Abstract
Alkaline phosphatase (ALP) plays an essential role in the regulation of tissue mineralization, and its activity is highly heritable. Guided by genetic associations discovered in a murine model, we hypothesized a role for rare coding variants in determining serum ALP level and bone mineral density (BMD) in humans. We sequenced the coding regions of the ALP gene (ALPL) in men with low and normal serum ALP activity levels. Single-nucleotide ALPL variants, including 19 rare nonsynonymous variants (minor allele frequency <1%), were much more frequent among the low ALP group (33.8%) than the normal group (1.4%, p = 1 × 10−11). Within the low ALP group, men with a rare, nonsynonymous variant had 11.2% lower mean serum ALP (p = 3.9 × 10−4), 6.7% lower BMD (p = 0.03), and 11.1% higher serum phosphate (p = 0.002) than those without. In contrast, common nonsynonymous variants had no association with serum ALP, phosphate, or BMD. Multiple rare ALPL coding variants are present in the general population, and nonsynonymous coding variants may be responsible for heritable differences in mineralization and thus BMD.
Keywords: OSTEOPOROSIS, HEREDITY, GENETICS, ALKALINE PHOSPHATASE
Introduction
Osteoporosis is a complex trait determined by both genetic and environmental factors. Twin and family studies have demonstrated moderate to high heritability of several bone traits, such as peak bone mineral density (BMD), femoral neck geometry, and bone turnover.(1) Yet despite substantial progress in identifying genetic variants conferring increased risk of low BMD, the heritability of osteoporosis remains largely unexplained. Only a small proportion of the variation in BMD can be explained by genetic variations thus far discovered in human populations.(2) Animal research can elucidate possible roles of genetic and environmental constituents in the regulation of bone mass that might otherwise be difficult to untangle. Approaches that combine human and nonhuman mammalian datasets and resources are likely to provide valuable insight into our understanding of the various genetic determinants of osteoporosis risk.(1)
Employing genome-wide quantitative trait loci (QTL) analysis in laboratory mice, we(3) and others(4–8) have linked a region on mouse chromosome 4 to measures of bone strength. In addition, a number of linkage-disequilibrium and genome-wide association (GWA) mapping studies have identified the corresponding homologous region in the human genome (1p36) as also containing DNA sequence polymorphisms associated with increased osteoporosis risk.(2,9–16) However, the genetic variation(s) residing within this region capable of influencing skeletal integrity are not yet known.
The gene for tissue-nonspecific alkaline phosphatase (TNAP) resides within the chromosome 4 QTL in mice and at 1p36 in humans. It is the product of the alkaline phosphatase (ALP) gene (ALPL; OMIM 171760), and its posttranscriptional modification into bone-specific alkaline phosphatase (BSAP) is essential for bone mineralization.(17) Mineralization provides mechanical resistance and occurs when calcium phosphate hydroxyapatite crystals are intercalated into the extracellular matrix of specialized connective tissues (eg, growth plate cartilage, bone, and dentin). ALP promotes mineralization by hydrolyzing pyrophosphate (an inhibitor of hydroxyapatite formation) into inorganic phosphate. The osteopenia and fractures that occur in mice genetically-deficient in Alpl (the murine homolog of ALPL) serve as persuasive evidence for a crucial role of TNAP in the development and mineralization of the murine skeleton.(18) Both animal (mouse, swine, baboon) and human studies have demonstrated substantial population variation in circulating levels of ALP, a portion of which appears to be hereditary.(6,8,19–24) The nature of the genetic factors influencing circulating ALP levels is uncertain, but a number of studies have detected strong associations between circulating ALP levels with the ALPL locus itself and another part of the variation (estimated to be less than 15% of the genetic variance) is attributed to the presumed presence or absence of the intestinal isoenzyme, associated with the ABO blood group polymorphism.(23–25) As is the case for BMD variation, much of the variation in ALP level remains unexplained.
While common ALPL sequence variations have been linked to circulating ALP levels in population samples, loss of function mutations in the ALPL gene also contribute to hypophosphatasia (HPP; OMIM 241500, 241510, 146300), a rare inborn error of mineral metabolism.(26) The disease is characterized by a global deficiency of ALP activity, inadequate hard tissue mineralization, and fractures. The clinical severity of HPP is widely variable and ranges from death in utero to mild dental complications. Mutation analyses of HPP subjects have identified over 200 mutations, of which approximately 80% are missense (for a complete listing the reader is referred to a database maintained by the SESEP Laboratory and the Human Molecular Genetics laboratory of the University of Versailles-Saint Quentin, Yvelines, France http://www.sesep.uvsq.fr/03_hypo_mutations.php).(27) In virtually every case, HPP appears to be a consequence of either a homozygous point mutation or a compound heterozygous genetic condition. The wide range of mutations is likely responsible for the variability in clinical presentation and disease severity.
The combined evidence from laboratory mouse experiments and clinical studies of patients with HPP led us to hypothesize that low-frequency coding variants in ALPL may also affect BMD in humans unaffected by HPP. For the studies reported here, we employed an interdisciplinary approach incorporating data from both mouse models and human studies. We aimed to discover coding variants in ALPL (among men in a population-based cohort) and its murine homolog, Alpl, and to test associations with serum ALP and BMD. In laboratory mice, we identified a coding sequence variant (L324P) in Alpl that was associated with reduced serum ALP, BMD, and bone strength in a panel of mouse strains. In elderly men with reduced serum ALP activity, we identified 25 single-nucleotide coding variants in ALPL, 22 of which had minor allele frequency (MAF)<1%. Seven of these variants can be considered novel because they have not been recorded in the hypophosphatasia or other single-nucleotide polymorphism (SNP) databases including the 1000 Genomes Project. We discovered that subjects bearing rare nonsynon-ymous coding variants in ALPL exhibit reduced mean serum ALP levels, elevated serum phosphate levels, and lower BMD. Thus, we describe an association between rare nonsynonymous variants in the gene encoding ALP with circulating enzyme activity and bone density in both mice and humans.
Materials and Methods
Animals
All mice used in these experiments were bred under identical conditions at the Portland VA Veterinary Medical Unit from breeding stock originally obtained from the Jackson Laboratory (Bar Harbor, ME, USA) no more than three generations prior to this work. At the time of weaning the mice were group housed (two to five animals per cage) and maintained with ad libitum water and laboratory rodent chow (Diet 5001: 23% protein, 10% fat, 0.95% calcium, and 0.67% phosphorus; PMI Feeds, Inc., St. Louis, MO, USA) in a 12-hour light/dark cycle (6:00 a.m. to 6:00 p.m.) at 21±2°C. Congenic mice with a 64 megabase (Mb) region of the DBA/2J (D2) genome introgressed onto a C57BL/6J (B6) genetic background were created by breeding mice heterozygous for markers flanking the chromosome 4 ALP QTL (D4Mit327 and D4Mit42) back to the B6 progenitor strain over 10 generations. At the conclusion of congenic development, we studied littermates originating from an F2 intercross between heterozygous congenic mice to insure that all differences in serum ALP activity originated exclusively from the genetic difference in the introgressed region. We studied all mice at 4 months of age. After an overnight fast, we euthanized the mice by CO2 inhalation and weighed them to the nearest 0.1 gm. Blood samples, spleen and left femur from each mouse carcass were harvested immediately. The spleens were frozen in liquid N2 and stored at −80°C for later extraction of genomic DNA using a salting-out method.(28) We wrapped the left femora in sterile gauze soaked in phosphate-buffered saline, and stored them frozen at ≤ −20°C for subsequent analyses. All procedures were approved by the VA Institutional Animal Care and Use Committee and performed in accordance with National Institutes of Health guidelines for the care and use of animals in research.
Murine skeletal phenotyping
We determined serum alkaline phosphatase activity by measuring the release of para-nitrophenol from para-nitrophenyl-phosphate spectrophotometrically and determined serum osteocalcin levels by radioimmunoassay (DRG International, Mountainside, NJ, USA). We determined whole femoral bone mineral density measurements by dual energy X-ray absorpti-ometry (PIXImus; GE-Lunar, Madison, WI, USA). We examined cortical femoral shaft bone geometry with a desktop X-ray microtomographic scanner (Model 1074; SkyScan, Aartselaar, Belgium). Images were analyzed with Optimas software (version 6.2; Media Cybernetics, Silver Spring, MD, USA). To determine femoral structural properties, the left femur was tested to failure by three-point bending on a high-resolution materials test apparatus (Model 4442; Instron Corp., Canton, MA, USA). Load and displacement data were recorded and failure load was determined using system software.
