Abstract
CRM1 is an export factor that together with its adaptor NMD3 transports numerous cargo molecules from the nucleus to cytoplasm through the nuclear pore. Previous studies have suggested that CRM1 and NMD3 are detected in the nucleolus. However, their localization with subnucleolar domains or participation in the activities of the nucleolus are unclear. We demonstrate here biochemically and using imaging analyses that CRM1 and NMD3 co-localize with nucleolar marker proteins in the nucleolus. In particular, their nucleolar localization is markedly increased by inhibition of RNA polymerase I (Pol I) transcription by actinomycin D or by silencing Pol I catalytic subunit, RPA194. We show that CRM1 nucleolar localization is dependent on its activity and the expression of NMD3, whereas NMD3 nucleolar localization is independent of CRM1. This suggests that NMD3 provides nucleolar tethering of CRM1. While inhibition of CRM1 by leptomycin B inhibited processing of 28S ribosomal (r) RNA, depletion of NMD3 did not, suggesting that their effects on 28S rRNA processing are distinct. Markedly, depletion of NMD3 and inhibition of CRM1 reduced the rate of pre-47S rRNA synthesis. However, their inactivation did not lead to nucleolar disintegration, a hallmark of Pol I transcription stress, suggesting that they do not directly regulate transcription. These results indicate that CRM1 and NMD3 have complex functions in pathways that couple rRNA synthetic and processing engines and that the rRNA synthesis rate may be adjusted according to proficiency in rRNA processing and export.
Keywords: rRNA synthesis, export, transcription, biogenesis, nucleolus
Introduction
The nucleolus is a membraneless structure with major function in ribosome biogenesis.1,2 Following transcription of 47S ribosomal (r) RNA precursor by RNA polymerase I (Pol I), the precursor undergoes extensive processing, modification and maturation to 18S and 28S rRNAs.3-6 The maturing rRNAs are assembled to 90S ribosome precursor and processed further to 40S and 60S pre-ribosomes. This process is mediated by over 150 participant proteins, occurs sequentially through dynamic protein exchange, and takes place in discrete nucleolar subdomains.7-10 The nucleolus consists of three subdomains, the fibrillar center (FC), the dense fibrillar component (DFC) and the granular component (GC).2 Various proteins are specifically associated with these domains and used to mark them. For example, upstream binding factor (UBF), a transcription factor required for rRNA synthesis, marks the FC, while fibrillarin (FBL), a component of rRNA modifying ribonucleoprotein complexes, marks the DFC.1 The GC is marked by the nucleophosmin (NPM), whose functions are associated with late maturation processes of the ribosome.1,2
The nucleolus has high dynamic bidirectional flow of proteins and RNA.11,12 Maturing 40S and 60S ribosomes are being shuttled from the nucleolus to the nucleus at high rates, but what propagates their movement is not fully understood. While some nucleolar proteins depend on GTP for their mobility, others do not.13 The propagation of ribosome movement may also take place by free diffusion.14 Quantitative proteomics has revealed extensive nucleolar dynamic responses to transcription stress and DNA damage.15-18 For example, following inhibition of Pol I by treatment with actinomycin D (ActD), the nucleoli condense and form so-called nucleolar cap structures, while simultaneously several GC proteins, such as nucleolin (NCL) and NPM, translocate to the nucleoplasm.12,18 The cessation of Pol I transcription leads to disassembly of maturing ribosomal particles.
The nuclear transport receptor CRM1 (chromosome region maintenance 1, XPO1) mediates export of a broad range of macromolecular cargos from the nucleus to the cytoplasm.19,20 The export activity depends on Ran GTPase that loads and unloads the cargos for their passage through the nuclear pores. The cargos or components of the cargoes normally contain a short leucine-rich nuclear export signal or other export signals of more complex 3-dimensional characters, such as folded domains.20 CRM1 exports also small nuclear RNAs, rRNAs, select mRNA and viral RNAs.19,20 Several adaptor proteins that contain RNA-binding motifs have been identified. In yeast CRM1 is involved in the transport of the large and small ribosome subunits to the cytoplasm, which requires the yeast Nmd3p export adaptor.7,21-23 While yeast has several export receptors for the ribosomes, like Mex67/Mtr2 and Arx1, vertebrate cells are dependent on the CRM1-NMD3 export complex.24,25 NMD3 is highly conserved and contains both nuclear export and nuclear localization sequences and presumably engages with its cargo in the nucleus.10,22 It binds stoichiometrically the 60S subunit.26 Following nuclear export of the pre-60S particle, 60S subunit is subjected to further maturation steps in the cytoplasm and release of NMD3.10,27 CRM1 may also be needed for the maturation of pre-40S in the cytoplasm by facilitating the nuclear export of the pre-18S rRNA.28 Remarkably, also Exportin-5 has been shown to export 60S subunit in a Ran-GTP-dependent manner, indicating that robust and overlapping pathways contribute to ribosome export.29
Accumulating evidence suggests that CRM1 is involved in transport between intranuclear structures and has functions unrelated to nuclear export.30,31 CRM1 affects shuttling of the U3 snoRNA from Cajal bodies to the nucleolus,30 binds snoRNA assembly complexes32 and influences the composition of the nucleoplasmic pre-snoRNP complexes.33 In addition, CRM1 interacts with kinetochores and centrosomes, suggesting that it participates in the organization of mitotic events.19 In a line of evidence obtained by inhibition of CRM1 by leptomycin B (LMB), a specific inhibitor of CRM1 function, in situ hybridization experiments suggested that both 28S and 18S rRNAs (or their precursors) accumulated in the nucleolus and in the nucleoplasm after LMB treatment.24 These findings indicate that CRM1 acts in diverse functions through its capacity to interact with multiple dynamic protein and RNA complexes.
CRM1 and NMD3 are detected in the nucleolar structures.24,25,34-37 Following ActD treatment, CRM1 is found in the proximity of the DFC protein FBL,35 which is surprising considering the extensive nucleolar disruption and displacement of major GC proteins to the nucleoplasm following transcription stress. However, in most of these studies, nucleolar protein co-localizations were not conducted. Conversely, as assessed by in situ hybridization, LMB increases the accrual of 28S in the nucleoli, suggesting that inhibition of CRM1 causes stalling in the ability to process and/or export rRNAs.24,38 Rev, a nucleolar HIV protein, increases CRM1 nucleolar association and retains its mobility.39 Finally, GFP-tagged NMD3 is localized to structures akin the nucleoli in LMB-treated cells,25 and when the NMD3 nuclear export sequence(s) is altered.24,25 These findings indicate that NMD3 may shuttle through the nucleolus in a CRM1-dependent manner.25
Collectively, the data suggests that CRM1 and NMD3 transit through the nucleolus. Given the apparent wide range of functions of CRM1 and NMD3, we were interested in investigating their nucleolar affiliations. We show here biochemically and using nucleolar marker protein co-localization analyses that inhibition of Pol I activity leads to substantial accumulation of CRM1 and NMD3 in the nucleolus. While the nucleolar accumulation of CRM1 is dependent on its activity, the accumulation of NMD3 is independent of CRM1. In contrast, we show that CRM1 nucleolar localization depends on NMD3, suggesting that NMD3 is requisite for CRM1 nucleolar tethering. Strikingly, both CRM1 and NMD3 affect ribosome biogenesis. Inhibition of CRM1 leads to a pronounced 18S processing defect while that of depletion of NMD3 does not. Yet, their inhibition leads to a prominent decrease in rRNA synthesis without causing disintegration of the nucleolus. These findings suggest that defects in rRNA processing and ribosome export feed back to suppress the rate of rRNA synthesis and hence, ribosome biogenesis.
