Abstract
The rare but recurrent RUNX1-USP42 fusion gene is the result of a t(7;21)(p22;q22) chromosomal translocation and has been described in 6 cases of acute myeloid leukemia (AML) and one case of refractory anemia with excess of blast. In the present study, we present the molecular genetic analysis and the clinical features of a t(7;21)(p22;q22)-positive AML case. PCR amplified two RUNX1-USP42 cDNA fragments but no reciprocal USP42-RUNX1 fragment indicating that the RUNX1-USP42 is the leukemogenic fusion gene. Sequencing of the two amplified fragments showed that exon 6 or exon 7 of RUNX1 (accession number NM_001754 version 3) was fused to exon 3 of USP42 (accession number NM_032172 version 2). The predicted RUNX1-USP42 fusion protein would contain the Runt homology domain (RHD), which is responsible for heterodimerization with CBFB and for DNA binding, and the catalytic UCH (ubiquitin carboxyl terminal hydroxylase) domain of the USP42 protein. The bone marrow cells in the present case also had a 5q deletion, and it was revealed that 5 out of the 8 reported cases (including the present case) with t(7;21)(p22;q22)/RUNX1-USP42 also had cytogenetic abnormalities of 5q. The fact that t(7;21) and 5q- occur together much more often than chance would allow seems to be unquestionable, although the pathogenetic connection between the two aberrations remains unknown.
Keywords: acute myeloid leukemia, cytogenetic, t(7;21)(p22;q22), 5q aberration, RUNX1, USP42, fusion gene
Introduction
It is now generally accepted that neoplastic disorders arise through the acquisition of genomic changes by suitably primed target cells (1). These somatic mutations are often cytogenetically visible in the form of balanced or unbalanced chromosome aberrations (1). Many hematologic malignancies are characterized by balanced chromosomal abnormalities resulting in chimeric genes of pathogenetic, diagnostic and prognostic importance (1). In fact, more than 200 different genes are now known to be rearranged through translocations in leukemias and leukemia-like disorders (1), with some genes being particularly promiscuous, having numerous partners in different fusions and disorders (1).
To date, the RUNX1 gene (previously AML1, CBFA2 in 21q22) has been shown to fuse in-frame with 23 different partner genes, encoding a structurally heterogeneous group of proteins, in acute myeloid and lymphoblastic leukemia (AML and ALL), chronic myeloid leukemia (CML; the fusion here occurs secondarily), and myelodysplastic syndromes (MDS) (2,3). Some of the fusions are common, such as the ETV6/RUNX1 [t(12;21)(p13;q22)] in ALL, RUNX1/RUNX1T1 (also known as AML1/ETO) [t(8;21)(q22;q22)] in AML and RUNX1/MECOM [t(3;21)(q26;q22)] in MDS, AML and CML in the blastic phase, whereas many of them have only been reported in single cases, i.e., they have not yet been shown to be recurrent (2,3). Whereas the prognostic impact of frequent RUNX1 fusions is well known, corresponding knowledge regarding infrequent chimeras is lacking (4,5). Considering that most treatment protocols are in part based on the presence of certain genetic changes in acute leukemias, it is of potential clinical value to obtain further information also about rare RUNX1 fusions, even in disease subgroups that cannot be treated with medications specifically directed against the leukemogenic defect. It is important to underscore that this may be the case also for rare pathogenetic mechanisms where information is gathered by the addition of single case reports, as recently exemplified by the story of the rare MLL/ARHGAP26 (GRAF) fusion in pediatric AML (6–8). For this reason, we here present the molecular genetic and clinical features of a case of AML with t(7;21)(p22;q22), a rare but recurrent chromosomal translocation that was first described in 2006 by Paulsson et al(9).
Materials and methods
Case history
The study was approved by the Regional Committee for Medical Research Ethics (REK Sør, http://helseforskning.etikkom.no), and written informed consent was obtained from the patient.
A 52-year-old woman was admitted to the hospital following a month of tiredness, sleepiness and symptoms of lower airway infection. She had been treated with antibiotics without any clinical improvement. Upon admission to the hospital, she had fever, severe anemia (hemoglobin 5.8 g/l), thrombocytopenia (116×109/l) and leukocytosis (34×109/l). A bone marrow aspirate showed >70% myeloblasts. The immunophenotypical features of the malignant cells confirmed the diagnosis of acute myeloblastic leukemia without differentiation. The myeloblasts expressed CD34, CD117, HLA-DR antigens, CD13, and partly CD11b, in addition to CD7 and CD56, but were negative for myeloperoxidase as well as B- and T-cell lineage markers. The clinical, blood sample and bone marrow findings (Fig. 1) were conclusive for acute myeloblastic leukemia without maturation (formerly FAB M0).