PCR genotyping
For construction of the congenic line, we genotyped mice using microsatellite markers from the MIT series by a method adapted from Dietrich and colleagues(29) and Serikawa and colleagues.(30) Markers were chosen with information from the Mouse Genome Database (www.nih.gov/science/models/mouse/resources/mgd.html). All primers were purchased from Research Genetics (Huntsville, AL, USA). We performed amplification on a Perkin-Elmer 9700 thermocycler (Perkin-Elmer Cetus, Branchburg, NJ, USA). Reaction mixture was 25 µL of total volume, consisting of approximately 150 ng of genomic DNA, 264 nM of both forward and reverse primers, 0.2 mM of each dNTP, 1 unit of Taq polymerase (Perkin-Elmer Cetus, Waltham, MA, USA), 2.5 µL of GeneAmp 10× PCR Buffer containing 100 mM Tris-HCl (pH 8.3), 15 mM MgCl2, 500 mM KCl, and 0.01% (wt/vol) gelatin. Thermal cycling included two 5-minute denaturation steps at 95°C then at 80°C, 40 cycles of 30 seconds at 94°C, 30 seconds at 53°C, 30 seconds at 72°C, and a final extension step for 10 minutes at 72°C. PCR products were separated on 4% agarose gels and visualized with ethidium bromide staining.
Alpl sequencing
We designed primers to amplify each exon plus at least 50 nucleotides of the adjacent introns. We carried out a PCR in a total volume of 10 µL including ~50 ng of DNA, 1× Amplitaq Gold PCR buffer, 1 mM MgCl2, 200 µM dNTPs, 0.5 U Taq DNA polymerase (Bioline USA Inc, Tauton, MA, USA) and 5 pmol of each primer using Applied Biosystems Gene Amp PCR System 9700 (Applied Biosystems, Carlsbad, CA, USA) and conditions: 95°C for 10 minutes, 32 cycles of 94°C for 30seconds, 59°C for 30 seconds, and 72°C for 30 seconds, followed by a further 5-minute extension at 72°C. We treated PCR products with ExoSAP-IT (USB, Cleveland, OH, USA) to enzymatically remove unincorporated primers and dNTPs, and then subjected them to thermal cycle sequencing using BigDye Terminator cycle sequencing reagents (Applied Biosystems, Carlsbad, CA, USA). The resulting fragments were analyzed on an Applied Biosystems 16-capillary 3130xl automated sequence analysis system, performed by the MMI DNA Analysis Core Facility at Oregon Health and Science University (OHSU). Sequences were compared by aligning to the genomic B6 sequence using MultAlin.(31) The sequencing was performed three times for each strain and the same sequencing results were found.
Human cohort
The Osteoporotic Fractures in Men (MrOS) Study enrolled 5994 participants from March 2000 through April 2002. Recruitment occurred at six U.S. clinical centers (Birmingham, AL, USA; Minneapolis, MN, USA; Palo Alto, CA, USA; Pittsburgh, PA, USA; Portland, OR, USA; and San Diego, CA, USA) and was accomplished primarily through mass mailings targeted to age-eligible men. Eligible participants were community-dwelling men who were at least 65 years of age, able to walk without assistance from another person, and who had not had bilateral hip replacements (in order to obtain a hip BMD measure). Details of the MrOS study design and recruitment have been published elsewhere.(32,33) Written informed consent was obtained from all participants, and the Institutional Review Board at each study site approved the study.
Subcohort for ALPL sequencing
We selected a subset of MrOS participants based on their serum ALP value to have direct ALPL sequencing of all coding exons(2–12) for the discovery of potentially functional single-nucleotide variants. We examined the distribution of baseline serum ALP values for men reporting non-Hispanic white race. To rule out high ALP due to liver or kidney disease, we excluded men with values of serum ALP that were >3 SD above the mean. We excluded participants reporting conditions or medication use that might alter serum ALP levels or BMD: those with a history of prostate cancer treatment with hormones or surgical orchiecto-my; and those currently using corticosteroids, antiandrogens, bisphosphonates, antithyroid agent, thyroid agent, Zafirlukast, or Montelukast. Within this cohort there were 74 men with ALP<40 U/L (range: 19.0–40.0 U/L) and 148 men with ALP closest to the mean value (range: 70–74 U/L) served as the control group.
Clinic visits
Participants completed self-administered questionnaires and attended a baseline clinic visit, when anthropometric measures were obtained and blood was drawn. We measured BMD using fan-beam DXA (QDR 4500W; Hologic Inc., Bedford, MA, USA). We asked participants to bring in all prescription medications used within the last 30 days. A computerized dictionary, based on the original Established Populations for Epidemiologic Studies of the Elderly (EPESE) coding system(34) was used to categorize medications. All recorded prescription medications were stored in an electronic medications inventory database (San Francisco Coordinating Center, San Francisco, CA, USA). Each medication was matched to its ingredient(s) based on the Iowa Drug Information Service (IDIS) Drug Vocabulary (College of Pharmacy, University of Iowa, Iowa City, IA, USA).(35)
Serum chemistry measures
We assayed baseline serum samples for albumin, creatinine, ALP, phosphate, electrolytes (chloride, sodium, potassium), calcium, alanine transaminase, high-density lipoprotein (HDL), low-density lipoprotein (LDL), and triglycerides. We conducted all assays at the Portland Veterans Administration Medical Center Clinical Lab using a Roche COBAS Integra 800 automated analyzer (Roche Diagnostics Corp., Indianapolis, IN, USA). The analyzer was calibrated daily in the clinical laboratory. One serum control, taken from one male volunteer, was included in each assay run. The interassay coefficient of variation for alkaline phosphatase was 2.71%. The estimated glomerular filtration rate (eGFR) was calculated from the Modification of Diet in Renal Disease (MDRD) study equation using serum creatinine and age (terms in the equation for female and African American were not required for this analysis of non-Hispanic white men).
DNA extraction
We collected blood samples from MrOS participants at baseline and frozen. Genomic DNA of all individuals with available blood samples who consented to genetic studies was extracted at the University of Pittsburgh using the Flexigene protocol (Qiagen, Valencia, CA, USA).
ALPL sequencing
We amplified the exons and flanking intronic sequences of ALPL by PCR and treated them with recombinant exonuclease I and shrimp alkaline phosphatase (ExoSAP-IT; USB, Cleveland, OH, USA). We sequenced both strands of each product on an ABI 3730 automated sequencer with BigDye Terminator cycle sequencing reagents (Applied Biosystems, Carlsbad, CA, USA). The sequences of the oligonucleotides used for sequencing are available on request. All sequence variants identified were verified by manual inspection of the chromatograms, and all missense changes were confirmed by an independent resequen-cing reaction. Separate pyrosequencing reactions and/or custom TaqMan® (Applied Biosystems, Carlsbad, CA, USA) sequencing reactions were also employed to further verify the presence of sequence variants. Standardized nomenclature was used to report the variants counting from the first (ATG initiation) codon, according to the recommendations of the Human Genome Variation Society.
Analytic methods
We calculated genotype proportions and MAFs. For the analysis of exonic single-nucleotide ALPL variants in the sequencing study, we tested the difference in proportions of participants with each variant in the low ALP and normal ALP groups using Fisher’s exact test. Because many variants were found in very few subjects, categories of exonic variants were generated: (1) any variant, (2) variants with MAF < 1% in the normal ALP group, and (3) variants with MAF ≥ 1% in the normal ALP group. Proportions of men in these categories were compared between low and normal ALP groups using the χ2test.
For the associations with BMD, serum ALP and phosphate in the low ALP group, we used collapsed variables of (1) synonymous variants, (2) common (MAF ≥ 1%) nonsynonymous variants, and (3) rare (MAF<1%) nonsynonymous variants. Regression analyses were used to determine the association between each of these three categories and each of the three phenotypes. Base models were adjusted for age, weight, and study site. Potential confounders of the associations were examined by assessing whether their addition to the linear regression model altered the coefficient for the genotype by ≥ 5%. Variables considered included age, eGFR, history of prostate cancer, use of various medications, serum calcium, glucose, and creatinine. Analyses were performed using SAS 9.2 (SAS Institute, Inc., Cary, NC, USA).
