SUMMARY
Computer-aided detection (CADe) and diagnosis (CAD) has been a rapidly growing, active area of research in medical imaging. Machine leaning (ML) plays an essential role in CAD, because objects such as lesions and organs may not be represented accurately by a simple equation; thus, medical pattern recognition essentially require “learning from examples.” One of the most popular uses of ML is the classification of objects such as lesion candidates into certain classes (e.g., abnormal or normal, and lesions or non-lesions) based on input features (e.g., contrast and area) obtained from segmented lesion candidates. The task of ML is to determine “optimal” boundaries for separating classes in the multidimensional feature space which is formed by the input features. ML algorithms for classification include linear discriminant analysis (LDA), quadratic discriminant analysis (QDA), multilayer perceptrons, and support vector machines (SVM). Recently, pixel/voxel-based ML (PML) emerged in medical image processing/analysis, which uses pixel/voxel values in images directly, instead of features calculated from segmented lesions, as input information; thus, feature calculation or segmentation is not required. In this paper, ML techniques used in CAD schemes for detection and diagnosis of lung nodules in thoracic CT and for detection of polyps in CT colonography (CTC) are surveyed and reviewed.
Keywords: machine learning in medical imaging, computer-aided diagnosis, classification, pixel-based machine learning, lung nodule, colorectal polyp, CT colonography
1. Introduction
CAD [1, 2] has been a rapidly growing, active area of research in medical imaging. CAD is defined as detection and/or diagnosis made by a radiologist/physician who takes into account the computer output as a “second opinion” [2]. Evidence suggests that CAD can help improve the diagnostic performance of radiologists/physicians in their image interpretations [3–6]. Consequently, many investigators have participated and developed CAD schemes such as those for detection of lung nodules in chest radiographs (also known as chest x-rays; CXRs) [7–10] and in thoracic CT [11–14], those for detection of microcalcifications/masses in mammography [15], breast MRI [16], and breast US [17], and those for detection of polyps in CTC [18–21].
A CADe scheme of lesions in medical images generally consists of two major components: (1) identification of lesion candidates and (2) classification of the identified candidates into lesions or non-lesions. Segmentation of the organ of interest is the first necessary step before the identification of lesion candidates. The development of the first component, the identification of lesion candidates, generally aims at obtaining a high sensitivity level, because the sensitivity lost in this step cannot be recovered in the later step. The second component, the classification of the identified candidates, is very important, because it determines the final performance of a CAD scheme. The development of the second component aims at removing as many non-lesions (i.e., false-positive (FP) detections in the first step) as possible while minimizing the removal of lesions (i.e., true-positive detections in the first step). Minimizing FPs is very important, because a large number of FPs could adversely affect the clinical application of CADe. A large number of FPs is likely to confound the radiologist’s task of image interpretation and thus lower radiologist efficiency. In addition, radiologists may lose their confidence in CADe as a useful tool. The evaluation of the standalone performance of a developed CAD scheme is the last step of CAD development, and the evaluation of radiologists’ performance with the use of the developed CAD scheme is the important last step in CAD research.
ML plays a very important role in a CAD scheme, because tasks on medical images in a CAD scheme require “learning from examples (or data).” Objects in medical images such as lesions and organs may be too complex to be represented accurately by a simple equation. Modeling of such complex objects often requires a number of parameters that have to be determined by examples or data. For example, a lung nodule is generally modeled as a solid sphere, but there are nodules of various shapes and nodules with internal inhomogeneities, such as spiculated nodules and ground-glass nodules. A polyp in the colon is modeled as a bulbous object, but there are also polyps which have a flat shape [22, 23]. Thus, CAD schemes need “learning from examples or data” to determine a number of parameters in a complex model. ML has been used in the second major step of a CAD scheme, i.e., classification of identified lesion candidates into certain classes (e.g., abnormal or normal, lesions or non-lesions, and malignant or benign) based on input features (e.g., contrast, area, and circularity) obtained from segmented lesion candidates (This class of ML is referred to as feature-based ML, or simply as a classifier). The task of ML here is to determine “optimal” boundaries for separating classes in the multidimensional feature space which is formed by the input features.
ML algorithms for classification include LDA, QDA, multilayer perceptron (one of the most popular artificial neural network (ANN) models) [24], and support vector machines [25]. Such ML algorithms have been applied to lung nodule detection in CXR [26] and thoracic CT [12, 27], classification of lung nodules into benign or malignant in CXR [28] and thoracic CT [29], and polyp detection in CTC [18, 30]. Recently, as available computational power has increased dramatically, PML emerged in medical image processing/analysis which uses pixel/voxel values in images directly, instead of features calculated from segmented regions, as input information; thus, feature calculation or segmentation is not required. PML has also been used in the classification of the identified lesion candidates in CAD schemes.
In this paper, ML techniques used in CAD schemes for detection and diagnosis of lung nodules in CT and for detection of polyps in CTC are surveyed and reviewed. Survey papers for CAD in thoracic CT have been published, including one for lung image analysis in CT with emphasis on a comprehensive survey for computer analysis of the lungs [31], one for CAD in thin-section CT [32], one for CAD in CT with emphasis on CAD performance [33], one for CAD in CT with emphasis on performance comparisons with clinical aspects [34], and one for CAD in both thoracic CT and CTC with emphasis on a methodological overview of major steps in CAD schemes [35]. This present paper focuses on surveys and comparisons of ML techniques in CADe and CADx schemes in thoracic CT and CTC.
2. Classes of Classification Techniques in CAD
There are three classes of classification techniques that have been developed and used in CAD schemes: feature-based classifiers (or feature-based ML), PML, and non-ML-based methods that are defined as methods that do not use ML techniques, such as a procedure that uses a geometrical relationship in a non-learning way. Non-ML methods are not surveyed in this paper.
2.1 Feature-based Classifiers
When an ML algorithm is used for classification, it is generally called a classifier. A standard classification approach based on a classifier such as a multilayer perceptron is illustrated in Fig. 1. First, target lesions are segmented by using a segmentation method. Next, features are extracted from the segmented lesions. Features may include morphologic (or shape-based), gray-level-based (including histogram-based), and texture features. Then, extracted features are entered as input to an ML model such as a multilayer perceptron [24]. The ML model is trained with sets of input features and correct class labels. A class label of 1 is assigned to the corresponding output unit when a training sample belongs to a certain class (e.g., class A), and 0 is assigned to the other output units (e.g., classes B, C, etc.). In the case of two-class classification, one output unit instead of two output units is often used with the output value 0 being class A, and 1 being class B. After training, the class of the unit with the maximum value is determined to be the corresponding class to which an unknown sample belongs. For details of feature-based classifiers, refer to one of many textbooks in pattern recognition such as [24, 25, 36, 37].
Fig. 1.
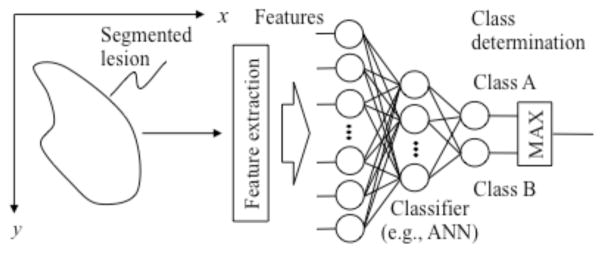
Feature-based ML (feature-based classifier) for classification of a detected and segmented lesion.
There are several important issues to be considered in the design of ML techniques: generalization, over-fitting, curse of dimensionality, training data annotation, and feature selection.
Generalization in ML is the ability of a trained ML model to perform on unseen cases. The generalization performance of the ML model is estimated by using cases in a test database, which is often lower than the performance for training cases. How to design an ML model with a high generalization performance is an important topic, which is closely related to the over-fitting issue. If an ML model is trained with only a small number of cases, the generalization ability will be lower, because the ML model may fit only the training cases. This is known as over-training (or over-fitting”) [38]. Over-fitting occurs when the number of training cases is too small to determine parameters in the ML model sufficiently. For achieving a high generalization performance, a large number of training cases, e.g., 400–800 cases, is generally required for an ANN in a CADs scheme [39]. For detailed information, please refer to the literature such as [37, 40].
How to estimate the generalization performance with a finite number of testing cases is an important topic as well. To estimate the generalization performance better, resampling schemes such as leave-one-out cross validation, N-fold cross validation, and bootstrapping are often employed [40]. The curse of dimensionality [41] is referred to as the following phenomenon: As the dimensionality of the input feature space for a ML model increases subject to the number of input features, the number of training samples required for the ML model increases exponentially. For detailed information, please refer to the literature such as [40]. To avoid the curse of dimensionality, feature selection and/or dimensionality reduction techniques are often utilized.
Annotating training cases is also an important topic, because the annotation is expensive or time-consuming when the number of training cases is large. There are methods for reducing the annotation labor or annotation itself in the general ML field, but the quality of annotation (or determining “gold standard”) is more important in the CAD research area. In order for the study to be clinically meaningful, “gold standard” annotations (or labels) have to be determined by using more reliable/accurate examinations, e.g., the “gold standard” for lung nodule presence in screening CT should be established by using their confirmation in upper-level follow-up examinations such as diagnostic CT or high-resolution CT (HRCT).
Feature selection has long been an active research topic in ML, because it is one of the main factors that determine the performance of a classifier. It avoids the curse of dimensionality by reducing the input dimension to the classifier. In general, many features are extracted from segmented lesions as the classifier input. Not all of the features, however, would be useful for a classifier to distinguish between lesions and non-lesions, because some of them might be highly correlated with each other or redundant; some of them may not be strongly associated with the given classification task. For designing a classifier with high performance, it is crucial to select “effective” features. In the field of CADe research, one of the most popular feature selection methods is a stepwise feature selection based on Wilks’ lambda. The method has been applied in various CADe schemes because of its simplicity [12, 29, 42]. The Wilks’ lambda criterion is good for LDA, but not necessarily for nonlinear classifiers. One of the most widely used deterministic feature selection methods is sequential forward or backward floating selection (SFFS or SBFS) [43]. SBFS was used for selection of input features for ANNs [44, 45]. SFFS was used for feature selection combined with various classifiers such as Naïve Bayes, SVMs, and AdaBoost [46] in different CADe schemes. Recently, Xu and Suzuki proposed SFFS coupled with an SVM for selection of the most relevant features that maximize the area under the receiver-operating-characteristic (ROC) curve (AUC) [47].