Figure 1.
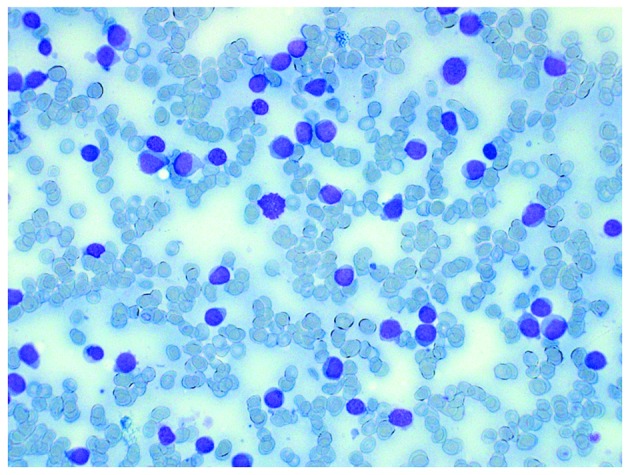
Bone marrow smear of the patient taken at diagnosis. A monomorphous image of blasts which are small and with a scarse cytoplasm is evident. Giemsa staining at magnification ×400.
The patient was transferred to the regional hospital and standard induction chemotherapy was administered. Following complete remission five months later, she received an allogenic bone marrow transplant with reduced conditioning (preferred because of complications during initial therapy) from a sibling donor. The patient is, at the time of the preparation of this manuscript, still in remission with a good chimerism status nine months after the primary diagnosis, although she is now being treated for complications due to graft vs. host disease and cytomegalovirus reactivation.
G-banding and fluorescence in situ hybridization (FISH)
Bone marrow cells were cytogenetically investigated by standard methods. Chromosome preparations were made from metaphase cells of a 24-h culture, G-banded using Leishman’s stain and karyotyped according to ISCN 2009 guidelines (10). As part of our standard cytogenetic diagnosis of AML patients, interphase FISH analyses of bone marrow cells were performed with the Cytocell Multiprobe AML/MDS panel (Cytocell, http://www.cytocell.co.uk/) searching for −5/del(5q), PML/RARα, del(17p) (TP53), AML1/ETO, trisomy 8, −7/del(7q), CBFβ/MYH11 and del(20q). The del(5q) probe contains the probe for the EGR1 gene in 5q31.1 labeled in red as well as a control probe at 5p15.31 flanking the marker D5S30 labeled in green. Fluorescent signals were captured and analyzed using the CytoVision system (Applied Imaging, Newcastle, UK).
PCR analyses
Total RNA (1 μg) was reverse-transcribed in a 20-μl reaction volume using iScript Advanced cDNA Synthesis kit for RT-qPCR according to the manufacturer’s instructions (Bio-Rad). cDNA corresponding to 50 ng total RNA was used as the template in subsequent PCR assays. The 25-μl PCR volume contained 12.5 μl Premix Ex Taq™ DNA Polymerase Hot Start version (Takara), 1 μl of diluted cDNA, and 0.2 μM of each of the forward and reverse primers. For the detection of the RUNX1-USP42 fusion transcript, the forward RUNX1–765F (GGATGTTCCAGATGGCACTCTGG) and the reverse USP42–562R (ACGTCCCCAGGATTACTGAGTGCC) primers were used. For the amplification of a possible USP42-RUNX1 fusion transcript, the primers USP42–116F (CAGAAT CAGCCTGGCAGCTCCGA) and RUNX1–1489R (GCCGA CATGCCGATGCCGAT) were used. The PCR was run on a C-1000 Thermal cycler (Bio-Rad) with an initial denaturation at 94°C for 30°sec, followed by 35 cycles of 7°sec at 98°C, 2°min at 68°C, and a final extension for 5°min at 68°C. PCR products (4 μl) were stained with GelRed (Biotium), analyzed by electrophoresis through 1.0% agarose gel and photographed. The remaining PCR products were excised from the gel, purified using the Qiagen Gel extraction kit (Qiagen), and cloned to the pCR4-TOPO vector using TOPO TA cloning kits for sequencing (Invitrogen). Colonies were sequenced at GATC Biotech (Germany, http://www.gatc-biotech.com/en/home.html). The BLAST software (http://www.ncbi.nlm.nih.gov/BLAST/) was used for computer analysis of sequence data.