Results
Murine studies
Our genome-wide QTL analysis in C57BL/6J (B6) and DBA/2J (D2) laboratory mice linked a region on mouse chromosome 4 (Ch4) harboring Alpl to variation in both circulating ALP activity and BMD.(3,6,8) The D2 allele in this Ch4 region was associated with both greater ALP activity and increased BMD when compared to mice bearing the B6 allele in this same Ch4 region. To pursue these QTL findings, a B6 background congenic mouse with a 64-Mb region of chromosome 4 replaced by the corresponding region of the D2 genome was generated (B6.D2.Ch4). The introgressed region of chromosome 4 extended from 86 to 151 Mb with the Alpl gene located at 137 Mb. Congenic B6.D2.Ch4 mice exhibited normal overall health and body size, and there were no differences in measured serum electrolytes, or in renal or hepatic function. However, as predicted from the initial QTL experiment, serum ALP activity was 58% lower in the B6 background strain than in the B6.D2.Ch4 congenic strain (60±5 IU/L versus 142 ±12 IU/L, p< 0.001). The reduction in ALP activity did not appear to reflect a generalized impairment in bone remodeling activity, as serum osteocalcin levels were similar between the two strains (249±23ng/mL versus 230±17ng/mL, p = not significant [n.s.]). Together with the increased serum ALP activity, congenic mice bearing the D2 allele also exhibited increased femoral cortical thickness, BMD, and resistance to fracture in comparison to the B6 background strain (Table 1).
Table 1.
Serum ALP and Skeletal Traits of B6 Background and B6.D2.Ch4 Congenic Mice
| B6 background (n=40; 20F/20M) | B6.D2.Ch4 congenic (n=39; 20F/19M) | p value | |
|---|---|---|---|
| Serum ALP activity (U/L) | 60±5 | 142±12 | <0.0001 |
| Femoral bone traits | |||
| BMD (mg/cm2) | 54.6±1.0 | 58.8±0.7 | <0.005 |
| Cortical thickness (µm) | 158±2 | 169±1 | <0.0005 |
| Ultimate failure load (N) | 14.5 ±0.5 | 17.3 ±0.8 | <0.01 |
ALP=alkaline phosphatase; BMD= bone mineral density; F = female; M=male.
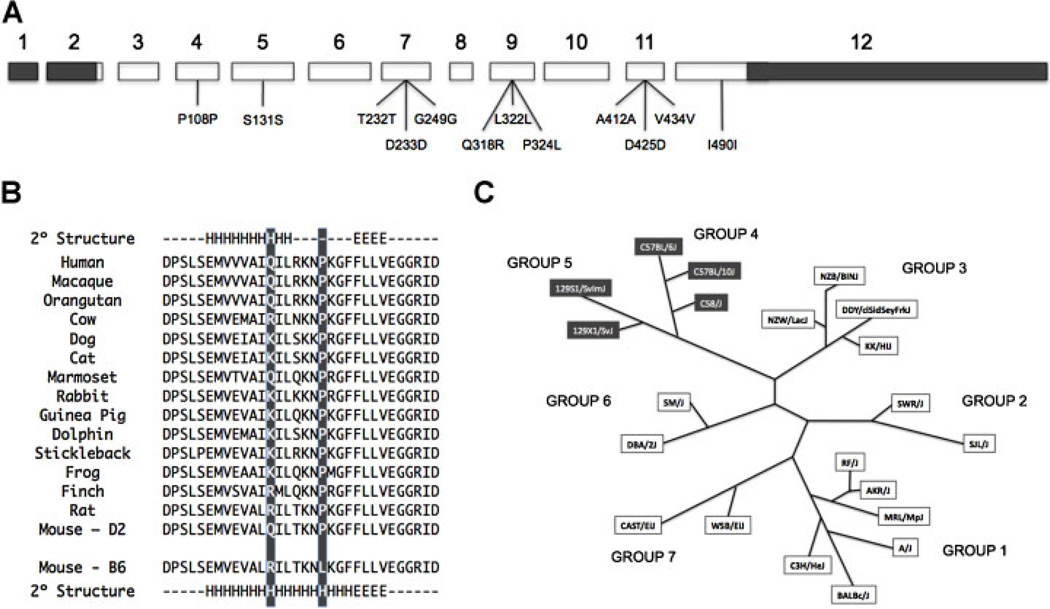
Because steady-state Alpl mRNA levels in hepatic and skeletal tissues were similar between the congenic and background mice (data not shown), coding sequence variation was pursued as an explanation for the observed genotype-dependent differences in circulating ALP enzyme activity and bone mass. A total of 12 sequence variations between the B6 and D2 alleles were identified (Fig. 1A). The majority of these differences were synonymous, but two changes residing in exon 9 did result in coding differences. One variant resulted in an amino acid substitution with arginine in the B6 allele and glutamine in the D2 allele at codon 318 (Gln318Arg) whereas the other variant resulted in an amino acid sequence substitution with leucine in the B6 allele and proline in the D2 allele at codon 324 (Leu324Pro). Although there is recognized evolutionary variability at the Gln318Arg site, the proline at position 324 is conserved in the ALP protein structure of all vertebrates in which sequence is available (Fig. 1B). Secondary structure modeling predicts this amino acid resides in a region of the enzyme corresponding to an alpha-helix (NNPredict(36)). When present, the proline prematurely disrupts this alpha-helix region. The evolutionary constancy of this proline residue and therefore the shortened alpha-helical region strongly suggest an important functional role for this amino acid or region of the enzyme.
Fig. 1.
Coding sequence variations in Alpl between B6 and D2 mice. (A) Exonic structure of Alpl. Open boxed regions indicate coding exons and 5′- and 3′-untranslated regions are represented by filled boxes. A total of 12 sequence variations between the B6 and D2 alleles were identified; two of which, residing within exon 9, are predicted to result in amino acid differences (Q318R and P324L). (B) Comparative sequence analysis of exon 9 of the homologous genes encoding tissue nonspecific ALP. There is evolutionary variability at the Gln318Arg site, but the proline corresponding to position 324 in murine Alpl is conserved in all vertebrates in which sequence is available, with the exception of the B6 inbred strain. Secondary structure modeling was performed with NNPredict.(28) “H,” a predicted helix element; “E,” a predicted beta strand element, or “-,” a predicted turn element. When present, the proline is predicted to prematurely disrupt an alpha-helix region. (C) Exon 9 of Alpl was sequenced from 21 laboratory mouse strains representing the seven phylogenetic divisions of the laboratory mouse family tree. The B6 haplotype (Gln 318/Leu 324) was limited to strains assigned to Groups 4 and 5 of the murine phylogenetic tree (highlighted), while the D2 haplotype was observed in the remaining five groups.
We next resequenced exon 9 of the Alpl gene from a variety of inbred strains representing the various phylogenetic divisions of the laboratory mouse family tree as constructed by Petkov and colleagues.(37) As shown in Figure 1C, the B6 haplotype (Gln 318/Leu 324) was limited to strains assigned to Groups 4 and 5 of the murine phylogenetic tree. Given the critical role that ALP plays in skeletal mineralization, we hypothesized that the Alpl exon 9 mutations could contribute to strain differences in serum ALP activity and measures of bone integrity. We tested this proposal in silico with the Mouse Phenome Database (MPD; http://www.jax.org/phenome), an open source, web-based repository of phenotypic data on commonly used and genetically diverse inbred strains of mice.(38) Mirroring the differences we observed between our B6 background and D2 congenic mice, those strains of mice bearing the B6 (Gln318/ Leu324) haplotype (B6, C57BL/10J, C57BLKS/J, C57L/J, C58/J and 129S1/SvImJ) exhibited reduced serum ALP activity, femoral BMD, cortical thickness, and failure load as compared to those strains of mice bearing the D2 (Arg318/Pro324) haplotype (A/J, BALB/cByJ, C3H/HeJ, CAST/EiJ, D2, NZB/BINJ, NZW/LacJ, SJL/J, SM/J and SWR/J) (Table 2).
Table 2.
