Abstract
Summary
Despite success with BRAFV600E–inhibitors, therapeutic responses in patients with metastatic melanoma are short-lived because of the acquisition of drug resistance. We identified a mechanism of intrinsic multi-drug resistance based on the survival of a tumor cell subpopulation. Treatment with various drugs, including cisplatin and vemurafenib, uniformly leads to enrichment of slow-cycling, long-term tumor-maintaining melanoma cells expressing the H3K4-demethylase JARID1B/KDM5B/PLU-1. Proteome-profiling revealed an upregulation in enzymes of mitochondrial oxidative-ATP-synthesis (OXPHOS) in this subpopulation. Inhibition of mitochondrial respiration blocked the emergence of the JARID1Bhigh subpopulation and sensitized melanoma cells to therapy, independent of their genotype. Our findings support a two-tiered approach combining anti-cancer agents that eliminate rapidly proliferating melanoma cells with inhibitors of the drug-resistant slow-cycling subpopulation.
Introduction
Malignant melanoma is a heterogeneous tumor of neuroectodermal origin that can be cured if excised at an early stage; however, once disseminated to distant organs, the median survival of patients drops below 9 months. All major cancer therapies have failed to persistently increase melanoma patients survival rates. This failure is attributed to different resistance mechanisms including increased DNA repair, overexpression of anti-apoptotic proteins such as BCL-2, and drug efflux pumps such as ABCB5 (Chen et al., 2009). Despite encouraging response rates seen with the BRAF inhibitor vemurafenib in BRAFV600E–positive individuals, relapses occur after a median duration of ~5 months following initial tumor shrinkage (Sosman et al., 2012). Several groups have identified various mechanisms of acquired therapy resistance in BRAF-targeted melanomas. Surviving melanomas re-activate pivotal networks such as the MAPK and PI3K pathways, e.g. through activation of PDGFR-β, CRAF, and IGFR1 (Nazarian et al., 2010; Villanueva et al. 2010), dimerization of aberrantly spliced BRAF isoforms (Poulikakos et al., 2011) or by secondary genetic events such as genomic amplification of COT, NRAS mutations, or the MEK1C121S mutation (Johannessen et al., 2010; Wagle et al., 2011). However, even among functionally and genetically heterogeneous tumors, common and intrinsic survival mechanisms exist. Cytotoxic agents including conventional chemotherapy, targeted therapy, or radiation most effectively eliminate rapidly dividing cells (Blagosklonny, 2005). This concept is gaining prominence in the field of cancer research with several studies reporting the enrichment of quiescent cancer stem cells after chemotherapy. For example, slow-cycling glioblastoma cells survive temozolomide treatment (Chen et al., 2012). Accordingly, if cell subpopulations can act as major drivers for tumor maintenance and mediate universal therapeutic resistance, approaches that target this phenotype could increase the efficacy of treatment regimens and reduce the risk of melanoma relapses.
We have recently demonstrated that even within highly proliferative melanomas, there is a slow-cycling cell subpopulation that is identifiable by the expression of the histone 3 K4 demethylase JARID1B (Roesch et al., 2005; Roesch et al., 2010). JARID1B (KDM5B/PLU-1/RBP2-H1) is a member of the highly conserved family of jumonji/ARID1 H3K4 demethylases which are involved in tissue development, cancer, and stem cell biology (Christensen et al., 2007; Dey et al., 2008; Yamane et al., 2007). The JARID1Bhigh slow-cycling subpopulation is required for the continuous tumor growth of melanoma; however, it does not follow a unidirectional cancer stem cell hierarchy. The JARID1Bhigh phenotype is temporarily distinct, dynamic, and can be acquired depending on the microenvironmental context (Roesch et al., 2010). In this respect, another study indicates that other types of cancers, such as breast or lung cancer, also harbor populations of quiescent cells that are drug resistant, and whose phenotypes can switch dynamically (Sharma et al., 2010). Interestingly, in the latter study slow-cycling cells were characterized by the expression of the chromatin-remodeling factor JARID1A, a homologue of JARID1B.
Considering JARID1B’s role in continuous tumor maintenance (Roesch et al., 2010), we asked, (1) whether the subpopulation of JARID1Bhigh slow-cycling melanoma cells displays lower drug susceptibility compared to the bulk of tumor cells, and (2) whether this resistant phenotype can be pharmacologically eradicated.
Results
Cytotoxic treatment of melanoma cells results in the enrichment of a surviving JARID1Bhigh slow-cycling subpopulation
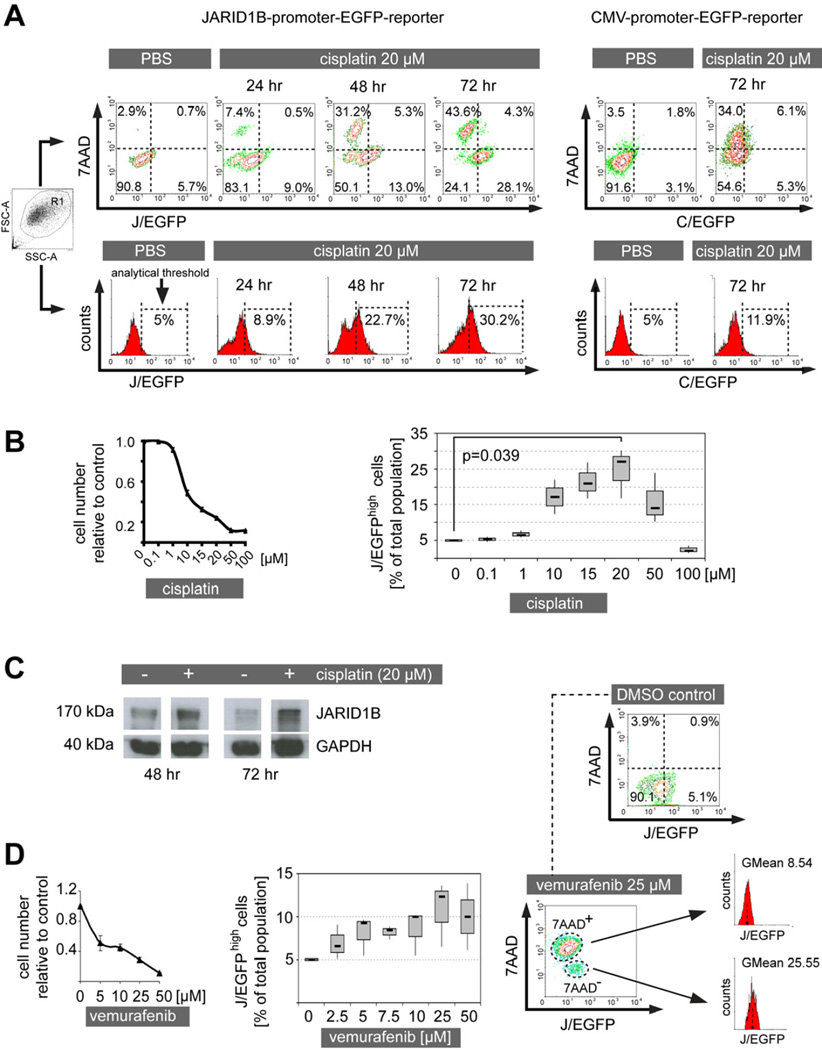
To assess whether slow-cycling melanoma cells show differences in the therapeutic response compared to the rapidly proliferating bulk, we treated melanoma cultures with various anti-cancer drugs. To discriminate between slowly and rapidly cycling subpopulations, we used a JARID1B-promoter-EGFP-reporter construct as previously described (Roesch et al., 2010). This fluorescence-based model allows both the detection of slow-cycling melanoma cells based on their expression of JARID1B and monitoring of the dynamic nature of the JARID1B phenotype while cells are alive and interacting with their microenvironment. Prior in vitro and in vivo expression studies confirmed the correlation between the endogenous expression of JARID1B and the JARID1B-promoter-induced expression of EGFP (abbreviated J/EGFP) using an analytical threshold set to the maximum 5% of the fluorescence signal (Figure S1A and Roesch et al., 2010). Cells with an EGFP signal above this threshold show the highest endogenous JARID1B levels and, thus, can be reliably selected and analyzed. Two genetically different melanoma cell lines, WM3734 and 1205Lu, were stably transduced with this reporter construct and named WM3734JARID1Bprom-EGFP and 1205LuJARID1Bprom-EGFP.
When the cell lines were treated with cisplatin at a highly toxic concentration (20 µM) for 24, 48, and 72 hr, a gradual separation of the total population emerged into live, 7AAD− and dead, 7AAD+ subpopulations (Figure 1A). With increasing incubation time, 7AAD− cells were incrementally enriched for J/EGFPhigh cells whereas 7AAD+ cells were relatively J/EGFPlow. Control cells stably expressing a CMV-promoter-EGFP-reporter construct failed to show such a separation/enrichment pattern. While the total number of cells was constantly decreasing with rising cisplatin doses, the relative number of J/EGFPhigh cells was significantly increasing in the drug resistant population (Figure 1B, Figure S1B). For example, up to 30.2% J/EGFPhigh cells were seen following 20 µM of cisplatin addition, compared to the 5% seen in vehicle controls. Immunoblotting confirmed a ~3-fold enrichment for endogenous JARID1B protein after densitometric normalization to GAPDH (Figure 1C). Vemurafenib also failed to eradicate all melanoma cells and enriched for J/EGFPhigh cells; however, to a lesser extent than seen for cisplatin (Figure 1D). Interestingly, even when high concentrations of vemurafenib were applied, a distinct subpopulation of cells remained alive (7AAD−) and displayed a ~3-fold higher fluorescence intensity for J/EGFP (GMean=25.55) than the population of vemurafenib-sensitive (7AAD+) cells (GMean=8.54, Figure 1D). The surviving melanoma cells did not change in their mutational status of BRAF as confirmed by DNA sequencing before and after treatment (not shown). In a previous publication, we have excluded the acquisition of secondary mutations for a large panel of diverse melanoma cell lines rendered resistant to BRAF inhibitors. We have tested 451Lu, Mel1617, WM983B, WM902B, SKMel28 (parental and resistant pairs, several passages) for mutations in BRAF, NRAS, TP53, PTEN, KIT, MEK1, MEK2, AKT1, CDKN2A, and CDK4 (Villanueva et al., 2010).