Results
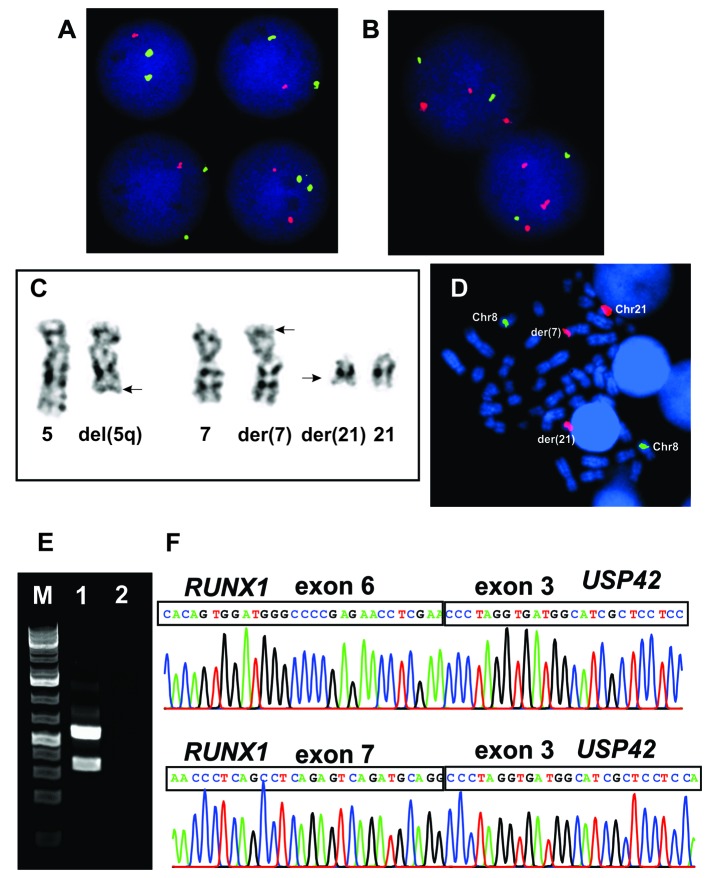
The G-banding analysis showed del(5)(q31) in all 15 cells analyzed which was confirmed by interphase FISH (Fig. 2A and C). The AML1/ETO probe (RUNX1/RUNX1T1) showed abnormal signals, i.e., splitting of the RUNX1 probe was observed in the majority of interphase nuclei examined in spite of no cytogenetically visible rearrangement of this chromosome arm (Fig. 2B). In the same experiment two metaphase cells were found which demonstrated that part of the RUNX1 probe was unexpectedly located on 7p22 (Fig. 2C and D). Other FISH analyses detected no PML/RARα, del(17p), trisomy 8, −7/del(7q), CBFβ/MYH11 or del(20q). Therefore, the whole karyotype was: 46,XX,del(5)(q31)[15].nuc ish (EGR1×1)[196/206],(ETOx2,AML1×3)[186/222].ish t(7;21)(p22;q22) (AML1+;AML1+)[2] (Fig. 1).
Figure 2.
Cytogenetic, FISH and PCR analyses. (A) Interphase FISH with del(5q) probe. The EGR1 probe (in 5q31) is labeled in red and the control probe mapped in 5p15.31 is labeled in green. Three nuclei had one red signal suggesting a hemizygous deletion of the EGR1 gene. All four nuclei had two green signals of the control probe. (B) Interphase FISH with the AML1/ETO probe. The AML1 probe (RUNX1) is labeled in red and the ETO probe (RUNX1T1) is labeled in green. Both nuclei had two green signals which suggest that the RUNX1T1 gene was not rearranged. Both nuclei had three red signals which suggest than one RUNX1 locus was rearranged. (C) Partial karyotype showing chromosome aberrations del(5q), der(7)t(7;21)(p22;q22), and der(21)t(7;21)(p22;q22) together with the corresponding normal homologues; breakpoint positions are indicated by arrows. (D) FISH on metaphase spread using the AML1/ETO probe. Green signals (ETO probe) are observed only on chromosomes 8 (normal RUNX1T1). Part of the AML1 probe (RUNX1) is located on 7p22. (E) cDNA fragment amplification of RUNX1-USP42 (lane 1) using the primers RUNX1–765F and USP42–562R. PCR with primers USP42–116F and RUNX1–1489R did not amplify any cDNA fragment (lane 2). M, 1 kb DNA ladder. (F) Partial sequence chromatograms of the two amplified RUNX1-USP42 fragments showing that exon 6 of RUNX1 is fused to exon 3 of USP42 and that exon 7 of RUNX1 is fused to exon 3 of USP42.