Skeletal Traits of Murine Strains With Either the B6 or D2 Alpl Exon 9 Haplotype
| MPD citation | B6-Haplotype strains | D2-Haplotype strains | p value | |
|---|---|---|---|---|
| Serum ALP activity (U/L) | (n = 77; 28F/36M) | (n=122; 68F/54M) | ||
| Ref. (60) | 76±4 | 85±2 | <0.02 | |
| Femoral bone traits | (n=64; 37F/40M) | (n=102; 47F/55M) | ||
| BMD (mg/cm2) | Ref. (61) | 63.0 ±5.0 | 69.1 ±6.0 | <0.001 |
| Cortical thickness (µm) | Ref. (62) | 221±3 | 253±5 | <0.001 |
| Ultimate failure load (N) | Ref. (63) | 23.2±0.6 | 27.4±0.7 | <0.001 |
Data derived from MPD. For serum ALP, activity data were available for the following B6-haplotype strains: 129S1/SvImJ, B6, C57BL/10J, C57BLKS/J, C57L/J; and D2-haplotype strains: A/J, BALB/cByJ, C3H/HeJ, CAST/EiJ, D2, NZW/LacJ and SJL/J. For the femoral bone traits data were available for the following B6-haplotype strains: 129S1/SvImJ, B6, C58/J; and D2-haplotype strains: BALB/cJ, C3H/HeJ, NZB/BINJ, SJL/J, and SWR/J.
ALP=alkaline phosphatase; BMD=bone mineral density; F=female; M=male; MPD=Mouse Phenome Database.
Human studies
Based on the evidence from murine experiments of an association between nonsynonymous coding polymorphisms in Alpl and variations in serum ALP, BMD, and bone mineral strength, we extended our studies to a well-characterized cohort of community-dwelling elderly men. We sequenced the 11 coding regions (exons 2–12, including intron-exon boundaries) of ALPL in a subset of white men in the MrOS cohort. To increase the probability of identifying functionally relevant ALPL variants, we selected 74 individuals with serum ALP below the clinical cut point for normal (<40 U/L, n=74)(39) and 148 with serum ALP levels nearest the median of the population distribution (72 U/L). A total of 25 sequence variants were identified, with 21 resulting in nonsynonymous changes in amino acid coding and 22 occurring at MAF<1% in the normal ALP group (Table 3; Supplementary Table 1). Seven of these variants were not previously reported in 1000 Genomes or SNP databases. In the low ALP group, 60.8% of men had any of the 25 variants, and in the normal ALP group, 37.1% had at least one of the variants (p for χ2 =6×10−4). The difference in frequency between groups was minimal for variants with MAF ≥ 1% (37.8% in those with low ALP and 33.1% in those with normal, p for χ2 = 0.49). However, a far greater proportion of men in the low ALP group had a rare variant (36.5%) than did men in the normal ALP group (3.4%, p for Fisher’s exact test = 1.6 × 10−10).
Table 3.
Single-Nucleotide Variants Detected by Sequencing ALPL in Groups of Men Selected for Low or Normal Serum ALP Activity, Osteoporotic Fractures in Men (MrOS) Study
| Group 1, selected for low ALP (n=74) | Group 2, selected for normal ALP (n=148) | ||||
|---|---|---|---|---|---|
| ALP (U/L) | |||||
| Mean (SD) | 34.2 (4.9) | 71.8 (1.1) | |||
| Range | 19.0–39.0 | 70.0–74.0 | |||
| Phosphate (mg/dL) | 3.33 (0.48) | 3.20 (0.48) | |||
| n (%) | n (%) | pa | |||
| Any variant (25 variants) | 45 (60.8%) | 65 (37.1%) | 0.0006 | ||
| Any variant with MAF ≥ 1% (3 variants) | 28(37.8%) | 49 (33.1%) | 0.49 | ||
| Any variant with MAF<1% (22 variants | 27(36.5%) | 5 (3.4%) | 1.6 × 10−10c | ||
| Variants detectedb | n (%) | n (%) | MAF (%) | pc | |
| Synonymous (four variants) | 20 (27.0) | 32 (21.6) | NA | 0.40 | |
| S110S | T>C | 2 (2.7) | 1 (0.7) | 0.3% | 0.26 |
| P292P | AA | 58 (78.4) | 118 (79.7) | 11.1% | 0.89 |
| AG | 15 (20.3) | 27 (18.2) | |||
| GG | 1 (1.4) | 3 (2.0) | |||
| T366T | C>G | 1 (1.4) | 0 (0.0) | d | 0.30 |
| A514A | T>G | 1 (3.2) | 2 (1.4) | 0.7% | >0.99 |
| Nonsynonymous, common (two variants) | 23 (31.1) | 45 (30.4) | NA | >0.99 | |
| Y263He | TT | 61 (82.4) | 119(80.4) | 10.1% | 0.90 |
| TC | 13 (17.6) | 28 (18.9) | |||
| CC | 0 (0.0) | 1 (0.7) | |||
| V522A | TC | 11 (14.9) | 22 (14.9) | 7.5% | >0.99 |
| Nonsynonymous, rare (19 variants) | 25 (33.8) | 2 (1.4) | NA | 9.9 × 10−12 | |
| T134He | A>C | 1 (1.4) | 0 (0.0) | d | 0.33 |
| E146Ke | G>A | 2 (2.7) | 0 (0.0) | d | 0.11 |
| R152H | G>A | 5 (6.8) | 2 (1.7) | 0.9% | 0.05 |
| S181Le | C>T | 1 (1.4) | 0 (0.0) | d | 0.33 |
| M192Te | T>C | 2 (2.7) | 0 (0.0) | d | 0.11 |
| R246Se | G>T | 1 (1.4) | 0 (0.0) | d | 0.33 |
| G249Ve | G>T | 1 (1.4) | 0 (0.0) | d | 0.33 |
| R272Ce | C>T | 2 (2.7) | 0 (0.0) | d | 0.11 |
| T273Me | C>T | 2 (2.7) | 0 (0.0) | d | 0.11 |
| F327Ce | T>C | 1 (1.4) | 0 (0.0) | d | 0.33 |
| R357Le | G>T | 1 (1.4) | 0 (0.0) | d | 0.33 |
| R391Ce | C>T | 2 (2.7) | 0 (0.0) | d | 0.11 |
| N417S | A>G | 2 (2.7) | 0 (0.0) | d | 0.11 |
| R428Qe | G>A | 1 (1.4) | 0 (0.0) | d | 0.33 |
| Y436Se | A>C | 1 (1.4) | 0 (0.0) | d | 0.33 |
| A443V | C>T | 1 (1.4) | 0 (0.0) | d | 0.33 |
| V461I | A>G | 1 (1.4) | 0 (0.0) | d | 0.33 |
| H482Ne | C>A | 1 (1.4) | 0 (0.0) | d | 0.33 |
| G491Re | G>A | 2 (2.7) | 0 (0.0) | d | 0.11 |
ALP = alkaline phosphatase; MAF=minor allele frequency; NA= not applicable.
For χ2, compared to the group with no variant of the same category.
All variants were detected as heterozygotes, except for P292P and Y263H.
For Fisher’s exact test.
Minor allele not detected in the “normal ALP” group.
Sequence variant resulting in a nonconservative amino acid substitution.
