Abstract
In the cytoplasm, actin filaments form crosslinked networks that enable eukaryotic cells to transport cargo, change shape, and move. Actin is also present in the nucleus but, in this compartment, its functions are more cryptic and controversial. If we distill the substantial literature on nuclear actin down to its essentials, we find four, recurring, and more-or-less independent, claims: (1) crosslinked networks of conventional actin filaments span the nucleus and help maintain its structure and organize its contents; (2) assembly or contraction of filaments regulates specific nuclear events; (3) actin monomers moonlight as subunits of chromatin remodeling complexes, independent of their ability to form filaments; and (4) modified actin monomers or oligomers, structurally distinct from canonical, cytoskeletal filaments, mediate nuclear events by unknown mechanisms. We discuss the evidence underlying these claims and as well as their strengths and weaknesses. Next, we describe our recent work, in which we built probes specific for nuclear actin and used them to describe the form and distribution of actin in somatic cell nuclei. Finally, we discuss how different forms of nuclear actin may play different roles in different cell types and physiological contexts.
Keywords: cytoskeleton, exportin 6, importin 9, nuclear actin, nuclear myosins, oocyte
Introduction and Scope
In the cytoplasm, actin filaments form crosslinked networks that enable eukaryotic cells to transport cargo, change shape, and move. Actin is also present in the nucleus1 but, in this compartment, its functions are more cryptic and controversial. If we distill the substantial literature on nuclear actin down to its essentials, we find four, recurring, and more-or-less independent, claims (Fig. 1): (1) crosslinked networks of conventional actin filaments span the nucleus and help maintain its structure and organize its contents; (2) assembly or contraction of filaments regulates specific nuclear events; (3) actin monomers moonlight as subunits of chromatin remodeling complexes, independent of their ability to form filaments; and (4) modified actin monomers or oligomers, structurally distinct from canonical, cytoskeletal filaments, mediate nuclear events by unknown mechanisms. We discuss the evidence underlying these claims and as well as their strengths and weaknesses. Next, we describe our recent work, in which we built probes specific for nuclear actin and used them to describe the form and distribution of actin in somatic cell nuclei. Finally, we discuss how different forms of nuclear actin may play different roles in different cell types and physiological contexts.
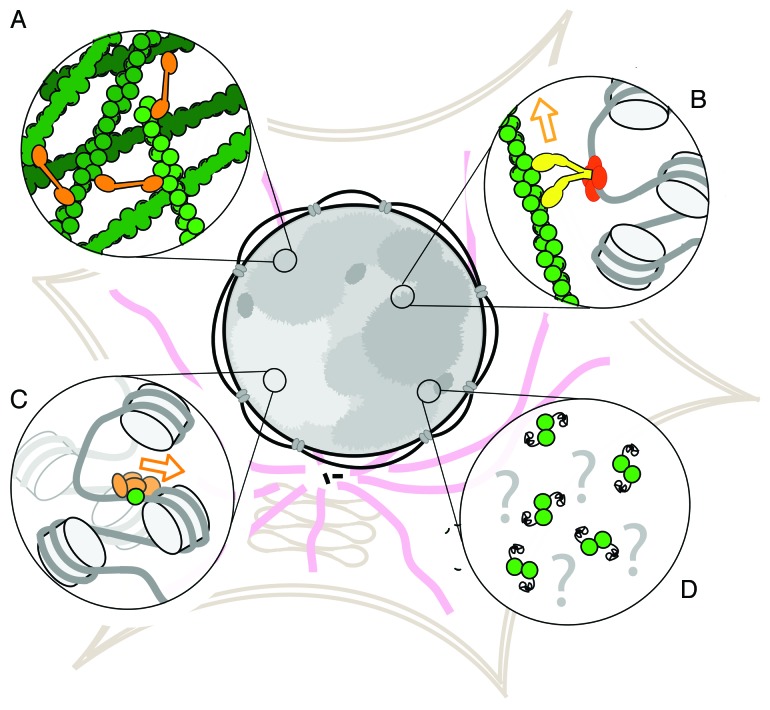
Figure 1. Proposed forms of actin in the nucleus. (A) Crosslinked networks of conventional actin filaments, (B) linear filaments that support motor-based transport, (C) monomers that regulate chromatin-based protein complexes and (D) non-canonical, modified actin monomers or oligomers.