Results
CRM1 and NMD3 accumulate in the nucleolus after inhibition of Pol I activity
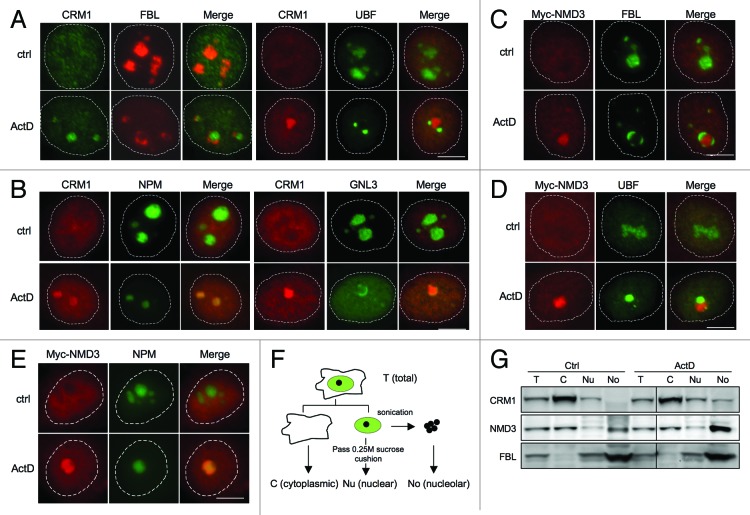
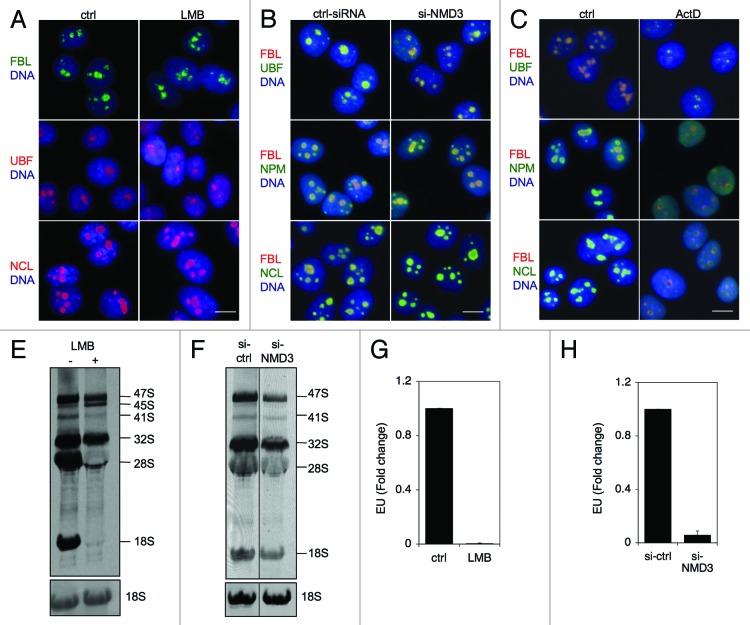
Although previous studies indicated that CRM1 is detected in the nucleolus, only limited co-localization assays with nucleolar marker proteins have been conducted (FBL).35 To verify the nucleolar localization of CRM1, we treated HeLa cells with ActD and co-stained CRM1 with proteins known to localize to different subnucleolar domains. These included FBL, denoting the DFC, UBF denoting the FC and two GC proteins nucleostemin (GNL3) and NPM. As shown by co-immunofluorescence staining, the nucleolus was reorganized following ActD treatment, and CRM1 localized to the remnants of the condensed GC and became surrounded by the FC and DFC proteins UBF and FBL, respectively (Fig. 1A and B). Similarly, we conducted co-staining analyses for NMD3. As several NMD3 antibodies tested failed to produce NMD3-specific staining in immunofluorescence analysis, we used ectopic expression of Myc-NMD3 and treated the cells with ActD. Cells that expressed high levels of Myc-NMD3 displayed diffuse nucleoplasmic and cytoplasmic expression (Fig. S1). However, in cells with low Myc-NMD3 expression, Myc-NMD3 was predominantly detected in the nucleoli (Fig. 1C and D). This implies that the nucleolar association of Myc-NMD3 is dependent on its relevant stoichiometric interactions masked by unphysiological levels of expression. As shown in Figure 1C and D, Myc-NMD3 was surrounded by FBL and UBF following ActD treatment and localized to the remnants of the condensed GC, which was suggested by its co-localization with NPM-ECGFP (Fig. 1E). Thus, the localization of Myc-NMD3 was consistent with that of CRM1.
Figure 1. CRM1 and Myc-NMD3 accumulate in the nucleolus. (A) HeLa cells were treated with ActD (50 ng/ml) for 3 h as indicated. The cells were co-stained for FBL and CRM1 or for UBF and CRM1 as shown. (B) Cells expressing NPM-ECGFP were treated with ActD followed by staining for CRM1, or HeLa cells were treated with ActD followed by co-staining for GNL3 and CRM1. (C and D) HeLa cells transfected with Myc-NMD3 expression vector were treated as in (A) and co-stained for Myc-NMD3 and FBL (C) or Myc-NMD3 and UBF (D). (E) HeLa cells stably expressing NPM-ECGFP were transfected with Myc-NMD3, treated as in (A) and stained for Myc-NMD3. Scale bars, 10 µm. (F) Protocol of cellular fractionation to purify cytoplasmic, C; nuclear, Nu and nucleolar, No fractions. Note that the nuclear fraction contains also the nucleoli. T denotes total cellular extract. (G) Cells were treated with Act D (50 ng/ml) for 3 h followed by fractionation according to the scheme in (F), followed by western blot analysis for CRM1 and NMD3. Forty micrograms of protein from each fraction was loaded. FBL was used as a nucleolar marker.
To confirm the subcellular localizations of the respective proteins, we performed cellular fractionation (Fig. 1F) that enabled the detection of changes in endogenous protein levels by biochemical means. For this purpose we used a TRIzol-based isolation technique for consecutive isolation of RNA, DNA and protein that we have developed.40 This allowed comparative isolation of the different macromolecular entities from the same specimens. We have previously shown that the method facilitates highly efficient recovery of the nucleolar proteins unobtainable using other techniques.40 As shown in Figure 1G, CRM1 was predominantly cytoplasmic and hardly detectable in the nucleolar fraction, whereas following ActD-treatment, its expression in the nucleolar fraction increased. We then assessed NMD3 distribution and asked whether ActD treatment affects NMD3 subcellular localization. NMD3 expression was also substantially increased in the nucleolar fraction (Fig. 1G). FBL served as a nucleolar marker protein and loading control.