PCR amplification using the RUNX1–765F and USP42–562R primers generated two RUNX1-USP42 fragments of 500- and 300-bp in size whereas PCR with primers USP42–116F and RUNX1–1489R did not amplified any cDNA fragment (Fig. 2E). Sequencing of the two amplified fragments showed that, in the 300-bp fragment, exon 6 of RUNX1 (accession number NM_001754 version 3) was fused to exon 3 of USP42 (accession number NM_032172 version 2), whereas in the 500-bp long fragment exon 7 of RUNX1 was fused to exon 3 of USP42 (Fig. 2F).
Discussion
The cryptic t(7;21)(p22;q22) chromosomal translocation was first described in a 7-year-old boy with AML-M0 together with aberrant expression of T-lymphocyte-associated markers (9). The translocation was an unexpected finding after FISH had been performed using whole chromosome painting probes for chromosome 7 while screening pediatric leukemias for the t(7;12)(q36;p13) translocation (9). In the present study, we also detected the t(7;21) unexpectedly as a result of our standard cytogenetic diagnosis of AML patients using interphase FISH analyses of bone marrow cells and searching for −5/del(5q), PML/RARα, del(17p), AML1/ETO, trisomy 8, −7/del(7q), CBFβ/MYH11 and del(20q). The finding of a split RUNX1 probe in 186 of 222 interphase nuclei examined triggered further investigations which led to the detection of the t(7;21). Notably, AML with t(7;21) seems to be associated with AML-M0 [our patient was also undifferentiated AML-M0] or myelomonocytic differentiation. One patient was found to present with MDS RAEB-2 (11,12). Since the AML M0 subset makes up <5% of all AMLs (14), it seems likely that the 7;21-translocation is unusual in leading to this particular undifferentiated myeloid leukemia in a dysproportionate number of cases (admittedly, they may also display monocytic differentiation according to a few reports). These leukemias are immunophenotypically characterized by the aberrant expression of CD7 as well as CD56 (present case and 5 previously reported cases) (9,12,13).
Subsequent molecular genetic investigations of the bone marrow cells showed that the result of the t(7;21)(p22;q22) is the fusion of USP42 (on 7p22) and RUNX1 (on 21q22) to generate a RUNX1-USP42 chimera (9). Including the present case, a RUNX1-USP42 chimera has now been found in 8 cases while the reciprocal USP42-RUNX1 was noted in 5 (9,11–13). These findings suggest that RUNX1-USP42 is the leukemogenic fusion. The incidence has hitherto been higher in males (n=6) than in females (n=2) and all but one patient (the first described case) were adults (age >30 years) (9,11–13).
Although recurrent, the RUNX1-USP42 fusion seems to be rare. Paulsson et al(9) screened 35 additional AML cases, Foster et al(11) screened 100 AML/MDS with normal karyotypes, and Giguére and Hebert (13) examined 95 leukemias without finding additional cases of RUNX1-USP42. Jeandidier et al(12) studied 397 AML cases and found only 3 cases with RUNX1-USP42 fusion. An interesting observation was that all 3 had additional 5q abnormalities resulting in loss of material from that chromosome arm. In total, 6 out of 8 cases (the present one included) with the t(7;21)(p22;q22)/RUNX1-USP42 fusion had cytogenetically visible changes of 5q resulting in loss of material (9,11–13). In the series presented by Jeandidier et al(12), 3 out of 35 leukemias with 5q- had the t(7;21)(p22;q22)/RUNX1-USP42 fusion gene (8.5%). In only one case was there direct evidence as to whether 5q- or t(7;21) was the primary cytogenetic abnormality. The patient described by Paulsson et al(9) had only the t(7;21) at the primary diagnosis (detected as RUNX1-USP42 fusion), whereas a 5q- occurred secondarily as an additional anomaly in a later sample.