Evidence for Nuclear Actin Networks
Nuclear actin was first identified in germinal vesicles of amphibian oocytes, where it is present at high concentration (> 100 µM).2 The high concentrations of actin in germinal vesicles is probably due to the fact that most oocytes do not express actin’s main nuclear export factor, Exportin-6.3 There has been much disagreement over the state of this actin: whether it is mostly monomeric or filamentous.4 Several early studies reported the existence of an actin filament-rich “nuclear gel” in germinal vesicles isolated from frog (Xenopus laevis) oocytes in magnesium- and potassium-containing buffers.5,6 Others reported that germinal vesicles when isolated under oil contain almost no filamentous actin.4,7
Recent experiments on intact oocytes are similarly contradictory. In Xenopus oocytes, images of phalloidin-stained cells from the Görlich laboratory, as well as high-resolution scanning electron micrographs, support the idea that actin filaments form a nucleoplasmic mesh required for maintenance of germinal vesicle integrity.8,9 Live cell imaging of actin in starfish (Patiria miniata) oocytes, however, seems to imply that the majority of actin in these germinal vesicles is monomeric and, upon nuclear envelope breakdown, polymerizes into a contractile mesh that ensnares chromosomes and facilitates their congression.10,11
Evidence that Actin Filaments Play a Role in Nuclear Processes
Very early studies in Xenopus and salamanders (Pleurodeles watlii) suggested that actin filaments associate with lampbrush chromosomes and are required for transcription.12-14 This proposal was based largely on the observation that actin antibodies microinjected into germinal vesicles inhibited both chromosome condensation and transcription. This hypothesis was not followed up for many years, but recent work in Xenopus has revived the idea that nuclear actin plays a role in oocyte gene expression. In an effort to understand the mechanisms of induced pluripotency, the Gurdon lab transferred differentiated C2C12 nuclei into Xenopus oocytes, and found that overexpression of non-polymerizing actin mutants inhibits induction of the pluripotency gene Oct4. Conversely, these authors also found that overexpression of polymer-stabilizing actin mutants enhances Oct4 induction.15 Whether nuclear actin filaments act directly on Oct4 transcription has yet to be determined. It has been subsequently reported that treatment of Xenopus, and avian oocytes (Gallus gallus domesticus, Coturnix coturnix japonicas, Fringilla coelebs) with the actin-depolymerizing drugs cytochalasin D and latrunculin A results in chromosome collapse and global inhibition of transcription.7 Thus, the inhibition of Oct4 induction may stem from a more general phenomenon.
In somatic cells, the functions and forms of nuclear actin are even less clear and even more hotly debated. We can find no good estimates in the literature for the concentration of actin in somatic cell nuclei but, because somatic cells do express actin’s nuclear export factor, Exportin-6, it is certainly much lower than in germinal vesicles. Although phalloidin does not label any filaments in somatic nuclei, the study of nuclear actin dynamics suggests that several forms of somatic nuclear actin may exist. Using fluorescence recovery after photobleaching (FRAP) and fluorescence correlation spectroscopy (FCS), McDonald and colleagues16 measured two different mobilities for GFP-tagged actin in somatic cell nuclei (0.009 µm2/sec and > 0.06 µm2/sec). It is unclear, however, whether the difference in measured mobilities arises from differences between the two techniques used to characterize molecular mobility or whether it reflects the existence of both filamentous and monomeric nuclear actin pools.
Several studies suggest that nuclear actin is required for optimal transcription in somatic cells. As in the oocyte systems, injecting anti-actin antibodies into HeLa cell nuclei appears to inhibit RNA polymerases I and II, and reducing the amount of nuclear actin by inhibiting nuclear import represses transcription globally.17-19 Some of these effects might be due to the role of actin monomers in chromatin remodeling complexes (see below), but actin filaments have also been proposed to play a role. Ye et al.20 performed the most rigorous test of this hypothesis to date using both pharmacological agents and actin mutants, with mixed results. The authors found that treating HEK293T cells or nuclear extracts with inhibitors of actin assembly (latrunculin B and cytochalasin D), decreases pre-rRNA synthesis, but found that this effect could not be reproduced by overexpressing non-polymerizing actin mutants. It is possible that this inconsistency results from high doses and long incubation times required for pharmacological inhibition, which may trigger stress responses that alter global transcriptional activity. Ye et al. also observed discrepancies when they compared the effects of actin filament stabilizing drugs (jasplakinolide and phalloidin) with the effects of filament-stabilizing actin mutants (S14C and V159N). The former has no effect on pre-rRNA levels, while the latter causes pre-rRNA levels to rise. Based on these results, the direct involvement of actin filaments in transcription remains an open question (Fig. 2).
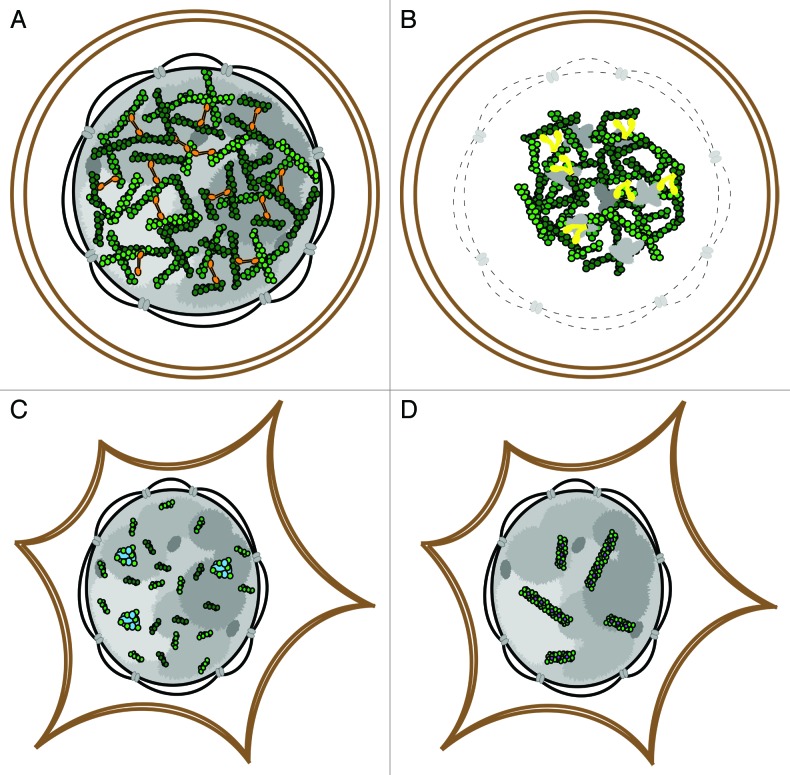
Figure 2. A comparison of known nuclear actin architectures. Oocyte germinal vesicles contain cross-linked actin networks (A) that undergo myosin-based contraction following nuclear envelope breakdown (B). In somatic nuclei, short actin filaments and a functional monomeric pool are present (C) and reorganize into cofilin-actin rods (D) following environmental stress.