Figure 1. Cytotoxic treatment results in the relative enrichment of therapy resistant JARID1Bhigh melanoma cells.
(A) WM3734 melanoma cells were persistently treated with cisplatin and analyzed for J/EGFP expression by flow cytometry at the indicated time points (left). Dead cells were detected by 7AAD staining. Control cells were analyzed for C/EGFP expression (right). Depicted is one representative of at least three independently performed experiments. (B) MTT cytotoxicity assay of WM3734 cells treated with increasing concentrations of cisplatin (left). Relative percentage of the J/EGFPhigh subpopulation under escalating cisplatin treatment (right). Both panels show results from 72 hr of treatment. (C) Immunoblot of JARID1B protein expression in pre- and post-cisplatin treated WM3734 cells at the indicated time points. (D) Cytotoxicity assay and quantitation of the J/EGFPhigh subpopulation after treatment of WM3734 cells for 72 hr with escalating vemurafenib concentrations (left and middle). Comparison of the J/EGFP expression levels of 7AAD+ and 7AAD cells (right). Error bars in the cytotoxicity assays represent SD. See also Figure S1.
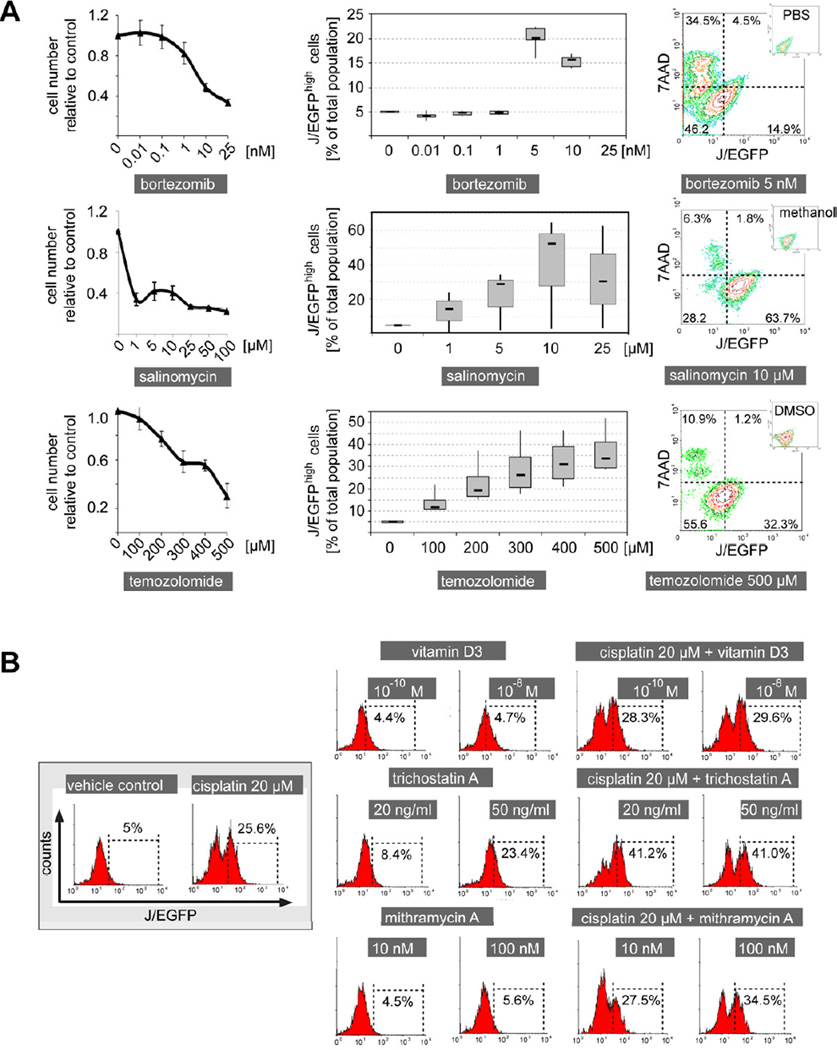
Interestingly, similar results were observed for other anti-cancer drugs such as bortezomib and temozolomide (Figure 2A). We also note that a number of cells with relatively low J/EGFP expression can survive various drug treatments indicating other potential operating mechanisms of phenotypic resistance. In contrast to its proposed effect on cancer stem cells (Gupta et al., 2009), the potassium ionophore salinomycin also showed a strong enrichment of J/EGFPhigh cells. To rule out an unspecific bias in our model, we tested vitamin D3 as a compound with a different anti-cancer mechanism (pro-differentiation; Figure 2B). Here, no significant change in J/EGFPhigh cells was seen. Next, we tested compounds that are expected to diminish the slow-cycling phenotype such as the HDAC inhibitor trichostatin A (Sharma et al., 2010) or the Sp1 inhibitor mithramycin A (Sp1 binding sites within the JARID1B promoter are indicative of a possible regulatory potential); however, both compounds failed to reduce the J/EGFPhigh subpopulation (Figure 2B). Similarly, leflunomide which was suggested to interfere with developmental processes in melanocytes and melanoma growth (White et al., 2011), did not reduce the J/EGFPhigh subpopulation in our model (Figure S2).
Figure 2. The JARID1Bhigh subpopulation of melanoma cells is resistant to a broad panel of anti-cancer drugs.
(A) Cytotoxicity assays of WM3734 cells after 72 hr of treatment with the indicated anti-cancer drugs (left). Flow cytometric determination of the relative percentage of the J/EGFPhigh subpopulation under these treatments (middle). Typical examples for the corresponding drug-associated distribution of 7AAD and J/EGFP signals (right). (B) Frequency of J/EGFPhigh WM3734 cells as measured by flow cytometry after incubation with the indicated compounds over 72 hr. Error bars represent SD. See also Figure S2.
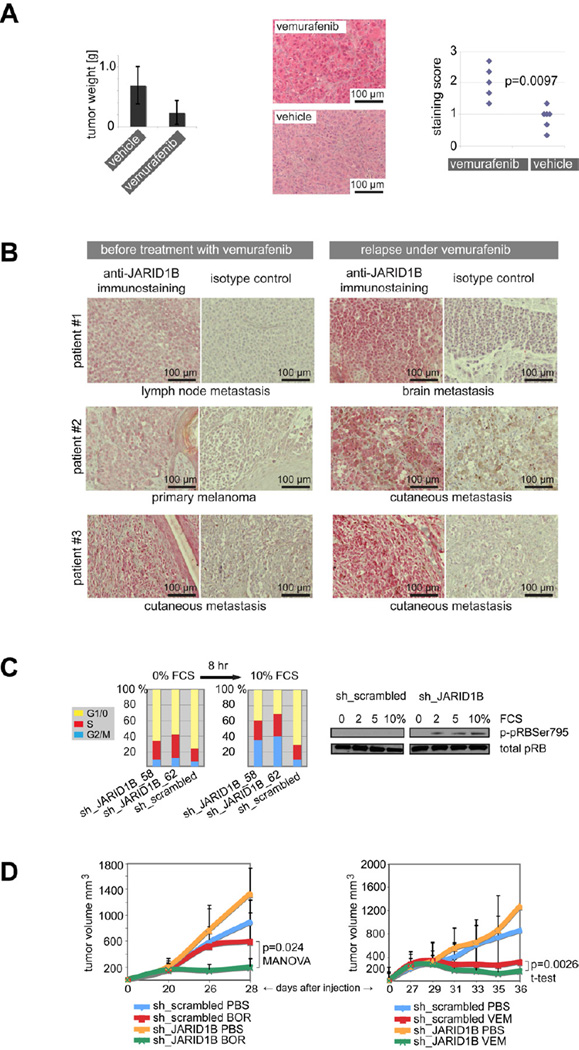
The in vivo relevance of our observations was confirmed in WM3734 cells xenografted to NSG mice and treated with vemurafenib (Figure 3A). While tumors decreased in size due to treatment, the number and staining intensity of resistant JARID1Bhigh cells significantly increased. Importantly, a clear increase in JARID1B expressing cells could be observed in 3 out of 4 matched pairs of patients’ melanomas that relapsed under vemurafenib (Figure 3B). Together, these data indicate a universal resistance of the slow-cycling JARID1Bhigh melanoma subpopulation against various anti-cancer agents.
Figure 3. The JARID1Bhigh subpopulation is enriched in melanomas resisting therapy and knockdown of JARID1B leads to increased drug sensitivity.