2.2. Pixel/voxel-Based Machine Learning (PML)
Recently, as available computational power has increased dramatically, pixel/voxel-based ML (PML) [35] emerged in medical image processing/analysis, which uses pixel/voxel values in images directly instead of features calculated from segmented regions as input information; thus, feature calculation or segmentation is not required. Because PML can avoid errors caused by inaccurate feature calculation and segmentation, the performance of PML can potentially be better for subtle/complex lesions than that of common feature-based classifiers.
There are three classes of PMLs: neural filters [48–50] (including neural edge enhancers [51, 52]), convolution neural networks (NNs) [53–57] (including shift-invariant NNs [58, 59]), and massive-training artificial neural networks (MTANNs) [19, 60–63] (including multiple MTANNs [12, 49, 50, 60, 64, 65], a mixture of expert MTANNs [20, 66], a multi-resolution MTANN [61], a Laplacian eigenfunction MTANN (LAP-MTANN) [67], a massive-training support vector regression (MTSVR), and a massive-training Gaussian process regression [68]). For details of the architectures and applications of PMLs in medical imaging, refer to a survey paper on PMLs [35].
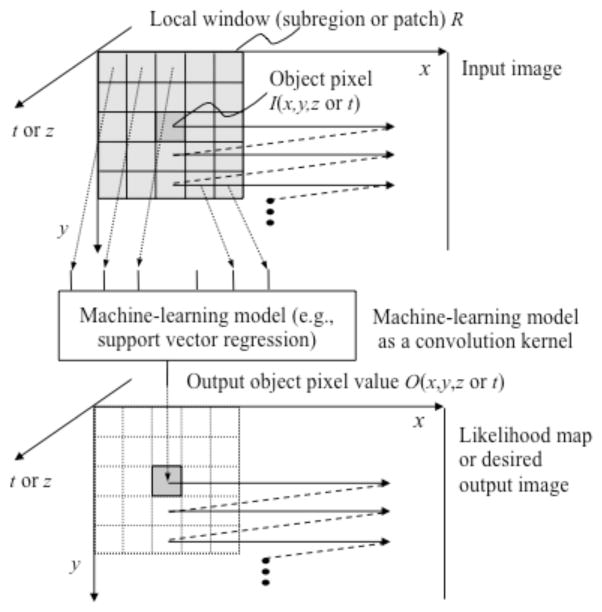
By extending the neural filter and the neural edge enhancer, two-dimensional (2D) MTANNs [11], which are a class of a PML based on an ANN regression model, have been developed for accommodating the task of distinguishing a specific opacity from other opacities in medical images. The MTANN learns the relationship between input images and corresponding “teaching” images (i.e., ideal or desired images) to distinguish lesions from non-lesions (i.e., FPs). The MTANN is trained with a massive number of subregions/subvolumes extracted from input images together with teaching pixels; hence the term “massive training”. The architecture of an MTANN is shown in Fig. 2. A 2D MTANN consists of a linear-output multilayer ANN regression model, which is capable of operating on voxel data directly [52],[51]. The MTANN is trained with input images/volumes and the corresponding “teaching” images/volumes for enhancement of a specific pattern and suppression of other patterns. The input to the MTANN consists of voxel values in a sub-region/volume (local window or patch) extracted from an input image/volume. The class of MTANNs has been used for classification, such as FP reduction in CAD schemes for detection of lung nodules in CXR [7] and CT [5, 11, 12], distinction between benign and malignant lung nodules in CT [65], and FP reduction in a CAD scheme for polyp detection in CTC [19, 20, 66–68]. The MTANNs have also been applied to pattern enhancement and suppression such as separation of bones from soft tissue in CXR [61, 62], and enhancement of lung nodules in CT [63].
Fig. 2.
Architecture of an MTANN (a class of PML) consisting of an ML regression model (e.g., linear-output ANN regression and support-vector regression) with sub-region (local window or patch) input and single-pixel output.
3. Classification in CAD of the Thorax
Lung cancer continues to rank as the leading cause of cancer deaths in America and other nations such as Japan. The number of lung cancer deaths in each year is greater than the combined number of breast, colon, and prostate cancer deaths in the United States [69]. Because CT is more sensitive than CXR in the detection of small nodules and of lung carcinoma at an early stage [70–72], lung cancer screening programs are being investigated in the United States [73], Japan [70, 71], and other countries with low-dose (LD) helical CT as the screening modality. Evidence suggests that early detection of lung cancer may allow more timely therapeutic intervention and thus a more favorable prognosis for the patient [71, 74]. Helical CT, however, generates a large number of images that must be read by radiologists/physicians. This may lead to “information overload” for the radiologists/physicians. Furthermore, radiologists/physicians may miss some cancers during interpretation of CT images. Therefore, a CAD scheme for detection of lung nodules in LDCT images has been investigated as a useful tool for lung cancer screening.
Classification is a major component in CAD schemes for detection and diagnosis of lung nodules in CT. CAD schemes for detection of lung nodules in thoracic CT (i.e., CADe) generally consists of two major steps: (1) identification of nodule candidates, followed by (2) classification of the identified nodule candidates into nodules or non-nodules (i.e., normal anatomic structures). The second major step in a CADe scheme aims at classification of the nodule candidates identified in the first step into nodules or non-nodules, whereas a CAD scheme for diagnosis (often abbreviated as CADx) aims at classification of the detected nodules (either by a computer or by a radiologist) into benign or malignant nodules.
3.1. Detection of lung nodules
Technical developments of the classification step in CADe schemes for detection of lung nodules in CT are summarized in Table 1. In 1994, Giger et al. [75] developed a CADe scheme for detection of lung nodules in CT. In 1999, Armato et al. [13, 27] extended the method to include 3D feature analysis, a rule-based scheme, and LDA for classification. They evaluated the performance of their scheme with a leave-one-out cross-validation (LOO) test. Kanazawa et al. [76] employed a rule-based scheme with features for classification in their CADe scheme. Gurcan et al. [77] employed a rule-based scheme based on 2D and 3D features, followed by LDA for classification. Lee et al. [78] employed a simpler approach which is a rule-based scheme based on 13 features for classification. Suzuki et al. [60] developed a PML technique called an MTANN for reduction of a single source of FPs and a multiple MTANN scheme for reduction of multiple sources of FPs that had not been removed by LDA. This MTANN approach did not require a large number of training cases: the MTANN was able to be trained with 10 positive and 10 negative cases [79–81], whereas feature-based classifiers generally require 400–800 training cases [79–81]. Arimura et al. [12] employed a rule-based scheme followed by LDA or by the MTANN [60] for classification. Farag et al. [82] developed a template-modeling approach that uses level sets for classification. Ge et al. [83] incorporated 3D gradient field descriptors and ellipsoid features in LDA for classification. Matsumoto et al. [84] employed LDA with 8 features for classification. Yuan et al. [85] tested a commercially available CADe system (ImageChecker CT, R2 Technology, CA). Bi et al. [86] developed an asymmetric cascade of classifiers for classification. Pu et al. [87] developed a scoring method based on the similarity distance of medial axis-like shapes for classification. Retico et al. [88] used the MTANN approach (i.e., a PML technique) for classification. Ye et al. [89] used a rule-based scheme followed by a weighted SVM for classification. Golosio et al. [90] used a fixed-topology ANN for classification, and they evaluated their CADe scheme with a publicly available database from the Lung Image Database Consortium (LIDC) [91]. Murphy et al. [92] used a k-nearest-neighbor classifier for classification. Tan et al. [93] developed a feature-selective classifier based on a genetic algorithm and ANNs for classification. Messay et al. [94] developed a sequential forward selection process for selecting the optimum features for LDA and QDA. Riccard et al. [95] used a heuristic approach based on geometric features, followed by an SVM for classification. Other than the development of CADe schemes, Rao et al. [96] performed an observer performance study with a CADe scheme. Thus, various approaches have been proposed for the classification component in CADe schemes. There are large variations in the performance of CADe schemes: sensitivities ranged from 70–94% with 0.7–64.1 FPs per case. It is difficult to say which CADe scheme performs better because of different databases and testing methods used, without a direct comparison. Some studies used thick-slice CT, and others used thin-slice CT. Some studies used nodules missed by radiologists, and some used nodules detected by radiologists. Evaluation of a CAD scheme with missed cases would be desirable, because the CAD scheme is likely to help radiologists more with such cases. Some studies used screening CT, some used diagnostic CT, and some used HRCT. Testing with screening CT cases would be more appropriate, given the purpose of CADe schemes. Some studies used an LOO test, some used an independent test, and some used N-fold cross-validation. Each testing method has its own advantages and limitations. For detailed information, please refer to the literature [39, 40, 97]. Since the current sensitivity and FP rate of CADe schemes are not high enough compared to radiologists’ performance, further developments of techniques to improve the performance would be necessary. In addition, more studies on the proof of the usefulness of CADe such as observer performance studies and clinical trials would be beneficial in the field.
Table 1.