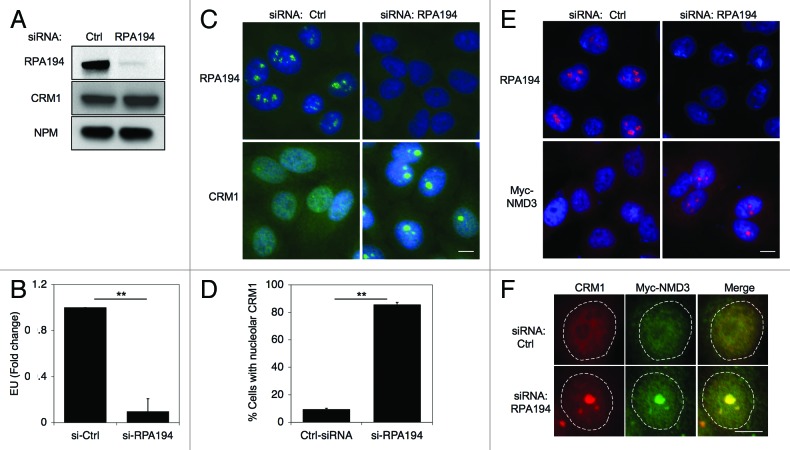
ActD is a DNA-binding drug and a transcription inhibitor capable of blocking the synthesis of rRNA by Pol I in animal cells at 50–100-fold lower concentrations than those required for inhibition of Pol II- and Pol III-mediated transcription.41 To obtain further evidence that the localization changes are caused by the inhibition of Pol I and not due to DNA damage (ActD is a strong DNA intercalating agent) or inhibition of the other polymerases, we silenced the expression of the large catalytic subunit of RNA Pol I, RPA194. The efficiency of silencing of RPA194 was verified by RPA194 immunofluorescence and western blotting, and loss of nascent RNA synthesis was verified using RNA Click-IT assay (Fig. 2A and B). Based on these assays RPA194 silencing led to its over 90% decrease and profound repression of Pol I activity. Under these conditions, immunofluorescence assay showed that CRM1 was present in nucleoli in over 80% of HeLa cells (Fig. 2C and D). Despite the change in localization, the expression of CRM1 protein was invariable (Fig. 2A). Similarly, RPA194 silencing led to an increase in Myc-NMD3 nucleolar localization, detectable in cells that expressed low levels of Myc-NMD3 (Fig. 2E). Lastly, co-staining for CRM1 and Myc-NMD3 in the RPA194 silenced cells showed their extensive co-staining (Fig. 2F).
Figure 2. Depletion of RPA194 increases CRM1 and NMD3 nucleolar localization. (A) HeLa cells were transfected with control or RPA194 targeting siRNAs and incubated for 48 h, followed by western analysis for RPA194 and CRM1, and NPM was used as a loading control. (B) rRNA synthesis was determined using EU incorporation. Control and RPA194 siRNA transfected cells were incubated for 1 h with EU. EU incorporation was determined by quantitative image analysis. Data represents means ± SD, n = 2 experiments. ** p < 0.01. (C) RPA194 depleted cells or control siRNA transfected cells were immunostained for RPA194 and CRM1. Scale bar, 10 µm. (D) Quantification of number of cells with CRM1 nucleolar accumulation from (C). Data represents means ± SD, n = 2 experiments. ** p < 0.01. (E) Cells were transfected with siRNAs as indicated, followed by transfection with Myc-NMD3 expression vector and incubation for 24 h. Cells were fixed and stained for RPA194 and Myc-NMD3. Scale bar, 10 µm. (F) Cells treated as in (E) were co-stained for CRM1 and Myc-NMD3. CRM1, red; Myc-NMD, green; Merge, yellow. Scale bar, 10 µm.
To further determine whether inhibition of other RNA polymerases could affect nucleolar accumulation of CRM1, HeLa cells were treated with Pol II inhibitor α-amanitin for three hours and cells were stained for CRM1 (Fig. S2). There was no change in CRM1 localization or expression following α-amanitin treatment. These experiments suggest that CRM1 nucleolar accumulation is a specific effect of inhibition of Pol I. In the following experiments, we used ActD to investigate the nucleolar effects of Pol I inhibition on CRM1.
NMD3 nucleolar association is independent of CRM1
The interaction between CRM1-NMD3 is dependent on CRM1-RanGTP activity.24 Having confirmed that the inhibition of Pol I results in nucleolar accumulation of CRM1 and NMD3, we asked whether their accumulation is dependent on CRM1 and its nuclear export activity. To test this, we treated HeLa cells with ActD in the presence of the CRM1 specific inhibitor LMB, and compared with cells treated with either ActD or LMB alone. As shown in Figure 3A and B, concomitant treatment of cells with ActD and LMB led to the loss of CRM1 nucleolar accumulation, suggesting that CRM1 nucleolar localization, prompted by inhibition of Pol I transcription, depends on its nuclear export activity.
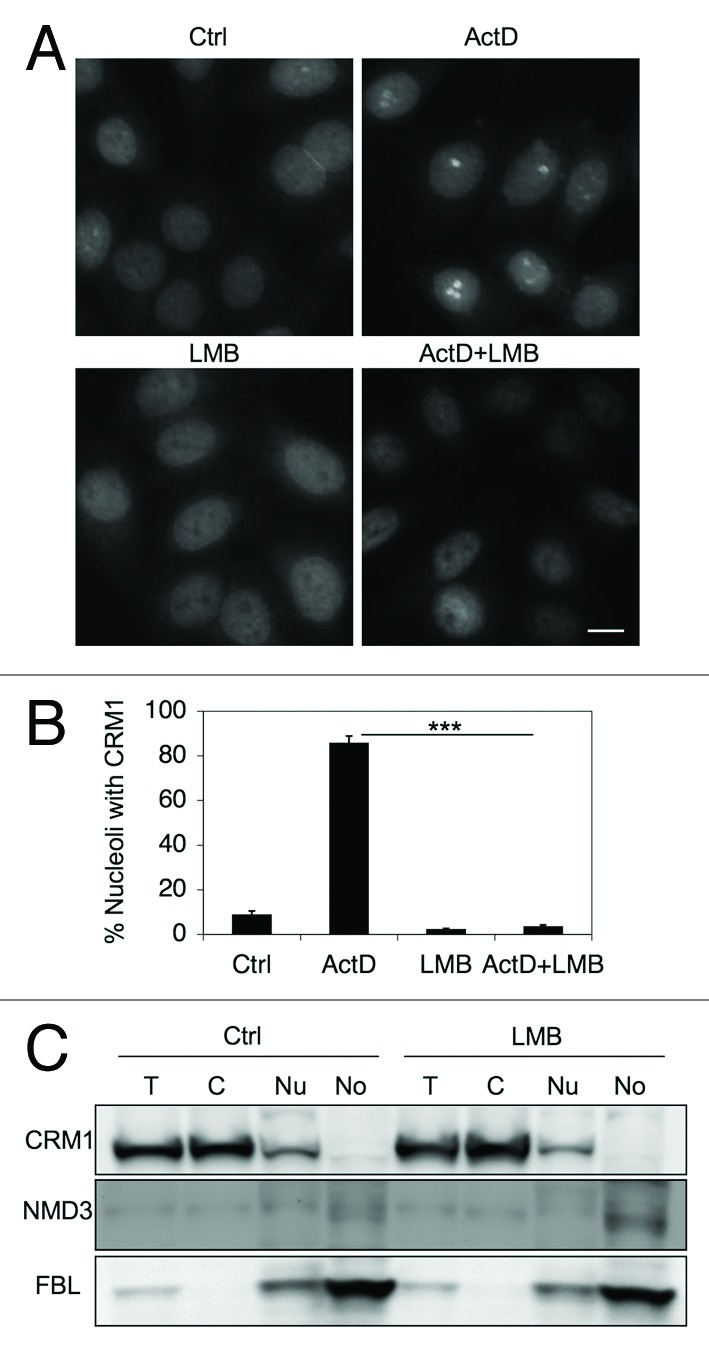
Figure 3. CRM1 nucleolar localization depends on its activity. (A) HeLa cells were treated with ActD (40 nM) or LMB (20 ng/ml) or their combination for 3 h, fixed and stained for CRM1. Scale bar, 10 µm. (B) Quantification of cells with CRM1 nucleolar accumulation in A. Data represents means ± SD, n = 2 experiments. *** p < 0.001. (C) Cells were treated with LMB (20 ng/ml) for 12 h followed by cellular fractionation and isolation of proteins as in Figure 1F. Protein loading was adjusted and analyzed by western blotting for CRM1, NMD3 and FBL.
