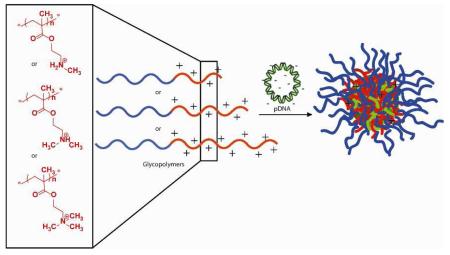
Abstract
A series of nine poly(2-deoxy-2-methacrylamido glucopyranose)-b-poly(methacrylate amine) diblock copolycations The cationic block was varied in length and in the degree of methyl group substitution (secondary, tertiary, quaternary) on the pendant amine in an effort to optimize the structure and activity for plasmid DNA delivery. Upon a thorough kinetic study of polymerization for each polymer, the glycopolymers were prepared with well-controlled Mn and Ð. The binding and colloidal stability of the polymer-pDNA nanocomplexes at different N/P ratios and in biological media has been investigated using gel electrophoresis and light scattering techniques. The toxicity and transfection efficiency of the polyplexes has been evaluated with Hep G2 (human liver hepatocellular carcinoma) cells; several polymers displayed excellent delivery and toxicity profiles justifying their further development for in vivo gene therapy.
Cationic polymers have been explored heavily in recent years as a promising modality for gene delivery.1,2 Many exciting advances have been made with polymeric delivery vehicles; yet more work is needed to further advance these systems towards the clinic.1,2 The anionic nature of polynucleotides [i.e., pDNA, small interfering RNA (siRNA), microRNA, and oligodeoxynucleotides] enables the electrostatic complexion with cationic polymers giving rise to nanoscale complexes termed polyplexes.3
Ideally, polymeric vehicles would efficiently bind nucleic acids to form colloidally stable polyplexes that have the ability to circulate in the blood, facilitate endocytosis into the targeted tissue, and ultimately release their cargo once inside the cell. In addition, the polyplexes must exhibit low cellular and immunotoxicity and avoid rapid clearance by the reticuloendothelial system. Polyplexes are typically formulated to encompass a slight net positive charge. This has been shown to increase endocytosis relative to the uptake of naked nucleic acid strands, which are often repelled by the negatively charged cell membrane.1e, f The polymer vehicles also promote in situ physical protection for the nucleic acid cargo against enzymatic degradation.3 The inclusion of a biocompatible component, most notably poly(ethylene glycol) (PEG), has been reported to increase the stability of polyplexes in biological media.4 PEG has also been reported to diminish the exposure of a cationic shell, reducing aggregation with proteins in the blood.4 However, recent research revealed that this system contained drawbacks; particularly that PEG may induce an immunological response that results in accelerated clearance of the polyplexes after multiple injections.1b, 2b, 5
Recently, our labs have reported the synthesis of a new generation of block-copolycations, poly(2-deoxy-2-methacrylamido glucopyranose), that was copolymerized with poly(aminoethylmethacrylamide) P(MAG-b-AEMA) as the cationic component and explored for its gene delivery capabilities.2b,d When polyplexed with pDNA, this series of polymer vehicles prevented colloidal aggregation in both salt and serum-containing cell culture media.2b,d Polyplexes formed with those systems have been shown to exhibit low toxicity, as well as provide potential sites for functionalization along the polymer backbone though the hydroxyl groups of the sugars.2b,6 Glycopolymers are also advantageous in their ability to promote specific biological interactions such targeting to specific tissues.2b, 6 While excellent cellular uptake of these polyplexes was reported, the efficiency of pDNA expression and siRNA-mediated gene knockdown was highly dependent on the length of the charge block. The shorter charge block length (21 repeat units) only yielded pDNA expression, and the longer charge block length (48 repeat units) only displayed siRNA-mediated gene knockdown. It was hypothesized that the short block was needed to fully release pDNA for gene expression, and the longer charged block was needed to fully complex and protect short strands of siRNA from degradation.
