another trans-acting factor for the TCS. Protein family members often bind to the same cis-elements. For example, the CPE in the cyclin B1 3′ UTR has trans-acting factors of CPEB and CPEB4, and the pumilio binding element binds PUF family proteins, of which there are three in vertebrates [35–37].
Like Zar2, Zar1 binds to the TCS in the Wee1 3′ UTR (Section 3.6). However, Zar1 has much higher affinity for RNA than Zar2 as determined by titrations of Zar proteins in EMSAs (Section 3.7). If Zar1 recognizes the same TCS-containing mRNAs as Zar2, but with higher affinity, and there are equivalent amounts of Zar1 and Zar2 in immature oocytes (Section 3.4), this could mean that Zar1 could out-compete Zar2 for binding to maternal mRNAs. However, we already know that endogenous Zar2 binds to endogenous Wee1 and Mos mRNAs in immature oocytes [9] arguing against Zar2 being out-competed. It could be that Zar1 and Zar2 are differentially localized within the oocyte and therefore they do not compete for the same mRNAs. For example both Zar1 and Zar2 (Zar1-like) have been found in P-body-like mRNP complexes [1, 38] suggesting that they are not homogenously distributed in the cytoplasm. Alternatively, because Zar1 and Zar2 expression peaks are at different stages of oogenesis this could indicate that they might have access to different mRNAs at these stages.
Alternatively, the mRNAs that we have been using for in vitro studies may not be the best RNAs to distinguish between Zar1 and Zar2 target mRNAs. When the binding of Zar1 and Zar2 to the Wee1 and Mos 3′ UTRs (Fig. 6) is analyzed, there is evidence that suggests that Zar1 and Zar2 sequence recognition is slightly different. Zar1 binds to Wee1 3′ UTR more strongly than the Mos 3′ UTR. In contrast, Zar2 binds the two 3′ UTRs with similar affinity. Moreover, mutation of the twelve characteristic amino acids in Zar2 to those found in Zar1 resulted in a stronger binding to Wee1 than to Mos. It should be noted that these differences are quite subtle, but there is enough of a difference to suggest that Zar1 and Zar2 may have different sequence specificities. Extrapolation of this data leads to the proposal that Zar1 and Zar2 may target different mRNAs. Indeed, within protein family members there can be differential RNA binding characteristics with respect to the exact sequence of the cis element. Pumilio binding elements have a core conserved sequence, but variation in the 3′ sequence confers specificity for different PUF proteins [39]. Future studies that compare HITS-CLIP [40] from Zar1 and Zar2 will help distinguish between Zar1 and Zar2 binding the same or different repertoires of maternal mRNAs. Even though this study shows the similarities between Zar1 and Zar2, it should be noted that Zar1 and Zar2 are not redundant. For example, Zar1 knockout mice are infertile [3], so Zar2 does not compensate for the loss of Zar1 in mouse eggs and/or embryos. It is tempting to speculate that Zar1 regulates the translation of a maternal RNA that is required for activation of the zygotic genome that Zar2 does not.
In another alternative scenario, Zar2 may require accessory factors to increase binding affinity in the oocyte that are missing in in vitro studies. A recent study explored how ternary complexes could determine RNA-binding specificities [41]. Analyzing proteins that co-purify with Zar1 and Zar2 will address this possibility. Identification of these proteins will also help in the understanding of how Zar proteins regulate translation.
4.3. There are two members of the Zygote arrest family
Murine Zar1 was the original zygote arrest family member [3]. Since then, Zar family proteins have acquired the names Zar1-like, Zar1-like protein-like, Zar2 and Xzar2 ([1,2,6,29] and Table 1). From these names, it is not clear if there are multiple members of the Zar family, or if there is no nomenclature established. When we aligned Zar sequences we found that they fell into two groups only, that we prefer to be called Zar1 and Zar2 (Sections 3.1, 3.3). The two Zar family members are conserved from fish to mammals. Other evidence that supports the contention that there are only two Zar family members is their highly conserved synteny that is maintained from fish to mammals (Section 3.2). Slain2, Slc10a4, Fryl and Fry can all be used to distinguish between Zar1 and Zar2 in all vertebrates including fish. Brca2 can distinguish Zar2 in vertebrates higher than fish. Brca2 is found in fish, but it is not in the same gene context as in higher vertebrates. Fish are also harder to classify by amino acid conservation: only six positions can distinguish between Zar1 and Zar2 in fish, whereas twelve positions distinguish between Zar1 and Zar2 in higher vertebrates. The only exception to this conserved synteny that we have found is mouse, where it is conceivable that a gene duplication event leads to gradual degeneration of the original Zar1 (now the pseudogene) rather than the copy. We propose that some of the sequences noted in Table 1 are renamed to reflect their sequence homology and synteny. As noted by others [29], we also did not see Zar-like sequences in Drosophila or C. elegans. However, recent additions to genomic databases include sequences from the chordates, amphioxus and Ciona, and from the mollusk, Pacific oyster. These animals have genes that show some homology to the Zar family, albeit at low levels. As these sequences do not contain the conserved amino acids that distinguish between Zar1 and Zar2, we would like to propose the name Zar-like for sequences in these animals (in preparation).