Classifications component in CADe schemes for detection of lung nodules in CT.
| Study | Database | Classifier/Method | Performance |
|---|---|---|---|
| Giger et al. [75] | Thick-slice diagnostic CT scans of 8 patients with 47 nodules | Comparison of geometric features | Sensitivity of 94% with 1.25 FPs per case |
| Armato et al. [13, 27] | Thick-slice (10 mm) diagnostic CT scans of 43 patients with 171 nodules | Rule-based scheme and LDA with 9 2D and 3D features | Sensitivity of 70% with 42.2 FPs per case in an LOO test |
| Kanaza wa et al. [76] | Thick-slice (10 mm) screening CT scans of 450 patients with 230 nodules | Rule-based scheme | Sensitivity of 90% |
| Gurcan et al. [77] | Thick-slice (2.5–5 mm) diagnostic CT scans of 34 patients with 63 nodules | Rule-based scheme and LDA with 6 2D and 3D features | Sensitivity of 84% with 74.4 FPs per case in an LOO test |
| Lee et al. [78] | Thick-slice (10 mm) diagnostic CT scans of 20 patients with 98 nodules | Rule-based scheme and LDA with 13 features | Sensitivity of 72% with 30.6 FPs per case |
| Suzuki et al. [60] | Thick-slice (10 mm) screening LDCT scans of 63 patients with 71 nodules with solid, part-solid and non-solid patterns, including 66 cancers | Multiple MTANNs with pixel values in a 9×9 subregion (local window or patch) as input | Sensitivity of 80.3% with 4.8 FPs per case in a validation test |
| Arimura et al. [12] | 106 thick-slice (10 mm) screening LDCT scans of 73 patients with 109 cancers with solid, part-solid and non-solid patterns | Rule-based scheme followed by multiple MTANNs with pixel values in a 9×9 subregion as input (or LDA with Wilks’ lambda stepwise feature selection) | Sensitivity of 83% with 5.8 FPs per case in a validation test (or an LOO test for LDA) |
| Farag et al. [82] | Thin-slice (2.5 mm) screening LDCT scans of 16 patients with 119 nodules and 34 normal patients | Template modeling approach using level sets | Sensitivity of 93.3% with an FP rate of 3.4% |
| Ge et al. [83] | 82 thin-slice (1.0–2.5 mm) CT scans of 56 patients with 116 solid nodules | LDA with Wilks’ lambda stepwise feature selection from 44 features | Sensitivity of 80% with 14.7 FPs per case in an LOO test |
| Matsumoto et al. [84] | Thick-slice (5 or 7 mm) diagnostic CT scans of 5 patients (4 of which used contrast media) with 50 nodules | LDA with 8 features | Sensitivity of 90% with 64.1 FPs per case in an LOO test |
| Yuan et al. [85] | Thin-slice (1.25 mm) CT scans of 150 patients with 628 nodules | ImageChecker CT LN-1000 by R2 Technology | Sensitivity of 73% with 3.2 FPs per case in an independent test |
| Bi et al. [86] | HRCT scans of 86 patients with 48 nodules | Asymmetric cascade of classifiers with column generation boosting feature selection | Sensitivity of 88% with 0.7 FPs per case in a validation test |
| Pu et al. [87] | Thin-slice (2.5 mm) screening CT scans of 52 patients with 184 nodules including 16 non-solid nodules | Scoring method based on the similarity distance combined with a marching cube algorithm | Sensitivity of 81.5% with 6.5 FPs per case |
| Retico et al. [88] | Thin-slice (1 mm) screening CT scans of 39 patients with 102 nodules | Voxel-based neural approach (MTANN) with pixel values in a subvolume as input | Sensitivities of 80–85% with 10–13 FPs per case |
| Ye et al. [89] | Thin-slice (1 mm) screening CT scans of 54 patients with 118 nodules including 17 non-solid nodules | Rule-based scheme followed by a weighted SVM with 15 features | Sensitivity of 90.2% with 8.2 FPs per case in an independent test |
| Golosio et al. [90] | Thin-slice (1.5–3.0 mm) CT scans of 83 patients with 148 nodules that one radiologist detected from LIDC database | Fixed-topology ANN with 42 features from multithreshold ROI | Sensitivity of 79% with 4 FPs per case in an independent test |
| Murphy et al. [92] | Thin-slice screening CT scans of 813 patients with 1,525 nodules | k-nearest-neighbor classifier with features selected from 135 features | Sensitivity of 80 with 4.2 FPs per case in an independent test |
| Tan et al. [93] | Thin-slice CT scans of 125 patients with 80 nodules that 4 radiologists agreed from LIDC database | Feature-selective classifier based on a genetic algorithm and ANNs with 45 initial features | Sensitivity of 87.5% with 4 FPs per case in an independent test |
| Messay et al. [94] | Thin-slice CT scans of 84 patients with 143 nodules from LIDC database | LDA and QDA with feature selection from 245 features | Sensitivity of 83% with 3 FPs per case in a 7-fold cross-validation test |
| Riccardi et al. [95] | Thin-slice CT scans of 154 patients with 117 nodules that 4 radiologists agreed on from LIDC database | Heuristic approach and SVM with maximum-intensity projection data from a volume of interest | Sensitivity of 71% with 6.5 FPs per case in a 2-fold cross-validation test |
3.2 Diagnosis of lung nodules
Although CT has been shown to be sensitive to the detection lung nodules, it may be difficult for radiologists to distinguish between benign and malignant nodules on LDCT images. In a screening program with LDCT in New York, 88% (206/233) of suspicious lesions were found to be benign on follow-up examinations [72]. In a screening program in Japan, only 83 (10%) among 819 scans with suspicious lesions were diagnosed to be cancer cases [98]. According to recent findings at the Mayo Clinic, 2,792 (98.6%) of 2,832 nodules detected by a multidetetor CT were benign, and 40 (1.4%) nodules were malignant [99]. Thus, a large number of benign nodules were found with CT; follow-up examinations such as HRCT and/or biopsy were performed on these patients. Therefore, CADx schemes for distinction between benign and malignant nodules in LDCT would be useful for reducing the number of “unnecessary” follow-up examinations.
A number of researchers developed CADx schemes for this task, which distinguish malignant nodules from benign nodules automatically and/or determine the likelihood of malignancy for the detected nodules. The performance of the schemes was generally evaluated by means of ROC analysis [100], because this task is a two-class classification. The AUC [101] was often used as a performance index. Studies on the development of CADx schemes for distinction between malignant and benign lung nodules in CT are summarized in Table 2. In 1998, Kawata et al. [102] described the calculation of nodule features for the purpose of distinction between malignant and benign nodules. In 1999, McNitt-Gray et al. [103] developed a classification scheme based on LDA for distinction between malignant and benign nodules in HRCT. They achieved a correct classification rate of 90.3% for a database of 17 malignant and 14 benign nodules. Matsuki et al. [104] used an ANN with subjective features determined by radiologists for classification between 99 malignant and 56 benign nodules in HRCT and achieved an AUC value of 0.951. Aoyama et al. [42] used LDA for distinction between malignant and benign nodules in thick-slice screening LDCT. They achieved an AUC value of 0.846 for a database of 73 patients with 76 primary cancers and 342 patients with 413 benign nodules. Mori et al. [105] developed a classification scheme for distinction between malignant and benign nodules in contrast-enhanced (CE) CT by using LDA with 3 features (i.e., attenuation, shape index, and curvedness value). Shah et al. [106] employed different classifiers such as logistic regression and QDA with features selected from a group of 31 by using stepwise feature selection based on the Akaike information criterion. Their scheme with logistic regression achieved an AUC value of 0.92 in the distinction between 19 malignant and 16 benign nodules in thin-slice CE-CT. Suzuki et al. [65] developed a PML technique called a multiple MTANN scheme for the classification task. They achieved an AUC value of 0.88 for thick-slice screening LDCT scans of 73 patients with 76 primary cancers and 342 patients with 413 benign nodules. Iwano et al. [107] achieved a sensitivity of 76.9% and a specificity of 80% with their scheme based on LDA with 2 features in their evaluation of HRCT images of 52 malignant and 55 benign nodules. Way et al. [108] incorporated nodule surface features into their classification based on LDA or an SVM, and they achieved an AUC value of 0.857 in the classification of 124 malignant and 132 benign nodules in 152 patients. Chen et al. [109] employed an ANN ensemble to classify 19 malignant and 13 benign nodules, and they achieved an AUC value of 0.915. Lee et al. [110] developed a two-step supervised learning scheme combining a genetic algorithm with a random subspace method, and they achieved an AUC value of 0.889 in the classification between 62 malignant and 63 benign nodules. Other than CADx approaches, Kawata et al. [111] developed a content-based image retrieval approach to provide radiologists with similar images for improving their diagnostic performance in distinction between benign and malignant nodules. Kawata et al. [112] also developed quantitative classification measures that correlate with pathologic characteristics of lung cancer and patients’ prognosis. Thus, various approaches to CADx schemes have been proposed. The database size varied in different studies, from 31–489. Generally achieving high performance for a large database is challenging, because it is likely to contain more variations of nodules. CT scans in the databases included screening LDCT, standard diagnostic CT, and HRCT. Diagnosis of lung nodules on LDCT images would be the most challenging due to a low image quality. Most studies used an LOO test. There are variations in the performance of CADx schemes: AUC values ranged from 0.846–0.951. Once again, it is difficult to say which CADx scheme performs better without a direct comparison. Since the current performance of CADx schemes would be close to or comparable to radiologists’ performance, more studies on the proof of the usefulness of CADx such as observer performance studies and clinical trials would be beneficial in the field.
Table 2.
Classification between malignant and benign nodules (CADx) for thoracic CT.
| Study | Database | Classifier/Method | Performance |
|---|---|---|---|
| McNitt-Gray et al. [103] | HRCT scans of 17 malignant and 14 benign nodules | LDA with stepwise feature selection | Correct classification rate of 90.3% |
| Matsuki et al. [104] | HRCT scans of 99 malignant and 56 benign nodules | ANN with 16 radiologists’ subjective features and 7 clinical data | AUC value of 0.951 in an LOO test |
| Aoyama et al. [113] | Thick-slice (10 mm) screening LDCT scans of 76 malignant and 413 benign nodules | LDA with Wilks’ lambda stepwise feature selection | AUC of 0.846 in an LOO test |
| Mori et al. [105] | Thin-slice (2 mm) CE-CT scans of 35 malignant and 27 benign nodules | LDA with 3 features | AUC of 0.91 and 1.0 with non-CE CT and CE-CT, respectively, in an LOO test |
| Shah et al. [106] | Thin-slice (≤ 3 mm) CE-CT scans of 19 malignant and 16 benign nodules | Logistic regression or QDA with stepwise feature selection from 31 features | AUC of 0.92 with QDA in an LOO test |
| Suzuki et al. [65] | Thick-slice (10 mm) screening LDCT scans of 76 malignant and 413 benign nodules | Multiple MTANNs with pixel values in a 9×9 subregion as input | AUC of 0.88 in an LOO test |
| Iwano et al. [107] | HRCT (0.5–1 mm slice) scans of 52 malignant and 55 benign nodules | LDA with 2 features | Sensitivity of 76.9% and a specificity of 80% |
| Way et al. [108] | CT scans of 124 malignant and 132 benign nodules in 152 patients | LDA or SVM with stepwise feature selection | AUC of 0.857 in an LOO test |
| Chen et al. [109] | CT (slice thickness of 2.5 or 5 mm) scans of 19 malignant and 13 benign nodules | ANN ensemble with selected features | AUC of 0.915 in an LOO test |
| Lee et al. [110] | Thick-slice (5 mm) CT scans of 62 malignant and 63 benign nodules | GA-based feature selection and a random subspace method | AUC value of 0.889 in an LOO test |
4. Classification in CADe of the Colon
4.1. CADe for detection of polyps in CTC
Colorectal cancer is the second leading cause of cancer deaths in the United States [114]. Evidence suggests that early detection and removal of polyps (i.e., precursors of colorectal cancer) can reduce the incidence of colorectal cancer [115]. CTC, also known as virtual colonoscopy, is a technique for detecting colorectal neoplasms by use of CT scans of the colon. The diagnostic performance of CTC in detecting polyps, however, remains uncertain due to a propensity for perceptual errors in detection of polyps. CADe of polyps has been investigated to address that issue with CTC [116]. CADe has the potential to improve radiologists’ diagnostic performance in the detection of polyps. A number of investigators have developed automated or semi-automated CADe schemes for the detection of polyps in CTC [117–120].