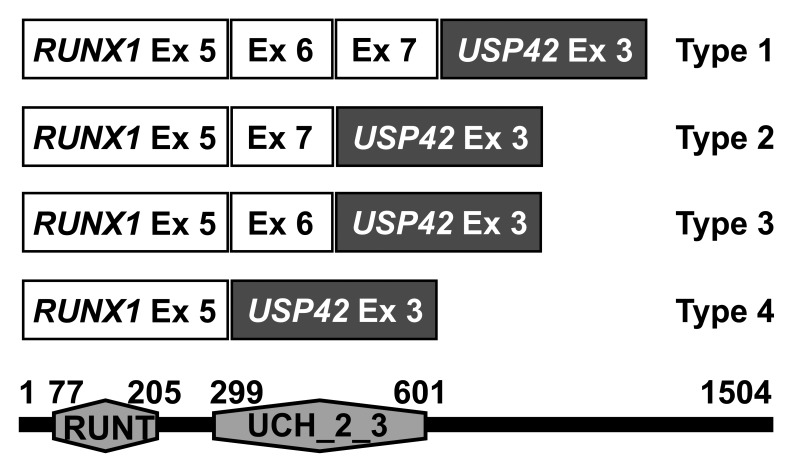
There are 4 types of RUNX1-USP42 chimeric transcripts, defined here as types 1 to 4 (Fig. 3). The RUNX1-USP42 type 1 was described as that in which exon 7 of RUNX1 is fused to exon 3 of USP42(9,11). The type 2 was defined as the transcript in which exon 7 of RUNX1 is fused to exon 3 of USP42 but exon 6 of RUNX1 is spliced out from the fusion (9,11). In the type 3 fusion transcript, exon 6 of RUNX1 is fused to exon 3 of USP42. In type 4, finally, exon 5 of RUNX1 is fused to exon 3 of USP42(11). In all RUNX1-USP42 fusion transcripts, the predicted fusion protein would be expected to contain the Runt homology domain (RHD) which is responsible for heterodimerization with CBFB and DNA binding, and the catalytic UCH (ubiquitin carboxyl terminal hydroxylase) domain of the USP42 protein (9,11). The function of this fusion protein and its cellular consequences leading to leukemia are unknown. It might exert its leukemogenic effect as other RUNX1 fusions do in which the RHD is retained but the transactivation domain of RUNX1 is removed, i.e., by acting as a dominant-negative inhibitor of wild-type RUNX1 in transcription activation (3). In fact, these RUNX1 fusions mimic the RUNX1α variant which has higher affinity to DNA binding but suppresses the transcription activation of RUNX1β (3). RUNX1-USP42 might also affect the regulation of TP53 since the USP42 interacts and deubiqitinates this protein (15). More information concerning the cellular function of the normal USP46 is clearly needed in order to understand the role of RUNX1-USP42 fusions in leukemias.
Figure 3.
Diagram showing the four known variants of the RUNX1-USP42 chimeric transcripts and the predicted RUNX1-USP42 protein of 1504 amino acids which corresponds to the type 1 chimeric transcript. The protein retains the RUNT domain of RUNX1 (position 77–205) and the ubiquitin carboxyl-terminal hydrolases family 2 domain (UCH_2_3; position 299–601).
Acknowledgements
The authors thank Kristin Andersen and Silje Bringsrud for their technical help. The present study was supported by grants from the Norwegian Cancer Society and the South-Eastern Norway Regional Healthy Authority.