4.4. Conclusions
This study supports the contention that Zygote arrest proteins regulate mRNA translation. The repertoire of target mRNAs and how they interact with the ribosome warrant further study. Although Zar proteins have generally similar molecular functions in the translational regulation of maternal mRNAs, they may have different roles in early development.
Supplementary Material
Acknowledgments
We thank Ms. Ling Liu for excellent technical assistance in the cloning of Zar1. We would like to thank Dr. Angus MacNicol, University of Arkansas for Medical Sciences, AR; Dr. Jeff Coller, Case Western Reserve University, OH; and Dr. Nancy Standart, University of Cambridge, UK for reagents. We also thank Dr. Aaron Johnson (Dept. Integrative Biology, UCD) and Spring 2013 Principles of Research class (UCD Biol 5705), for critical reading of this manuscript, as well as RNA Club at University of Colorado Denver Anschutz Medical Campus for intellectual and technical support. This work was funded in part by ACS RSG 0804401, NIH P20 GM104325, and University of Colorado Denver start up funds to AC. The DNA samples were sequenced by the University of Colorado Cancer Center DNA Sequencing and Analysis Core (http://DNASequencingCore.ucdenver.edu), which is supported by an NIH/NCI Cancer Center Core Support Grant (P30 CA046934).
Abbreviations
- CPE
cytoplasmic polyadenylation element
- CPEB
cytoplasmic polyadenylation element binding protein
- EMSA
electrophoretic mobility shift assay
- MBE
Musashi binding element
- TCS
Translational Control Sequence
- UTR
untranslated region
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbagrm.2013.06.001.
Contributor Information
Tomomi M. Yamamoto, Email: tomomi.yamamoto@ucdenver.edu.
Jonathan M. Cook, Email: jonathan.cook@ucdenver.edu.
Cassandra V. Kotter, Email: cassandra.kotter@ucdenver.edu.
Jefferson D. Knight, Email: jefferson.knight@ucdenver.edu.
Amanda Charlesworth, Email: amanda.charlesworth@ucdenver.edu.
References
- 1.Hu J, Wang F, Zhu X, Yuan Y, Ding M, Gao S. Mouse ZAR1-like (XM_359149) colocalizes with mRNA processing components and its dominant-negative mutant caused two-cell-stage embryonic arrest. Dev Dyn. 2010;239:407–424. doi: 10.1002/dvdy.22170. [DOI] [PubMed] [Google Scholar]
- 2.Nakajima Y, Okamoto H, Kubo T. Expression cloning of Xenopus zygote arrest 2 (Xzar2) as a novel epidermalization-promoting factor in early embryos of Xenopus laevis. Genes Cells. 2009;14:583–595. doi: 10.1111/j.1365-2443.2009.01291.x. [DOI] [PubMed] [Google Scholar]
- 3.Wu X, Viveiros MM, Eppig JJ, Bai Y, Fitzpatrick SL, Matzuk MM. Zygote arrest 1 (Zar1) is a novel maternal-effect gene critical for the oocyte-to-embryo transition. Nat Genet. 2003;33:187–191. doi: 10.1038/ng1079. [DOI] [PubMed] [Google Scholar]
- 4.Wu X, Wang P, Brown CA, Zilinski CA, Matzuk MM. Zygote arrest 1 (Zar1) is an evolutionarily conserved gene expressed in vertebrate ovaries. Biol Reprod. 2003;69:861–867. doi: 10.1095/biolreprod.103.016022. [DOI] [PubMed] [Google Scholar]
- 5.Uzbekova S, Roy-Sabau M, Dalbies-Tran R, Perreau C, Papillier P, Mompart F, Thelie A, Pennetier S, Cognie J, Cadoret V, Royere D, Monget P, Mermillod P. Zygote arrest 1 gene in pig, cattle and human: evidence of different transcript variants in male and female germ cells, Reprod. Biol Endocrinol. 2006;4:12. doi: 10.1186/1477-7827-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sangiorgio L, Strumbo B, Brevini TA, Ronchi S, Simonic T. A putative protein structurally related to zygote arrest 1 (Zar1), Zar1-like, is encoded by a novel gene conserved in the vertebrate lineage. Comp Biochem Physiol B Biochem Mol Biol. 2008;150:233–239. doi: 10.1016/j.cbpb.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 7.Michailidis G, Argiriou A, Avdi M. Expression of chicken zygote arrest 1 (Zar1) and Zar1-like genes during sexual maturation and embryogenesis. Vet Res Commun. 2010;34:173–184. doi: 10.1007/s11259-010-9343-z. [DOI] [PubMed] [Google Scholar]