(A) NSG mice xenografted with WM3734 melanoma cells were treated with vemurafenib or vehicle (n=7). Tumor weights were assessed after 11 days of treatment (left). Tumor residues were immunostained for JARID1B expression (middle) and nuclear staining signals of each tumor were scored at 40x magnification in 3 representative fields of vision (right, two-sided Wilcoxon two-sample test). (B) Immunstaining of JARID1B in matched pairs of melanoma samples before and relapse under vemurafenib treatment from three melanoma patients. (C) Two different JARID1B knockdown clones (sh_JARID1B_58 and 62) of WM35 melanoma cells and the sh_scrambled control were starved for 4 days at 0% FCS and subsequently stimulated by 10% FCS for 8 hr. The bar graph shows one example of five independent experiments of propidium iodide-based cell cycle analysis (left). The immunoblot shows pRB phosphorylation (right). (D) NSG mice were xenotransplanted with WM3734 melanoma cells stably knocked down for JARID1B (sh_JARID1B) or with control cells (sh_scrambled). Each mouse received 104 cells (n=8). After tumors reached an average volume of ~200mm3, mice were treated with bortezomib (left, BOR), vemurafenib (right, VEM), or vehicle (PBS). Tumor volumes were measured at the indicated time points. Error bars represent SD. See also Figure S3.
Knockdown of JARID1B leads to increased in vivo sensitivity to anti-melanoma treatment
Previous studies suggested that the abrogation of the slow-cycling phenotype by JARID1B-specific knockdown leads in vitro and in vivo to increased cell proliferation followed by melanoma exhaustion (Roesch et al., 2010). To quantify the shifts in cell cycle progression depending on the presence or absence of JARID1B, we performed cell cycle experiments in WM35 melanoma cells stably knocked down for JARID1B (Figure 3C and S3). WM35 were chosen because changes in cell cycle of this low tumorigenic cell line could be better assessed compared to lines with high doubling rate. Influences of cell densities and growth factors were considered. Cells were seeded at different densities and serum-starved at 0% FCS for 4 days to synchronize the cell cycles. Under these conditions, knockdown of JARID1B led to an increase in S phase cells when compared to the control. Following stimulation with 2%, 5%, and 10% FCS over 4, 8, 16, and 24 hr, JARID1B knockdown cells shifted to G2/M. The most consistent changes in the cell cycle were seen after 8 hr of 10% FCS stimulation. Across five independently performed experiments, we saw an average decrease in G1 phase cells in JARID1B knocked down cells by ~20% [ranging from ~5% to 40% (the latter experiment is depicted in Figure 3C)], while S phase cells increased by ~6% (~2% to 12%), and G2/M phase cells by ~14% (~1% to 31%). As predicted by previous studies showing JARID1B/pRB-dependent cell cycle control (Roesch et al., 2006), JARID1B knockdown was accompanied by an increase in pRB phosphorylation at the JARID1B-specific phosphorylation site Ser795 (Figure 3C). Consequently, we asked if the inhibition of JARID1B could affect the therapeutic responsiveness of melanoma cells to anti-cancer drugs while releasing them from their slow-cycling phenotype. Indeed, stable knockdown of JARID1B in WM3734 melanoma cells xenografted to NSG mice led to increased cell proliferation accompanied by a significant sensitization to different treatments (Figure 3D). JARID1B knockdown plus bortezomib showed an additive effect, improving the moderate anti-melanoma effect of bortezomib to an almost complete stasis of tumor growth. When we combined JARID1B knockdown with vemurafenib, which by itself dramatically prevents melanoma growth, we still observed a decrease in the average tumor volume below that of the treatment-starting volume. Because of the biological variability of tumor sizes this effect was not significant in the MANOVA, but an analysis of the change in the logarithm of volume per unit time by t-test had significantly different rates. In sum, these results support the hypothesis that targeting the slow-cycling cell phenotype can sensitize melanoma to anti-cancer approaches including chemotherapeutics and molecular targeted drugs.
Dynamics of the therapy-resistant JARID1Bhigh phenotype
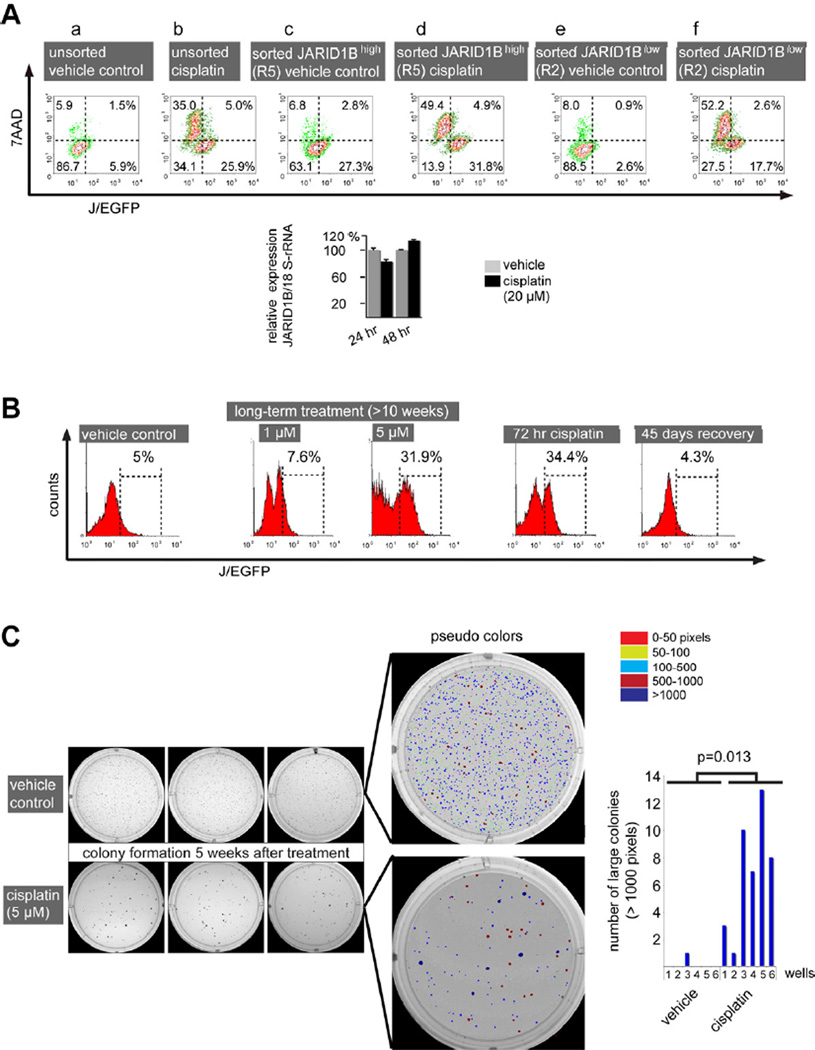
According to our previous observations, the progeny of both J/EGFPhigh and J/EGFPlow cells can dynamically interconvert into the opposing phenotype, indicating a flexible system that is dependent on the microenvironmental context (Roesch et al., 2010). Thus, we asked (1) if the newly developing progenies of initially sorted J/EGFPhigh or J/EGFPlow cells show a differential drug response and (2) if the observed enrichment of J/EGFPhigh cells after anti-cancer treatment results from selection of J/EGFPhigh cells or induction of J/EGFPhigh expression. In regular culture, spontaneous phenotype interconversion usually becomes visible within a few days after FACS sorting of the two populations. For example, 69.9% of FACS-sorted J/EGFPhigh cells turned negative 72 hr after separation (Figure 4Ac, upper left plus lower left quadrant), while in total 3.5% of the progeny of sorted J/EGFPlow cells became J/EGFPhigh (Figure 4Ae). When sorted cells were treated with 20 µM cisplatin for 72 hr, we saw no substantial enrichment for J/EGFPhigh cells in the progeny of initially J/EGFPhigh cells indicating that no additional JARID1B-expression cells were induced (summarized upper and lower right quadrants, Figure 4Ac vs. 4Ad). Interestingly, most newly developed J/EGFPlow cells became 7AAD+ under cisplatin treatment (Figure 4Ac vs. d, upper and lower left quadrants). The apparent preference for JARID1B induction in the progeny of J/EGFPlow cells (Figure 4Ae vs. f) was counteracted by an increased rate of cisplatin-induced cell death in cells derived from the J/EGFPlow population. Manual counts revealed a significantly lower number of surviving cells in the progeny of sorted J/EGFPlow cells compared to the progeny of J/EGFPhigh cells [p<0.05 for 5 µM and 10 µM cisplatin (data not shown)]. Together with the results from consecutive measurements of JARID1B mRNA under cisplatin treatment that did not show transcriptional upregulation (Figure 4A), we concluded that the major mechanism for the enrichment of J/EGFPhigh cells following therapy is a selection of pre-existent JARID1Bhigh slow-cycling cells. However, to a lesser extent JARID1B induction may also occur, e.g. shifting cells with intermediate JARID1B expression to higher levels.
Figure 4. Dynamics of the therapy resistant JARID1Bhigh phenotype.
(A) WM3734JARID1Bprom-EGFP cells were FACS-sorted according to their J/EGFP expression and the isolated subfractions were cultured as adherent monolayers over night. After subsequent treatment for 72 hr, the frequencies of J/EGFPhigh and J/EGFPlow cells were again quantified by flow cytometry. Dead cells were detected by 7AAD staining. JARID1B mRNA expression was quantified by QPCR. (B) Flow cytometric detection of J/EGFPhigh WM3734 cells after prolonged incubation with different concentrations of cisplatin and after drug recovery. (C) Colony forming assay of WM3734 cells that underwent cytotoxic pulse treatment prior to seeding. Cells were treated for 72 hr with cisplatin or vehicle. Surviving cells were subsequently embedded into soft agar (2000 cells/well). Colony sizes and numbers were digitally quantified and color-coded with dark blue colonies representing large colonies above 1000 pixels. Shown are results from two independent experiments, each performed in triplicate. Error bars represent SD.