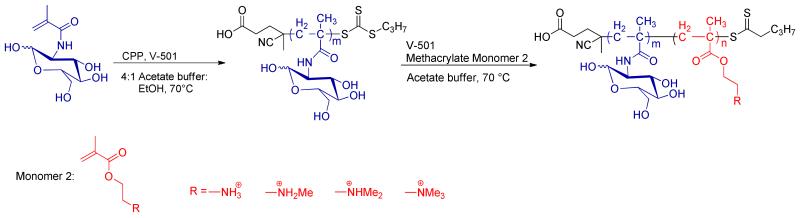
To further understand the role of charge type and block length of polymer gene delivery vehicles, a family of copolymers was generated comprising polyMAG and polymethacrylates of various block lengths bearing secondary, tertiary, and quaternary amine functionalities. To address the inability of the acrylamide-containing P(MAG-b-AEMA) to release its genetic material, polymethacrylates were investigated. In general, methacrylates are more susceptible to hydrolysis than acrylamides, which may result in the destabilization of the polyplexes and allow for the release of genetic material.7d Polyplex cytotoxicity and transfection efficiency were then measured to identify the optimal glycopolymer structure for future in vivo gene delivery studies. The methacrylate monomers investigated include aminoethylmethacrylate (AEMT), N-methyl aminoethylmethacrylate (MAEMT), N,N-dimethyl aminoethylmethacrylate (DMAEMT), and N,N,N-trimethylammoniumethylmethacrylate (TMAEMT). It was expected that differing the amine groups and block lengths would create diverse delivery characteristics (including binding, release, internalization, toxicity) due to the number and nature of the cationic amino/ammonium presented at the electrostatic binding site. Exploring the structural aspects of the cationic block of the glycopolymers may identify a polymer that balances high transfection efficiency with low toxicity. It should be noted that polymers containing PDMAEMT charge groups have been investigated in the past for nucleic acid delivery and some structural differences have been explored for PDMAEMT and PTMAEMT.4a, 7 To our knowledge however, no full systematic study has been explored for block-coglycopolymers containing PMAEMT, PDMAEMT, and PTMAEMT. The synthesis of this polymer series is presented in Scheme 1 and characterization is presented in Table 1.
Scheme 1.
Preparation of the P(MAG-b-methacrylate) nucleic acid delivery vehicles via RAFT polymerization that contain blocks of a methacrylate monomer with pendant primary amine (PAEMT), secondary amine (PMAEMT), tertiary amine (PDMAEMT), and quaternary ammonium groups (PTMAEMT). At the pH of biological media (7.2-7.4), the some of the amines along the backbone exhibit a positive charge, which facilitates binding and compaction of polynucleotides.
Table 1.
Molecular Weight Data for P(MAG-b-Methacrylate) containing the indicated charge block.
| Polymer | Mn (kDa) |
Ð | MAG DP |
Methacrylate DP |
|---|---|---|---|---|
| PMAEMT-1 | 18 | 1.02 | 51 | 30 |
| PMAEMT-2 | 20 | 1.02 | 51 | 42 |
| PMAEMT-3 | 27 | 1.05 | 51 | 76 |
| PDMAEMT-1 | 20 | 1.02 | 56 | 32 |
| PDMAEMT-2 | 24 | 1.02 | 56 | 53 |
| PDMAEMT-3 | 28 | 1.03 | 56 | 71 |
| PTMAEMT-1 | 25 | 1.29 | 57 | 33 |
| PTMAEMT-2 | 29 | 1.06 | 57 | 48 |
| PTMAEMT-3 | 36 | 1.12 | 57 | 72 |
The monomers AEMT, MAEMT, DMAEMT and TMAEMT were either purchased from Polysciences (AEMT) or Aldrich (DMAEMT) or synthesized as detailed in the Supporting Information material. The PMAG macro chain transfer agent (CTA) was prepared by polymerizing MAG in a mixed solvent of acetate buffer (pH 5.2)/ethanol (4:1, v:v) in the presence of 4-cyano-4-(propylsulfanylthiocarbonyl) sulfanylpentanoic acid (CPP) and 4,4′-azobis(4-cyanopentanoic acid) (V501) according to a previously published procedure.2b Cationic block copolymers of P(MAG-b-methacrylate) were prepared using reversible addition-fragmentation chain transfer (RAFT) polymerization. The block lengths of each rendered copolymer were achieved by conversion rate rather than by simple monomer to CTA ratios to avoid an increase in the dispersity index (Ð). The kinetic rates of the polymerizations were determined with NMR spectrometry, using the conversion rate of the methacrylate monomers to calculate cationic polymer block lengths (SI, Figures S2-S5). The resulting polymers were purified by dialysis against water and then lyophilized yielding solids of the final structures. As indicated by the GPC traces (SI, Figures S7 and S8), the Mn and Ð for all of the obtained polymers were low, meaning the polymerizations were well controlled. Detailed characterization information for each polymer can be found in Table 1. As shown, three classes of polymers were created that contained PMAG blocks of similar length and charged polymethacrylate blocks (containing secondary, tertiary, and quaternary amines) of three different yet comparable lengths. This study allowed for a systematic comparison of the effects of amine type and block length on the pDNA delivery capabilities of these systems in vitro.