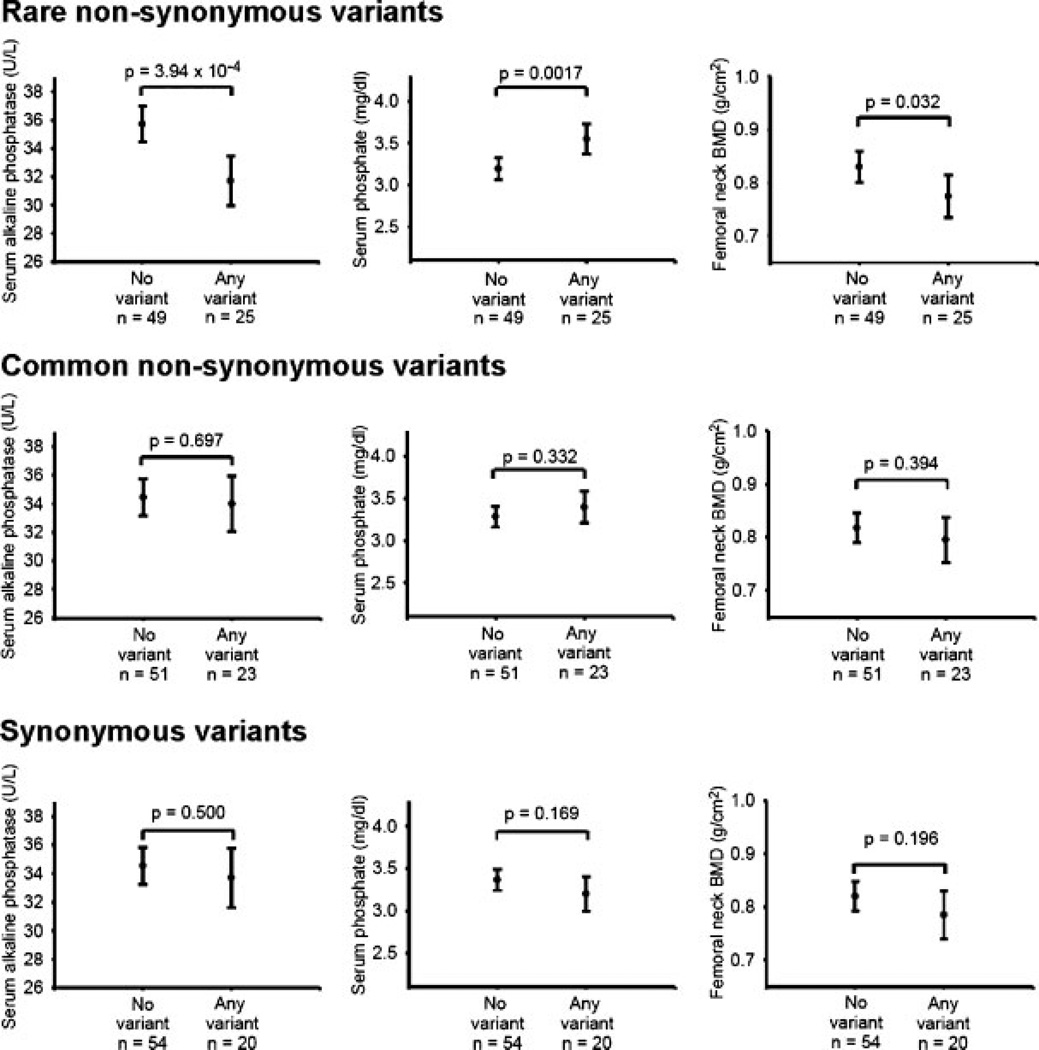
To test the hypothesis that nonsynonymous rare variants in ALPL were associated with physiological consequences, we examined the group of men in the low ALP group, in which there was a sufficient frequency of rare variants to explore this hypothesis. Beyond direct measure of the circulating enzyme activity, other clinical markers of defective ALP action include increased urinary phosphoethanolamine (PEA) and increased serum phosphate and pyridoxal 5’-phosphate (PLP) levels.(17,40) Neither PEA excretion nor PLP levels were measured in the MrOS cohort, but fasting serum phosphate levels were available for analysis. Although each individual variant occurred in only one, two, or five of the men, 25 men (33.8%) in the low ALP group had at least one rare variant. Therefore, the association with this combined category of ‘‘any rare, nonsynonymous variant’’ was examined for serum ALP, serum phosphate, femoral neck BMD, and whole-body BMD. Men with a rare, nonsynonymous variant had 11.2% lower mean serum ALP (p = 3.9× 10−4), 6.7% lower femoral neck BMD (p = 0.03), 6.6% lower whole-body BMD (p = 0.007), and 11.1% higher serum phosphate (p = 0.002) than those without such a variant. In contrast, serum ALP, phosphate or BMD did not differ according to the presence of a common nonsynonymous variant or a synonymous variant (Fig. 2).
Fig. 2.
Those subjects with rare (MAF < 1% in the normal ALP group) nonsynonymous ALPL variants exhibited lower serum ALP activity, higher serum phosphate concentrations, and lower femoral neck BMD values. However, there were no statistically significant differences in serum ALP, phosphate, or BMD between participants with and without synonymous or common (MAF>1% in the normal ALP group) nonsynonymous variants. Plots show means and 95% confidence intervals, adjusted for age, weight, and clinic site, in the MrOS group selected for low serum ALP (n = 74).
Discussion
By combining data from mouse models and human studies, we uncovered associations between rare variants in the ALP gene sequence (murine Alpl and human ALPL) and both circulating ALP enzyme activity and bone density. These associations contribute to our understanding of heritable differences in BMD and fracture risk. Our findings of rare ALPL polymorphisms that are related to variation in relevant skeletal traits points to the need for carefully designed studies to determine their population level impact and to discover additional rare variants in skeletal mineralization genes. Moreover, the finding of rare variants in the ALPL gene and their association with clinically important phenotypes has implications for the significance of rare alleles in the determination of complex human traits.
Our studies in mouse models, and similar studies by others, strongly suggest that substantial variation in alkaline phospha-tase activity, bone density and bone strength are the result of polymorphisms in the ALP gene. A review of the findings from multiple murine experiments suggests that the phenotype-genotype association that links ALP activity and measures of bone strength to a chromosome 4 QTL is dependent on the presence or absence of Alpl exon 9 coding variations in the strains of laboratory mice examined. In published QTL experiments involving B6 and D2(6,8) or B6 and C3H/HeJ(41) in which the chromosome 4 (Alpl) locus has been implicated, Alpl exon 9 coding variations (Gln318Arg and Leu324Pro) exist between the two mouse strains. In contrast, when sequence variation is absent at this site (eg, between MRL/MpJ and SJL/J mice), no linkage to chromosome 4 has been identified.(21) A similar observation is made when murine linkage analyses for BMD and/ or bone strength traits are reviewed. The chromosome 4 region was identified in experimental intercrosses using strains divergent for the Alpl haplotype—B6 × C3H/HeJ(4,5,7) and B6 × D2(3,8)—whereas this region was not implicated when strains bearing the same Alpl haplotype were employed — MRL × SJL,(42) NZB/B1NJ × RF/J,(43) SM/J × NZB/BlNJ,(44) or B6 × 129S1/SvImJ.(45) Certainly, other nearby genetic variants in strong linkage with the exon 9 Alpl variants could also be responsible. However, the strong concordance of results from multiple laboratories employing a variety of genetically distinct mouse strains provides compelling circumstantial evidence for an underlying role for these particular coding variations in Alpl in determining strain-dependent differences in ALP activity and measures of skeletal integrity. The results of our studies of a congenic strain support the hypothesis that genetic variation on chromosome 4 is responsible for alterations in ALP activity, bone density, and bone strength. Together, these results provided a strong rationale for exploring genetic variation in the human gene and its association with similar human phenotypes.
This is the first study to describe multiple rare variants in ALPL detected in a healthy cohort of older men designed to be representative of the U.S. population. Of the 25 sequence variants in ALPL identified here, seven had not previously been reported and are unlikely to have been discovered in this cohort if we had not sequenced those men with the lowest serum ALP. With these variants identified, future work can focus on determining the prevalence of such variants in larger samples of men and in women and on the proportion of variation in BMD that they explain in the population. While the current study included sequence variants in coding regions only, additional sequencing of intronic regions and of regulatory regions may reveal additional variants that affect gene transcription or stability of the protein.
Although it is generally accepted that the genetic basis of osteoporosis—among other common chronic diseases—is likely the consequence of multiple genetic factors working in aggregate, there is considerable debate over the importance of genetic variations that occur commonly versus those that rarely occur. The first model suggests that complex traits are likely caused by alleles common in the population with modest phenotypic effects—the common disease, common variant (CDCV) hypothesis. An alternative model argues that multiple rare DNA sequence variations, each with relatively large individual effects, are major contributors to genetic susceptibility to common diseases—the common disease, rare variant (CDRV) hypothesis.
Based on the CDCV hypothesis, numerous linkage-disequilibrium and GWA mapping studies have identified the genomic region harboring ALPL (1p36) as containing common genetic variants responsible for altering circulating ALP activity.(23,24) serum phosphorus,(46) and osteoporosis risk.(2,9–16,47) However, the genetic variation(s) residing within this region capable of influencing these traits are not yet known. The rare variant hypothesis suggests direct sequencing of targeted subsets of the population may be the optimal way to identify alleles associated with complex phenotypes. Although the relative frequency of variants that contribute to common diseases continues to be a subject of intense speculation,(48) recent population studies of plasma triglycerides,(49) HDL,(50) and LDL(51) cholesterol levels, as well as type 1 diabetes risk,(52) blood pressure variation,(53) and colorectal adenoma development(54) indicate that rare variants are important for disease susceptibility and should not be ignored in genetic studies of complex traits. Still, a major stumbling block at this juncture is determining for which gene(s) it is appropriate to commit the substantial resources required for sequencing efforts. As demonstrated in this report, we believe that animal models can be instrumental in pinpointing “high-value” genes for such intensive scrutiny in human populations.