When cells were long-term challenged with cisplatin, the J/EGFPhigh subpopulation remained constantly elevated over several weeks in a dose-dependent manner. The cisplatin-induced elevation of J/EGFPhigh cells was fully reversible following drug withdrawal (Figure 4B). In line with the concept of dynamic phenotype switching (Roesch et al., 2010), drug-resistant cells that are enriched for JARID1B can regain the original cell distribution and J/EGFP-ratios after drug removal. Moreover, cisplatin-surviving JARID1B-enriched cells gave rise to fewer but significantly larger, i.e. rapidly growing, 3D colonies in soft agar compared to untreated cells (Figure 4C). This observation is reminiscent of earlier clonogenicity and single cell dilution assays showing that untreated FACS-isolated J/EGFPhigh cells give rise to a rapidly-proliferating progeny with elevated repopulation capacity when compared to JARID1Blow cells (Roesch et al., 2010).
Bioenergetic metabolism plays an important role in the survival of slow-cycling melanoma cells
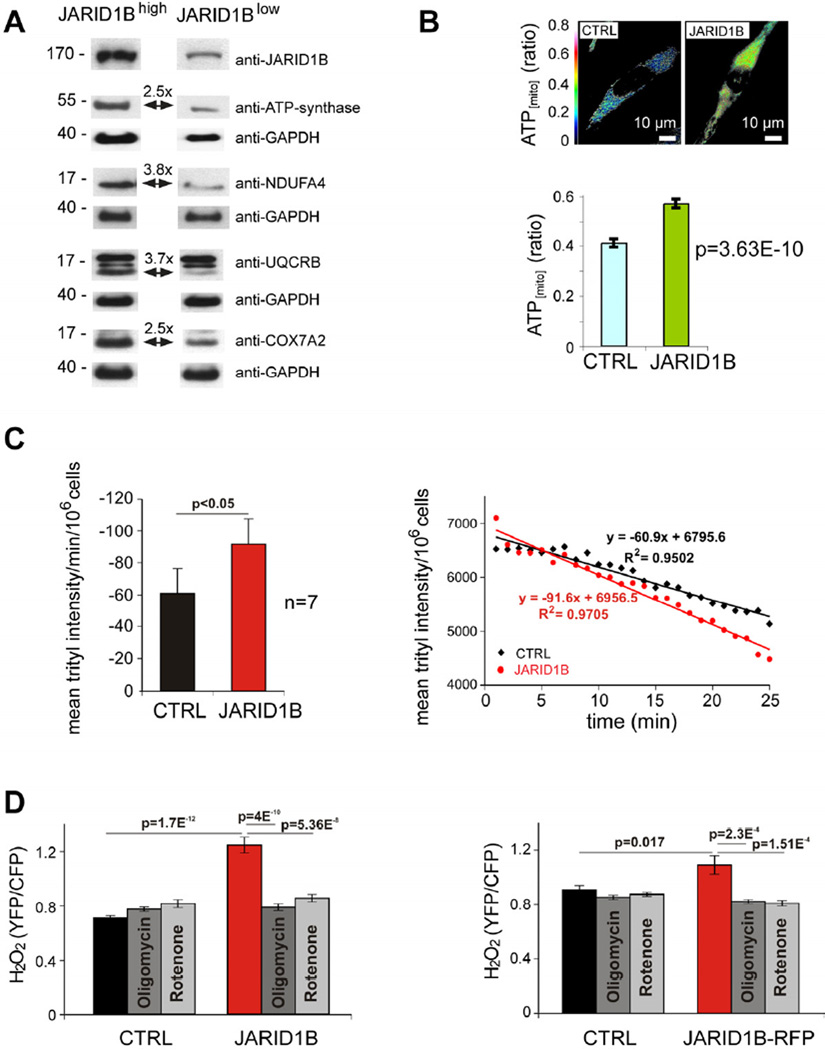
We next designed experiments to unravel druggable targets that are specifically activated in slow-cycling melanoma cells. Since global gene expression screening with cDNA microarrays did not provide significant hit candidates in FACS-sorted J/EGFPhigh vs. J/EGFPlow or JARID1B knockdown vs. control cells (not shown), we performed quantitative proteome profiling. Following our established protocols for isolation of slow-cycling melanoma cells (Roesch et al., 2010), we co-stained WM3734JARID1Bprom-EGFP cells with the PKH26 dye, then FACS-isolated the label-retaining and J/EGFPhigh subpopulation vs. non-label-retaining J/EGFPlow cells after a period of 4 weeks of cell divisions. Prior to sorting, the cells were grown as melanospheres to attain a more stringent J/EGFP phenotype as previously reported (Roesch et al., 2010). Tryptic digests of the cell lysates were analyzed by liquid chromatography-mass spectrometry/mass spectrometry. Label-free computational quantitation identified 423 proteins with significant expression changes between label-retaining J/EGFPhigh cells and non-label-retaining J/EGFPlow cells with a p-value <0.001. Unexpectedly, neither common cancer target nor drug-resistance-related protein could be detected by this approach. Instead, we found a differential regulation of a number of mitochondrial proteins with particular functions in bioenergetic metabolism (Table S1). Many of the upregulated proteins play a major role in cell respiratory electron transport (oxidative phosphorylation, OXPHOS) such as NADH dehydrogenase (complex I of the mitochondrial electron transport chain), ubiquinol cytochrome c reductase (complex III), cytochrome c oxidase (complex IV), and ATP-synthase. In contrast, glycolytic hexokinase I and II were downregulated. Immunoblotting of lysates from J/EGFP-sorted cells confirmed upregulation of ATP synthase, NADH dehydrogenase, ubiquinol-cyctochrome C reductase-binding protein, and cytochrome c oxidase in J/EGFPhigh cells (Figure 5A). WM3734 cells transiently overexpressing JARID1B had a higher mitochondrial energy production compared to the mock control (Figure 5B). Accordingly, JARID1B-overexpressing WM3734 cells consumed ~ 50% more oxygen than control cells (Figure 5C). Treatment with different concentrations of oligomycin and rotenone severely reduced the oxygen consumption by up to 94%, indicating a major role for mitochondria in this process (not shown). Because high mitochondrial electron transfer activity can lead to increase in superoxide and hydrogen peroxide (H2O2) production, we analyzed the H2O2 level. 60–72 hours after transfection, JARID1B-overexpressing cells generated 76% more H2O2 than control cells (Figure 5D). The H2O2 increase was completely abolished by treatment with oligomycin or rotenone. These results suggest that redox signaling may play an important role for JARID1Bhigh cells. Furthermore, high oxygen consumption can ultimately lead to hypoxic conditions, which is known to favor the JARID1Bhigh slow-cycling phenotype (Xia et al., 2009; Roesch et al., 2010).
Figure 5. Elevated mitochondrial bioenergy metabolism in the JARID1Bhigh subpopulation.
(A) Immunoblotting of mitochondrial respiratory enzymes in FACS-sorted J/EGFPhigh and J/EGFPlow WM3734 cells. Fold changes were normalized to GAPDH. (B) Relative mitochondrial ATP levels in control (CTRL) or JARID1B-transfected WM3734 cells were measured using the ATP sensor protein ATEAM. Representative ATEAM ratio images (top) and summarized ATEAM ratio values in control (n=72) and JARID1B-transfected (n=47) cells (bottom). (C) Mean oxygen consumption of control- (black) or JARID1B-transfected (red) WM3734 cells presented as decline of trityl radical intensity per minute per 106 cells (left). Mean oxygen concentration in presence of control or JARID1B-transfected WM3734 cells presented as trityl intensity per 106 cells. Each data point is an average of 7 independent experiments. (D) Relative H2O2 levels were measured in JARID1B-transfected WM3734 cells using the H2O2 sensor protein Hyper. Columns represent averaged Hyper ratio values of control- (black, n=71) or JARID1B-transfected cells (red, n=48). To test the role of mitochondria as source of H2O2, cells were treated with 1 µM oligomycin (dark gray) or 1 µM rotenone (light gray). Numbers of cells averaged were: CTRL plus oligomycin (n=50), CTRL plus rotenone (n=42), JARID1B plus oligomycin (n=45) and JARID1B plus rotenone (n=52). In a confirmation experiment with WM3734 cells transfected with pCAGGS-JARID1B-IRES-RFP (right), positively transfected cells were selected for analysis based on the expression of RFP. Error bars represent SEM. See also Table S1.