References
- 1.Heim S, Mitelman F. Cancer Cytogenetics. 3rd edition. John Wiley & Sons, Inc; Hoboken, NJ: 2009. [Google Scholar]
- 2.Abe A, Katsumi A, Kobayashi M, et al. A novel RUNX1-C11orf41 fusion gene in a case of acute myeloid leukemia with a t(11;21)(p14;q22) Cancer Genet. 2012;205:608–611. doi: 10.1016/j.cancergen.2012.10.001. [DOI] [PubMed] [Google Scholar]
- 3.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Ferec C, De Braekeleer M. RUNX1 translocations and fusion genes in malignant hemopathies. Future Oncol. 2011;7:77–91. doi: 10.2217/fon.10.158. [DOI] [PubMed] [Google Scholar]
- 4.Cho EK, Bang SM, Ahn JY, et al. Prognostic value of AML 1/ ETO fusion transcripts in patients with acute myelogenous leukemia. Korean J Intern Med. 2003;18:13–20. doi: 10.3904/kjim.2003.18.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gandemer V, Chevret S, Petit A, et al. Excellent prognosis of late relapses of ETV6/RUNX1-positive childhood acute lymphoblastic leukemia: lessons from the FRALLE 93 protocol. Haematologica. 2012;97:1743–1750. doi: 10.3324/haematol.2011.059584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borkhardt A, Bojesen S, Haas OA, et al. The human GRAF gene is fused to MLL in a unique t(5;11)(q31;q23) and both alleles are disrupted in three cases of myelodysplastic syndrome/acute myeloid leukemia with a deletion 5q. Proc Natl Acad Sci USA. 2000;97:9168–9173. doi: 10.1073/pnas.150079597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panagopoulos I, Kitagawa A, Isaksson M, Morse H, Mitelman F, Johansson B. MLL/GRAF fusion in an infant acute monocytic leukemia (AML M5b) with a cytogenetically cryptic ins(5;11)(q31;q23q23) Genes Chromosomes Cancer. 2004;41:400–404. doi: 10.1002/gcc.20097. [DOI] [PubMed] [Google Scholar]
- 8.Wilda M, Perez AV, Bruch J, et al. Use of MLL/GRAF fusion mRNA for measurement of minimal residual disease during chemotherapy in an infant with acute monoblastic leukemia (AML-M5) Genes Chromosomes Cancer. 2005;43:424–426. doi: 10.1002/gcc.20182. [DOI] [PubMed] [Google Scholar]
- 9.Paulsson K, Bekassy AN, Olofsson T, Mitelman F, Johansson B, Panagopoulos I. A novel and cytogenetically cryptic t(7;21)(p22;q22) in acute myeloid leukemia results in fusion of RUNX1 with the ubiquitin-specific protease gene USP42. Leukemia. 2006;20:224–229. doi: 10.1038/sj.leu.2404076. [DOI] [PubMed] [Google Scholar]
- 10.Schaffer LG, Slovak ML, Campbell LJ. ISCN 2009: an International System for Human Cytogenetic Nomenclature; Karger, Basel. 2009. [Google Scholar]
- 11.Foster N, Paulsson K, Sales M, et al. Molecular characterisation of a recurrent, semi-cryptic RUNX1 translocation t(7;21) in myelodysplastic syndrome and acute myeloid leukaemia. Br J Haematol. 2010;148:938–943. doi: 10.1111/j.1365-2141.2009.08039.x. [DOI] [PubMed] [Google Scholar]
- 12.Jeandidier E, Gervais C, Radford-Weiss I, et al. A cytogenetic study of 397 consecutive acute myeloid leukemia cases identified three with a t(7;21) associated with 5q abnormalities and exhibiting similar clinical and biological features, suggesting a new, rare acute myeloid leukemia entity. Cancer Genet. 2012;205:365–372. doi: 10.1016/j.cancergen.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Giguere A, Hebert J. Microhomologies and topoisomerase II consensus sequences identified near the breakpoint junctions of the recurrent t(7;21)(p22;q22) translocation in acute myeloid leukemia. Genes Chromosomes Cancer. 2011;50:228–238. doi: 10.1002/gcc.20848. [DOI] [PubMed] [Google Scholar]
- 14.Bennett J, Catovsky D, Daniel M, Flandrin G, Galton D, Gralnick H, Sultan C. Proposals for the classification of the acute leukaemias. French-American-British (FAB) co-operative group. Br J Haematol. 1976;33:451–458. doi: 10.1111/j.1365-2141.1976.tb03563.x. [DOI] [PubMed] [Google Scholar]
- 15.Hock AK, Vigneron AM, Carter S, Ludwig RL, Vousden KH. Regulation of p53 stability and function by the deubiquitinating enzyme USP42. EMBO J. 2011;30:4921–4930. doi: 10.1038/emboj.2011.419. [DOI] [PMC free article] [PubMed] [Google Scholar]