All resulting P(MAG-b-MAEMT), P(MAG-b-DMAEMT) and P(MAG-b-TMAEMT) polymers given in Table 1 exhibited high water solubility. It should be noted that the P(MAG-b-AEMT) polymers (the primary amine charge center) were prepared with a similar procedures, but could only be dissolved in acidic aqueous conditions. This decrease in water solubility limited characterization of the molecular weight for these polymers to GPC analysis in an acidic mobile phase. Comparing the molecular weight of the crude products obtained before dialysis to that obtained after dialysis revealed a dramatic increase in Mn and Ð for the final products (S.I., Figures S:7). Previous reports of polymers prepared using this methacrylate monomer with a pendant primary amine revealed that the ester bonds are susceptible to nucleophilic attack,8 suggesting that the observed increase in the Mn and Ð are due to crosslinking among different macromolecules, likely as the result of both intra- and inter-molecular amidation from the nucleophilic primary amines attacking the labile ester carbonyls. Considering that the architectures of the P(MAG-b-AEMT) have changed to significantly decrease the solubility in neutral aqueous conditions, all PAEMT polymers were excluded from in vitro investigations.
To investigate the association of the glycopolycations with pDNA, polyplex formulations were run via gel electrophoresis to determine the optimal N/P ratios [the number ratio of pendant amines (N) on the diblock copolymer to the phosphates (P) on the nucleic acid backbone] for binding and complex formation (SI, Figure S9). As the supporting material indicates, binding of pDNA with P(MAG-b-MAEMT) and P(MAG-b-TMAEMT) is observed at N/P values of 2 or 3 and above, but P(MAG-b-DMAEMT) polymers do not bind pDNA until reaching an N/P value of 5 (SI, Figure S9). To explain the difference in binding capabilities among the three different types of polymers, it is hypothesized that the lower binding capabilities of the P(MAG-b-DMAEMT) structures could be due to the difference in pKa of that charge center as compared to the secondary amine derivatives. The quaternary ammonium derivatives, P(MAG-b-TMAEMT)s, possess a stronger ion strength, which could result in very stable polyplex formation. In gel shift assays of polymer-pDNA binding with these systems, tight binding at N/P ratios below 4 was observed (in fact, ethidium bromide was not able to intercalate to stain the pDNA in these polyplexes indicating tight binding strength with the tetraalkylammonium cationic units). Tight binding could result in decreased pDNA release and gene expression.