Targeting the mitochondrial respiratory chain inhibits the JARID1Bhigh subpopulation and reveals long-term effects against melanoma cells
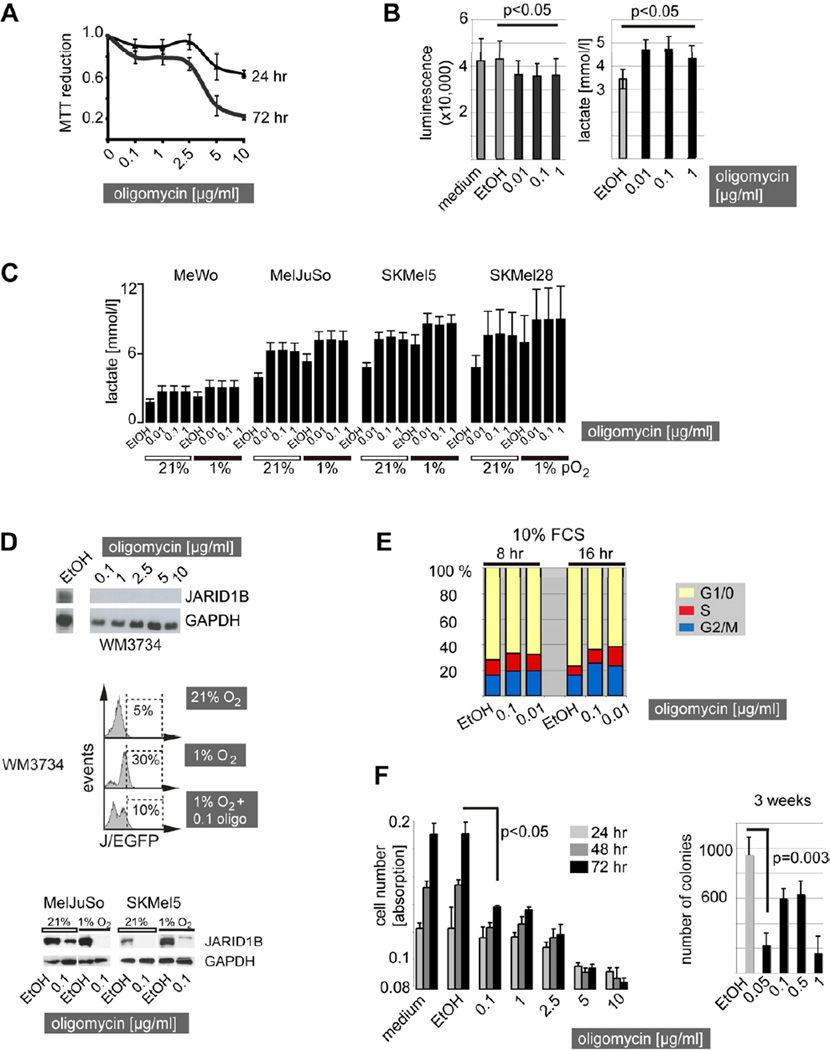
Inhibition of the mitochondrial respiratory chain has been reported to exert tumor suppressive effects in different types of cancers including neuroectodermal glioblastoma cells (Hao et al., 2010). Since enzymes of almost all respiratory chain complexes were found differentially regulated in slow-cycling J/EGFPhigh cells (Table S1), we explored representative inhibitory tool compounds such as (i) the ATP-synthase inhibitors oligomycin and Bz-423, (ii) the NADH dehydrogenase (complex I) inhibitors rotenone and phenformin, and (iii) antimycin A which inhibits the ubiquinol cytochrome c reductase in a series of preliminary cytotoxicity assays (not shown). Here, ATP-synthase and complex I inhibitors revealed the most prominent results; thus, these were selected for subsequent studies. High concentrations of oligomycin were reported to exert unspecific effects in eukaryotes due to the uncoupling of the respiratory chain (Charton et al., 2004). Therefore, MTT assays were used to estimate the minimal concentration of oligomycin that could cause mitochondrial uncoupling in our cell lines since reduction of the tetrazolium compound MTT is dependent on intact mitochondrial electron transport (Grazette et al., 2004). We observed an abrupt decrease in MTT reduction above an oligomycin concentration of 2.5–5 µg/ml independent of the incubation time and the genetic background of all cell lines tested (Figure 6A, depicted are WM3734 as an example). These results are in line with oligomycin concentrations that were reported by others to specifically block oxidative phosphorylation (Hao et al., 2010). Thus, we considered oligomycin concentrations below 2.5 µg/ml to work through the specific inhibition of the ATP-synthase and not through mitochondrial uncoupling.
Figure 6. Inhibition of the mitochondrial ATP synthase inhibits the JARID1Bhigh subpopulation and reveals long-term melanoma cell suppressive effects.
(A) MTT reduction curve of regularly cultured WM3734 melanoma cells after oligomycin treatment. (B) Total cellular ATP levels were luminometrically determined in WM3734 cells after 24 hr of treatment with oligomycin at the indicated concentrations (left). Lactate production was measured in WM3734 cell culture supernatants after 24 hr of oligomycin. Ethanol (EtOH, diluted 1:1000) was used as vehicle control. Shown are means of three independently performed experiments. (C) Lactate measurement in a panel of genetically diverse melanoma cell lines under normoxic vs. hypoxic culture conditions after 24 hr of treatment. (D) Immunoblot of JARID1B protein expression in WM3734 cells treated with oligomycin for 72 hr at the indicated concentrations (top). Frequency of J/EGFPhigh WM3734 cells under 72 hr of hypoxia vs. hypoxia plus oligomycin (oligo) treatment as determined by flow cytometry (middle). Immunoblots of endogenous JARID1B expression in MelJuSo and SKMel5 melanoma cells under hypoxia and hypoxia plus oligomycin treatment (bottom). (E) WM3734 cells were starved at 0% FCS and then released by 10% FCS for 8 and 16 hr. The bar graph shows one example of three independent experiments of propidium iodide-based cell cycle analysis. Dead cells (SubG1) were gated out. (F) Short-term effects of increasing concentrations of oligomycin on total WM3734 cell numbers as determined by crystal violet assays (left). Long-term growth reduction as measured by 3D soft agar colony formation assays (bi-daily treatment, right). Error bars represent SD. See also Figure S4.
We next asserted that low-dose oligomycin still leads to functional and significantly measurable effects such as a decrease in ATP production or increase in lactate secretion in WM3734 cells (Figure 6B), and subsequently confirmed this for a panel of genetically different melanoma lines under normoxic and hypoxic conditions (Figure 6C). The detected changes in ATP and lactate were comparable to those measured by others for a number of different cell types (Hao et al., 2010). Interestingly, ATP-synthase inhibition was accompanied by a complete loss of endogenously expressed JARID1B protein after 72 hr of oligomycin treatment (Figure 6D). Also, the enrichment of JARID1Bhigh cells usually seen after hypoxic treatment was inhibited by low-dose oligomycin.
Due to the evolutionary link between non-proliferative and OXPHOS-dependent cell phenotypes (Berridge et al., 2010; Jose et al., 2011; Vander Heiden et al., 2009), we next asked if inhibition of mitochondrial ATP-production also influences the cell cycle of melanoma cells. In fact, even very low-doses of oligomycin (0.01 µg/ml) caused a measurable decrease in the percentage of cells in G1/0 phase and an increase in the percentage of cells in S- and G2/M-phase compared to vehicle controls. To optimize the readout, cells were serum-starved for 4 days and subsequently stimulated with FCS for 8 and 16 hr (Figure 6E). After 24 hr, cell cycle phases were again equal in oligomycin and vehicle control-treated cell groups (not shown). Despite the increase in proliferative cell cycle phases, we found a relative decrease in overall cell numbers after oligomycin treatment compared to controls (Figure 6F). A likely explanation for this paradox is that simultaneously to (or after) the acceleration of the cell cycle, a considerable portion of the cells is undergoing cell death as indicated by independently performed 7AAD staining and subsequent flow cytometric analysis (e.g. ~20% dead cells after 72 hr of 0.1 µg/ml oligomycin, Figure S4A). Short-term analysis of oligomycin’s effect on total cell numbers showed that the cell population slightly expands at low doses (Figure 6F, left panel), whereas high doses starting from 5 µg/ml caused persisting or decreasing cell numbers, possibly due to highly toxic mitochondrial uncoupling. The full effect of the anti-tumor potential of low-dose oligomycin became visible in long-term colony formation assays. Here, a significant reduction in colony growth was seen after 3 weeks of treatment (Figure 6F, right panel), which is reminiscent of the results previously reported for JARID1B knockdown (Roesch et al., 2010). Short-term treatment of WM3734 cells with the ATP-synthase inhibitor Bz-423 and the complex I inhibitors rotenone and phenformin confirmed these findings (Figure S4B, S4C, and S4D). In conclusion, this suggests that pharmacological targeting of the mitochondrial respiratory chain can inhibit the JARID1Bhigh slow-cycling subpopulation and reveal long-term suppressive effects in melanoma cells.