Ectopic expression of actin mutants has suggested that nuclear filaments might facilitate long-range transport of activated genes, and the identification of numerous nuclear myosin motor proteins has made this idea quite attractive.21-26 In Dundr et al.27 overexpression of a nucleus-targeted, non-polymerizing actin mutant (R62D) in HeLa cells inhibited long-range (2–3 micron) movement of an array of activated U2 snRNA minigenes to Cajal bodies. Chuang et al.28 observed a similar effect on the motion of a transgenic DHFR gene in CHO cells in the presence of a nucleus-targeted G13R actin mutant, also defective in polymerization.
Several reports have proposed an indirect role for nuclear actin in regulating the activity of the actin-binding transcriptional regulator, MRTF (myotonin-related transcription factor). Briefly, MRTF binds monomeric actin via three short sequences, called RPEL domains. Actin monomers occupying these sites block access of importins to MRTF’s nuclear localization sequence. Decreases in cytoplasmic concentrations of monomeric actin, caused by bursts of polymerization, decrease the occupancy of the RPEL domains and drive MRTF into the nucleus. Once MRTF is inside the nucleus, however, nuclear actin monomers can still bind the RPEL domains and inhibit transcriptional activity. Serum stimulation of starved cells produces a burst of actin assembly in the cytoplasm sufficient to drive MRTF into the nucleus.29,30 Based on a set of persuasive experiments, Baarlink et al.31 found that serum stimulation additionally causes a burst of actin polymerization in the nucleus sufficient to enable MRTF to carry out its program of transcriptional activation. These authors used a nucleus-targeted version of a fluorescent probe, Lifeact, to directly observe the transient burst of nuclear actin filament assembly triggered by serum stimulation. In this mechanism it is not nuclear actin filaments per se that promote transcription but the polymerization-linked depletion of nuclear actin monomers.
Significantly, Baarlink et al. demonstrated that serum-induced nuclear actin filament assembly requires the activity of formin-family actin nucleators inside the nucleus. In addition to formins and myosins, a growing number of actin-associated proteins have been found in the nucleus, including: cofilin,19,32 α-actinin,33 filamin,34 and coronin 2A.35 Some of these proteins appear to translocate from the cytoplasm to the nucleus in response to environmental stimuli and may regulate cofilin-actin rods, nuclear actin bundles that form in a range of cell types following external stress. Early work identified actin rods in nuclei of several species of Dictyostelium following treatment with DMSO.36,37 Subsequent studies in various model systems have shown that nuclear actin rod formation is also induced by heat shock, ATP depletion and oxidative stress.32,38,39 Little is understood of the physiological role of nuclear actin rods, though many reports have strongly implicated nuclear cofilin as a major regulatory factor.40,41
The Role of Monomeric Actin in Chromatin-Remodeling Complexes
It is now a widely accepted fact that conventional actin monomers, along with several actin-related proteins (Arps), form conserved subunits of complexes that modify and remodel chromatin from yeast to human.42 These include the INO80, SWR1 and SNI/SNF-type chromatin-remodeling complexes and the NuA4 histone acetyltransferase complex.43-46 These multi-component, molecular machines produce dynamic changes in chromatin architecture, gene transcription, and DNA repair. The role of actin and nuclear Arps in these complexes is unknown, but some early studies suggested that, similar to the Arp2/3 complex, they might nucleate conventional actin filaments.47 More recent work, including structural studies of the Arp subunits, argues against this idea.48 Interestingly, the activity of chromatin remodelers can be reconstituted in vitro from partial complexes or sub-complexes lacking actin and/or Arps, suggesting that they are required for regulation in vivo.49,50
Non-Canonical Actin Monomers and Oligomers in the Nucleus
After years of work failed to produce convincing images of canonical actin filaments in nuclei of unstressed, somatic cells, some investigators proposed that nuclear actin self-associates into non-filamentous oligomers—possibly related to the anti-parallel, “lower” dimers that can be generated in vitro by chemical crosslinking.51 Evidence for the existence of such non-canonical structures in cells comes almost exclusively from the use of antibodies raised against chemically crosslinked actin. Furthermore, detecting these antibodies in the nucleus requires specialized fixation and extraction methods, making it difficult to judge their physiological relevance. Recently, Hofmann et al.52 discovered a pool of SUMO-ylated actin, which appears to be restricted to the nucleus. This post-translational modification may play a role in nuclear retention of actin but its role in nuclear function is still a mystery.