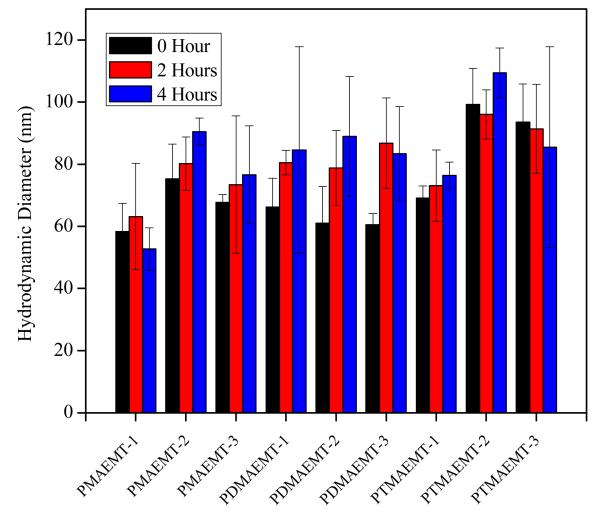
Among the advantages for the cationic PolyMAG-based glycopolymers is their biocompatibility and stability.1c, 6 Colloidal stability of the polyplexes dispersed in Opti-MEM were examined by monitoring the change their diameter using dynamic light scattering (DLS) at time intervals of 0, 2, and 4 hours at N/P ratios of 5 and 15 (Figure 1 and SI, Figure S10). It was observed that the sizes of the polyplexes stayed within a range of 50-110 nm, and were, in general, stable over a period of 4 hours. Polyplex sizes revealed by TEM (SI, Figure S12) are typically smaller than those shown by light scattering techniques (DLS measures hydrodynamic radius). However, the TEM images showed that the polyplexes were uniform in size. Measurement of zeta potentials for the polyplexes was performed at N/P ratios of 5 and 15 (SI Figure S11); the obtained values were between 15-25 mV for N/P 5 and within 25-40 mV for N/P 15 (typically observed for this class of polyplexes).
Figure 1.
Hydrodynamic diameters for polyplexes made from pDNA and the P(MAG-b-methacrylate) containing the charge block indicated above at N/P ratio of 15 measured in Opti-MEM.
The primary goal for this series of polymers is to screen the in vitro delivery efficacy to select candidates for murine screening in vivo for pDNA delivery applications. To this end, the pDNA delivery capabilities of these polymers were examined in cultured Hep G2 (human liver hepatocellular carcinoma) cells in two media conditions, both serum-free Opti-MEM and DMEM containing 10% serum. To probe the effect of amine structure and block length on transfection efficiency, pDNA encoding the luciferase reporter gene was used (SI, Figure S13) for polymers containing blocks of PMAEMT, PDMAEMT, and PTMAEMT at N/P ratios of 2, 5, 10, 15, 20, and 30. It was observed in both Opti-MEM and DMEM that an increase in the N/P ratio for most polymers did not show a significant increase in luciferase gene expression. However, the luciferase expression results did show a general trend when considering the amine type. According to this assay, the transfection efficiency decreased slightly as the amount of methyl groups on the amine increased (secondary > tertiary > quaternary), suggesting that the PMAEMT containing polymers should be further optimized and examined for nucleic acid delivery. It was also of interest to further examine transfection efficiency between the three different types of amines at one amine length, which is discussed below.
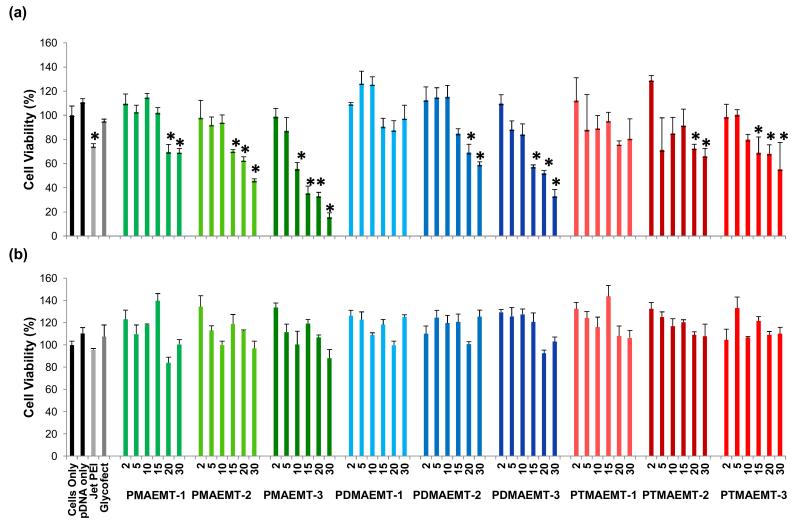
HepG2 cell viability was determined 48 hours post transfection with the glycopolymer/pDNA polyplexes via an MTT assay (Figure 2) in both Opti-MEM and DMEM for the P(MAG-b-methacrylate)s containing the charge block PMAEMT, PDMAEMT, and PTMAEMT at N/P ratios of 2, 5, 10, 15, 20, and 30. In general, the cell viability decreased with an increase in the N/P ratio in the presence of serum-free Opti-MEM. Most of the polymers were not toxic to cells up to an N/P ratio of about 10, though some polymers, most notably, those containing PMAEMT-2 and -3 and PDMAEMT -3, showed higher levels of toxicity at and above an N/P of 15. This result suggested that for the secondary and tertiary amine types, increased amine length increases toxicity at higher N/P ratios. In DMEM, all of the polymers showed low toxicity (~90% cell viability), even at higher N/P ratios.