We then sought to address how similar treatments affect NMD3. Western blotting analysis of subcellular fractions of LMB-treated HeLa cells indicated that NMD3 expression was only slightly increased in the nucleolar fraction (Fig. 3C). FBL served as a loading control for the nucleolar fraction. The changes in CRM1 and NMD3 localization did not result in changes of expression of the respective proteins (Fig. S3). Hence, in contrast to Trotta et al.25 but similarly to Thomas et al.,24 we could not detect substantial accumulation of NMD3 in the nucleoli following LMB-treatment alone. We then used Myc-NMD3 expression to detect its subcellular localization. In repeated experiments we observed only few (≤ 5%) cells with nucleolar Myc-NMD3 expression, although, as expected, LMB-treatment led to the nucleoplasmic accumulation of Myc-NMD3 (Fig. S1).
We hence used RPA194 silencing to address this further. Again, depletion of RPA194 increased nucleolar accumulation of Myc-NMD3 but LMB treatment alone was without effect (Fig. 4A). However, in contrast to CRM1, NMD3 retained its nucleolar presence after inhibition of both Pol I and CRM1 (Fig. 4A). To further assess the dependency of NMD3 on CRM1, we used expression of a NMD3 mutant where three leucines critical for CRM1 recognition are mutated to alanine (Myc-NMD3-NES).24,25 Myc-NMD3-NES mutant accumulated in the nucleoli in RPA194-depleted cells and the accumulation was unaffected in LMB-treated cells (Fig. 4B and C). This suggested that CRM1 activity was required for its nucleolar accumulation, but that NMD3 accrual was independent of CRM1.
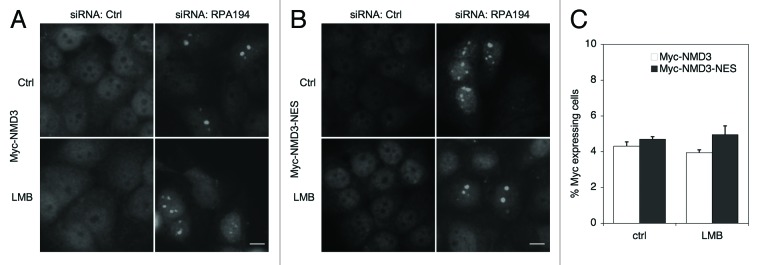
Figure 4. Myc-NMD3 nucleolar localization is independent of CRM1. (A and B) Cells were depleted of RPA194 using RNAi, transfected with (A) Myc-NMD3 or (B) Myc-NMD3-NES expression vectors and treated with LMB as shown for 4 h. Cells were fixed and stained for Myc-NMD3 using Myc-epitope tag antibody. Scale bar, 10 µm. (C) Quantification of nucleolar expression of Myc-tagged NMD3 and Myc-NMD3-NES. The percentage of cells with nucleolar expression is shown. Data represents means ± SD, n = 2 experiments.
CRM1 nucleolar localization depends on NMD3
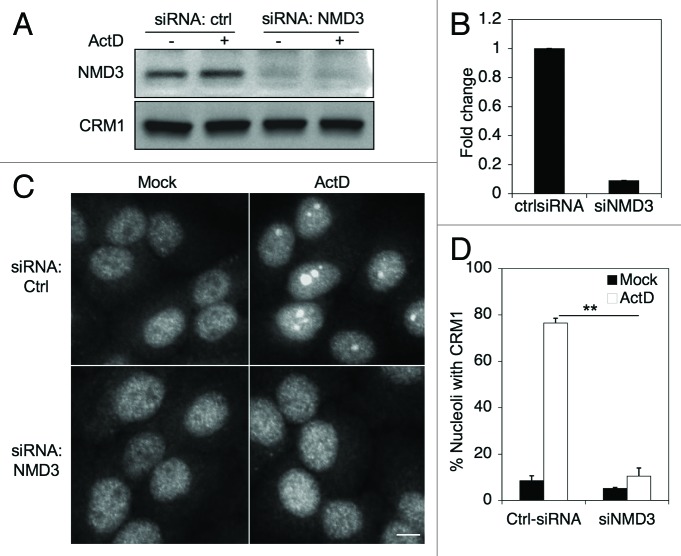
The function of NMD3 as the CRM1 adaptor suggests that CRM1 nucleolar localization could depend on NMD3. To test this we silenced NMD3 in HeLa cells using siRNA, and subsequently treated the cells with ActD. Depletion of NMD3 was verified by western blotting and qRT-PCR, and did not affect CRM1 expression (Fig. 5A and B). However, in comparison to cells treated with control siRNAs, silencing of NMD3 reduced nucleolar CRM1 accumulation by ActD treatment by approximately 80% (Fig. 5C and D). This indicated that CRM1 nucleolar localization depends on NMD3.
Figure 5. CRM1 nucleolar accumulation is dependent on NMD3. (A) Cells were transfected with control or NMD3 targeting siRNAs and incubated for 48 h, followed by Western analysis for NMD3 and CRM1. (B) Quantitative PCR for the expression of NMD3 transcript. qPCR was normalized to GAPDH. (C) Cells were silenced for NMD3 as in A and treated with ActD (50 ng/ml) for 3 h and stained for CRM1. Scale bars, 10 µm. (D) Quantification of cells with CRM1 nucleolar accumulation in (C). Data represents means ± SD, n = 2 experiments. **, p < 0.001.