Major Obstacles and Recent Progress
Although compelling evidence places actin inside the nucleus, we know little about its nuclear functions and the mechanisms by which it carries them out. High concentrations of actin in the cytoplasm make it difficult to see the trace amounts in somatic cell nuclei. Also, since the actin cytoskeleton participates in many different cellular processes it is difficult to attribute the effects of actin mutants or pharmacological agents to a specific nuclear pool. To make progress the field must develop new methods to: (1) perturb the concentration, localization, and/or architecture of actin inside the nucleus without disrupting actin in the cytoplasm, and (2) visualize nuclear actin structures in living cells.
Recent work on the nucleo-cytoplasmic trafficking of actin has provided tools that may enable us to manipulate nuclear actin concentrations without perturbing cytoplasmic actin. In Stüven et al.,3 the Görlich lab identified Exportin-6 (XPO6) as a factor that appears to specifically export profilin-bound actin and a small set of actin-associated proteins out of the nucleus. The Vartianen group closed the import-export loop by identifying Importin-9 (IPO9) as the nuclear import factor for actin.19 Using RNAi, these authors demonstrated that nuclear actin levels could be tuned by altering the ratios of import and export. These discoveries open a door to more decisive mechanistic studies and they would be further enhanced by development of additional tools, such as nucleus-targeted actin monomer-sequestering proteins or depolymerization factors.
A final key to solving the puzzles posed by nuclear actin would be provided by reliable methods to image actin-containing nuclear structures in live cells. Conventional techniques for labeling cellular actin are problematic. The signal from fluorescent derivatives of phalloidin or fluorescent actin fusion proteins is dominated by the contribution from the cytoplasm. In addition, fusion of fluorescent proteins to actin renders them poor substrates for some actin regulators. Formin-family proteins, already implicated in generating nuclear actin filaments in response to serum stimulation31 for example, cannot nucleate or elongate filaments using GFP-actin. “Nucleus-specific” actin antibodies label punctate structures in the nucleus, but these results are highly dependent upon cell type and fixation conditions and cannot be used for live-cell imaging. Such immunofluorescence experiments are also unable to discriminate between monomeric and filamentous actin.
To characterize nuclear actin architectures in somatic cells, we designed nucleus-specific actin reporters consisting of an EGFP-tagged actin binding domain (ABD) fused to a strong nuclear localization signal (NLS). We tested a range of ABDs that represent multiple classes of well-characterized actin-binding proteins specific for either actin filaments or monomers. When expressed in human U2OS cells, the majority of the ABDs we tested exhibited one of two flaws: (1) they failed to detect actin structures in the cytoplasm, or (2) they clearly perturbed the nuclear actin pool and created artifactual nuclear actin structures.
One monomer-specific probe, the RPEL1 ABD from MAL, had neither of these defects. Similar to our EGFP-NLS control construct, RPEL1-EGFP-NLS (R1EN) has a diffuse nucleoplasmic localization and is enriched in nucleoli. R1EN is also enriched in nuclear speckles, a nuclear body involved in pre-mRNA processing events. Mutation of residues required for binding actin (R81D/R82D) abolished the enrichment in nuclear speckles, suggesting that monomeric actin is present in nuclear speckles. This result is consistent with a previously proposed role for nuclear actin in RNA processing.
Of the filament-specific ABDs tested, only two efficiently labeled cytoplasmic actin filaments in vivo: the actin binding peptide, Lifeact, and a truncation of the human actin-binding protein utrophin, Utr261. When targeted to the nucleus, however, both probes induced formation of large, bundled actin networks. In the case of Lifeact, this is likely due to the peptide’s high affinity for monomeric actin, which probably increases the nuclear concentration of actin. Since we could find no easy way to improve Lifeact’s specificity for filaments, we did not pursue it further. Utr261 is highly specific for filaments, but potently induces actin-filament assembly and bundling in vitro. We suspect that this bundling activity arises from self-association of Utr261 into a dimer that contains several non-overlapping actin-binding sites capable of crosslinking filaments and forming bundles. To create a utrophin ABD variant that binds actin filaments without bundling them, we made several truncation mutants of Utr261 and expressed them with EGFP-NLS tags. One of these truncations, Utr230-EGFP-NLS (Utr230-EN) retained the ability to bind filaments in the cytoplasm and, in the nucleus, localized to punctate structures of a relatively fixed size (< 500 nm) scattered throughout the nucleoplasm.
The number and size of nuclear actin particles does not vary with the expression level of Utr230-EN, indicating that they are native structures rather than aggregates or artifacts of overexpression. Additional lines of evidence argue that these structures contain actin filaments: (1) they cannot be detected by Utr230-EN mutants deficient in actin binding; (2) they can be stained by phalloidin (but only after cytoplasmic actin structures are depolymerized by latrunculin B); and (3) their presence in the nucleoplasm can be abolished by knockdown of importin-9. These controls provide compelling evidence that Utr230-EN provides an accurate picture of actin filament distribution in somatic cell nuclei.