Figure 2.
Cell viability as measured via MTT assay with HepG2 cells transfected with polyplexes formed at N/P ratios of 2, 5, 10, 15, 20, and 30 with pDNA and all the P(MAG-b-methacrylate) containing the charge blocks of PMAEMT, PDMAEMT, and PTMAEMT polymers in (a) serum free Opti-MEM and (b) DMEM with 10% serum. Error bars are representative of the standard deviation of analyzed data from three replicates. All measurements found to be statistically significant (p<0.05) as compared to cells only are marked with an asterisk.
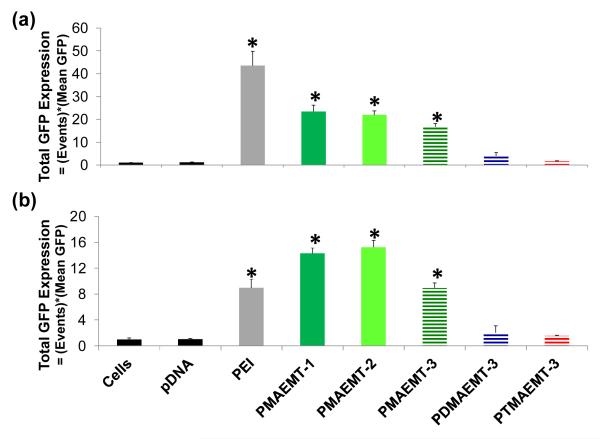
These large experiments allowed for the selection of a smaller group of polymers to further test for transfection efficiency and cellular internalization via flow cytometry analysis. This smaller group of polymers was selected to be an orthogonal study and include the three different amine types containing comparable charge block lengths, PMAEMT-3, PDMAEMT-3, and PTMAEMT-3. As the secondary amine derivatives yielded the highest gene expression in the preliminary screen, PMAEMT-1 and -2 were also included to further investigate the effect of amine length on transfection efficiency. All additional tests were performed by formulating the polyplexes at an N/P ratio of 15. To further probe transfection efficiency in a quantitative manner, Green Fluorescent Protein (eGFP) was used as the reporter gene and the number of cells positive for GFP expression was analyzed using flow cytometry. It should be noted that the difference in the data between the luciferase and GFP assays are due to the nature of how gene expression is detected. The luciferase assays only detect the bulk gene expression in the culture as relative light units (RLUs). However, the eGFP expression assay is more quantitative in that both the number of cells expressing the transgene and the average gene expression per cell is determined. In both serum-free Opti-MEM and DMEM containing 10% (by volume) fetal bovine serum, transfection efficiency decreased as methyl substitution of the amine increased (Figure 3, horizontal striped bars), which agreed with the luciferase expression data. When examining the effect of amine length (Figure 3, green bars), PMAEMT-1 and -2 showed similar transfection efficiency in both serum free Opti-MEM and DMEM containing 10% serum, while the polymer containing the longest amine length, PMAEMT-3, exhibited significantly lower GFP expression (p value<0.05) in DMEM containing 10% serum than in serum free Opti-MEM. This result possibly indicates that a longer amine block could inhibit pDNA release to promote gene expression.
Figure 3.
Total GFP expression (number of events measured multiplied by mean GFP measured) of Hep G2 cells transfected with polyplexes formed at an N/P ratio of 15 with a GFP reporter pDNA gene and the P(MAG-b-methacrylate) polymers containing the charge blocks PMAEMT-1,-2, and -3, PDMAEMT-3, and PTMAEMT-3 in (a) serum free Opti-MEM and (b) DMEM containing 10% serum. Error bars are representative of the standard deviation of analyzed data from three replicates. All measurements found to be statistically significant (p<0.05) as compared to cells only are marked with an asterisk.