CRM1 and NMD3 affect rRNA biogenesis
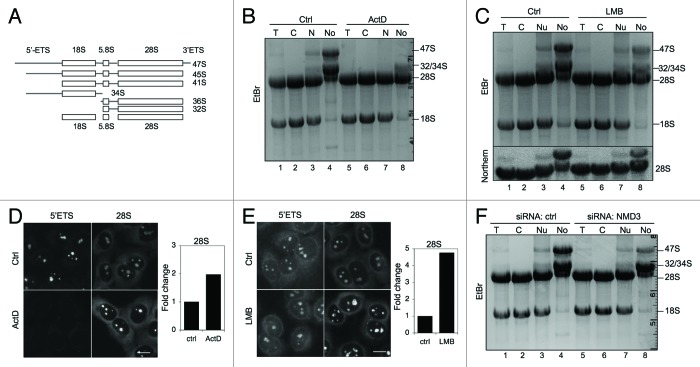
The observed nucleolar localization of CRM1 and NMD3 begs the question as to whether the CRM1-NMD3 complex is involved in the transport of the ribosomal subunits from the nucleolus or has other functions in relation to ribosome biogenesis. Furthermore, CRM1 has earlier been implicated in the processing of 18S rRNA.28 To monitor the presence of the rRNAs and their processing (schematically depicted in Fig. 6A) in the subcellular compartments, we extracted total RNAs from cellular fractions (nucleolus, nucleus, cytoplasm and total) purified from mock, ActD and LMB-treated cells. The RNA fractions correspond to protein fractions analyzed in Figures 1G and 3C. Agarose gel electrophoresis of the RNA samples showed that the nucleolar RNA content was unique and that ActD-treatment, as expected, abolished the synthesis of the 47S pre-rRNA (Fig. 6B, compare lane 4 to lane 8). In contrast, ActD-treatment enhanced a band corresponding to 28S rRNA in the nucleolar fraction (Fig. 6B, compare lane 4 to lane 8). Similarly, an analysis of LMB-treated cells showed that a band corresponding to 28S rRNA was increased in the nucleolar fraction while 47S precursor rRNA decreased (Fig. 6C, compare lane 4 to lane 8). We further conducted Northern hybridization for 28S rRNA, which confirmed the identity and expression of the 28S rRNA (Fig. 6C, bottom panel; Fig. S4). Thus, both ActD and LMB treatments increased the level of 28S rRNA in the nucleolus.
Figure 6. Alterations in nucleolar rRNAs following ActD and LMB treatments and NMD3 silencing. (A) Schematic presentation of rRNA processing. (B) RNA gel analysis of total RNA of cellular fractions treated with ActD (50 ng/ml) for 3 h. RNA was extracted from the samples in Figure 1G. EtBr staining. The migration of rRNAs is indicated to the right. (C) Cells were treated with LMB (20 ng/ml) for 12 h followed by cellular fractionation and isolation of RNA. The RNA gel was stained with EtBr (top panel). Subsequently, the RNA agarose gel was hybridized to digoxigenin-labeled 28S LNA probe (Northern, bottom panel). RNA was extracted from the samples in Figure 3C. (D) Cells were treated with ActD followed by in situ hybridization using probes detecting 5′ETS and 28S rRNAs. Quantification of the image analysis (right panel). Scale bar, 10 µm. (E) Cells were treated with LMB followed by in situ hybridization using probes detecting 5′ETS and 28S rRNA. Quantification of the image analysis (right panel). Scale bar, 10 µm. (F) RNA gel analysis of total RNA of cellular fractions from cells transfected with control or NMD3 siRNAs. EtBr staining.
To further assess the presence of rRNAs in cellular subfractions derived from cells treated with ActD and LMB, we performed in situ hybridization using probes for short-lived 5′ETS and 28S rRNA. After ActD treatment, the 5′ETS was no longer detectable, as attributed to its degradation by ActD and lack of new rRNA synthesis,42 while 28S rRNA signal increased by 2-fold in the nucleoli (Fig. 6D). In contrast, LMB-treatment did not noticeably change 5′ETS hybridization signal, while 28S rRNA increased by over 4-fold (Fig. 6E). Thus, the in situ hybridization is consistent with the increase of 28S rRNA in the nucleolar RNA fraction. Finally, we assessed the effect of NMD3 silencing on rRNA expression and localization. As shown in Figure 6F, NMD3 depletion did not affect the presence of 28S rRNA in the nucleolus, but caused a noticeable decrease in the expression of the pre-47S rRNA.
The decrease in pre-47S rRNA indicated that the inhibition of CRM1 and depletion of NMD3 could affect rRNA synthesis. If transcriptional stress, i.e., inhibition of Pol I transcription is involved, these treatments should be evidenced by nucleolar segregation. However, neither treatment affected nucleolar integrity as assessed by localization of FBL, UBF and NCL as compared with their relocalization in response to ActD (Fig. 7A–C). To analyze further the synthesis and processing of newly synthesized rRNA we conducted metabolic labeling of rRNA using 3H-uridine incorporation. Cells were first treated with LMB for 4 h and then labeled with 3H-uridine for the last 2 h, followed by extraction of RNA. LMB-treatment led to abrogation of 18S rRNA maturation in accordance with the study by Rouquette et al.28 (Fig. 7D). In addition, we observed a decrease in 47S rRNA precursor and defective processing of the 41/45S rRNA precursor affecting also maturation of 28S rRNA (Fig. 7D). Silencing of NMD3 decreased the level of pre-47S rRNA precursor and those of 18S and 28S rRNA slightly (Fig. 7E). To investigate the effects of LMB and NMD3 depletion on rRNA synthesis we analyzed nascent nucleolar RNA synthesis using EU incorporation in LMB-treated and NMD3-silenced cells. To provide representative comparison to NMD3 silencing, the LMB-treatment was conducted for 14 h. The analysis showed a prominent decrease in uridine incorporation to nascent RNA after both treatments (Fig. 7F and G). These results suggest that CRM1 and NMD3 significantly impact rRNA biogenesis.
Figure 7. Inhibition of CRM1 and NMD3 affects rRNA synthesis. (A, B and C) Immunofluorescence analysis of the indicated nucleolar proteins following treatment of cells with LMB for 14 h (A), NMD3 silencing (B) or treatment of the cells with ActD for 3 h (C). Scale bars, 10 µm. (D) Cells were treated with LMB for 4 h and labeled for the last two hours with 3H-uridine. RNA was isolated and separated by agarose gel electrophoresis. Autoradiogram is shown. (E) Cells were transfected with or without NMD3 silencing siRNAs and metabolic labeling of rRNA was conducted as in (D). Position of rRNAs are indicated. Total 18S rRNA as stained by EtBr is shown at the bottom. (F and G) Cells were treated with LMB for 13 h as in (D) or NMD3 was silenced as in (E) and cells were incubated for the last hour with EU. EU incorporation was detected, cells were imaged and nucleolar EU signals were quantified of over 200 cells in each assay. n = 2. Error bars, SD. Data represents means ± SD, n = 2 experiments.
Discussion
We validate here by biochemical and imaging analyses that CRM1 and NMD3 localize to nucleolar structures. CRM1 nucleolar localization was mediated by NMD3 expression, required CRM1 activity and was greatly increased when rRNA transcription was inhibited. Under these conditions, CRM1 and NMD3 co-localized to the remnants of GC, the subdomain where late rRNA processing takes place. This suggests that inhibition of Pol I transcription, which leads to depletion of maturing rRNAs, also leads to accrual of CRM1 and NMD3 to this location. These findings are consistent with NMD3 acting as also a nucleolar adaptor for CRM1. Our data are in accordance with that of Thomas et al.24 that inhibition of CRM1 leads to nucleolar accrual of 28S RNA and/or its precursors. This supports the notion that the CRM1-NMD3 complex engages with its cargo during the ribosome biosynthesis in the nucleolus and that they are needed for the nucleolar processing of the rRNAs. Finally, we show that inactivation of CRM1 and NMD3 lead to reduction in the synthesis of rRNA. This finding suggests that rRNA export function effectively feeds back to titrate the rate of rRNA synthesis.