We used colocalization assays to address proposed functions of actin filaments in the nucleus. Surprisingly, we found no strong overlap between nuclear actin filaments and markers for numerous nuclear processes, including chromatin remodeling, transcription, and RNA processing. In fact, DAPI staining and markers for chromatin indicated that actin filaments are generally excluded from chromatin-rich regions of the nucleus. These experiments appear to rule out direct participation of actin filaments in many RNA- and chromatin-based processes. It is, however, still possible that actin filaments interact with subsets of genes in chromatin-poor regions.
What do our results say about actin-based cargo transport within the nucleus? Actin-based transport can occur in one of two ways: (1) by motor proteins moving along actin filament tracks, and (2) by actin filaments polymerizing to generate propulsive forces. The first mechanism seems unlikely given the small size (< 500 nm) of the nuclear actin filaments and the fact that “directed” motions observed in the nucleus can span several microns. To address the second mechanism, we tracked nuclear actin filaments in live cells using confocal microscopy and calculated the speed correlation index (SCI) of the resulting trajectories. Briefly, the SCI is a measure of how persistently a particle moves in a given direction. It compares the direction of motion at each time point with the direction at subsequent times. High SCI values across several frames of a particle trajectory indicate directed motion. Our SCI calculations for nuclear actin filaments demonstrated that these signatures of directed motion were entirely absent, ruling out a role for actin in directing intranuclear transport.
Curiously, the SCI values we measured indicate the opposite of persistent, directed motion: actin filaments are more likely to “backtrack” along their trajectories than to diffuse randomly. The velocity autocorrelation function (VCF), a parameter similar to the SCI that specifically identifies “backtracking” events and oscillations, supports this observation. The most likely physical interpretation of this result is that the actin filaments in the nucleoplasm are embedded in a medium that provides an elastic force that “pushes back.”
Similar “backtracking” behavior has been observed and modeled for particles embedded in viscoelastic networks, including individual genes within chromosomes and monomers within larger cytoskeletal polymers. The actin filaments in the nucleus are too small and dispersed to create such a viscoelastic mesh on their own, so the source of the elastic force must be something else. Because actin is excluded from chromatin-rich regions of the nucleus, it is unlikely to be chromatin, and may instead be a protein-based mesh similar to the proposed nuclear “matrix.” One possibility is that short, nuclear actin filaments serve as scaffolds that locally organize nuclear proteins within a viscoelastic mesh.
In addition to providing evidence that actin interacts with a viscoelastic medium in the nucleoplasm, our particle tracking analysis also revealed multiple mobility populations of nuclear actin filaments. They majority of filaments formed three overlapping populations with diffusion coefficients between 0.06 and 0.08 µm2/sec. The motion of these particles was consistent with simple diffusion, with α values approaching 1. A smaller subset of filaments moves much more slowly and subdiffusively, with average diffusion coefficients of 0.015 and 0.04 µm2/sec and α values approaching 0.67. This lower α value of is consistent with those calculated for particles embedded in viscoelastic media (~0.7), suggesting that a subset of actin filaments interacts strongly with low-mobility elements in the medium.53,54 The multiple mobility populations also indicate that nuclear actin filaments exist in multiple, distinct pools.
The size and spatial distribution of actin filaments we observe in somatic cell nuclei argue strongly against many of their proposed functions. We found no evidence for an actin filament meshwork spanning the nucleus and, in our work as well as that of other groups, inhibition of actin import has no obvious effect on the morphology or integrity of the nucleus. Our work does not address the existence of noncanonical, oligomeric actin structures in the nucleus.
Our monomer-specific actin reporter R1-EN suggests that actin monomers may have a role in RNA processing at nuclear speckles, but there may be other actin monomer populations that cannot be detected using R1-EN because they are not enriched above background in an obvious nuclear landmark. Determining the extent to which nuclear actin monomers participate in RNA processing and other nucleoplasmic processes will require additional molecular tools.
Moving Forward
Tools to visualize and perturb nuclear actin are beginning to advance our understanding, but more and better tools are needed. Utrophin- and Lifeact-based probes revealed populations of nuclear actin filaments, but neither probe should be considered a universal in vivo actin reporter. Endogenous actin binding proteins, for example, can block access of probes to their binding sites on the filament and the probes themselves can interfere with normal actin dynamics. Development and characterization of a suite of probes with different affinities and binding sites could help uncover distinct pools of filamentous actin with distinct functions.
Additional agents to perturb nuclear actin without affecting cytoplasmic actin are also needed. The ability to block nuclear import and export of actin is an important first step, but it is necessary to note that IPO9 and XPO6 may have additional unknown client proteins whose loss also affects nuclear processes. Identifying these client proteins will improve our ability to interpret the results of knockdown of these transport factors. In addition, the study of nuclear actin would benefit greatly from agents that block assembly of nuclear actin filaments or stabilize them against disassembly, enabling more detailed mechanistic studies. One strategy for identifying these agents would be to target actin-perturbing proteins to the nucleus. Another approach would be to modulate expression of endogenous regulators of nuclear actin dynamics. Some formin-family proteins, for example, are already thought to promote nuclear actin assembly but identification of other regulators will provide more knobs to turn.