4.2. Classification component in CADe schemes
Major sources of non-polyps (i.e., FPs) remaining after the first step in CADe schemes include haustral folds, residual stool, rectal tubes, the ileocecal valve, and extra-colonic structures such as the small bowel and stomach. Technical developments of the classification step in CADe schemes for detection of polyps in CTC are summarized in Table 3. Many investigators employed feature-based classifiers in the second component of CADe schemes. Gokturk et al. [121] employed an SVM with histogram input that is used as a shape signature for classification. Näppi et al. developed a classification method based on volumetric features [122]. Acar et al. [123] used edge-displacement fields to model the changes in consecutive crosssectional views of CTC data and QDA for classifcation. Jerebko et al. [30] used a multilayer perceptron to classify polyp candidates in their CADe scheme and improved the performance by incorporating a committee of multilayer perceptrons [124] and a committee of SVMs [125]. Wang et al. [126] developed a classification method based on LDA with internal features (geometric, morphologic, and textural) of polyps. Suzuki et al. [127] developed a PML technique called a 3D MTANN by extending the structure of a 2D MTANN [11] to process 3D volume data in CTC. They removed FPs due to rectal tubes by using a single 3D MTANN [127] and multiple sources of FPs by developing and using a mixture of expert 3D MTANNs [20]. Li et al. [128] developed a classification method based on an SVM classifier with wavelet-based features. Wang et al. [129] improved the SVM performance by using nonlinear dimensionality reduction (i.e., a diffusion map and locally linear embedding). Yao at al. [130] employed a topographic height map for calculating features for an SVM classifier.. Suzuki et al. [66] tested a CADe scheme with MTANNs (i.e., a PML technique) on polyps that had been “missed” by radiologists [131] in a multicenter clinical trial [132]. Suzuki et al. [67] also improved the efficiency of the MTANN approach by incorporating principal-component analysis-based and Laplacian eigenmap-based dimension reduction techniques. Xu and Suzuki [68] showed that other nonlinear regression models such as support vector and nonlinear Gaussian process regression models instead of the ANN regression model could be used as the core model in the MTANN framework. Zhou et al. [133] developed projection features for an SVM classifier. Wang et al. [134] improved the performance of a CAD scheme by adding statistical curvature features in multiple-kernel learning. Multiple kernel learning is a recent topic in SVM research.
Table 3.
Classification components in CADe schemes for detection of polyps in CT colonography
| Study | Database | Classifier/Met hod | Performance |
|---|---|---|---|
| Gokturk et al. [121] | CTC data (2.5–3.0 mm collimation) of 48 patients in either supine or prone, containing 40 polyps (2–15 mm) | SVM with high-dimensional histograms used as shape signature | Sensitivity of 100% (95%) with a specificity of 0.69 (0.74) [14.3 FPs/patient (12.0 FPs/patient)] |
| Näppi et al. [122]. | CTC data (5 mm collimation) of 40 patients in both supine and prone, including 12 polyps in 11 patients | LDA or QDA with 54 volumetric features (9 statistics of 6 features) | By-patient (by-polyp) sensitivity of 100% (95%) with 2.4 FPs/patient |
| Acar et al. [123] | CTC data (2.5–3.0 mm collimation) of 48 patients in either supine or prone, containing 40 polyps (2–15 mm) | QDA with edge-displacement fields | Sensitivity of 100% (95%) with a specificity of 0.47 (0.56) |
| Jerebko et al. [30] | CTC data (5 mm collimation) of 40 patients in both supine and prone, including 29 polyps (3–25 mm) in 20 patients | A multilayer perceptron with 4 features selected from 17 features | Sensitivity of 90% with a specificity of 95% with 32 FPs/patient |
| Jerebko et al. [124] | CTC data (5 mm collimation) of 40 patients in both supine and prone, including 21 polyps (5–25 mm) | A committee of multilayer perceptrons with 12 features | Sensitivity of 82.9% with a specificity of 95.3% with 5.4 FPs/patient |
| Jerebko et al. [125] | CTC data (5 mm collimation) of 40 patients in both supine and prone, including 21 polyps (5–25 mm) | A committee of SVMs with 9 selected features | Sensitivity of 86.7% for larger polyps (≥10mm) and 75% for other polyps with 3 FPs/patient in a independent test |
| Wang et al. [126] | CTC data (5 mm collimation) of 153 patients in both supine and prone, including 61 polyps (4–30 mm) in 45 patients | LDA with internal features of polyps | Sensitivity of 100% and 100% for larger polyps (≥10mm) and other polyps with 4 and 6.9 FPs/patient, respectively |
| Suzuki et al. [127] | CTC data (1.25–5 mm collimation) of 73 patients in both supine and prone, including 28 polyps (5–25 mm) in 15 patients | Bayesian ANN and a single 3D MTANN with voxel values in a 7×7×7 subvolume as input | By-polyp (by-patient) sensitivity of 96.4% (100%) with 2.1 FPs/patient in an LOO test of the classification part |
| Suzuki et al. [20] | CTC data (1.25–5 mm collimation) of 73 patients in both supine and prone, including 28 polyps (5–25 mm) in 15 patients | Bayesian ANN and a mixture of expert 3D MTANNs with voxel values in a 7×7×7 subvolume as input | By-polyp (by-patient) sensitivity of 96.4% (100%) with 1.1 FPs/patient in an LOO test of the classification part |
| Li et al. [128] | CTC data of 44 patients containing 45 polyps (6–9 mm) | SVM classifier with wavelet-based features | Sensitivity of 71% with 5.4 FPs/patient in a 4-fold cross-validation test of the classification part |
| Wang et al. [129] | CTC data (1.25–2.5 mm collimation) of 791 patients in both supine and prone, including 123 polyps (6–9 mm) and 25 polyps (≥10mm) | SVM with nonlinear dimensionality reduction (i.e., diffusion map and locally linear embedding) | Sensitivity of 83% for polyps (6–9 mm) with 9 FPs/patient |
| Yao at al. [130] | CTC data (1.25–2.5 mm collimation) of 792 patients in both supine and prone, including 226 polyps (> 6 mm) | SVM classifier with features from a topographic height map | Sensitivity of 93% and 76% for larger polyps (≥10 mm) and other polyps with 1.2 and 3.1 FPs/patient, respectively, in a 10-fold cross-validation test of the classification part |
| Suzuki et al. [66] | CTC data (1.25–5 mm collimation) of 24 patients in both supine and prone, including 23 polyps (6–25 mm) and a mass (35 mm), that had been “missed” by radiologists in a multicenter clinical trial [132] | Bayesian ANN and a mixture of expert 3D MTANNs with voxel values in a 7×7×7 subvolume as input | By-polyp (by-patient) sensitivity of 96.4% (100%) with 1.1 FPs/patient in an LOO test of the classification part |
| Zhou et al. [133] | CTC data (1.25–5.0 mm collimation) of 325 patients in supine and/or prone, including 347 polyps and masses (5–60 mm) | SVM classifier with projection features | By-polyp sensitivity of 93.1% and 80.6% for larger polyps (≥10mm) and other polyps with 1.9 and 5.2 FPs/patient, respectively |
| Wang et al. [134] | CTC data (1.25–2.5 mm collimation) of 66 patients in supine and/or prone, including 96 polyps | Multiple-kernel learning with statistical curvature and 18 geometric features | Sensitivity of 83% with 5 FPs/patient in an LOO test of the classification part |
5. Summary
In this paper, ML techniques used in CAD schemes for the thorax and colon have been surveyed. These CAD schemes included CADe and CADx of lung nodules in thoracic CT and CADe of polyps in CTC. The second of the two major components of most CAD schemes, i.e., the classification of lesion candidates, used ML techniques. There are three classes of classification techniques used in CAD schemes: feature-based ML, PML, and non-ML methods. Feature-based ML is the most popular technique in the classification step. Various ML models have been used in this class, including LDA, a multilayer perceptron, an SVM, an ML ensemble, and multiple-kernel learning. Feature selection is an important step for maximizing the performance of a feature-based ML technique, and thus it was often used. The most popular feature selection method in CAD is stepwise feature selection with Wilks’ lambda for linear classifiers such as LDA. Recently, feature selection for nonlinear classifiers has been studied. The most recent development is SFFS under the maximum AUC criterion coupled with an SVM. Recently, PML emerged and used for removal of FPs that had not been removed by feature-based ML. An MTANN is a representative PML model, and there are variations of the MTANNs, including a mixture of expert MTANNs, MTSVR, and Lap-MTANNs. Thus, many investigators have been studying ML in CAD, which indicates the importance of ML in this field. Most CAD schemes employ feature-based MLs that had originally been developed and established in the pattern recognition field. On the other hand, MTANNs were born in the medical imaging field. Evidence demonstrated that PML including MTANNs was effective for improving the performance of CAD schemes. It is hoped that this survey will be useful for researchers in understanding the past studies and the current status of ML in CAD, and in advancing the research area of ML in CAD. It is also hoped to see more original ML techniques/models created in the CAD field.
Acknowledgments
The author is grateful to Ms. E. Lanzl for improving the manuscript. This work was partly supported by NIH R01CA120549, S10 RR021039 and P30 CA14599. CAD and ML technologies developed at University of Chicago have been licensed to companies including R2 Technology (Hologic), Riverain Medical, Median Technologies, Mitsubishi Space Software, General Electric, and Toshiba. It is the policy of University of Chicago that investigators disclose publicly actual or potential significant financial interests that may appear to be affected by research activities.