Wang and colleagues(55) have recently demonstrated that rare variants can create synthetic association signals in GWA studies by occurring relatively more frequently in association with one of the alleles of a common tag SNP, therefore explaining a situation in which a common SNP (or SNPs) seem to confer risk for a complex trait. Although our study identified many rare ALPL variants, we have not yet genotyped them in larger cohorts to determine whether the frequency of these variants is high enough to make an association study feasible. In the future, genotyping for all possible rare variants would allow us to collapse the results into a single ‘‘rare variant’’ category and estimate the amount of variation in ALP and/or BMD attributable to the existence of rare variants in the population. Confirmation of our findings with multiple other ALPL variants and in other populations, would support the importance of sequencing in candidate gene regions or in whole exomes to detect rare coding variants that are more likely to provide evidence of causal links to heritable outcomes than those reported for common tag SNPs.
Establishing the functional consequences of the ALPL variants described here will require extensive experimental work. The variants we report are located in various domains of the TNAP structure (eg, in the active site, crown domain, homodimer interface, and calcium binding site), and deleterious mutations have been described in each of these regions.(56) Yet it seems probable that rare nonsynonymous coding variants identified by resequencing will be more likely to result in functional changes than the common variants with smaller effects detected by GWA studies. In this study, rare nonsynonymous variants were always present in the heterozygous state, suggesting they might be functioning through a dominant negative mechanism (eg, the activity of the wild type monomer is inhibited by the mutated monomer when both the normal and the mutated ALP protein dimerize(52)). Support for this possibility can be derived from previous site-directed in vitro mutagenesis studies on subjects with mild HPP, indicating that a majority of tested heterozygous ALPL mutations exhibit a dominant negative effect.(57–59) It is intriguing to consider the possibility that a mild genetic form of HPP (ie, heterozygous inheritance of a single ALPL missense mutation) might explain some proportion of adult-onset osteoporosis.
Supplementary Material
Acknowledgments
The Osteoporotic Fractures in Men (MrOS) Study is supported by National Institutes of Health funding; the following institutes provide support: the National Institute of Arthritis and Musculo-skeletal and Skin Diseases (NIAMS), the National Institute on Aging (NIA), the National Center for Research Resources (NCRR), and NIH Roadmap for Medical Research under the following grant numbers: U01 AR45580, U01 AR45614, U01 AR45632, U01 AR45647, U01 AR45654, U01 AR45583, U01 AG18197, U01-AG027810, and UL1 RR024140. Dr. Nielson is also supported by the Office of Research on Women’s Health and the National Institute of Child Health and Human Development, Building Interdisciplinary Research Careers in Women’s Health (BIRCWH) grant number HD043488-08. Dr. Klein’s research effort is supported by the National Institute of Arthritis, Musculoskeletal and Skin Diseases (AR44659) and the VA Medical Research Service. Dr. Zmuda’s research effort is supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases (R01-AR051124) and by the National Institute on Aging (R01-AG033618).
Footnotes
Additional Supporting Information may be found in the online version of this article.
Disclosures
All authors state that they have no conflicts of interest.
Authors’ roles: CMN conceived of the study, analyzed, and interpreted the human data and wrote the manuscript. JMZ directed the human DNA extraction, and contributed to data analysis and manuscript preparation. EAL, under the direction of RFK, carried out the murine skeletal phenotyping and data analysis. ASC and WJW, under the direction of RFK, performed the DNA sequencing and data analysis. ESO directed the MrOS cohort study, assisted in the interpretation of data, and contributed to manuscript revision. RFK conceived of the study, designed the murine experiments, analyzed and interpreted the data, and wrote the manuscript.
References
- 1.Ralston SH, Uitterlinden AG. Genetics of osteoporosis. Endocr Rev. 2010;31(5):629–662. doi: 10.1210/er.2009-0044. [DOI] [PubMed] [Google Scholar]
- 2.Rivadeneira F, Styrkarsdottir U, Estrada K, Halldorsson BV, Hsu YH, Richards JB, Zillikens MC, Kavvoura FK, Amin N, Aulchenko YS, Cupples LA, Deloukas P, Demissie S, Grundberg E, Hofman A, Kong A, Karasik D, van Meurs JB, Oostra B, Pastinen T, Pols HA, Sigurdsson G, Soranzo N, Thorleifsson G, Thorsteinsdottir U, Williams FM, Wilson SG, Zhou Y, Ralston SH, van Duijn CM, Spector T, Kiel DP, Stefansson K, Ioannidis JP, Uitterlinden AG. Twenty bone-mineral-density loci identified by large-scale meta-analysis of genome-wide association studies. Nat Genet. 2009;41(11):1199–1206. doi: 10.1038/ng.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klein RF, Carlos AS, Vartanian KA, Chambers VK, Turner EJ, Phillips TJ, Belknap JK, Orwoll ES. Confirmation and fine mapping of chromosomal regions influencing peak bone mass in mice. J Bone Miner Res. 2001;16(11):1953–1961. doi: 10.1359/jbmr.2001.16.11.1953. [DOI] [PubMed] [Google Scholar]
- 4.Beamer WG, Shultz KL, Donahue LR, Churchill GA, Sen S, Wergedal JR, Baylink DJ, Rosen CJ. Quantitative trait loci for femoral and lumbar vertebral bone mineral density in C57BL/6J and C3H/HeJ inbred strains of mice. J Bone Miner Res. 2001;16(7):1195–1206. doi: 10.1359/jbmr.2001.16.7.1195. [DOI] [PubMed] [Google Scholar]
- 5.Farber CR, van Nas A, Ghazalpour A, Aten JE, Doss S, Sos B, Schadt EE, Ingram-Drake L, Davis RC, Horvath S, Smith DJ, Drake TA, Lusis AJ. An integrative genetics approach to identify candidate genes regulating BMD: combining linkage, gene expression, and association. J Bone Miner Res. 2009;24(1):105–116. doi: 10.1359/JBMR.080908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foreman JE, Blizard DA, Gerhard G, Mack HA, Lang DH, Van Nimwe-gen KL, Vogler GP, Stout JT, Shihabi ZK, Griffith JW, Lakoski JM, McClearn GE, Vandenbergh DJ. Serum alkaline phosphatase activity is regulated by a chromosomal region containing the alkaline phos-phatase 2 gene (Akp2) in C57BL/6J and DBA/2J mice. Physiol Genomics. 2005;23(3):295–303. doi: 10.1152/physiolgenomics.00062.2005. [DOI] [PubMed] [Google Scholar]
- 7.Koller DL, Schriefer J, Sun Q, Shultz KL, Donahue LR, Rosen CJ, Foroud T, Beamer WG, Turner CH. Genetic effects for femoral biomechanics, structure, and density in C57BL/6J and C3H/HeJ inbred mouse strains. J Bone Miner Res. 2003;18(10):1758–1765. doi: 10.1359/jbmr.2003.18.10.1758. [DOI] [PubMed] [Google Scholar]
- 8.Lang DH, Sharkey NA, Mack HA, Vogler GP, Vandenbergh DJ, Blizard DA, Stout JT, McClearn GE. Quantitative trait loci analysis of structural and material skeletal phenotypes in C57BL/6J and DBA/2 second-generation and recombinant inbred mice. J Bone Miner Res. 2005;20(1):88–99. doi: 10.1359/JBMR.041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ermakov S, Toliat MR, Cohen Z, Malkin I, Altmuller J, Livshits G, Nurnberg P. Association of ALPL and ENPP1 gene polymorphisms with bone strength related skeletal traits in a Chuvashian population. Bone. 2010;46(5):1244–1250. doi: 10.1016/j.bone.2009.11.018. [DOI] [PubMed] [Google Scholar]