In the 1990s and 2000s the use of fluorescent cytoskeletal proteins, photobleaching, photoactivation, and speckle microscopy, along with the development of pharmacological agents such as latrunculin, jasplakinolide, and blebbistatin, drove fundamental advances in our understanding of cytoplasmic actin dynamics. This has shaped our modern views of how cells move, how they interact with their environment, and how they divide. Now, with new tools for visualizing and perturbing nuclear actin, the nucleus is poised for a similar advance. The nucleus looks to be an important new frontier for cytoskeletal discovery and these discoveries may shape the way we understand how cells regulate gene expression and make fundamental decisions regarding their fate.
Submitted
07/12/2013
Accepted
07/30/2013
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/25960
References
- 1.de Lanerolle P, Serebryannyy L. Nuclear actin and myosins: life without filaments. Nat Cell Biol. 2011;13:1282–8. doi: 10.1038/ncb2364. [DOI] [PubMed] [Google Scholar]
- 2.Clark TG, Merriam RW. Diffusible and bound actin nuclei of Xenopus laevis oocytes. Cell. 1977;12:883–91. doi: 10.1016/0092-8674(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 3.Stüven T, Hartmann E, Görlich D. Exportin 6: a novel nuclear export receptor that is specific for profiling actin complexes. EMBO J. 2003;22:5298–5240. doi: 10.1093/emboj/cdg565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gall JG, Wu Z. Examining the contents of isolated Xenopus germinal vesicles. Methods. 2010;51:45–51. doi: 10.1016/j.ymeth.2009.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clark TG, Rosenbaum JL. An actin filament matrix in hand-isolated nuclei of X. laevis oocytes. Cell. 1979;18:1101–8. doi: 10.1016/0092-8674(79)90223-X. [DOI] [PubMed] [Google Scholar]
- 6.Gounon P, Karsenti E. Involvement of contractile proteins in the changes in consistency of oocyte nucleoplasm of the newt Pleurodeles waltlii. J Cell Biol. 1981;88:410–21. doi: 10.1083/jcb.88.2.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maslova A, Krasikova A. Nuclear actin depolymerization in transcriptionally active avian and amphibian oocytes leads to collapse of intranuclear structures. Nucleus. 2012;3:300–11. doi: 10.4161/nucl.20393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiseleva E, Drummond SP, Goldberg MW, Rutherford SA, Allen TD, Wilson KL. Actin- and protein-4.1-containing filaments link nuclear pore complexes to subnuclear organelles in Xenopus oocyte nuclei. J Cell Sci. 2004;117:2481–90. doi: 10.1242/jcs.01098. [DOI] [PubMed] [Google Scholar]
- 9.Bohnsack MT, Stüven T, Kuhn C, Cordes VC, Görlich D. A selective block of nuclear actin export stabilizes the giant nuclei of Xenopus oocytes. Nat Cell Biol. 2006;8:257–63. doi: 10.1038/ncb1357. [DOI] [PubMed] [Google Scholar]
- 10.Lénárt P, Bacher CP, Daigle N, Hand AR, Eils R, Terasaki M, Ellenberg J. A contractile nuclear actin network drives chromosome congression in oocytes. Nature. 2005;436:812–8. doi: 10.1038/nature03810. [DOI] [PubMed] [Google Scholar]
- 11.Mori M, Monnier N, Daigle N, Bathe M, Ellenberg J, Lénárt P. Intracellular transport by an anchored homogeneously contracting F-actin meshwork. Curr Biol. 2011;21:606–11. doi: 10.1016/j.cub.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 12.Karsenti E, Gounon P, Bornens M. Immunocytochemical study of lampbrush chromosomes: presence of tubulin and actin. Biol Cell. 1978;31:210–24. [Google Scholar]
- 13.Rungger D, Rungger-Brändle E, Chaponnier C, Gabbiani G. Intranuclear injection of anti-actin antibodies into Xenopus oocytes blocks chromosome condensation. Nature. 1979;282:320–1. doi: 10.1038/282320a0. [DOI] [PubMed] [Google Scholar]
- 14.Scheer U, Hinssen H, Franke WW, Jockusch BM. Microinjection of actin-binding proteins and actin antibodies demonstrates involvement of nuclear actin in transcription of lampbrush chromosomes. Cell. 1984;39:111–22. doi: 10.1016/0092-8674(84)90196-X. [DOI] [PubMed] [Google Scholar]
- 15.Miyamoto K, Pasque V, Jullien J, Gurdon JB. Nuclear actin polymerization is required for transcriptional reprogramming of Oct4 by oocytes. Genes Dev. 2011;25:946–58. doi: 10.1101/gad.615211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald D, Carrero G, Andrin C, de Vries G, Hendzel MJ. Nucleoplasmic β-actin exists in a dynamic equilibrium between low-mobility polymeric species and rapidly diffusing populations. J Cell Biol. 2006;172:541–52. doi: 10.1083/jcb.200507101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hofmann WA, Stojiljkovic L, Fuchsova B, Vargas GM, Mavrommatis E, Philimonenko V, Kysela K, Goodrich JA, Lessard JL, Hope TJ, et al. Actin is part of pre-initiation complexes and is necessary for transcription by RNA polymerase II. Nat Cell Biol. 2004;6:1094–101. doi: 10.1038/ncb1182. [DOI] [PubMed] [Google Scholar]