- 8.Brevini TA, Cillo F, Colleoni S, Lazzari G, Galli C, Gandolfi F. Expression pattern of the maternal factor zygote arrest 1 (Zar1) in bovine tissues, oocytes, and embryos. Mol Reprod Dev. 2004;69:375–380. doi: 10.1002/mrd.20140. [DOI] [PubMed] [Google Scholar]
- 9.Charlesworth A, Yamamoto TM, Cook JM, Silva KD, Kotter CV, Carter GS, Holt JW, Lavender HF, Macnicol AM, Ying Wang Y, Wilczynska A. Xenopus laevis zygote arrest 2 (zar2) encodes a zinc finger RNA-binding protein that binds to the translational control sequence in the maternal Wee1 mRNA and regulates translation. Dev Biol. 2012;369:177–190. doi: 10.1016/j.ydbio.2012.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farley BM, Ryder SP. Regulation of maternal mRNAs in early development. Crit Rev Biochem Mol Biol. 2008;43:135–162. doi: 10.1080/10409230801921338. [DOI] [PubMed] [Google Scholar]
- 11.Colegrove-Otero LJ, Minshall N, Standart N. RNA-binding proteins in early development. Crit Rev Biochem Mol Biol. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- 12.MacNicol MC, MacNicol AM. Developmental timing of mRNA translation–integration of distinct regulatory elements. Mol Reprod Dev. 2010;77:662–669. doi: 10.1002/mrd.21191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pique M, Lopez JM, Foissac S, Guigo R, Mendez R. A combinatorial code for CPE-mediated translational control. Cell. 2008;132:434–448. doi: 10.1016/j.cell.2007.12.038. [DOI] [PubMed] [Google Scholar]
- 14.Radford HE, Meijer HA, de Moor CH. Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochim Biophys Acta. 2008;1779:217–229. doi: 10.1016/j.bbagrm.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Villalba A, Coll O, Gebauer F. Cytoplasmic polyadenylation and translational control. Curr Opin Genet Dev. 2011;21:452–457. doi: 10.1016/j.gde.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 16.Murakami MS, Vande Woude GF. Analysis of the early embryonic cell cycles of Xenopus; regulation of cell cycle length by Xe-wee1 and Mos. Development. 1998;125:237–248. doi: 10.1242/dev.125.2.237. [DOI] [PubMed] [Google Scholar]
- 17.Charlesworth A, Welk J, MacNicol AM. The temporal control of Wee1 mRNA translation during Xenopus oocyte maturation is regulated by cytoplasmic polyadenylation elements within the 3′-untranslated region. Dev Biol. 2000;227:706–719. doi: 10.1006/dbio.2000.9922. [DOI] [PubMed] [Google Scholar]
- 18.Murakami MS, Copeland TD, Vande Woude GF. Mos positively regulates Xe-Wee1 to lengthen the first mitotic cell cycle of Xenopus. Genes Dev. 1999;13:620–631. doi: 10.1101/gad.13.5.620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murakami MS, Moody SA, Daar IO, Morrison DK. Morphogenesis during Xenopus gastrulation requires Wee1-mediated inhibition of cell proliferation. Development. 2004;131:571–580. doi: 10.1242/dev.00971. [DOI] [PubMed] [Google Scholar]
- 20.Wang YY, Charlesworth A, Byrd SM, Gregerson R, MacNicol MC, MacNicol AM. A novel mRNA 3′ untranslated region translational control sequence regulates Xenopus Wee1 mRNA translation. Dev Biol. 2008;317:454–466. doi: 10.1016/j.ydbio.2008.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagata N, Daar I, Oskarsson M, Showalter SD, Vande Woude GF. The product of the mos proto-oncogene as a candidate “initiator” for oocyte maturation. Science. 1989;245:643–646. doi: 10.1126/science.2474853. [DOI] [PubMed] [Google Scholar]
- 22.Dupre A, Jessus C, Ozon R, Haccard O. Mos is not required for the initiation of meiotic maturation in Xenopus oocytes. EMBO J. 2002;21:4026–4036. doi: 10.1093/emboj/cdf400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sagata N, Watanabe N, Vande Woude GF, Ikawa Y. The c-mos proto-oncogene product is a cytostatic factor responsible for meiotic arrest in vertebrate eggs. Nature. 1989;342:512–518. doi: 10.1038/342512a0. [DOI] [PubMed] [Google Scholar]