Biography
Kenji Suzuki received his Ph.D. degree in information engineering from Nagoya University in 2001. From 1993 to 2001, he worked at Hitachi Medical Corporation, and then Aichi Prefectural University as faculty. In 2001, he joined Department of Radiology at University of Chicago. Since 2006, he has been Assistant Professor of Radiology, Medical Physics, and Cancer Research Center there.
References
- 1.Giger ML, Suzuki K. Computer-Aided Diagnosis (CAD) In: Feng DD, editor. Biomedical Information Technology. Academic Press; 2007. pp. 359–374. [Google Scholar]
- 2.Doi K. Current status and future potential of computer-aided diagnosis in medical imaging. Br J Radiol. 2005;78 (1):S3–S19. doi: 10.1259/bjr/82933343. [DOI] [PubMed] [Google Scholar]
- 3.Chan HP, Sahiner B, Helvie MA, Petrick N, Roubidoux MA, Wilson TE, Adler DD, Paramagul C, Newman JS, Sanjay-Gopal S. Improvement of radiologists’ characterization of mammographic masses by using computer-aided diagnosis: an ROC study. Radiology. 1999 Sep;212(3):817–827. doi: 10.1148/radiology.212.3.r99au47817. [DOI] [PubMed] [Google Scholar]
- 4.Li F, Aoyama M, Shiraishi J, Abe H, Li Q, Suzuki K, Engelmann R, Sone S, Macmahon H, Doi K. Radiologists’ performance for differentiating benign from malignant lung nodules on high-resolution CT using computer-estimated likelihood of malignancy. AJR Am J Roentgenol. 2004 Nov;183(5):1209–1215. doi: 10.2214/ajr.183.5.1831209. [DOI] [PubMed] [Google Scholar]
- 5.Li F, Arimura H, Suzuki K, Shiraishi J, Li Q, Abe H, Engelmann R, Sone S, MacMahon H, Doi K. Computer-aided detection of peripheral lung cancers missed at CT: ROC analyses without and with localization. Radiology. 2005 Nov;237(2):684–690. doi: 10.1148/radiol.2372041555. [DOI] [PubMed] [Google Scholar]
- 6.Suzuki K, Hori M, McFarland E, Friedman AC, Rockey DC, Dachman AH. Can CAD help improve the performance of radiologists in detection of difficult polyps in CT colonography?. Proceedings of RSNA Annual Meeting; Chicago, IL. 2009. p. 872. [Google Scholar]
- 7.Suzuki K, Shiraishi J, Abe H, MacMahon H, Doi K. False-positive reduction in computer-aided diagnostic scheme for detecting nodules in chest radiographs by means of massive training artificial neural network. Academic Radiology. 2005 Feb;12(2):191–201. doi: 10.1016/j.acra.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 8.van Ginneken B, ter Haar Romeny BM, Viergever MA. Computer-aided diagnosis in chest radiography: a survey. IEEE Trans Med Imaging. 2001 Dec;20(12):1228–1241. doi: 10.1109/42.974918. [DOI] [PubMed] [Google Scholar]
- 9.Giger ML, Doi K, MacMahon H. Image feature analysis and computer-aided diagnosis in digital radiography. 3. Automated detection of nodules in peripheral lung fields. Med Phys. 1988 Mar-Apr;15(2):158–166. doi: 10.1118/1.596247. [DOI] [PubMed] [Google Scholar]
- 10.Chen S, Suzuki K, MacMahon H. A computer-aided diagnostic scheme for lung nodule detection in chest radiographs by means of two-stage nodule-enhancement with support vector classification. Med Phys. 2011;38:1844–1858. doi: 10.1118/1.3561504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki K, Armato SG, Li F, Sone S, Doi K. Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low-dose CT. Med Phys. 2003 Jul;30(7):1602–1617. doi: 10.1118/1.1580485. [DOI] [PubMed] [Google Scholar]
- 12.Arimura H, Katsuragawa S, Suzuki K, Li F, Shiraishi J, Sone S, Doi K. Computerized scheme for automated detection of lung nodules in low-dose computed tomography images for lung cancer screening. Acad Radiol. 2004 Jun;11(6):617–629. doi: 10.1016/j.acra.2004.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Armato SG, 3rd, Giger ML, Moran CJ, Blackburn JT, Doi K, MacMahon H. Computerized detection of pulmonary nodules on CT scans. Radiographics. 1999 Sep-Oct;19(5):1303–1311. doi: 10.1148/radiographics.19.5.g99se181303. [DOI] [PubMed] [Google Scholar]
- 14.Armato SG, 3rd, Li F, Giger ML, MacMahon H, Sone S, Doi K. Lung cancer: performance of automated lung nodule detection applied to cancers missed in a CT screening program. Radiology. 2002 Dec;225(3):685–692. doi: 10.1148/radiol.2253011376. [DOI] [PubMed] [Google Scholar]
- 15.Chan HP, Doi K, Galhotra S, Vyborny CJ, MacMahon H, Jokich PM. Image feature analysis and computer-aided diagnosis in digital radiography. I. Automated detection of microcalcifications in mammography. Med Phys. 1987 Jul-Aug;14(4):538–548. doi: 10.1118/1.596065. [DOI] [PubMed] [Google Scholar]
- 16.Gilhuijs KG, Giger ML, Bick U. Computerized analysis of breast lesions in three dimensions using dynamic magnetic-resonance imaging. Med Phys. 1998 Sep;25(9):1647–1654. doi: 10.1118/1.598345. [DOI] [PubMed] [Google Scholar]
- 17.Drukker K, Giger ML, Metz CE. Robustness of computerized lesion detection and classification scheme across different breast US platforms. Radiology. 2005 Dec;237(3):834–840. doi: 10.1148/radiol.2373041418. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida H, Nappi J. Three-dimensional computer-aided diagnosis scheme for detection of colonic polyps. IEEE Trans Med Imaging. 2001 Dec;20(12):1261–1274. doi: 10.1109/42.974921. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki K, Yoshida H, Nappi J, Dachman AH. Massive-training artificial neural network (MTANN) for reduction of false positives in computer-aided detection of polyps: Suppression of rectal tubes. Medical Physics. 2006;33(10):3814–3824. doi: 10.1118/1.2349839. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki K, Yoshida H, Nappi J, Armato SG, 3rd, Dachman AH. Mixture of expert 3D massive-training ANNs for reduction of multiple types of false positives in CAD for detection of polyps in CT colonography. Med Phys. 2008 Feb;35(2):694–703. doi: 10.1118/1.2829870. [DOI] [PubMed] [Google Scholar]
- 21.Summers RM, Beaulieu CF, Pusanik LM, Malley JD, Jeffrey RB, Jr, Glazer DI, Napel S. Automated polyp detector for CT colonography: feasibility study. Radiology. 2000 Jul;216(1):284–290. doi: 10.1148/radiology.216.1.r00jl43284. [DOI] [PubMed] [Google Scholar]
- 22.Lostumbo A, Wanamaker C, Tsai J, Suzuki K, Dachman AH. Comparison of 2D and 3D views for evaluation of flat lesions in CT colonography. Acad Radiol. 2010 Jan;17(1):39–47. doi: 10.1016/j.acra.2009.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Lostumbo A, Suzuki K, Dachman AH. Flat lesions in CT colonography. Abdom Imaging. 2010 Oct;35(5):578–583. doi: 10.1007/s00261-009-9562-3. [DOI] [PubMed] [Google Scholar]
- 24.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323:533–536. [Google Scholar]
- 25.Vapnik VN. The Nature of Statistical Learning Theory. Springer-Verlag; Berlin: 1995. [Google Scholar]
- 26.Shiraishi J, Li Q, Suzuki K, Engelmann R, Doi K. Computer-aided diagnostic scheme for the detection of lung nodules on chest radiographs: localized search method based on anatomical classification. Med Phys. 2006 Jul;33(7):2642–2653. doi: 10.1118/1.2208739. [DOI] [PubMed] [Google Scholar]
- 27.Armato SG, 3rd, Giger ML, MacMahon H. Automated detection of lung nodules in CT scans: preliminary results. Med Phys. 2001 Aug;28(8):1552–1561. doi: 10.1118/1.1387272. [DOI] [PubMed] [Google Scholar]
- 28.Aoyama M, Li Q, Katsuragawa S, MacMahon H, Doi K. Automated computerized scheme for distinction between benign and malignant solitary pulmonary nodules on chest images. Med Phys. 2002 May;29(5):701–708. doi: 10.1118/1.1469630. [DOI] [PubMed] [Google Scholar]
- 29.Aoyama M, Li Q, Katsuragawa S, Li F, Sone S, Doi K. Computerized scheme for determination of the likelihood measure of malignancy for pulmonary nodules on low-dose CT images. Med Phys. 2003 Mar;30(3):387–394. doi: 10.1118/1.1543575. [DOI] [PubMed] [Google Scholar]
- 30.Jerebko AK, Summers RM, Malley JD, Franaszek M, Johnson CD. Computer-assisted detection of colonic polyps with CT colonography using neural networks and binary classification trees. Medical Physics. 2003 Jan;30(1):52–60. doi: 10.1118/1.1528178. [DOI] [PubMed] [Google Scholar]
- 31.Sluimer I, Schilham A, Prokop M, van Ginneken B. Computer analysis of computed tomography scans of the lung: a survey. IEEE Trans Med Imaging. 2006 Apr;25(4):385–405. doi: 10.1109/TMI.2005.862753. [DOI] [PubMed] [Google Scholar]
- 32.Li Q. Recent progress in computer-aided diagnosis of lung nodules on thin-section CT. Comput Med Imaging Graph. 2007 Jun-Jul;31(4–5):248–257. doi: 10.1016/j.compmedimag.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fraioli F, Serra G, Passariello R. CAD (computed-aided detection) and CADx (computer aided diagnosis) systems in identifying and characterising lung nodules on chest CT: overview of research, developments and new prospects. Radiol Med. 2010 Apr;115(3):385–402. doi: 10.1007/s11547-010-0507-2. [DOI] [PubMed] [Google Scholar]