- 10.Huang QY, Li GH, Kung AW. Multiple osteoporosis susceptibility genes on chromosome 1p36 in Chinese. Bone. 2009;44(5):984–988. doi: 10.1016/j.bone.2009.01.368. [DOI] [PubMed] [Google Scholar]
- 11.Karasik D, Myers RH, Hannan MT, Gagnon D, McLean RR, Cupples LA, Kiel DP. Mapping of quantitative ultrasound of the calcaneus bone to chromosome 1 by genome-wide linkage analysis. Osteoporos Int. 2002;13(10):796–802. doi: 10.1007/s001980200110. [DOI] [PubMed] [Google Scholar]
- 12.Lee YH, Rho YH, Choi SJ, Ji JD, Song GG. Meta-analysis of genome-wide linkage studies for bone mineral density. J Hum Genet. 2006;51(5):480–486. doi: 10.1007/s10038-006-0390-9. [DOI] [PubMed] [Google Scholar]
- 13.Streeten EA, Beck TJ, O’Connell JR, Rampersand E, McBride DJ, Takala SL, Pollin TI, Uusi-Rasi K, Mitchell BD, Shuldiner AR. Autosome-wide linkage analysis of hip structural phenotypes in the Old Order Amish. Bone. 2008;43(3):607–612. doi: 10.1016/j.bone.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Willaert A, Van Pottelbergh I, Zmierczak H, Goemaere S, Kaufman JM, De Paepe A, Coucke P. A genome-wide linkage scan for low spinal bone mineral density in a single extended family confirms linkage to 1p36.3. Eur J Hum Genet. 2008;16(8):970–976. doi: 10.1038/ejhg.2008.31. [DOI] [PubMed] [Google Scholar]
- 15.Xiao P, Shen H, Guo YF, Xiong DH, Liu YZ, Liu YJ, Zhao LJ, Long JR, Guo Y, Recker RR, Deng HW. Genomic regions identified for BMD in a large sample including epistatic interactions and gender-specific effects. J Bone Miner Res. 2006;21(10):1536–1544. doi: 10.1359/jbmr.060717. [DOI] [PubMed] [Google Scholar]
- 16.Zhang YP, Deng FY, Chen Y, Pei YF, Fang Y, Guo YF, Guo X, Liu XG, Zhou Q, Liu YJ, Deng HW. Replication study of candidate genes/loci associated with osteoporosis based on genome-wide screening. Osteoporos Int. 2009;21(5):785–795. doi: 10.1007/s00198-009-1014-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Orimo H. The mechanism of mineralization and the role of alkaline phosphatase in health and disease. J Nippon Med Sch. 2010;77(1):4–12. doi: 10.1272/jnms.77.4. [DOI] [PubMed] [Google Scholar]
- 18.Fedde KN, Blair L, Silverstein J, Coburn SP, Ryan LM, Weinstein RS, Waymire K, Narisawa S, Millan JL, MacGregor GR, Whyte MP. Alkaline phosphatase knock-out mice recapitulate the metabolic and skeletal defects of infantile hypophosphatasia. J Bone Miner Res. 1999;14(12):2015–2026. doi: 10.1359/jbmr.1999.14.12.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kalousdian S, Fabsitz R, Havlik R, Christian J, Rosenman R. Heritability of clinical chemistries in an older twin cohort: the NHLBI Twin Study. Genet Epidemiol. 1987;4(1):1–11. doi: 10.1002/gepi.1370040102. [DOI] [PubMed] [Google Scholar]
- 20.Reiner G, Clemens N, Fischer R, Kohler F, Berge T, Hepp S, Willems H. Mapping of quantitative trait loci for clinical-chemical traits in swine. Anim Genet. 2009;40(1):57–64. doi: 10.1111/j.1365-2052.2008.01804.x. [DOI] [PubMed] [Google Scholar]
- 21.Srivastava AK, Masinde G, Yu H, Baylink DJ, Mohan S. Mapping quantitative trait loci that influence blood levels of alkaline phos-phatase in MRL/MpJ and SJL/J mice. Bone. 2004;35(5):1086–1094. doi: 10.1016/j.bone.2004.07.011. [DOI] [PubMed] [Google Scholar]
- 22.Whitfield JB, Martin NG. Determinants of variation in plasma alkaline phosphatase activity: a twin study. Am J Hum Genet. 1983;35(5):978–986. [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, Vollenweider P, Stirnadel H, Johnson T, Bergmann S, Beckmann ND, Li Y, Ferrucci L, Melzer D, Hernandez D, Singleton A, Scott J, Elliott P, Waeber G, Cardon L, Frayling TM, Kooner JS, Mooser V. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet. 2008;83(4):520–528. doi: 10.1016/j.ajhg.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamatani Y, Matsuda K, Okada Y, Kubo M, Hosono N, Daigo Y, Nakamura Y, Kamatani N. Genome-wide association study of hema-tological and biochemical traits in a Japanese population. Nat Genet. 2010;42(3):210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 25.Walker BA, Eze LC, Tweedie MC, Evans DA. The influence of ABO blood groups, secretor status and fat ingestion on serum alkaline phosphatase. Clin Chim Acta. 1971;35(2):433–444. doi: 10.1016/0009-8981(71)90218-x. [DOI] [PubMed] [Google Scholar]
- 26.Whyte MP. Physiological role of alkaline phosphatase explored in hypophosphatasia. Ann N Y Acad Sci. 2010;1192:190–200. doi: 10.1111/j.1749-6632.2010.05387.x. [DOI] [PubMed] [Google Scholar]
- 27.Mornet E, Beck C, Bloch-Zupan A, Girschick H, Le Merrer M. Clinical utility gene card for: hypophosphatasia. Eur J Hum Genet. 2011;19:163. doi: 10.1038/ejhg.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buck KJ, Metten P, Belknap JK, Crabbe JC. Quantitative trait loci involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17(10):3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dietrich W, Katz H, Lincoln SE, Shin HS, Friedman J, Dracopoli NC, Lander ES. A genetic map of the mouse suitable for typing intraspe-cific crosses. Genetics. 1992;131(2):423–447. doi: 10.1093/genetics/131.2.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serikawa T, Kuramoto T, Hilbert P, Mori M, Yamada J, Dubay CJ, Lindpainter K, Ganten D, Guenet JL, Lathrop GM, et al. Rat gene mapping using PCR-analyzed microsatellites. Genetics. 1992;131(3):701–721. doi: 10.1093/genetics/131.3.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16(22):10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blank JB, Cawthon PM, Carrion-Petersen ML, Harper L, Johnson JP, Mitson E, Delay RR. Overview of recruitment for the osteoporotic fractures in men study (MrOS) Contemp Clin Trials. 2005;26(5):557–568. doi: 10.1016/j.cct.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 33.Orwoll E, Blank JB, Barrett-Connor E, Cauley J, Cummings S, Ensrud K, Lewis C, Cawthon PM, Marcus R, Marshall LM, McGowan J, Phipps K, Sherman S, Stefanick ML, Stone K. Design and baseline characteristics of the osteoporotic fractures in men (MrOS) study—a large observational study of the determinants of fracture in older men. Contemp Clin Trials. 2005;26(5):569–585. doi: 10.1016/j.cct.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 34.Cornoni-Huntley J, Ostfeld AM, Taylor JO, Wallace RB, Blazer D, Berkman LF, Evans DA, Kohout FJ, Lemke JH, Scherr PA, et al. Established populations for epidemiologic studies of the elderly: study design and methodology. Aging (Milano) 1993;5(1):27–37. doi: 10.1007/BF03324123. [DOI] [PubMed] [Google Scholar]
- 35.Pahor M, Chrischilles EA, Guralnik JM, Brown SL, Wallace RB, Carbonin P. Drug data coding and analysis in epidemiologic studies. Eur J Epidemiol. 1994;10(4):405–411. doi: 10.1007/BF01719664. [DOI] [PubMed] [Google Scholar]
- 36.Kneller DG, Cohen FE, Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990;214(1):171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- 37.Petkov PM, Ding Y, Cassell MA, Zhang W, Wagner G, Sargent EE, Asquith S, Crew V, Johnson KA, Robinson P, Scott VE, Wiles MV. An efficient SNP system for mouse genome scanning and elucidating strain relationships. Genome Res. 2004;14(9):1806–1811. doi: 10.1101/gr.2825804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grubb SC, Maddatu TP, Bult CJ, Bogue MA. Mouse phenome database. Nucleic Acids Res. 2009;37:D720–D730. doi: 10.1093/nar/gkn778. Database issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaplan MM. Alkaline phosphatase. N Engl J Med. 1972;286(4):200–202. doi: 10.1056/NEJM197201272860407. [DOI] [PubMed] [Google Scholar]