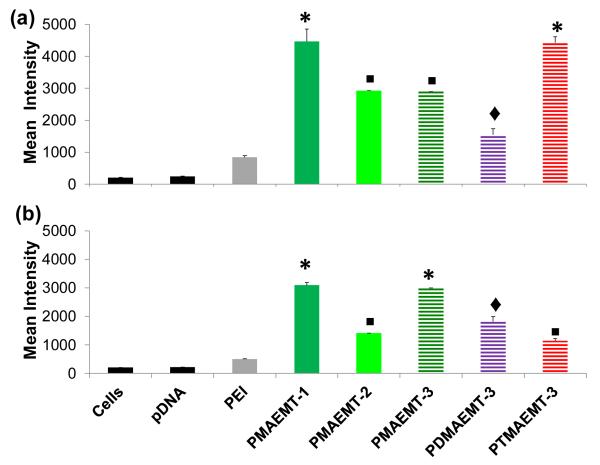
To further understand the effect of amine type and length, the cellular uptake profile was determined with Cy5-labeled pDNA using flow cytometry by measuring the number of cells positive for Cy5 fluorescence (Figure 4). For PMAEMT-1,-2 and -3, the Cy5 intensity is lower in DMEM than in Opti-MEM, agreeing with the lower GFP expression in DMEM as compared with Opti-MEM (green bars, Figure 3 and 4). Though PDMAEMT-3 showed lower Cy5 intensity than the polyplexes containing PMAEMT in Opti-MEM, this polymer showed significant Cy5 intensity as compared to cells only and pDNA. However, PDMAEMT-3 did not show significant GFP expression above background levels (blue striped bars, Figure 3 and 4), indicating trouble in release for the polymer vehicles containing the tertiary and quaternary ammonium charge centers (meaning either polyplex release from lysosomes and/or pDNA release from polyplexes). In Opti-MEM, PTMAEMT-3 showed Cy5 intensity similar to PMAEMT-1, but no significant GFP expression above background (red striped bars, Figure 3 and 4). In DMEM, PTMAEMT-3 showed the least Cy5 intensity among the polymers, but still showed significantly higher Cy5 intensity than cells only. We speculate that this disparate result could indicate that the “hard” quaternary ammonium charge on the PTMAEMT polyplexes could promote strong binding to the cell surface in the absence of serum (Opti-MEM), which would trigger high cell internalization. However, in the presence of serum in the culture media (DMEM), the charged serum proteins could adsorb onto the polyplexes in a strong manner, thus dampening cell surface interaction and uptake. To this end, GFP expression in DMEM of polyplexes with PTMAEMT-3 was not significantly different from cells only.
Figure 4.
Mean Cy5 intensity of HepG2 cells. Cells transfected with polyplexes were formed at an N/P ratio of 15 with Cy5-labeled pDNA and the P(MAG-b-methacrylate) structure containing the charge blocks PMAEMT-1,-2, and -3, PDMAEMT-3, and PTMAEMT-3 in (a) serum free Opti-MEM and (b) DMEM containing 10% serum. Error bars are representative of the standard deviation of analyzed data from three replicates. All measurements were found to be statically significant (p<0.05) as compared to cells only and pDNA are marked with an asterisk, a square, or a diamond. Measurements that share the same symbol are not significant different (p<0.05) from each other.