Inhibition of the mitochondrial respiratory chain overcomes intrinsic multi-drug resistance of melanoma
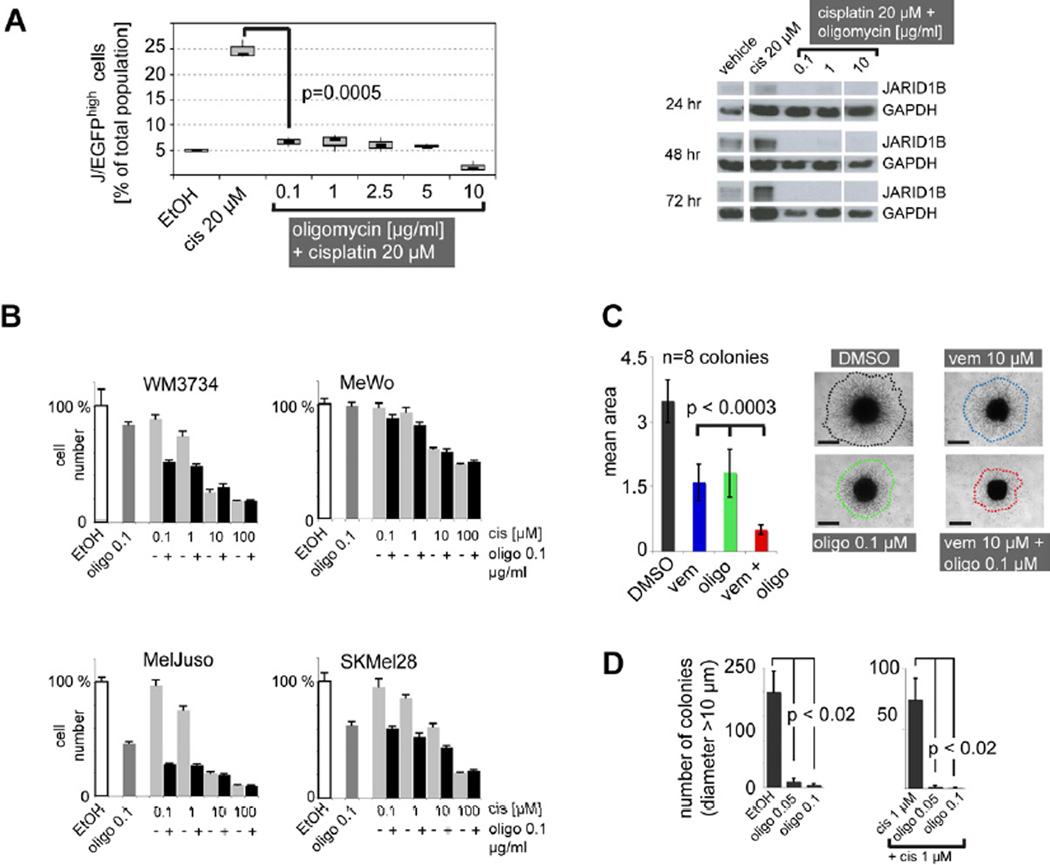
We asked if inhibition of the mitochondrial respiration could counteract the intrinsic multi-drug resistance mediated by slow-cycling melanoma cells. Flow-cytometric quantitation of J/EGFP levels in WM3734 cells (Figure 7A) and 1205LU cells (Figure S5A) revealed a significant blockade of the JARID1Bhigh subpopulation when cells were treated with the combination of cisplatin plus oligomycin. Similar results were seen for Bz-423, which also acts as an ATP-synthase inhibitor (Figure S5B). Even treatment with extremely low doses of oligomycin (0.001 µg/ml,Figure S5C) still blocked the enrichment of J/EGFPhigh cells. At 0.001 µg/ml, single agent oligomycin did not induce substantial cell death as measured by 7AAD flow cytometry in WM3734 cells (Figure S4A); however, when combined with cisplatin the overall number of dead cells strongly increased (Figure S5C). Immunoblots revealed a complete loss of endogenous JARID1B under oligomycin co-treatment overcoming the cisplatin-induced enrichment of JARID1B protein after 24, 48, and 72 hr (Figure 7A). Cytotoxicity assays confirmed that the combination with oligomycin sensitizes melanoma cells to cisplatin treatment, but depending on the cell line used the contribution of single agent oligomycin to cell killing can vary (Figure 7B). MeWo cells, the melanoma cell line with the lowest oligomycin response in lactate secretion (Figure 6C), were not sensitized to cisplatin indicating that the combination with oligomycin is not universally cytotoxic but instead, responses match our mitochondrial activity-based hypothesis. The tumor suppressive potential of combinatorial ATP-synthase inhibition was further demonstrated in 3D collagen-embedded spheroids of 1205Lu cells, a model which more readily mimics the melanoma microenvironment. The combination of oligomycin plus vemurafenib more potently inhibited cell invasion into collagen than each drug alone (Figure 7C, p<0.0003). Notably, treatment of WM3734 cells with a single pulse of low-dose oligomycin plus cisplatin over 72 hr substantially reduced long-term growth in soft agar and the number of 3D-colonies remained at almost undetectable levels even after 5 weeks of culture in drug-free medium (Figure 7D). This effect reached statistical significance with p<0.02 when large colonies above a diameter of 10 µm were counted. Consistent with the observation that significantly larger colonies grew from cisplatin pre-treated cells (that were enriched for the J/EGFPhigh subpopulation, Figure 4C), these results indicate that (i) adding mitochondrial inhibitors to common anti-cancer treatment significantly diminishes the number of cells with high repopulation and invasion potential and (ii) that even a relatively short-term treatment can yield a sustained anti-tumor effect.
Figure 7. The combination of anti-cancer agents plus inhibition of the mitochondrial ATP-synthase overcomes the enrichment of the JARID1Bhigh subpopulation in vitro.
(A) Relative percentage of the J/EGFPhigh subpopulation under combinatorial treatment of WM3734 cells for 72 hr with cisplatin (cis) and oligomycin as determined by flow cytometry (left). Ethanol (EtOH) was used as vehicle control. Immunoblots of the endogenous JARID1B protein expression of WM3734 cells under co-treatment with oligomycin at the indicated time points (right). (B) Cytotoxicity assays of melanoma cell lines after 72 hr of co-treatment with cisplatin (cis) and oligomycin (oligo). (C) Three-dimensional growth/invasion/survival assay of 1205Lu cells treated with vemurafenib (vem) and oligomycin (oligo) for 96 hr. Colony sizes were quantified digitally based on the relative number of pixels. Scale bar represents 500 µm. (D) Soft agar colony forming assay of WM3734 cells after 72-hr pulse treatment with oligomycin (left) and oligomycin plus cisplatin (right). Colonies were counted manually after 5 weeks of growth. Results shown are from one of two independent experiments. Error bars represent SD. See also Figure S5.
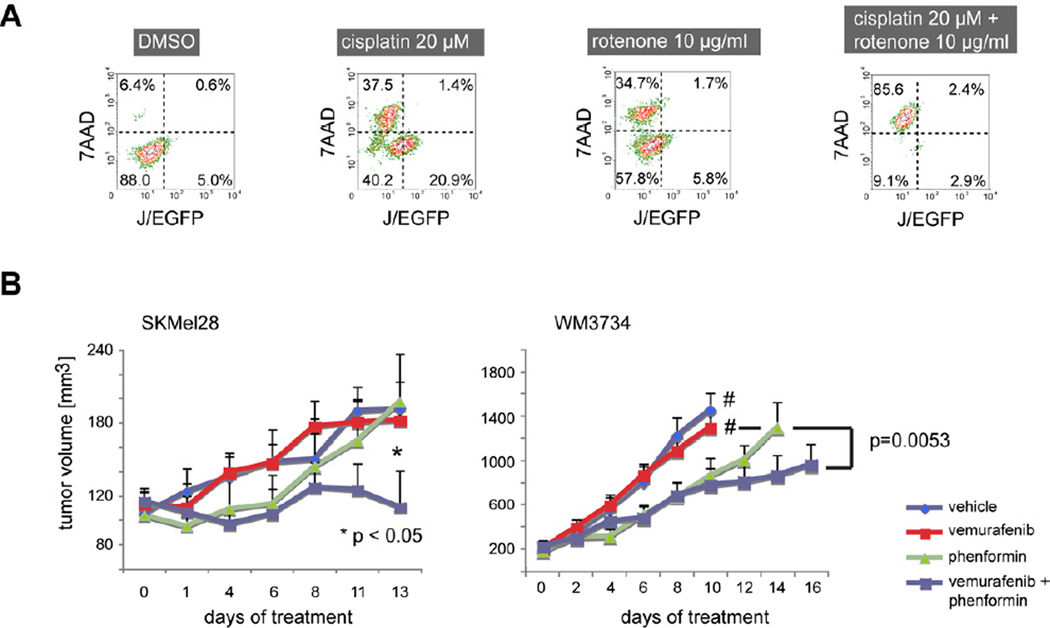
Mitochondrial inhibition as a combinatorial strategy against cancer drug-induced enrichment of slow-cycling JARID1Bhigh melanoma cells was further confirmed for the complex I inhibitors rotenone and phenformin. Phenformin is used in the clinics as an anti-diabetic drug. Both compounds potently blocked the emergence of JARID1Bhigh cells during anti-melanoma treatment (Figure 8A, Figure S6A–Figure S6D). Immunoblots and immunohistochemistry of phenformin-treated cells confirmed that complex I inhibition affects the endogenous JARID1B phenotype in vitro and in vivo (Figure S6C and S6D). Similar to the combinations of bortezomib or vemurafenib with JARID1B gene knockdown shown in Figure 3D, the mitochondrial inhibition by phenformin improved the tumor suppressive potential of vemurafenib in xenotransplanted melanomas in vivo (Figure 8B and Figure S6D). Statistical overall significance was shown by MANOVA (p=0.0263). Significant differences between the combination and single treatments were seen after adjusting for multiple hypothesis testing (combination vs. vemurafenib was p=0.022, vs. phenformin p=0.010, and vs. control p=0.010). Altogether, these findings suggest that multi-resistant, slow-cycling melanoma cells are susceptible to the inhibition of the mitochondrial respiratory chain and that the oxidative energy metabolism could be a prime target to overcome intrinsic drug resistance in melanoma.
Figure 8. The effect of anti-cancer agents is increasedin vitroandin vivoby the combination with complex I inhibitors.
(A) Flow cytometric pattern of 7AAD and J/EGFP signals in cultured WM3734 cells after 72 hr of treatment with cisplatin and rotenone. (B) SCID beige mice were xenotransplanted with 105 SKMel28 human melanoma cells (left, n=7 per treatment group, n=6 in vehicle group), NSG mice with 5×104 WM3734 cells (right, n=8 per group). SKMel28 tumors were treated when an average volume of ~120 mm3 was reached, WM3734 tumors at an average volume of 200 mm3. Tumor volumes were assessed every other day. Groups marked with # were terminated earlier due to excessive tumor growth. Error bars represent SEM. See also Figure S6.