- 18.Philimonenko VV, Zhao J, Iben S, Dingová H, Kyselá K, Kahle M, Zentgraf H, Hofmann WA, de Lanerolle P, Hozák P, et al. Nuclear actin and myosin I are required for RNA polymerase I transcription. Nat Cell Biol. 2004;6:1165–72. doi: 10.1038/ncb1190. [DOI] [PubMed] [Google Scholar]
- 19.Dopie J, Skarp KP, Rajakylä EK, Tanhuanpää K, Vartiainen MK. Active maintenance of nuclear actin by importin 9 supports transcription. Proc Natl Acad Sci U S A. 2012;109:E544–52. doi: 10.1073/pnas.1118880109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye J, Zhao J, Hoffmann-Rohrer U, Grummt I. Nuclear myosin I acts in concert with polymeric actin to drive RNA polymerase I transcription. Genes Dev. 2008;22:322–30. doi: 10.1101/gad.455908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pestic-Dragovich L, Stojiljkovic L, Philimonenko AA, Nowak G, Ke Y, Settlage RE, Shabanowitz J, Hunt DF, Hozak P, de Lanerolle P. A myosin I isoform in the nucleus. Science. 2000;290:337–41. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 22.Vreugde S, Ferrai C, Miluzio A, Hauben E, Marchisio PC, Crippa MP, Bussi M, Biffo S. Nuclear myosin VI enhances RNA polymerase II-dependent transcription. Mol Cell. 2006;23:749–55. doi: 10.1016/j.molcel.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 23.Lindsay AJ, McCaffrey MW. Myosin Vb localises to nucleoli and associates with the RNA polymerase I transcription complex. Cell Motil Cytoskeleton. 2009;66:1057–72. doi: 10.1002/cm.20408. [DOI] [PubMed] [Google Scholar]
- 24.Cameron RS, Liu C, Mixon AS, Pihkala JP, Rahn RJ, Cameron PL. Myosin16b: The COOH-tail region directs localization to the nucleus and overexpression delays S-phase progression. Cell Motil Cytoskeleton. 2007;64:19–48. doi: 10.1002/cm.20162. [DOI] [PubMed] [Google Scholar]
- 25.Pranchevicius MC, Baqui MM, Ishikawa-Ankerhold HC, Lourenço EV, Leão RM, Banzi SR, dos Santos CT, Roque-Barreira MC, Espreafico EM, Larson RE. Myosin Va phosphorylated on Ser1650 is found in nuclear speckles and redistributes to nucleoli upon inhibition of transcription. Cell Motil Cytoskeleton. 2008;65:441–56. doi: 10.1002/cm.20269. [DOI] [PubMed] [Google Scholar]
- 26.Salamon M, Millino C, Raffaello A, Mongillo M, Sandri C, Bean C, Negrisolo E, Pallavicini A, Valle G, Zaccolo M, et al. Human MYO18B, a novel unconventional myosin heavy chain expressed in striated muscles moves into the myonuclei upon differentiation. J Mol Biol. 2003;326:137–49. doi: 10.1016/S0022-2836(02)01335-9. [DOI] [PubMed] [Google Scholar]
- 27.Dundr M, Ospina JK, Sung MH, John S, Upender M, Ried T, Hager GL, Matera AG. Actin-dependent intranuclear repositioning of an active gene locus in vivo. J Cell Biol. 2007;179:1095–103. doi: 10.1083/jcb.200710058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuang CH, Carpenter AE, Fuchsova B, Johnson T, de Lanerolle P, Belmont AS. Long-range directional movement of an interphase chromosome site. Curr Biol. 2006;16:825–31. doi: 10.1016/j.cub.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 29.Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–52. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- 30.Mouilleron S, Guettler S, Langer CA, Treisman R, McDonald NQ. Molecular basis for G-actin binding to RPEL motifs from the serum response factor coactivator MAL. EMBO J. 2008;27:3198–208. doi: 10.1038/emboj.2008.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baarlink C, Wang H, Grosse R. Nuclear actin network assembly by formins regulates the SRF coactivator MAL. Science. 2013;340:864–7. doi: 10.1126/science.1235038. [DOI] [PubMed] [Google Scholar]
- 32.Iida K, Matsumoto S, Yahara I. The KKRKK sequence is involved in heat shock-induced nuclear translocation of the 18-kDa actin-binding protein, cofilin. Cell Struct Funct. 1992;17:39–46. doi: 10.1247/csf.17.39. [DOI] [PubMed] [Google Scholar]
- 33.Kumeta M, Yoshimura SH, Harata M, Takeyasu K. Molecular mechanisms underlying nucleocytoplasmic shuttling of actinin-4. J Cell Sci. 2010;123:1020–30. doi: 10.1242/jcs.059568. [DOI] [PubMed] [Google Scholar]