This data combined with the GFP expression data suggests that polyplexes from the polymers in this study are efficiently internalized by the cell. However, the relatively low percentage of cells expressing GFP for polyplexes formed with the polymers containing the PDMAEMT and PTMAEMT blocks (Figure 3), suggests that pDNA is not efficiently released. As mentioned, this may be a result of a higher binding affinity at N/P 15 for the polyplexes formulated with the PDMAEMT and PTMAEMT polymers, thus not allowing pDNA release for gene expression. Also, the low overall GFP expression and the high percentage of cells containing Cy5 for PDMAEMT may be attributed to the high stability of this polymethacrylate against hydrolysis as suggested by van de Wetering, et. al.7d,e PMAEMT-3 also exhibits a lower GFP expression compared to PMAEMT-1 and -2, but shows roughly the same amount of Cy5 positive cells, suggesting that the longer amine block may also hinder the release of the pDNA from polyplexes in the cytoplasm of the cell. Another possible explanation for high uptake and low expression of polyplexes may be an inefficient endosomal/lysosomal escape and trafficking to the nucleus. Previous reports demonstrated that polymers containing monomers with a pKa between 5.0 and 7.0 increase transfection efficiency.9 Determination of the pKa’s of these polymers is of high interest to explore the possibility of low endosomal escape.
The binding of polycations and polyanions (here pDNA) are entropically driven due to counterion release, and in this study, it is typically related to the structure of the cationic moieties in polymers. These results demonstrate how varying the methyl substitution on the cationic amine moieties in polymers with comparable amine block lengths can significantly alter the ability of the polymer to both transport pDNA into the cell and subsequently release pDNA after internalization and delivery. This ultimately results in a substantial change in transfection efficiency. As the results indicate, the glycopolymers containing the charge block PMAEMT (pendant secondary amines) exhibit toxicities similar or less than Jet PEI, superior cellular internalization, and higher luciferase and eGFP expression compared to the other polyamines studied.
In summary, a comparable series of glycopolymers exhibiting well-defined molecular weights has been created through the investigation and application of polymer kinetics. Different bioassays were utilized to probe the gene delivery capabilities of polyplexes made from pDNA and this glycopolymer series. In general, the P(MAG-b-MAEMT) polymers exhibited increased delivery efficiency as compared to polymers with more highly substituted amines along with relatively low cytotoxicity profiles. In vivo work is currently ongoing with this polymer series to determine structure-activity relationships and will be reported in due course.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. Nilesh Ingle, Department of Chemistry, University of Minnesota, for consistent support to this work, and the Institute of Technology Characterization Facility, University of Minnesota, a member of the NSF-funded Materials Research Facilities Network, for assistance in TEM imaging. We also thank the National Institutes of Health (NIH) Director’s New Innovator Award Program (DP2OD006669-01) for partial financial support of this project. Also, this work was supported partially by the National Science Foundation through the University of Minnesota MRSEC under the Award Number DMR-0819885.
Funding Sources
NIH DP2OD006669-02
NSF MRSEC DMR-0819885
Footnotes
ASSOCIATED CONTENT