Nmd3p binds to the maturing large ribosome subunit in the nucleus which following cytoplasmic transit and maturation leads to its release.21,23,25,26 Prior reports using GFP-tagged NMD3 have proposed that NMD3 localizes to the nucleolus and that this might be the initial site of NMD3 engagement with the ribosome subunits. However, analyses of the presence of human NMD3 in the nucleolus have lacked the use of nucleolar markers or biochemical purification of the nucleoli. Our data show by various methods that human NMD3 locates to the nucleolus in HeLa cells, and that the extent of this localization is influenced by Pol I activity. NMD3 and CRM1 displayed extensive nucleolar co-localization in cells when Pol I activity was inhibited. However, they lack co-staining with nucleolar proteins associated with Pol I transcription and those that are typically observed in nucleolar cap structures following transcription stress. Their localization and co-staining was consistent with localization in the remnants of the GC, which is remarkable considering the extensive relocation of nucleolar GC proteins and pre-60S ribosome processing nucleolar proteins during transcription stress.12,15,17 This suggests that CRM1 and NMD3 stalling could be due to lack of protein binding partners, rRNA, or both. The localization of CRM1 in the nucleolus was dependent on its activity and that of NMD3. This shows that NMD3 is the key factor that supports the nucleolar association and mediates CRM1 nucleolar tethering.
The data documented here are compatible with the binding of NMD3 with the maturing large ribosome subunit already in the nucleolus. However, in contrast to Trotta et al.,25 we did not observe retention of endogenous or Myc-NMD3 in the nucleolus when CRM1 activity was inhibited. This difference may reflect properties of the expression tags (Myc vs. GFP) used that can affect the mobility, interaction or stoichiometric binding of the fusion proteins. The nucleolar tethering of Myc-NMD3 following Pol I transcription block indicates that its transit rate depends on the ongoing rRNA synthesis. Given that in Pol I-inhibited cells NMD3 was detected in the nucleolus also when CRM1 activity was inhibited and that Myc-NMD3 export mutant also accrued in the nucleolus, suggests that the nucleolar entry of CRM1 and NMD3 are independent. They also appear to have independent and distinct activities related to rRNA processing. Depletion of NMD3 did not lead to obvious rRNA processing defects, whereas CRM1 inhibition abrogates the early processing of 28S rRNA, and possibly by interfering with U3 nucleolar transport, that of maturation of 18S rRNA.28,30 The maturation of pre-ribosome particles depends on successive, but transient interactions of several non-ribosomal proteins.7,8 Stoichiometric interaction of NMD3 with 60S subunit and its binding interface with the 28S rRNA suggests that NMD3 supports the quality control of rRNA at an early stage which is independent of its association with the CRM1 export complex.
We show here using multiple assays that depletion of NMD3 and blocking CRM1 activity leads to inhibition of rRNA synthesis. This was evident in assays measuring nascent RNA synthesis and production of the 47S rRNA precursor. The decrease in rRNA synthesis did not lead to nucleolar segregation, which, during Pol I transcription stress, is evident by extensive structural changes of the nucleolus and localization changes of nucleolar proteins. Since these changes are predominantly observed when Pol I transcription is directly affected,18,43 it seems likely that neither CRM1 nor NMD3 are involved with Pol I transcription initiation or elongation. The absence of nucleolar reorganization following LMB-treatment is in agreement with an earlier report by Muro et al.36 Interestingly, interfering with 28S processing by depletion of Pescadillo/WDR12/Bop1 complex proteins does not affect pre-47S rRNA synthesis, suggesting that maturation defects alone do not necessarily feed back to transcription.44,45 The rate of Pol I transcription is driven by external signaling events that regulate posttranslationally Pol I pre-initiation complex assembly on rDNA.46 Thus, the attenuation of rRNA synthesis following depletion of NMD3 and inhibition of CRM1 activity is suggestive that the decrease in RNA processing, ribosome transport and/or maturation provides a negative feedback to the transcriptional machinery. This could take place through abrogation of the ubiquitous signaling pathways positively supporting Pol I transcription. Alternatively, it is plausible that CRM1/NMD3 interfere directly with the Pol I transcription holocomplex. However, they do not demonstrate co-localization with factors (UBF, FBL) associated with transcription or early rRNA maturation indicating that this is unlikely. While there is increasing understanding of co-transcriptional regulation of RNA processing, the capability of the RNA export activity to impact rRNA synthesis has not previously been described and will need further studies. Hence the data provides a context that couples the export activity with the synthesis of its most abundant cargo.
Materials and Methods
Cell culture and reagents
HeLa cervical adenocarcinoma cells (CCL-2, ATCC) were maintained in DMEM supplemented with 10% FCS. All cell culture reagents were obtained from Invitrogen. Stable NPM-EGCFP expressing HeLa cells were established as in ref. 36. The following drugs were used; α-amanitin (A2263, Sigma), Actinomycin D (ActD, A1410, Sigma) and leptomycin B (LMB, 50–230–5970, Calbiochem).
Expression plasmids
Myc-NMD3 expression plasmid (RC203044) was purchased from Origene. Myc-NMD-NES was constructed by site-directed mutagenesis of L480, L484 and L487 to A (QuickChange II XL-site-Directed mutagenesis Kit, Agilent) and sequence-verified.
Immunofluorescenc
HeLa cells grow on glass coverslips were fixed with 3.7% paraformadehyde and permeabilized with 0.5% NP40. The following primary antibodies were used: mouse anti-CRM1 (611833, BD Transduction), mouse anti-RPA194 (SC48385, Santa Cruz Biotechnology), mouse anti-Myc tag (05–419, Millipore), rabbit anti-UBF (H-300, Santa Cruz Biotechnology), rabbit anti-nucleostemin (GNL3, H-270, Santa Cruz Biotechnology), rabbit anti-Myc tag (GTX29106, GeneTex) and rabbit anti-FBL (AB5821, Abcam). Antibodies were detected with secondary antibodies conjugated to Alexa 488 or 594 (Molecular Probes, Invitrogen) and nuclei were counterstained with Hoechst 33258 (H-21491, Sigma). Images were captured using Axioplan2 fluorescence microscope (Zeiss) equipped with AxioCam HRc CCD-camera and AxioVision 4.5 software using EC Plan-Neofluar 40 × /0.75 objective (Zeiss).