- 34.Loy CJ, Sim KS, Yong EL. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc Natl Acad Sci U S A. 2003;100:4562–7. doi: 10.1073/pnas.0736237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang W, Ghisletti S, Saijo K, Gandhi M, Aouadi M, Tesz GJ, Zhang DX, Yao J, Czech MP, Goode BL, et al. Coronin 2A mediates actin-dependent de-repression of inflammatory response genes. Nature. 2011;470:414–8. doi: 10.1038/nature09703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fukui Y. Intranuclear actin bundles induced by dimethyl sulfoxide in interphase nucleus of Dictyostelium. J Cell Biol. 1978;76:146–57. doi: 10.1083/jcb.76.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukui Y, Katsumaru H. Dynamics of nuclear actin bundle induction by dimethyl sulfoxide and factors affecting its development. J Cell Biol. 1980;84:131–40. doi: 10.1083/jcb.84.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pendleton A, Pope B, Weeds A, Koffer A. Latrunculin B or ATP depletion induces cofilin-dependent translocation of actin into nuclei of mast cells. J Biol Chem. 2003;278:14394–400. doi: 10.1074/jbc.M206393200. [DOI] [PubMed] [Google Scholar]
- 39.Kim JS, Huang TY, Bokoch GM. Reactive oxygen species regulate a slingshot-cofilin activation pathway. Mol Biol Cell. 2009;20:2650–60. doi: 10.1091/mbc.E09-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bernstein BW, Shaw AE, Minamide LS, Pak CW, Bamburg JR. Incorporation of cofilin into rods depends on disulfide intermolecular bonds: implications for actin regulation and neurodegenerative disease. J Neurosci. 2012;32:6670–81. doi: 10.1523/JNEUROSCI.6020-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Munsie LN, Desmond CR, Truant R. Cofilin nuclear-cytoplasmic shuttling affects cofilin-actin rod formation during stress. J Cell Sci. 2012;125:3977–88. doi: 10.1242/jcs.097667. [DOI] [PubMed] [Google Scholar]
- 42.Farrants AO. Chromatin remodelling and actin reorganization. FEBS Lett. 2008;585:2041–50. doi: 10.1016/j.febslet.2008.04.032. [DOI] [PubMed] [Google Scholar]
- 43.Cairns BR, Erdjument-Bromage H, Tempst P, Winston F, Kornberg RD. Two actin-related proteins are shared functional components of the chromatin-remodeling complexes RSC and SWI/SNF. Mol Cell. 1998;2:639–51. doi: 10.1016/S1097-2765(00)80162-8. [DOI] [PubMed] [Google Scholar]
- 44.Zhao K, Wang W, Rando OJ, Xue Y, Swiderek K, Kuo A, Crabtree GR. Rapid and phosphoinositol-dependent binding of the SWI/SNF-like BAF complex to chromatin after T lymphocyte receptor signaling. Cell. 1998;95:625–36. doi: 10.1016/S0092-8674(00)81633-5. [DOI] [PubMed] [Google Scholar]
- 45.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–4. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 46.Galarneau L, Nourani A, Boudreault AA, Zhang Y, Héliot L, Allard S, Savard J, Lane WS, Stillman DJ, Côté J. Multiple links between the NuA4 histone acetyltransferase complex and epigenetic control of transcription. Mol Cell. 2000;5:927–37. doi: 10.1016/S1097-2765(00)80258-0. [DOI] [PubMed] [Google Scholar]
- 47.Rando OJ, Zhao K, Janmey P, Crabtree GR. Phosphatidylinositol-dependent actin filament binding by the SWI/SNF-like BAF chromatin remodeling complex. Proc Natl Acad Sci U S A. 2002;99:2824–9. doi: 10.1073/pnas.032662899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schubert HL, Wittmeyer J, Kasten MM, Hinata K, Rawling DC, Héroux A, Cairns BR, Hill CP. Structure of an actin-related subcomplex of the SWI/SNF chromatin remodeler. Proc Natl Acad Sci U S A. 2013;110:3345–50. doi: 10.1073/pnas.1215379110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–53. doi: 10.1016/S1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 50.Szerlong H, Saha A, Cairns BR. The nuclear actin-related proteins Arp7 and Arp9: a dimeric module that cooperates with architectural proteins for chromatin remodeling. EMBO J. 2003;22:3175–87. doi: 10.1093/emboj/cdg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schoenenberger CA, Buchmeier S, Boerries M, Sütterlin R, Aebi U, Jockusch BM. Conformation-specific antibodies reveal distinct actin structures in the nucleus and the cytoplasm. J Struct Biol. 2005;152:157–68. doi: 10.1016/j.jsb.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 52.Hofmann WA, Arduini A, Nicol SM, Camacho CJ, Lessard JL, Fuller-Pace FV, de Lanerolle P. SUMOylation of nuclear actin. J Cell Biol. 2009;186:193–200. doi: 10.1083/jcb.200905016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weber SC, Spakowitz AJ, Theriot JA. Bacterial chromosomal loci move subdiffusively through a viscoelastic cytoplasm. Phys Rev Lett. 2010;104:238102. doi: 10.1103/PhysRevLett.104.238102. a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weber SC, Theriot JA, Spakowitz AJ. Subdiffusive motion of a polymer composed of subdiffusive monomers. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;82:011913. doi: 10.1103/PhysRevE.82.011913. b. [DOI] [PMC free article] [PubMed] [Google Scholar]