Supporting Information. All synthesis procedures, NMR spectra, GPC traces, gel electrophoresis images, light scattering and zeta potential data, TEM images and luciferase expression data are presented in electronic supporting information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.(a) Godbey WT, Wu KK, Mikos AG. J. Controlled Release. 1999;60:149–160. doi: 10.1016/s0168-3659(99)00090-5. [DOI] [PubMed] [Google Scholar]; (b) Ahmed M, Narain R. Prog. Polym. Sci. in Press. [Google Scholar]; (c) Zhang S, Zhao Y, Zhao B, Wang B. Bioconjugate Chem. 2010;21:1003–1009. doi: 10.1021/bc900261c. [DOI] [PubMed] [Google Scholar]; (d) Punam D. Nat. Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]; (e) Davis ME. Curr. Opin. Biotechnol. 2002;13:128–131. doi: 10.1016/s0958-1669(02)00294-x. [DOI] [PubMed] [Google Scholar]; (f) Grandinetti G, Smith AE, Reineke TM. Mol. Pharmaceutics. 2012;9:53–538. doi: 10.1021/mp200368p. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Park H-J, Yang F, Cho S-W. Adv. Drug Delivery Rev. 2012;64:40–52. doi: 10.1016/j.addr.2011.09.005. [DOI] [PubMed] [Google Scholar]; (h) Samal SK, Dash M, Van Vliebergh S, Kaplan DL, Chiellini E, van Blitterswijk C, Moroni L, Dubruel P. Chem. Soc. Rev. 2012;41:7147–7194. doi: 10.1039/c2cs35094g. [DOI] [PubMed] [Google Scholar]
- 2.(a) Anderson K, Sizovs A, Cortez M, Waldron C, Haddleton DM, Reineke TM. Biomacromolecules. 2012;13:2229–2239. doi: 10.1021/bm300471n. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Smith AE, Sizovs A, Grandinetti G, Xue L, Reineke TM. Biomacromolecules. 2011;12:3015–3022. doi: 10.1021/bm200643c. [DOI] [PubMed] [Google Scholar]; (c) Ingle NP, Malone B, Reineke TM. Trends in Biotechnol. 2011;29:443–453. doi: 10.1016/j.tibtech.2011.04.012. [DOI] [PubMed] [Google Scholar]; (d) Buckwalter DJ, Sizovs A, Ingle NP, Reineke TM. ACS Macro. Lett. 2012;1:609–613. doi: 10.1021/mz300081d. [DOI] [PubMed] [Google Scholar]; (e.) Lee C-C, Liu Y, Reineke TM. ACS Macro Lett. 2012;1:1388–1392. doi: 10.1021/mz300505t. [DOI] [PubMed] [Google Scholar]
- 3.(a) Borchard G. Adv. Drug Delivery Rev. 2001;52:145–150. doi: 10.1016/s0169-409x(01)00198-3. [DOI] [PubMed] [Google Scholar]; (b) Grigsby CL, Leong KW. J. R. Soc. Interface. 2010;7(Suppl 1):S67–S82. doi: 10.1098/rsif.2009.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Grimm D. Adv. Drug Delivery Rev. 2009;61:672–703. doi: 10.1016/j.addr.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 4.(a) Tang R, Palumbo RN, Nagarajan L, Krogstad E, Wang C. J. Controlled Release. 2010;142:229–237. doi: 10.1016/j.jconrel.2009.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Choi YH, Liu F, Kim J-S, Choi YK, Jong Sang P, Kim SW. J. Controlled Release. 1998;54:39–48. doi: 10.1016/s0168-3659(97)00174-0. [DOI] [PubMed] [Google Scholar]; (c) Dai J, Zou S, Pei Y, Cheng D, Ai H, Shuai X. Biomaterials. 2011;32:1694–1705. doi: 10.1016/j.biomaterials.2010.10.044. [DOI] [PubMed] [Google Scholar]
- 5.Knop K, Hoogenboom R, Fischer D, Schubert US. Angew. Chem. Int. Ed. 2010;49:6288–6308. doi: 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- 6.(a) Ting SRS, Chen G, Stenzel MH. Polym. Chem. 2010;1:1392–1412. [Google Scholar]; (b) Voit B, Appelhans D. Macromol. Chem. Phys. 2010;211:727–735. [Google Scholar]
- 7.(a) Li Z, Yin H, Zhang Z, Liu KL, Li J. Biomacromolecules. 2012;13:3162–3172. doi: 10.1021/bm300936x. [DOI] [PubMed] [Google Scholar]; (b) Agarwal S, Zhang Y, Maji S, Greiner A. Mater. Today. 2012;15:388–393. [Google Scholar]; (c) Zhu C, Zheng M, Meng F, Mickler FM, Ruthardt N, Zhu X, Zhong Z. Biomacromolecules. 2012;13:769–778. doi: 10.1021/bm201693j. [DOI] [PubMed] [Google Scholar]; (d) van de Wetering P, Zuldam NJ, van Steenbergen J, van der Houwen OAGJ, Underberg WJM, Hennink WE. Macromolecule. 1998;31:8063–8068. [Google Scholar]; (e) van de Wetering P, Moret EE, Schuurmans-Nteuwenbroek NME, van Steenbergen MJ, Hennink WE. Bioconjugate Chem. 1999;10:589–597. doi: 10.1021/bc980148w. [DOI] [PubMed] [Google Scholar]
- 8.He L, Read ES, Armes SP, Adams DJ. Macromolecules. 2007;40:4429–4438. [Google Scholar]
- 9.(a) Asokan A, Cho MJ. J. Pharm. Sci. 2002;91:903–913. doi: 10.1002/jps.10095. [DOI] [PubMed] [Google Scholar]; (b) Cho YW, Kim J, Park K. J. Pharm. Pharmacol. 2003;55:721–734. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.