- 34.Goo JM. A computer-aided diagnosis for evaluating lung nodules on chest CT: the current status and perspective. Korean journal of radiology: official journal of the Korean Radiological Society. 2011 Mar-Apr;12(2):145–155. doi: 10.3348/kjr.2011.12.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki K. Pixel-based Machine-Learning (PML) in Medical Imaging. International Journal of Biomedical Imaging, vol.2012, no. Special issue on machine learning in medical imaging. 2012:792079, 792018. doi: 10.1155/2012/792079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duda RO, Hart PE, Stork DG. Pattern Recognit. Wiley Interscience; Hoboken, NJ: 2001. [Google Scholar]
- 37.Bishop CM. Neural Networks for Pattern Recognition. Oxford University Press; New York: 1995. [Google Scholar]
- 38.Bishop CM. An example - character recognition. In: Bishop CM, editor. Neural Networks for Pattern Recognition. Oxford University Press; New York: 1995. pp. 1–4. [Google Scholar]
- 39.Chan HP, Sahiner B, Wagner RF, Petrick N. Classifier design for computer-aided diagnosis: effects of finite sample size on the mean performance of classical and neural network classifiers. Med Phys. 1999 Dec;26(12):2654–2668. doi: 10.1118/1.598805. [DOI] [PubMed] [Google Scholar]
- 40.Fukunaga K. Introduction to Statistical Pattern Recognition. Academic Press; San Diego: 1990. [Google Scholar]
- 41.Bellman RE. Adaptive control processes: a guided tour. Princeton University Press; Princeton, N.J: 1961. [Google Scholar]
- 42.Aoyama M, Li Q, Katsuragawa S, MacMahon H, Doi K. Automated computerized scheme for distinction between benign and malignant solitary pulmonary nodules on chest images. Med Phys. 2002 May;29(5):701–708. doi: 10.1118/1.1469630. [DOI] [PubMed] [Google Scholar]
- 43.Pudil P, Novovicova J, Kittler J. Floating search methods in feature selection. Pattern Recognition Letters. 1994;15:1119–1125. [Google Scholar]
- 44.Suzuki K. Determining the receptive field of a neural filter. J Neural Eng. 2004 Dec;1(4):228–237. doi: 10.1088/1741-2560/1/4/006. [DOI] [PubMed] [Google Scholar]
- 45.Suzuki K, Horiba I, Sugie N. A simple neural network pruning algorithm with application to filter synthesis. Neural Process Lett. 2001 Feb;13(1):43–53. [Google Scholar]
- 46.Takemura A, Shimizu A, Hamamoto K. Discrimination of Breast Tumors in Ultrasonic Images Using an Ensemble Classifier Based on the AdaBoost Algorithm With Feature Selection. IEEE Trans on Medical Imaging. 2010;29(3):598–609. doi: 10.1109/TMI.2009.2022630. [DOI] [PubMed] [Google Scholar]
- 47.Xu J, Suzuki K. Computer-aided detection of hepatocellular carcinoma in hepatic CT: False positive reduction with feature selection. IEEE International Symposium on Biomedical Imaging (IEEE ISBI); Chicago, IL. 2011. pp. 1097–1100. [Google Scholar]
- 48.Suzuki K, Horiba I, Sugie N, Ikeda S. Improvement of image quality of x-ray fluoroscopy using spatiotemporal neural filter which learns noise reduction, edge enhancement and motion compensation. Proc. Int. Conf. Signal Processing Applications and Technology (ICSPAT); Boston, MA. 1996. pp. 1382–1386. [Google Scholar]
- 49.Suzuki K, Horiba I, Sugie N, Nanki M. Neural filter with selection of input features and its application to image quality improvement of medical image sequences. IEICE Trans Inf Syst. 2002 Oct;E85-D(10):1710–1718. [Google Scholar]
- 50.Suzuki K, Horiba I, Sugie N. Efficient approximation of neural filters for removing quantum noise from images. IEEE Trans Signal Process. 2002 Jul;50(7):1787–1799. [Google Scholar]
- 51.Suzuki K, Horiba I, Sugie N. Neural edge enhancer for supervised edge enhancement from noisy images. IEEE Trans Pattern Anal Mach Intell. 2003 Dec;25(12):1582–1596. [Google Scholar]
- 52.Suzuki K, Horiba I, Sugie N, Nanki M. Extraction of left ventricular contours from left ventriculograms by means of a neural edge detector. IEEE Trans Med Imaging. 2004 Mar;23(3):330–339. doi: 10.1109/TMI.2004.824238. [DOI] [PubMed] [Google Scholar]
- 53.Lo SB, Lou SA, Lin JS, Freedman MT, Chien MV, Mun SK. Artificial convolution neural network techniques and applications for lung nodule detection. IEEE Trans Med Imaging. 1995;14(4):711–718. doi: 10.1109/42.476112. [DOI] [PubMed] [Google Scholar]
- 54.Lo SCB, Chan HP, Lin JS, Li H, Freedman MT, Mun SK. Artificial convolution neural network for medical image pattern recognition. Neural Networks. 1995;8(7–8):1201–1214. [Google Scholar]
- 55.Lin JS, Lo SB, Hasegawa A, Freedman MT, Mun SK. Reduction of false positives in lung nodule detection using a two-level neural classification. IEEE Trans Med Imaging. 1996;15(2):206–217. doi: 10.1109/42.491422. [DOI] [PubMed] [Google Scholar]
- 56.Lo SC, Li H, Wang Y, Kinnard L, Freedman MT. A multiple circular path convolution neural network system for detection of mammographic masses. IEEE Trans Med Imaging. 2002 Feb;21(2):150–158. doi: 10.1109/42.993133. [DOI] [PubMed] [Google Scholar]
- 57.Sahiner B, Chan HP, Petrick N, Wei D, Helvie MA, Adler DD, Goodsitt MM. Classification of mass and normal breast tissue: a convolution neural network classifier with spatial domain and texture images. IEEE Trans Med Imaging. 1996;15(5):598–610. doi: 10.1109/42.538937. [DOI] [PubMed] [Google Scholar]
- 58.Wei D, Nishikawa RM, Doi K. Application of texture analysis and shift-invariant artificial neural network to microcalcification cluster detection. Radiology. 1996 Nov;201:696–696. [Google Scholar]
- 59.Zhang W, Doi K, Giger ML, Wu Y, Nishikawa RM, Schmidt RA. Computerized detection of clustered microcalcifications in digital mammograms using a shift-invariant artificial neural network. Med Phys. 1994 Apr;21(4):517–524. doi: 10.1118/1.597177. [DOI] [PubMed] [Google Scholar]
- 60.Suzuki K, Armato SG, 3rd, Li F, Sone S, Doi K. Massive training artificial neural network (MTANN) for reduction of false positives in computerized detection of lung nodules in low-dose computed tomography. Med Phys. 2003 Jul;30(7):1602–1617. doi: 10.1118/1.1580485. [DOI] [PubMed] [Google Scholar]
- 61.Suzuki K, Abe H, MacMahon H, Doi K. Image-processing technique for suppressing ribs in chest radiographs by means of massive training artificial neural network (MTANN) IEEE Trans Med Imaging. 2006 Apr;25(4):406–416. doi: 10.1109/TMI.2006.871549. [DOI] [PubMed] [Google Scholar]
- 62.Oda S, Awai K, Suzuki K, Yanaga Y, Funama Y, MacMahon H, Yamashita Y. Performance of radiologists in detection of small pulmonary nodules on chest radiographs: effect of rib suppression with a massive-training artificial neural network. AJR Am J Roentgenol. 2009 Nov;193(5):W397–402. doi: 10.2214/AJR.09.2431. [DOI] [PubMed] [Google Scholar]
- 63.Suzuki K. A supervised ‘lesion-enhancement’ filter by use of a massive-training artificial neural network (MTANN) in computer-aided diagnosis (CAD) Phys Med Biol. 2009 Sep 21;54(18):S31–45. doi: 10.1088/0031-9155/54/18/S03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Suzuki K, Shiraishi J, Abe H, MacMahon H, Doi K. False-positive reduction in computer-aided diagnostic scheme for detecting nodules in chest radiographs by means of massive training artificial neural network. Acad Radiol. 2005 Feb;12(2):191–201. doi: 10.1016/j.acra.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki K, Li F, Sone S, Doi K. Computer-aided diagnostic scheme for distinction between benign and malignant nodules in thoracic low-dose CT by use of massive training artificial neural network. IEEE Trans Med Imaging. 2005 Sep;24(9):1138–1150. doi: 10.1109/TMI.2005.852048. [DOI] [PubMed] [Google Scholar]
- 66.Suzuki K, Rockey DC, Dachman AH. CT colonography: Advanced computer-aided detection scheme utilizing MTANNs for detection of “missed” polyps in a multicenter clinical trial. Med Phys. 2010;30:2–21. doi: 10.1118/1.3263615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suzuki K, Zhang J, Xu J. Massive-training artificial neural network coupled with Laplacian-eigenfunction-based dimensionality reduction for computer-aided detection of polyps in CT colonography. IEEE Trans Med Imaging. 2010 Nov;29(11):1907–1917. doi: 10.1109/TMI.2010.2053213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J, Suzuki K. Massive-training support vector regression and Gaussian process for false-positive reduction in computer-aided detection of polyps in CT colonography. Med Phys. 2011;38:1888–1902. doi: 10.1118/1.3562898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, Thun MJ. Cancer statistics, 2008. CA Cancer J Clin. 2008 Mar-Apr;58(2):71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 70.Kaneko M, Eguchi K, Ohmatsu H, Kakinuma R, Naruke T, Suemasu K, Moriyama N. Peripheral lung cancer: screening and detection with low-dose spiral CT versus radiography. Radiology. 1996 Dec;201(3):798–802. doi: 10.1148/radiology.201.3.8939234. [DOI] [PubMed] [Google Scholar]
- 71.Sone S, Takashima S, Li F, Yang Z, Honda T, Maruyama Y, Hasegawa M, Yamanda T, Kubo K, Hanamura K, Asakura K. Mass screening for lung cancer with mobile spiral computed tomography scanner. Lancet. 1998 Apr 25;351(9111):1242–1245. doi: 10.1016/S0140-6736(97)08229-9. [DOI] [PubMed] [Google Scholar]