- 24.Paris J, Richter JD. Maturation-specific polyadenylation and translational control: diversity of cytoplasmic polyadenylation elements, influence of poly(A) tail size, and formation of stable polyadenylation complexes. Mol Cell Biol. 1990;10:5634–5645. doi: 10.1128/mcb.10.11.5634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Charlesworth A, Wilczynska A, Thampi P, Cox LL, MacNicol AM. Musashi regulates the temporal order of mRNA translation during Xenopus oocyte maturation. EMBO J. 2006;25:2792–2801. doi: 10.1038/sj.emboj.7601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Charlesworth A, Ridge JA, King LA, MacNicol MC, MacNicol AM. A novel regulatory element determines the timing of Mos mRNA translation during Xenopus oocyte maturation. EMBO J. 2002;21:2798–2806. doi: 10.1093/emboj/21.11.2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacNicol MC, Pot D, MacNicol AM. pXen, a utility vector for the expression of GST-fusion proteins in Xenopus laevis oocytes and embryos. Gene. 1997;196:25–29. doi: 10.1016/s0378-1119(97)00171-6. [DOI] [PubMed] [Google Scholar]
- 28.Altschul SF. Amino acid substitution matrices from an information theoretic perspective. J Mol Biol. 1991;219:555–565. doi: 10.1016/0022-2836(91)90193-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Misra S, Sharma S, Agarwal A, Khedkar SV, Tripathi MK, Mittal MK, Chaudhuri G. Cell cycle-dependent regulation of the bi-directional overlapping promoter of human BRCA2/ZAR2 genes in breast cancer cells. Mol Cancer. 2010;9:50. doi: 10.1186/1476-4598-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.OhEigeartaigh SS, Armisen D, Byrne KP, Wolfe KH. Systematic discovery of unannotated genes in 11 yeast species using a database of orthologous genomic segments. BMC Genomics. 2011;12:377. doi: 10.1186/1471-2164-12-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jun J, Mandoiu II, Nelson CE. Identification of mammalian orthologs using local synteny. BMC Genomics. 2009;10:630. doi: 10.1186/1471-2164-10-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gilligan P, Brenner S, Venkatesh B. Fugu and human sequence comparison identifies novel human genes and conserved non-coding sequences. Gene. 2002;294:35–44. doi: 10.1016/s0378-1119(02)00793-x. [DOI] [PubMed] [Google Scholar]
- 33.Gray NK, Coller JM, Dickson KS, Wickens M. Multiple portionsof poly(A)-binding protein stimulate translation in vivo. EMBO J. 2000;19:4723–4733. doi: 10.1093/emboj/19.17.4723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheets MD, Fox CA, Hunt T, Vande Woude G, Wickens M. The 3′-untranslated regions of c-mos and cyclin mRNAs stimulate translation by regulating cytoplasmic polyadenylation. Genes Dev. 1994;8:926–938. doi: 10.1101/gad.8.8.926. [DOI] [PubMed] [Google Scholar]
- 35.Novoa I, Gallego J, Ferreira PG, Mendez R. Mitotic cell-cycle progression is regulated by CPEB1 and CPEB4-dependent translational control. Nat Cell Biol. 2010;12:447–456. doi: 10.1038/ncb2046. [DOI] [PubMed] [Google Scholar]
- 36.Igea A, Mendez R. Meiosis requires a translational positive loop where CPEB1 ensues its replacement by CPEB4. EMBO J. 2010;29:2182–2193. doi: 10.1038/emboj.2010.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quenault T, Lithgow T, Traven A. PUF proteins: repression, activation and mRNA localization. Trends Cell Biol. 2011;21:104–112. doi: 10.1016/j.tcb.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 38.Nakamura Y, Tanaka KJ, Miyauchi M, Huang L, Tsujimoto M, Matsumoto K. Translational repression by the oocyte-specific protein P100 in Xenopus. Dev Biol. 2010;344:272–283. doi: 10.1016/j.ydbio.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 39.Wickens M, Bernstein DS, Kimble J, Parker R. A PUF family portrait: 3′ UTR regulation as a way of life. Trends Genet. 2002;18:150–157. doi: 10.1016/s0168-9525(01)02616-6. [DOI] [PubMed] [Google Scholar]
- 40.Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, Darnell JC, Darnell RB. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Campbell ZT, Bhimsaria D, Valley CT, Rodriguez-Martinez JA, Menichelli E, Williamson JR, Ansari AZ, Wickens M. Cooperativity in RNA–protein interactions: global analysis of RNA binding specificity. Cell Rep. 2012;1:570–581. doi: 10.1016/j.celrep.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