Cellular fractionation
Nuclear (including the nucleoplasm and nucleolus) and cytoplasmic fractions were prepared essentially as in ref. 40. Cell pellets were resuspended in hypotonic buffer (10 mM Hepes, pH 7.9, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT) and incubated on ice for 5 min. Cell suspensions were homogenized with a Dounce tissue homogenizer (Wheaton) with frequent checks under a phase contrast microscope until 90% of the cells had burst. The nuclei were collected by centrifugation at 300 g for 5 min at 4°C. The supernatants were used for isolation of cytoplasmic fraction. Collected nuclei were purified by sucrose cushion centrifugation (0.35 M sucrose, 0.5 mM MgCl2). Cytoplasmic fractions were purified by centrifugation at 5,000 g for 10 min at 4°C. Nucleolar fractions were prepared by resuspending purified nuclear fractions in 0.35 M sucrose, 0.5 mM MgCl2 and sonicated using sonicator S-4000 (QSonica) fitted with a microtip. The sonicated suspension was checked under a phase contrast microscope until no intact nuclei were observed. Nucleoli were prepared by sucrose cushion centrifugation and collecting the fraction passing through the sucrose cushion as described before.15
RNA analyses
Total RNA was extracted from the different cellular compartments using TRIzol (15596–018, Invitrogen) reagent as described before.17 RNA concentrations were quantified using Nanodrop. Equal amounts of RNAs from the different cellular compartments were denatured in 2 × RNA loading buffer (AM8551, Ambion) and separated on 1% agarose gels. After electrophoresis the RNA was visualized by ethidium bromide staining.
In situ hybridization
In situ hybridization was performed essentially as described before.37
Quantitative RT-PCR
HeLa cells were treated with control siRNA and siRNA against NMD3. Total RNAs were extracted using TRIzol and quantified using Nanodrop (NanoDrop Products). cDNA was prepared using SuperScript® III Reverse Transcriptase (18080–093, Invitrogen) and random hexamers according to the manufacturer’s instructions. qPCR was performed using SYBR Green (AB-4162B, Atila Biosystem) and specific primer pairs for NMD3 and GAPDH. Amplification was conducted for 40 cycles at 94°C for 10 min each using an ABI7900 thermocycler (Applied Biosystems). The data were normalized against GAPDH. The primer sequences used were as follows. NMD3, forward: CTCCAAAACTGGCACAAAGCCTG, reverse: GTGACTCCAGAAAGTGCTCCCA. GAPDH, forward: GGTGATGGCATCTGAATGAA, reverse: CCCTTGGCATCAGTTTCTGT.
Ethynyl uridine-labeling
Cells were labeled with 1 mM ethynyl uridine (EU) for 1 h (E10345, Invitrogen). Cells were fixed and EU signal was detected using Click-iT RNA Alexa Fluor® 488 Imaging Kit (C10329, Invitrogen) according to manufacturer’s protocol. Image quantification was performed using FrIDA.47 Hue saturation and brightness range were defined and normalized to DNA. An average of > 200 cells was quantified from four fields for each experiment. P values were calculated using the Student two-tailed t test.
RNAi
HeLa cells were plated on coverslips or 6-well plates and transfected using Lipofectamine RNAiMAX (13778150, Invitrogen) with siRNAs (10 nM) either at the time of plating or the following day. The following siRNAs were used: RPA194 (S403, Ambion), NMD3 (SR309504, Origene).
Immunoblotting analyses
To obtain cellular lysates, cells were scraped, solubilized in RIPA buffer (50 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% NP-40, 0.1% sodium dodecyl sulfate, 1% sodium deoxycholate) supplemented with Complete Protease Inhibitor Tablets (11836153001, Roche) followed by centrifugation at 13,000 rpm for 20 min. Protein concentrations were determined using Bio-Rad Bradford protein assay (Bio-Rad). Equal amounts of protein were loaded onto 3–8% Tris-Acetate Nu-PAGE and transferred into nitrocellulose membrane (Trans-Blot, Transfer Medium, Bio-Rad). Immunoblotting was performed using mouse anti-NPM (32500, Zymed/Invitrogen), rabbit anti-NMD3 (SAB4200451, Sigma), mouse anti-RPA194 (SC48385, Santa Cruz Biotechnology), rabbit anti-FBL (AB5821, Abcam) antibodies followed by incubation with secondary antibodies conjugated to horseradish peroxidase, after which the signals were detected using enhanced chemiluminescence (RPN2106, ECL, Amersham Life Sciences).
Metabolic labeling
Cells were incubated with 3H-labeled uridine (NET367001MC, Perkin Elmer, final concentration 3.5 μCi/ml) for 2 h. RNA was extracted using TRIzol and RNA concentrations were measured with NanoDrop. Equal amounts of RNA was separated on 1% formaldehyde-agarose gel and transferred onto Nylon membranes (11209272001, Roche). The filter was cross-linked and sprayed with EN3HANCE (6NE970C, Perkin Elmer). Autoradiographs were developed 1 d later.
Supplementary Material
Acknowledgments
We thank members of Laiho lab at Helsinki and Hopkins for helpful discussion. This work was supported by NIH P30 CA006973 and Johns Hopkins University start-up funds.
Glossary
Abbreviations:
- ActD
actinomycin D
- CRM1
chromosomal maintenance 1
- FBL
fibrillarin
- FC
fibrillar center
- DFC
dense fibrillar component
- GC
granular component
- leptomycin B
LMB
- NCL
nucleolin
- NPM
nucleophosmin
- UBF
upstream binding factor
- qPCR
quantitative real-time PCR
- RNA polymerase I
Pol I
- ribosomal RNA
rRNA
Submitted
04/10/2013
Revised
05/21/2013
Accepted
06/10/2013
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplemental Materials
Supplemental materials may be found here: www.landesbioscience.com/journals/nucleus/article/25342.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/25342
References
- 1.Boisvert FM, van Koningsbruggen S, Navascués J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–85. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 2.Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3:a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fatica A, Tollervey D. Making ribosomes. Curr Opin Cell Biol. 2002;14:313–8. doi: 10.1016/S0955-0674(02)00336-8. [DOI] [PubMed] [Google Scholar]
- 4.Granneman S, Baserga SJ. Crosstalk in gene expression: coupling and co-regulation of rDNA transcription, pre-ribosome assembly and pre-rRNA processing. Curr Opin Cell Biol. 2005;17:281–6. doi: 10.1016/j.ceb.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 5.Russell J, Zomerdijk JC. The RNA polymerase I transcription machinery. Biochem Soc Symp. 2006;73:203–16. doi: 10.1042/bss0730203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haag JR, Pikaard CS. RNA polymerase I: a multifunctional molecular machine. Cell. 2007;131:1224–5. doi: 10.1016/j.cell.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 7.Nissan TA, Bassler J, Petfalski E, Tollervey D, Hurt E. 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. EMBO J. 2002;21:5539–47. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bassler J, Kallas M, Pertschy B, Ulbrich C, Thoms M, Hurt E. The AAA-ATPase Rea1 drives removal of biogenesis factors during multiple stages of 60S ribosome assembly. Mol Cell. 2010;38:712–21. doi: 10.1016/j.molcel.2010.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kressler D, Hurt E, Bassler J. Driving ribosome assembly. Biochim Biophys Acta. 2010;1803:673–83. doi: 10.1016/j.bbamcr.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 10.Panse VG, Johnson AW. Maturation of eukaryotic ribosomes: acquisition of functionality. Trends Biochem Sci. 2010;35:260–6. doi: 10.1016/j.tibs.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dundr M, Hoffmann-Rohrer U, Hu Q, Grummt I, Rothblum LI, Phair RD, et al. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–6. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 12.Hernandez-Verdun D. Nucleolus: from structure to dynamics. Histochem Cell Biol. 2006;125:127–37. doi: 10.1007/s00418-005-0046-4. [DOI] [PubMed] [Google Scholar]