Discussion
Despite recent discoveries in the area of therapy resistance in melanoma, we do not fully know how therapeutic escape arises and if it is the result of an evolutionary process under treatment or from the selection of pre-existing resistant subpopulations or a combination of both. Likely, even among functionally and genetically heterogeneous tumors, common mechanisms may exist that allow cells to survive in harsh conditions in general. Based on our experience with vemurafenib-resistant clones in vitro, we observed that functionally acquired resistance in cell lines that are initially vemurafenib-sensitive takes time to develop, but eventually leads to re-activation of the MAPK and PI3K pathways (Villanueva et al., 2010). Intrinsic resistance conferred by the slow-cycling cell phenotype could represent an additional and universal protection mechanism that helps cells survive the very first contact with various cytotoxic agents and provides cells with added survival time to establish longer-lasting secondary resistance mechanisms. A major challenge for successful therapy is that these slow-cycling cells can be present at any stage of the disease and that this phenotype can be dynamically acquired by other melanoma cells depending on the microenvironmental context, thus representing a ‘moving target’ (Roesch et al., 2005; Roesch et al., 2010).
Our findings indicate that the slow-cycling phenotype reflects a metabolic state that is characterized by high expression of mitochondrial bioenergetic enzymes relative to the rapidly proliferating tumor bulk population. Otto Warburg predicted that cancer cells generally rely on an increase in glucose uptake, the enhancement of glycolysis and lactate production along with the absence of oxidative respiration, despite the presence of sufficient oxygen concentrations (Warburg et al., 1924). He erroneously suggested defective mitochondria as a possible cause. However, it is becoming clear now that mitochondria in cancer cells are mostly intact and may play an important role in tumor growth (Caro et al., 2012; Weinberg et al., 2010). The cells’ decision to employ glycolysis or oxidative phosphorylation for their energy supply depends on multiple factors, including the cancer type and stage, the sequence of oncogenes activated which can directly influence mitochondrial metabolism, the current microenvironmental context, and also the rate of cell proliferation (Berridge et al., 2010; Jose et al., 2011; Vander Heiden et al., 2009). Our results support the general hypothesis that slow-cycling cells rely more on oxidative phosphorylation, whereas rapidly growing cells are characterized by high glycolysis (Berridge et al., 2010; Jose et al., 2011; Vander Heiden et al., 2009). As previously shown by others, the blockage of glycolysis by bromopyruvate treatment is only efficient on rapidly growing cells and barely useful to decrease the growth rate of tumor cells with slow proliferation (Jose et al., 2011). This observation together with ours indicates that cell cycling and energy metabolism represent interwoven biologic processes that holistically contribute to the phenotype of tumor cell subpopulations. With regard to intrinsic drug susceptibility, we found that the JARID1Bhigh slow-cycling phenotype is not responding to exogenous cell cycle influences such as serum starvation (data not shown). Since depletion of JARID1B does affect cell cycle phases, we would consider JARID1B expression to be a “driver” of the slow cycling phenotype rather than a “passenger”. Treatment with the multi-CDK inhibitor dinaciclib resulted in massive cell death rather than in proliferation arrest and, thus, passively selected for JARID1Bhigh cells as seen before for other anti-cancer agents (data not shown). We further suggest that consideration of single biologic features, such as cell cycling, without the metabolic contribution may not be sufficient to predict therapeutic response. For example, MeWo cells, which are known to have a high ability to metabolize glutamine in the (reversed) TCA cycle as an alternative energy source (Scott et al., 2011), responded least to cytotoxic killing and mitochondrial inhibition, although having a rather high proliferation rate (and low endogenous JARID1B expression, data not shown).
We cannot exclude the possibility that our initial proteome profiling strategy may have missed additional important factors contributing to the biology of slow-cycling cells. Therefore, we analyzed common molecular players of melanoma tumorigenesis by immunoblotting of sorted J/EGFPhigh vs. J/EGFPlow cells. We found no difference in MAPK activation (measured by pMEK and pERK levels) nor in the expression of the apoptosis markers BCL-2, MCL-1, and BIM or the cell cycle regulators CDK2, CDK4, and CDK9 (not shown). Interestingly, we found an increase in pAKT in J/EGFPhigh cells that correlates well with our previous finding that re-activation of PI3K signaling can overcome vemurafenib’s cytotoxicity in melanoma cells (Villanueva et al., 2010). The relevance of this finding with regard to therapeutic strategies comprising combinatorial inhibition of PI3K in slow-cycling cells will be the subject of future studies. Considering the heterogeneity of melanoma, we are currently delineating additional molecular mechanisms underlying resistance of the JARID1Bhigh slow-cycling phenotype. However, non-JARID1B-related phenotypic resistance is likely to occur and all potential resistance mechanisms could contribute to melanoma recurrence.
Our data confirm that slow-cycling melanoma cells of different genetic backgrounds are intrinsically resistant to various anti-cancer drugs and are characterized by the presence of several key enzymes of oxidative energy metabolism. Depletion of the JARID1Bhigh slow-cycling phenotype either by gene knockdown or by targeting of its bioenergetic metabolism sensitizes melanoma for a more pronounced and long-lasting therapeutic effect. Selective targeting of the slow-cycling subpopulation in combination with conventional tumor “debulking” strategies could help to eliminate a dramatically higher percentage of melanoma cells and significantly prolong the therapeutic response in patients with malignant melanoma.
Experimental Procedures
Melanoma cell lines, tissue, and vector constructs
Establishment and studying of non-commercial melanoma cell lines from human tumor samples was approved by the Internal Review Boards of the University of Pennsylvania School of Medicine and The Wistar Institute. Immunohistochemical staining of human melanoma samples was approved by the ethics committees of the University of Frankfurt and the Saarland University. Informed consents were obtained from all participating subjects. For details on cell culture techniques, see Supplemental Experimental Procedures. The consistency of cellular genotypes was confirmed by DNA fingerprinting at the Department of Human Genetics of The Saarland University Hospital. The lentiviral vector constructs for stable knockdown of JARID1B were purchased from Sigma. Details on lentiviral pLU-JARID1Bprom-EGFP-Blast, pLU-CMVprom-EGFP-Blast, and transient transfection with pBIND-RBP2H1(JARID1B) were previously described (Roesch et al., 2010; Roesch et al., 2008).
Detection of JARID1B and flow cytometry of J/EGFP signals
For immunoblotting and immunostaining rabbit polyclonal anti-JARID1B antibodies (Novus 22260002 and NB100-97821) were used following established protocols (Roesch et al., 2010). Quantitative real time RT-PCR was performed on a StepOnePlus instrument (Applied Biosystems) using self-designed primers or primers from the Harvard primer bank as described earlier (Roesch et al., 2010). Flow cytometric detection of JARID1B promoter-driven EGFP signals was performed using a FACSCanto II (BD Biosciences) as described in the supplements. Dead cells were excluded by staining for 7AAD. Fluorescence activated cell sorting was done on a Cytomation MoFlo (DakoCytomation) or a FACSAriaIII (BD Biosciences) instrument.
Cell proliferation and three-dimensional growth/invasion/survival assays
Cell proliferation was quantified by MTT and crystal violet assays according to standard protocols. Anchorage-independent colony formation was measured in 0.35% soft agar assays and quantified manually or digitally as described before using Image Pro Plus 7.0 (Roesch et al., 2010). Collagen-embedded three-dimensional melanoma spheroids were prepared and treated with inhibitors at the designated concentrations for 72 hours as previously described (Smalley et al., 2006). Following treatment, spheroids were imaged using a Nikon-300 inverted fluorescence microscope. To quantitate invasion into the matrix, borders were established around the invasive edge based on an ImageJ-defined cell density parameter and the same parameters were applied to all images.
Measurement of ATP and lactate
Total cellular ATP was measured using the ATP-based CellTiter-Glo kit (Promega) according to the manufacturer’s instructions. Each well on a 96-well plate was seeded with 2000 cells which were incubated with compounds at designated concentrations over varying time points (1, 2, 4, 24 hr). Luminescence was measured on a Tecan M200 Infinite plate reader. Mitochondrial ATP levels were determined using the mitochondrial ATP sensor protein ATEAM as reported (Imamura et al., 2009) and described in the supplements. For lactate measurement, 105 cells were plated per well on a 6-well plate and treated with compounds over 6, 24, or 72 hr. Cell culture supernatant (500 µl) was collected and centrifuged to remove any cellular debris. Subsequently, an aliquot of 100 µl was analyzed using Roche’s automated Modular Analytics system.
In vivo melanoma models
All animal experiments were performed in accordance with institutional and national guidelines and regulations. The protocols have been approved by the Wistar IACUC or the Saarland Veterinary Administration. NOD/LtSscidIL2Rγnull (NSG) and SCID beige mice were used for xenotransplantation. WM3734 or SKMel28 human melanoma cells were subcutaneously injected at a Matrigel®/culture medium ratio of 1:1. Mice were treated with (i) bortezomib every other day (20 µg per intraperitoneal injection) or (ii) with vemurafenib every 12 hr (100 mg/kg per oral gavage) or (iii), for combination studies, with vemurafenib (15 mg/kg) plus phenformin [150 mg/kg (Appleyard et al., 2012)] every 12 hr. Buffered saline or 0.1% Tween 80/0.5% methylcellulose served as vehicle controls. Treatment started at an average tumor volume of ~120–200 mm³. Tumor growth was measured every 2–3 days using a caliper. Tumor volumes were calculated according to the formulas V=WxDxH [mm³].