- 72.Henschke CI, McCauley DI, Yankelevitz DF, Naidich DP, McGuinness G, Miettinen OS, Libby DM, Pasmantier MW, Koizumi J, Altorki NK, Smith JP. Early Lung Cancer Action Project: overall design and findings from baseline screening. Lancet. 1999 Jul 10;354(9173):99–105. doi: 10.1016/S0140-6736(99)06093-6. [DOI] [PubMed] [Google Scholar]
- 73.Henschke CI, Yankelevitz DF, Naidich DP, McCauley DI, McGuinness G, Libby DM, Smith JP, Pasmantier MW, Miettinen OS. CT screening for lung cancer: suspiciousness of nodules according to size on baseline scans. Radiology. 2004 Apr;231(1):164–168. doi: 10.1148/radiol.2311030634. [DOI] [PubMed] [Google Scholar]
- 74.Heelan RT, Flehinger BJ, Melamed MR, Zaman MB, Perchick WB, Caravelli JF, Martini N. Non-small-cell lung cancer: results of the New York screening program. Radiology. 1984 May;151(2):289–293. doi: 10.1148/radiology.151.2.6324279. [DOI] [PubMed] [Google Scholar]
- 75.Giger ML, Bae KT, MacMahon H. Computerized detection of pulmonary nodules in computed tomography images. Invest Radiol. 1994 Apr;29(4):459–465. doi: 10.1097/00004424-199404000-00013. [DOI] [PubMed] [Google Scholar]
- 76.Kanazawa K, Kawata Y, Niki N, Satoh H, Ohmatsu H, Kakinuma R, Kaneko M, Moriyama N, Eguchi K. Computer-aided diagnosis for pulmonary nodules based on helical CT images. Comput Med Imaging Graph. 1998 Mar-Apr;22(2):157–167. doi: 10.1016/s0895-6111(98)00017-2. [DOI] [PubMed] [Google Scholar]
- 77.Gurcan MN, Sahiner B, Petrick N, Chan HP, Kazerooni EA, Cascade PN, Hadjiiski L. Lung nodule detection on thoracic computed tomography images: preliminary evaluation of a computer-aided diagnosis system. Med Phys. 2002 Nov;29(11):2552–2558. doi: 10.1118/1.1515762. [DOI] [PubMed] [Google Scholar]
- 78.Lee Y, Hara T, Fujita H, Itoh S, Ishigaki T. Automated detection of pulmonary nodules in helical CT images based on an improved template-matching technique. IEEE Trans Med Imaging. 2001 Jul;20(7):595–604. doi: 10.1109/42.932744. [DOI] [PubMed] [Google Scholar]
- 79.Suzuki K, Doi K. How can a massive training artificial neural network (MTANN) be trained with a small number of cases in the distinction between nodules and vessels in thoracic CT? Acad Radiol. 2005 Oct;12(10):1333–1341. doi: 10.1016/j.acra.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 80.Chan HP, Sahiner B, Wagner RF, Petrick N. Classifier design for computer-aided diagnosis: effects of finite sample size on the mean performance of classical and neural network classifiers. Med Phys. 1999 Dec;26(12):2654–2668. doi: 10.1118/1.598805. [DOI] [PubMed] [Google Scholar]
- 81.Sahiner B, Chan HP, Hadjiiski L. Classifier performance prediction for computer-aided diagnosis using a limited dataset. Med Phys. 2008 Apr;35(4):1559–1570. doi: 10.1118/1.2868757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Farag AA, El-Baz A, Gimelfarb G, El-Ghar MA, Eldiasty T. Quantitative nodule detection in low dose chest CT scans: new template modeling and evaluation for CAD system design. Medical image computing and computer-assisted intervention: MICCAI International Conference on Medical Image Computing and Computer-Assisted Intervention. 2005;8(Pt 1):720–728. doi: 10.1007/11566465_89. [DOI] [PubMed] [Google Scholar]
- 83.Ge Z, Sahiner B, Chan HP, Hadjiiski LM, Cascade PN, Bogot N, Kazerooni EA, Wei J, Zhou C. Computer-aided detection of lung nodules: false positive reduction using a 3D gradient field method and 3D ellipsoid fitting. Med Phys. 2005 Aug;32(8):2443–2454. doi: 10.1118/1.1944667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matsumoto S, Kundel HL, Gee JC, Gefter WB, Hatabu H. Pulmonary nodule detection in CT images with quantized convergence index filter. Med Image Anal. 2006 Jun;10(3):343–352. doi: 10.1016/j.media.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 85.Yuan R, Vos PM, Cooperberg PL. Computer-aided detection in screening CT for pulmonary nodules. AJR Am J Roentgenol. 2006 May;186(5):1280–1287. doi: 10.2214/AJR.04.1969. [DOI] [PubMed] [Google Scholar]
- 86.Bi J, Periaswamy S, Okada K, Kubota T, Fung G, Salganicoff M, Rao RB. Computer aided detection via asymmetric cascade of sparse hyperplane classifiers. Proc Proc of ACM SIGKDD. 2006:837–844. [Google Scholar]
- 87.Pu J, Zheng B, Leader JK, Wang XH, Gur D. An automated CT based lung nodule detection scheme using geometric analysis of signed distance field. Med Phys. 2008 Aug;35(8):3453–3461. doi: 10.1118/1.2948349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Retico A, Delogu P, Fantacci ME, Gori I, Preite Martinez A. Lung nodule detection in low-dose and thin-slice computed tomography. Computers in biology and medicine. 2008 Apr;38(4):525–534. doi: 10.1016/j.compbiomed.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 89.Ye X, Lin X, Dehmeshki J, Slabaugh G, Beddoe G. Shape-based computer-aided detection of lung nodules in thoracic CT images. IEEE transactions on bio-medical engineering. 2009 Jul;56(7):1810–1820. doi: 10.1109/TBME.2009.2017027. [DOI] [PubMed] [Google Scholar]
- 90.Golosio B, Masala GL, Piccioli A, Oliva P, Carpinelli M, Cataldo R, Cerello P, De Carlo F, Falaschi F, Fantacci ME, Gargano G, Kasae P, Torsello M. A novel multithreshold method for nodule detection in lung CT. Med Phys. 2009 Aug;36(8):3607–3618. doi: 10.1118/1.3160107. [DOI] [PubMed] [Google Scholar]
- 91.Armato SG, 3rd, McLennan G, McNitt-Gray MF, Meyer CR, Yankelevitz D, Aberle DR, Henschke CI, Hoffman EA, Kazerooni EA, MacMahon H, Reeves AP, Croft BY, Clarke LP. Lung image database consortium: developing a resource for the medical imaging research community. Radiology. 2004 Sep;232(3):739–748. doi: 10.1148/radiol.2323032035. [DOI] [PubMed] [Google Scholar]
- 92.Murphy K, van Ginneken B, Schilham AM, de Hoop BJ, Gietema HA, Prokop M. A large-scale evaluation of automatic pulmonary nodule detection in chest CT using local image features and k-nearest-neighbour classification. Med Image Anal. 2009 Oct;13(5):757–770. doi: 10.1016/j.media.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 93.Tan M, Deklerck R, Jansen B, Bister M, Cornelis J. A novel computer-aided lung nodule detection system for CT images. Med Phys. 2011 Oct;38(10):5630–5645. doi: 10.1118/1.3633941. [DOI] [PubMed] [Google Scholar]
- 94.Messay T, Hardie RC, Rogers SK. A new computationally efficient CAD system for pulmonary nodule detection in CT imagery. Med Image Anal. 2010 Jun;14(3):390–406. doi: 10.1016/j.media.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 95.Riccardi A, Petkov TS, Ferri G, Masotti M, Campanini R. Computer-aided detection of lung nodules via 3D fast radial transform, scale space representation, and Zernike MIP classification. Med Phys. 2011 Apr;38(4):1962–1971. doi: 10.1118/1.3560427. [DOI] [PubMed] [Google Scholar]
- 96.Rao RB, Bi J, Fung G, Salganicoff M, Obuchowski N, Naidich D. LungCAD: a clinically approved, machine learning system for lung cancer detection. Proc Proc ACM SIGKDD. 2007:1033–1037. [Google Scholar]
- 97.Sahiner B, Chan HP, Petrick N, Wagner RF, Hadjiiski L. Feature selection and classifier performance in computer-aided diagnosis: the effect of finite sample size. Med Phys. 2000 Jul;27(7):1509–1522. doi: 10.1118/1.599017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li F, Sone S, Abe H, MacMahon H, Armato SG, 3rd, Doi K. Lung cancers missed at low-dose helical CT screening in a general population: comparison of clinical, histopathologic, and imaging findings. Radiology. 2002 Dec;225(3):673–683. doi: 10.1148/radiol.2253011375. [DOI] [PubMed] [Google Scholar]
- 99.Swensen SJ, Jett JR, Hartman TE, Midthun DE, Sloan JA, Sykes AM, Aughenbaugh GL, Clemens MA. Lung cancer screening with CT: Mayo Clinic experience. Radiology. 2003 Mar;226(3):756–761. doi: 10.1148/radiol.2263020036. [DOI] [PubMed] [Google Scholar]
- 100.Metz CE. ROC methodology in radiologic imaging. Invest Radiol. 1986 Sep;21(9):720–733. doi: 10.1097/00004424-198609000-00009. [DOI] [PubMed] [Google Scholar]
- 101.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983 Sep;148(3):839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 102.Kawata Y, Niki N, Ohmatsu H, Kakinuma R, Eguchi K, Kaneko M, Moriyama N. Quantitative surface characterization of pulmonary nodules based on thin-section CT images. IEEE Transactions on Nuclear Science. 1998;45(4):2132–2138. [Google Scholar]
- 103.McNitt-Gray MF, Hart EM, Wyckoff N, Sayre JW, Goldin JG, Aberle DR. A pattern classification approach to characterizing solitary pulmonary nodules imaged on high resolution CT: preliminary results. Med Phys. 1999 Jun;26(6):880–888. doi: 10.1118/1.598603. [DOI] [PubMed] [Google Scholar]
- 104.Matsuki Y, Nakamura K, Watanabe H, Aoki T, Nakata H, Katsuragawa S, Doi K. Usefulness of an artificial neural network for differentiating benign from malignant pulmonary nodules on high-resolution CT: evaluation with receiver operating characteristic analysis. AJR Am J Roentgenol. 2002 Mar;178(3):657–663. doi: 10.2214/ajr.178.3.1780657. [DOI] [PubMed] [Google Scholar]