- 13.Pederson T, Tsai RY. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184:771–6. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Politz JC, Tuft RA, Pederson T. Diffusion-based transport of nascent ribosomes in the nucleus. Mol Biol Cell. 2003;14:4805–12. doi: 10.1091/mbc.E03-06-0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 16.Boisvert FM, Lam YW, Lamont D, Lamond AI. A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol Cell Proteomics. 2010;9:457–70. doi: 10.1074/mcp.M900429-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore HM, Bai B, Boisvert FM, Latonen L, Rantanen V, Simpson JC, et al. Quantitative proteomics and dynamic imaging of the nucleolus reveal distinct responses to UV and ionizing radiation. Mol Cell Proteomics. 2011;10:009241. doi: 10.1074/mcp.M111.009241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–27. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutten S, Kehlenbach RH. CRM1-mediated nuclear export: to the pore and beyond. Trends Cell Biol. 2007;17:193–201. doi: 10.1016/j.tcb.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Köhler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–73. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 21.Gadal O, Strauss D, Kessl J, Trumpower B, Tollervey D, Hurt E. Nuclear export of 60s ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p, that associates with the large subunit protein Rpl10p. Mol Cell Biol. 2001;21:3405–15. doi: 10.1128/MCB.21.10.3405-3415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson AW, Lund E, Dahlberg J. Nuclear export of ribosomal subunits. Trends Biochem Sci. 2002;27:580–5. doi: 10.1016/S0968-0004(02)02208-9. [DOI] [PubMed] [Google Scholar]
- 23.Ho JH, Kallstrom G, Johnson AW. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J Cell Biol. 2000;151:1057–66. doi: 10.1083/jcb.151.5.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas F, Kutay U. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J Cell Sci. 2003;116:2409–19. doi: 10.1242/jcs.00464. [DOI] [PubMed] [Google Scholar]
- 25.Trotta CR, Lund E, Kahan L, Johnson AW, Dahlberg JE. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 2003;22:2841–51. doi: 10.1093/emboj/cdg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sengupta J, Bussiere C, Pallesen J, West M, Johnson AW, Frank J. Characterization of the nuclear export adaptor protein Nmd3 in association with the 60S ribosomal subunit. J Cell Biol. 2010;189:1079–86. doi: 10.1083/jcb.201001124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo KY, Li Z, Bussiere C, Bresson S, Marcotte EM, Johnson AW. Defining the pathway of cytoplasmic maturation of the 60S ribosomal subunit. Mol Cell. 2010;39:196–208. doi: 10.1016/j.molcel.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouquette J, Choesmel V, Gleizes PE. Nuclear export and cytoplasmic processing of precursors to the 40S ribosomal subunits in mammalian cells. EMBO J. 2005;24:2862–72. doi: 10.1038/sj.emboj.7600752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wild T, Horvath P, Wyler E, Widmann B, Badertscher L, Zemp I, et al. A protein inventory of human ribosome biogenesis reveals an essential function of exportin 5 in 60S subunit export. PLoS Biol. 2010;8:e1000522. doi: 10.1371/journal.pbio.1000522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell. 2004;16:777–87. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 31.Verheggen C, Bertrand E. CRM1 plays a nuclear role in transporting snoRNPs to nucleoli in higher eukaryotes. Nucleus. 2012;3:132–7. doi: 10.4161/nucl.19266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, et al. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell. 2004;16:789–98. doi: 10.1016/j.molcel.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 33.Pradet-Balade B, Girard C, Boulon S, Paul C, Azzag K, Bordonné R, et al. CRM1 controls the composition of nucleoplasmic pre-snoRNA complexes to licence them for nucleolar transport. EMBO J. 2011;30:2205–18. doi: 10.1038/emboj.2011.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, et al. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–16. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ernoult-Lange M, Wilczynska A, Harper M, Aigueperse C, Dautry F, Kress M, et al. Nucleocytoplasmic traffic of CPEB1 and accumulation in Crm1 nucleolar bodies. Mol Biol Cell. 2009;20:176–87. doi: 10.1091/mbc.E08-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muro E, Gébrane-Younís J, Jobart-Malfait A, Louvet E, Roussel P, Hernandez-Verdun D. The traffic of proteins between nucleolar organizer regions and prenucleolar bodies governs the assembly of the nucleolus at exit of mitosis. Nucleus. 2010;1:202–11. doi: 10.4161/nucl.1.2.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Latonen L, Moore HM, Bai B, Jäämaa S, Laiho M. Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene. 2011;30:790–805. doi: 10.1038/onc.2010.469. [DOI] [PubMed] [Google Scholar]
- 38.Alavian CN, Politz JC, Lewandowski LB, Powers CM, Pederson T. Nuclear export of signal recognition particle RNA in mammalian cells. Biochem Biophys Res Commun. 2004;313:351–5. doi: 10.1016/j.bbrc.2003.11.126. [DOI] [PubMed] [Google Scholar]
- 39.Daelemans D, Costes SV, Lockett S, Pavlakis GN. Kinetic and molecular analysis of nuclear export factor CRM1 association with its cargo in vivo. Mol Cell Biol. 2005;25:728–39. doi: 10.1128/MCB.25.2.728-739.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai B, Laiho M. Efficient sequential recovery of nucleolar macromolecular components. Proteomics. 2012;12:3044–8. doi: 10.1002/pmic.201200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol. 1970;76:127–39. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- 42.Shcherbik N, Wang M, Lapik YR, Srivastava L, Pestov DG. Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Rep. 2010;11:106–11. doi: 10.1038/embor.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burger K, Mühl B, Harasim T, Rohrmoser M, Malamoussi A, Orban M, et al. Chemotherapeutic drugs inhibit ribosome biogenesis at various levels. J Biol Chem. 2010;285:12416–25. doi: 10.1074/jbc.M109.074211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grimm T, Hölzel M, Rohrmoser M, Harasim T, Malamoussi A, Gruber-Eber A, et al. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006;34:3030–43. doi: 10.1093/nar/gkl378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohrmoser M, Hölzel M, Grimm T, Malamoussi A, Harasim T, Orban M, et al. Interdependence of Pes1, Bop1, and WDR12 controls nucleolar localization and assembly of the PeBoW complex required for maturation of the 60S ribosomal subunit. Mol Cell Biol. 2007;27:3682–94. doi: 10.1128/MCB.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grummt I. Wisely chosen paths--regulation of rRNA synthesis: delivered on 30 June 2010 at the 35th FEBS Congress in Gothenburg, Sweden. FEBS J. 2010;277:4626–39. doi: 10.1111/j.1742-4658.2010.07892.x. [DOI] [PubMed] [Google Scholar]
- 47.Cornish T, Morgan J, Gurel B, De Marzo A. FrIDA: An open source framework for image data set analysis. Arch Pathol Lab Med. 2008;132:856. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.