Statistics
Multivariate analysis of variance (MANOVA) was used to evaluate differences between treatment groups; the change in the natural logarithm of volume over time was estimated for each animal from a linear regression and mean slopes were compared using a t-test. For the SKMel28 animal study, p-values were adjusted according to Hochberg, 1988. The two-sided Wilcoxon two-sample test was used when assumptions for the t-test were violated (Figure 3A), otherwise the Student’s t test was used. Box plots show the sample minimum (lower whisker), the lower quartile (bottom of the box), the median (line inside the box), the upper quartile (top of the box), and the sample maximum (upper whisker). A p value of less than 0.05 was considered significant. As software tools, SAS version 9.2 using Proc GLM and Microsoft Excel were used.
Supplementary Material
Highlights.
-
#1
slow-cycling melanoma cells expressing JARID1B show intrinsic multi-drug resistance
-
#2
slow-cycling melanoma cells show upregulation of OXPHOS
-
#3
inhibition of OXPHOS sensitizes slow-cycling cells to therapy
-
#4
OXPHOS inhibition can overcome drug resistance irrespective of melanoma genotypes
Significance.
Metastatic melanoma is a heterogeneous tumor of neuroectodermal origin with a median survival below 9 months. All chemotherapies, immunotherapies, and radiotherapies have failed to persistently increase survival. Despite the encouraging response rates following BRAFV600E–targeting, relapses occur after a median duration of ~5 months because melanoma cells acquire multiple resistance mechanisms. However, even among functionally and genetically heterogeneous tumors, common and intrinsic mechanisms exist that support the immediate survival of certain cells against the attack of cytotoxic agents. We have identified a subpopulation of slow-cycling tumor-maintaining melanoma cells with intrinsic phenotypic resistance to various therapies, irrespective of their mode of action. Targeting the slow-cycling subpopulation could increase the efficacy of current treatment regimens and reduce relapses.
Acknowledgments
We thank K Speicher and T Beer (Wistar Proteomics Facility), J Hayden and F Keeney (Wistar Microscopy Facility), JS Faust (Wistar Flow Cytometry Facility), R DelGiacco (Wistar Histology Facility), C Junker and R Kappl (Department Biophysics, Saarland University), and A Liu (The Saarland University Hospital, Department of Neurology) for technical support; S Kugler, H Palm, A Stark, G Rajan, and R Koerner for technical assistance. The animals were housed at Wistar’s Animal Facility and the Institute for Clinical and Experimental Surgery, Saarland University. Lactate measurement was done at the Department of Clinical Chemistry of The Saarland University Hospital. We also thank N Demaurex, H Imamura, and H Noji for providing the ATEAM probe, V Belousov for Hyper and B Niemeyer for the JARID1B pCAGGS-IRES-RFP construct. The research was funded by NIH grants CA25874, CA047159, CA10815, the Dr. Miriam and Scheldon G. Adelson Medical Research Foundation, and by the DFG grants SFB1027 project C4, SFB 894 (A1), R3577/2-1 and R3577/3-1.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflict of interest.
References
- Appleyard MV, Murray KE, Coates PJ, Wullschleger S, Bray SE, Kernohan NM, Fleming S, Alessi DR, Thompson AM. Phenformin as prophylaxis and therapy in breast cancer xenografts. Br J Cancer. 2012;106:1117–1122. doi: 10.1038/bjc.2012.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MV, Herst PM, Tan AS. Metabolic flexibility and cell hierarchy in metastatic cancer. Mitochondrion. 2010;10:584–588. doi: 10.1016/j.mito.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Blagosklonny MV. Why therapeutic response may not prolong the life of a cancer patient: selection for oncogenic resistance. Cell Cycle. 2005;4:1693–1698. doi: 10.4161/cc.4.12.2259. [DOI] [PubMed] [Google Scholar]
- Caro P, Kishan AU, Norberg E, Stanley IA, Chapuy B, Ficarro SB, Polak K, Tondera D, Gounarides J, Yin H, et al. Metabolic signatures uncover distinct targets in molecular subsets of diffuse large B cell lymphoma. Cancer Cell. 2012;22:547–560. doi: 10.1016/j.ccr.2012.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charton C, Ulaszewski S, da Silva Vieira MR, Henoux V, Claisse ML. Effects of oligomycins on adenosine triphosphatase activity of mitochondria isolated from the yeasts Saccharomyces cerevisiae and Schwanniomyces castellii. Biochem Biophys Res Commun. 2004;318:67–72. doi: 10.1016/j.bbrc.2004.03.185. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KG, Valencia JC, Gillet JP, Hearing VJ, Gottesman MM. Involvement of ABC transporters in melanogenesis and the development of multidrug resistance of melanoma. Pigment Cell Melanoma Res. 2009;22:740–749. doi: 10.1111/j.1755-148X.2009.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimtriou J, Wynder C. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol. 2008;17:5312–5327. doi: 10.1128/MCB.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grazette LP, Boecker W, Matsui T, Semigran M, Force TL, Hajjar RJ, Rosenzweig A. Inhibition of ErbB2 causes mitochondrial dysfunction in cardiomyocytes: implications for herceptin-induced cardiomyopathy. J Am Coll Cardiol. 2004;44:2231–2238. doi: 10.1016/j.jacc.2004.08.066. [DOI] [PubMed] [Google Scholar]
- Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Chang CP, Tsao CC, Xu J. Oligomycin-induced bioenergetic adaptation in cancer cells with heterogeneous bioenergetic organization. J Biol Chem. 2010;285:12647–12654. doi: 10.1074/jbc.M109.084194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochberg Y. A Sharper Bonferroni Procedure for Multiple Significance testing. Biometrika. 1988;75:800–803. [Google Scholar]
- Imamura H, Nhat KP, Togawa H, Saito K, Iino R, Kato-Yamada Y, Nagai T, Noji H. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proc Natl Acad Sci U S A. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannessen CM, Boehm JS, Kim SY, Thomas SR, Wardwell L, Johnson LA, Emery CM, Stransky N, Cogdill AP, Barretina J, et al. COT drives resistance to RAF inhibition through MAP kinase pathway reactivation. Nature. 2010;468:968–972. doi: 10.1038/nature09627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jose C, Bellance N, Rossignol R. Choosing between glycolysis and oxidative phosphorylation: a tumor’s dilemma? Biochim Biophys Acta. 2011;1807:552–561. doi: 10.1016/j.bbabio.2010.10.012. [DOI] [PubMed] [Google Scholar]
- Nazarian R, Shi H, Wang Q, Kong X, Koya RC, Lee H, Chen Z, Lee MK, Attar N, Sazegar H, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468:973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, Shi H, Atefi M, Titz B, Gabay MT, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E) Nature. 2011;480:387–390. doi: 10.1038/nature10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Becker B, Meyer S, Wild P, Hafner C, Landthaler M, Vogt T. Retinoblastoma-binding protein 2-homolog 1: a retinoblastoma-binding protein downregulated in malignant melanomas. Mod Pathol. 2005;18:1249–1257. doi: 10.1038/modpathol.3800413. [DOI] [PubMed] [Google Scholar]
- Roesch A, Becker B, Schneider-Brachert W, Hagen I, Landthaler M, Vogt T. Re-expression of the retinoblastoma-binding protein 2-homolog 1 reveals tumor-suppressive functions in highly metastatic melanoma cells. J Invest Dermatol. 2006;126:1850–1859. doi: 10.1038/sj.jid.5700324. [DOI] [PubMed] [Google Scholar]
- Roesch A, Fukunaga M, Schmidt E, Zabierowski S, Brafford P, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer. 2008;122:1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- Scott DA, Richardson AD, Filipp FV, Knutzen CA, Chiang GG, Ronai ZA, Osterman AL, Smith JW. Comparative metabolic flux profiling of melanoma cell lines: beyond the Warburg effect. J Biol Chem. 2011;286:42626–42634. doi: 10.1074/jbc.M111.282046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley KS, Haass NK, Brafford PA, Lioni M, Flaherty KT, Herlyn M. Multiple signaling pathways must be targeted to overcome drug resistance in cell lines derived from melanoma metastases. Mol Cancer Ther. 2006;5:1136–1144. doi: 10.1158/1535-7163.MCT-06-0084. [DOI] [PubMed] [Google Scholar]
- Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, McArthur GA, Hutson TE, Moschos SJ, Flaherty KT, et al. Survival in BRAF V600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva J, Vultur A, Lee JT, Somasundaram R, Fukunaga-Kalabis M, Cipolla AK, Wubbenhorst B, Xu X, Gimotty PA, Kee D, et al. Acquired resistance to BRAF inhibitors mediated by a RAF kinase switch in melanoma can be overcome by cotargeting MEK and IGF-1R/PI3K. Cancer Cell. 2010;18:683–695. doi: 10.1016/j.ccr.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagle N, Emery C, Berger MF, Davis MJ, Sawyer A, Pochanard P, Kehoe SM, Johannessen CM, Macconaill LE, Hahn WC, et al. Dissecting therapeutic resistance to RAF inhibition in melanoma by tumor genomic profiling. J Clin Oncol. 2011;29:3085–3096. doi: 10.1200/JCO.2010.33.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburg O, Posener K, Negelein E. Z Biochem. 1924;152:319. [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature. 2011;471:518–522. doi: 10.1038/nature09882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proc Natl Acad Sci U S A. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.