- 105.Mori K, Niki N, Kondo T, Kamiyama Y, Kodama T, Kawada Y, Moriyama N. Development of a novel computer-aided diagnosis system for automatic discrimination of malignant from benign solitary pulmonary nodules on thin-section dynamic computed tomography. J Comput Assist Tomogr. 2005 Mar-Apr;29(2):215–222. doi: 10.1097/01.rct.0000155668.28514.01. [DOI] [PubMed] [Google Scholar]
- 106.Shah SK, McNitt-Gray MF, Rogers SR, Goldin JG, Suh RD, Sayre JW, Petkovska I, Kim HJ, Aberle DR. Computer aided characterization of the solitary pulmonary nodule using volumetric and contrast enhancement features. Academic radiology. 2005 Oct;12(10):1310–1319. doi: 10.1016/j.acra.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 107.Iwano S, Nakamura T, Kamioka Y, Ikeda M, Ishigaki T. Computer-aided differentiation of malignant from benign solitary pulmonary nodules imaged by high-resolution CT. Comput Med Imaging Graph. 2008 Jul;32(5):416–422. doi: 10.1016/j.compmedimag.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 108.Way TW, Sahiner B, Chan HP, Hadjiiski L, Cascade PN, Chughtai A, Bogot N, Kazerooni E. Computer-aided diagnosis of pulmonary nodules on CT scans: improvement of classification performance with nodule surface features. Medical physics. 2009 Jul;36(7):3086–3098. doi: 10.1118/1.3140589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chen H, Xu Y, Ma Y, Ma B. Neural network ensemble-based computer-aided diagnosis for differentiation of lung nodules on CT images: clinical evaluation. Acad Radiol. 2010 May;17(5):595–602. doi: 10.1016/j.acra.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 110.Lee MC, Boroczky L, Sungur-Stasik K, Cann AD, Borczuk AC, Kawut SM, Powell CA. Computer-aided diagnosis of pulmonary nodules using a two-step approach for feature selection and classifier ensemble construction. Artificial intelligence in medicine. 2010 Sep;50(1):43–53. doi: 10.1016/j.artmed.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 111.Kawata Y, Niki N, Ohmatsu H, Moriyama N. Example-based assisting approach for pulmonary nodule classification in three-dimensional thoracic computed tomography images. Acad Radiol. 2003 Dec;10(12):1402–1415. doi: 10.1016/s1076-6332(03)00507-5. [DOI] [PubMed] [Google Scholar]
- 112.Kawata Y, Niki N, Ohmatsu H, Kusumoto M, Tsuchida T, Eguchi K, Kaneko M, Moriyama N. Quantitative classification based on CT histogram analysis of non-small cell lung cancer: correlation with histopathological characteristics and recurrence-free survival. Med Phys. 2012 Feb;39(2):988–1000. doi: 10.1118/1.3679017. [DOI] [PubMed] [Google Scholar]
- 113.Abe H, MacMahon H, Engelmann R, Li Q, Shiraishi J, Katsuragawa S, Aoyama M, Ishida T, Ashizawa K, Metz CE, Doi K. Computer-aided diagnosis in chest radiography: results of large-scale observer tests at the 1996–2001 RSNA scientific assemblies. Radiographics. 2003 Jan-Feb;23(1):255–265. doi: 10.1148/rg.231025129. [DOI] [PubMed] [Google Scholar]
- 114.Jemal A, Murray T, Ward E, Samuels A, Tiwari RC, Ghafoor A, Feuer EJ, Thun MJ. Cancer statistics, 2005. CA: A Cancer Journal for Clinicians. 2005 Jan-Feb;55(1):10–30. doi: 10.3322/canjclin.55.1.10. [DOI] [PubMed] [Google Scholar]
- 115.Winawer SJ, Fletcher RH, Miller L, Godlee F, Stolar MH, Mulrow CD, Woolf SH, Glick SN, Ganiats TG, Bond JH, Rosen L, Zapka JG, Olsen SJ, Giardiello FM, Sisk JE, Van Antwerp R, Brown-Davis C, Marciniak DA, Mayer RJ. Colorectal cancer screening: clinical guidelines and rationale. Gastroenterology. 1997 Feb;112(2):594–642. doi: 10.1053/gast.1997.v112.agast970594. [DOI] [PubMed] [Google Scholar]
- 116.Suzuki K, Dachman AH. Computer-aided diagnosis in CT colonography. In: Dachman AH, Laghi A, editors. Atlas of Virtual Colonoscopy. Springer; New York: 2011. pp. 163–182. [Google Scholar]
- 117.Yoshida H, Näppi J. Three-dimensional computer-aided diagnosis scheme for detection of colonic polyps. IEEE Trans Med Imaging. 2001 Dec;20(12):1261–1274. doi: 10.1109/42.974921. [DOI] [PubMed] [Google Scholar]
- 118.Summers RM, Johnson CD, Pusanik LM, Malley JD, Youssef AM, Reed JE. Automated polyp detection at CT colonography: feasibility assessment in a human population. Radiology. 2001 Apr;219(1):51–59. doi: 10.1148/radiology.219.1.r01ap0751. [DOI] [PubMed] [Google Scholar]
- 119.Paik DS, Beaulieu CF, Rubin GD, Acar B, Jeffrey RB, Jr, Yee J, Dey J, Napel S. Surface normal overlap: a computer-aided detection algorithm with application to colonic polyps and lung nodules in helical CT. IEEE Trans Med Imaging. 2004 Jun;23(6):661–675. doi: 10.1109/tmi.2004.826362. [DOI] [PubMed] [Google Scholar]
- 120.Kiss G, Van Cleynenbreugel J, Thomeer M, Suetens P, Marchal G. Computer-aided diagnosis in virtual colonography via combination of surface normal and sphere fitting methods. Eur Radiol. 2002 Jan;12(1):77–81. doi: 10.1007/s003300101040. [DOI] [PubMed] [Google Scholar]
- 121.Gokturk SB, Tomasi C, Acar B, Beaulieu CF, Paik DS, Jeffrey RB, Jr, Yee J, Napel S. A statistical 3-D pattern processing method for computer-aided detection of polyps in CT colonography. IEEE Transactions on Medical Imaging. 2001 Dec;20(12):1251–1260. doi: 10.1109/42.974920. [DOI] [PubMed] [Google Scholar]
- 122.Nappi J, Yoshida H. Automated detection of polyps with CT colonography: evaluation of volumetric features for reduction of false-positive findings. Acad Radiol. 2002 Apr;9(4):386–397. doi: 10.1016/s1076-6332(03)80184-8. [DOI] [PubMed] [Google Scholar]
- 123.Acar B, Beaulieu CF, Gokturk SB, Tomasi C, Paik DS, Jeffrey RB, Jr, Yee J, Napel S. Edge displacement field-based classification for improved detection of polyps in CT colonography. IEEE Transactions on Medical Imaging. 2002 Dec;21(12):1461–1467. doi: 10.1109/TMI.2002.806405. [DOI] [PubMed] [Google Scholar]
- 124.Jerebko AK, Malley JD, Franaszek M, Summers RM. Multiple neural network classification scheme for detection of colonic polyps in CT colonography data sets. Academic Radiology. 2003 Feb;10(2):154–160. doi: 10.1016/s1076-6332(03)80039-9. [DOI] [PubMed] [Google Scholar]
- 125.Jerebko AK, Malley JD, Franaszek M, Summers RM. Support vector machines committee classification method for computer-aided polyp detection in CT colonography. Academic Radiology. 2005 Apr;12(4):479–486. doi: 10.1016/j.acra.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 126.Wang Z, Liang Z, Li L, Li X, Li B, Anderson J, Harrington D. Reduction of false positives by internal features for polyp detection in CT-based virtual colonoscopy. Med Phys. 2005 Dec;32(12):3602–3616. doi: 10.1118/1.2122447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Suzuki K, Yoshida H, Nappi J, Dachman AH. Massive-training artificial neural network (MTANN) for reduction of false positives in computer-aided detection of polyps: Suppression of rectal tubes. Med Phys. 2006 Oct;33(10):3814–3824. doi: 10.1118/1.2349839. [DOI] [PubMed] [Google Scholar]
- 128.Li J, Van Uitert R, Yao J, Petrick N, Franaszek M, Huang A, Summers RM. Wavelet method for CT colonography computer-aided polyp detection. Med Phys. 2008 Aug;35(8):3527–3538. doi: 10.1118/1.2938517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang S, Yao J, Summers RM. Improved classifier for computer-aided polyp detection in CT colonography by nonlinear dimensionality reduction. Med Phys. 2008 Apr;35(4):1377–1386. doi: 10.1118/1.2870218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yao J, Li J, Summers RM. Employing topographical height map in colonic polyp measurement and false positive reduction. Pattern Recognition. 2009;42(6):1029–1040. doi: 10.1016/j.patcog.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Doshi T, Rusinak D, Halvorsen RA, Rockey DC, Suzuki K, Dachman AH. CT Colonography: False-Negative Interpretations. Radiology. 2007 Jul;244(1):165–173. doi: 10.1148/radiol.2441061122. [DOI] [PubMed] [Google Scholar]
- 132.Rockey DC, Paulson E, Niedzwiecki D, Davis W, Bosworth HB, Sanders L, Yee J, Henderson J, Hatten P, Burdick S, Sanyal A, Rubin DT, Sterling M, Akerkar G, Bhutani MS, Binmoeller K, Garvie J, Bini EJ, McQuaid K, Foster WL, Thompson WM, Dachman A, Halvorsen R. Analysis of air contrast barium enema, computed tomographic colonography, and colonoscopy: prospective comparison. Lancet. 2005 Jan 22–28;365(9456):305–311. doi: 10.1016/S0140-6736(05)17784-8. [DOI] [PubMed] [Google Scholar]
- 133.Zhu H, Liang Z, Pickhardt PJ, Barish MA, You J, Fan Y, Lu H, Posniak EJ, Richards RJ, Cohen HL. Increasing computer-aided detection specificity by projection features for CT colonography. Med Phys. 2010 Apr;37(4):1468–1481. doi: 10.1118/1.3302833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wang S, Yao J, Petrick N, Summers RM. Combining Statistical and Geometric Features for Colonic Polyp Detection in CTC Based on Multiple Kernel Learning. International journal of computational intelligence and applications. 2010 Jan 1;9(1):1–15. doi: 10.1142/S1469026810002744. [DOI] [PMC free article] [PubMed] [Google Scholar]