- 40.Chodirker BN, Evans JA, Seargeant LE, Cheang MS, Greenberg CR. Hyperphosphatemia in infantile hypophosphatasia: implications for carrier diagnosis and screening. Am J Hum Genet. 1990;46(2):280–285. [PMC free article] [PubMed] [Google Scholar]
- 41.Richman C, Kutilek S, Miyakoshi N, Srivastava AK, Beamer WG, Donahue LR, Rosen CJ, Wergedal JE, Baylink DJ, Mohan S. Postnatal and pubertal skeletal changes contribute predominantly to the differences in peak bone density between C3H/HeJ and C57BL/6J mice. J Bone Miner Res. 2001;16(2):386–397. doi: 10.1359/jbmr.2001.16.2.386. [DOI] [PubMed] [Google Scholar]
- 42.Masinde GL, Li X, Gu W, Wergedal J, Mohan S, Baylink DJ. Quantitative trait loci for bone density in mice: the genes determining total skeletal density and femur density show little overlap in F2 mice. Calcif Tissue Int. 2002;71(5):421–428. doi: 10.1007/s00223-001-1113-z. [DOI] [PubMed] [Google Scholar]
- 43.Wergedal JE, Sheng MH, Ackert-Bicknell CL, Beamer WG, Baylink DJ. Mouse genetic model for bone strength and size phenotypes: NZB/ B1NJ and RF/J inbred strains. Bone. 2002;31(6):670–674. doi: 10.1016/s8756-3282(02)00908-0. [DOI] [PubMed] [Google Scholar]
- 44.Ishimori N, Stylianou IM, Korstanje R, Marion MA, Li R, Donahue LR, Rosen CJ, Beamer WG, Paigen B, Churchill GA. Quantitative trait loci for BMD in an SM/J by NZB/BlNJ intercross population and identification of Trps1 as a probable candidate gene. J Bone Miner Res. 2008;23(9):1529–1537. doi: 10.1359/JBMR.080414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishimori N, Li R, Walsh KA, Korstanje R, Rollins JA, Petkov P, Pletcher MT, Wiltshire T, Donahue LR, Rosen CJ, Beamer WG, Churchill GA, Paigen B. Quantitative trait loci that determine BMD in C57BL/6J and 129S1/SvImJ inbred mice. J Bone Miner Res. 2006;21(1):105–112. doi: 10.1359/JBMR.050902. [DOI] [PubMed] [Google Scholar]
- 46.Kestenbaum B, Glazer NL, Kottgen A, Felix JF, Hwang SJ, Liu Y, Lohman K, Kritchevsky SB, Hausman DB, Petersen AK, Gieger C, Ried JS, Meitinger T, Strom TM, Wichmann HE, Campbell H, Hayward C, Rudan I, de Boer IH, Psaty BM, Rice KM, Chen YD, Li M, Arking DE, Boerwinkle E, Coresh J, Yang Q, Levy D, van Rooij FJ, Dehghan A, Rivadeneira F, Uitterlinden AG, Hofman A, van Duijn CM, Shlipak MG, Kao WH, Witteman JC, Siscovick DS, Fox CS. Common genetic variants associate with serum phosphorus concentration. J Am Soc Nephrol. 2010;21(7):1223–1232. doi: 10.1681/ASN.2009111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goseki-Sone M, Sogabe N, Fukushi-Irie M, Mizoi L, Orimo H, Suzuki T, Nakamura H, Orimo H, Hosoi T. Functional analysis of the single nucleotide polymorphism (787T>C) in the tissue-nonspecific alkaline phosphatase gene associated with BMD. J Bone Miner Res. 2005;20(5):773–782. doi: 10.1359/JBMR.041229. [DOI] [PubMed] [Google Scholar]
- 48.Schork NJ, Murray SS, Frazer KA, Topol EJCommon vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19(3):212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johansen CT, Wang J, Lanktree MB, Cao H, McIntyre AD, Ban MR, Martins RA, Kennedy BA, Hassell RG, Visser ME, Schwartz SM, Voight BF, Elosua R, Salomaa V, O’Donnell CJ, Dallinga-Thie GM, Anand SS, Yusuf S, Huff MW, Kathiresan S, Hegele RA. Excess of rare variants in genes identified by genome-wide association study of hypertrigly-ceridemia. Nat Genet. 2010;42(8):684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 51.Cohen JC, Pertsemlidis A, Fahmi S, Esmail S, Vega GL, Grundy SM, Hobbs HH. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103(6):1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ji W, Foo JN, O’Roak BJ, Zhao H, Larson MG, Simon DB, Newton-Cheh C, State MW, Levy D, Lifton RP. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40(5):592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fearnhead NS, Wilding JL, Winney B, Tonks S, Bartlett S, Bicknell DC, Tomlinson IP, Mortensen NJ, Bodmer WF. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proc Natl Acad Sci U S A. 2004;101(45):15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang K, Dickson SP, Stolle CA, Krantz ID, Goldstein DB, Hakonarson H. Interpretation of association signals and identification of causal variants from genome-wide association studies. Am J Hum Genet. 2010;86(5):730–742. doi: 10.1016/j.ajhg.2010.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mornet E. Hypophosphatasia. Best Pract Res Clin Rheumatol. 2008;22(1):113–127. doi: 10.1016/j.berh.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 57.Fauvert D, Brun-Heath I, Lia-Baldini AS, Bellazi L, Taillandier A, Serre JL, de Mazancourt P, Mornet E. Mild forms of hypophosphatasia mostly result from dominant negative effect of severe alleles or from compound heterozygosity for severe and moderate alleles. BMC Med Genet. 2009;10:51. doi: 10.1186/1471-2350-10-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lia-Baldini AS, Brun-Heath I, Carrion C, Simon-Bouy B, Serre JL, Nunes ME, Mornet E. A new mechanism of dominance in hypophosphatasia: the mutated protein can disturb the cell localization of the wild-type protein. Hum Genet. 2008;123(4):429–432. doi: 10.1007/s00439-008-0480-1. [DOI] [PubMed] [Google Scholar]
- 59.Lia-Baldini AS, Muller F, Taillandier A, Gibrat JF, Mouchard M, Robin B, Simon-Bouy B, Serre JL, Aylsworth AS, Bieth E, Delanote S, Freisinger P, Hu JC, Krohn HP, Nunes ME, Mornet E. A molecular approach to dominance in hypophosphatasia. Hum Genet. 2001;109(1):99–108. doi: 10.1007/s004390100546. [DOI] [PubMed] [Google Scholar]
- 60.Yuan R, Korstanje R. Mouse Phenome Database web site [Internet] Bar Harbor, ME, USA: The Jackson Laboratory; [Cited October 1, 2011]. Aging Study: Blood Chemistry for 32 Inbred Strains of Mice. MPD: 24411. Available from: http://phenome.jax.org/db/qp?rtn=views/measplot&brieflook=24411. [Google Scholar]
- 61.Donahue L, Beamer WG, Bogue MA, Churchill GA. Mouse Phenome Database web site [Internet] Bar Harbor, ME, USA: The Jackson Laboratory; [Cited October 1, 2011]. Characterization of Skeletal Geometry and Bone Strength in 10 Inbred Strains of Mice. MPD: 17203. Available from: http://phenome.jax.org/db/qp?rtn=views/measplot&brieflook=17203. [Google Scholar]
- 62.Donahue L, Beamer WG, Bogue MA, Churchill GA. Mouse Phenome Database web site [Internet] Bar Harbor, ME, USA: The Jackson Laboratory; [Cited October 1, 2011]. Characterization of Skeletal Geometry and Bone Strength in 10 Inbred Strains of Mice. MPD: 17217. Available from: http://phenome.jax.org/db/qp?rtn=views/measplot&brieflook=17217. [Google Scholar]
- 63.Donahue L, Beamer WG, Bogue MA, Churchill GA. Mouse Phenome Database web site [Internet] Bar Harbor, ME, USA: The Jackson Laboratory; [Cited October 1, 2011]. Characterization of Skeletal Geometry and Bone Strength in 10 Inbred Strains of Mice. MPD: 17208. Available from: http://phenome.jax.org/db/qp?rtn=views/measplot&brieflook=17208. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.