Abstract
Macropinocytosis is a highly conserved endocytic process by which extracellular fluid and its contents are internalized into cells via large, heterogeneous vesicles known as macropinosomes. Oncogenic Ras proteins have been shown to stimulate macropinocytosis but the functional contribution of this uptake mechanism to the transformed phenotype remains unknown1-3. Here we show that Ras-transformed cells utilize macropinocytosis to transport extracellular protein into the cell. The internalized protein undergoes proteolytic degradation, yielding amino acids including glutamine that can enter central carbon metabolism. Accordingly, the dependence of Ras-transformed cells on free extracellular glutamine for growth can be suppressed by the macropinocytic uptake of protein. Consistent with macropinocytosis representing an important route of tumor nutrient uptake, its pharmacological inhibition compromised the growth of Ras-transformed pancreatic tumor xenografts. These results identify macropinocytosis as a mechanism by which cancer cells support their unique metabolic needs and point to the possible exploitation of this process in the design of anti-cancer therapies.
To date, the induction of macropinocytosis by oncogenic Ras has been characterized in the setting of overexpressed proteins1-3. To determine whether stimulated macropinocytosis is a feature of cancer cells endogenously expressing oncogenic Ras, we analyzed fluid-phase uptake in human pancreatic and urinary bladder cancer cell lines harboring oncogenic Ras mutations and compared uptake to wild-type Ras-expressing cells originating from carcinomas of the same tissue type. Macropinosomes were visualized based on the ability of cells to internalize extracellular medium containing tetramethylrhodamine-labeled high molecular weight dextran (TMR-dextran), an established marker of macropinocytosis. Pancreatic adenocarcinoma-derived human MIA PaCa-2 cells, which are homozygous for the K-RasG12C allele4, displayed appreciably higher levels of TMR-dextran uptake compared to BxPC-3 cells, which express wild-type K-Ras (Fig. 1a, b)5. That the TMR-dextran labeling in the oncogenic Ras-expressing cells reflects uptake via macropinocytosis is indicated by the observation that uptake was inhibited in a dose-dependent manner by 5-[N-ethyl-N-isopropyl] amiloride (EIPA) (Fig. 1c, d), which has been shown to inhibit macropinosome formation without affecting other endocytic pathways6,7. Importantly, the knockdown of K-Ras led to an attentuation of macropinocytosis, confirming the dependence of this uptake mechanism on oncogenic Ras expression (Supplementary Fig. 1, a-d). This conclusion is further supported by the observation that bladder carcinoma-derived T24 cells, which are homozygous for the H-RasG12V allele8, exhibit increased levels of macropinocytosis than 5637 cells, which express wild-type H-Ras (Supplementary Fig. 2, a-c)9.
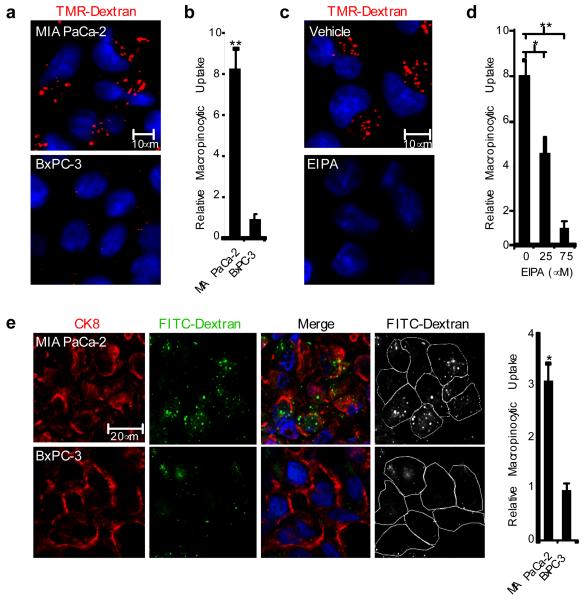
Figure 1. Oncogenic K-Ras-expressing pancreatic cancer cells display elevated levels of macropinocytosis both in culture and in vivo.
a, A macropinocytosis uptake assay utilizing TMR-dextran as a marker of macropinosomes (red) indicates that MIA PaCa-2 cells display elevated levels of macropinocytosis compared to BxPC-3 cells. DAPI staining (blue) identifies nuclei. b, Data are expressed as arbitrary units and are presented relative to the values obtained for BxPC-3 cells. c, Macropinocytic uptake in MIA PaCa-2 cells treated with either vehicle (DMSO) or 75 μM EIPA. d, Quantification of macropinocytic uptake in MIA PaCa-2 cells treated with 0, 25 or 75 μM EIPA. Data are presented relative to the values obtained for the 75 μM condition. e, Visualization and quantification of macropinocytosis in vivo. Representative images from sections of FITC-dextran (green) injected tumor xenografts stained with anti-CK8 (red). Cell boundaries (white outline) were delineated based on the CK8 staining. Data are presented relative to the values obtained for the BxPC-3 tumors. For all graphs, error bars indicate mean +/− SEM for n=3 independent experiments with at least 300 cells scored per experiment. Statistical significance was determined via t-test; *p<0.05, **p<0.01.
To examine whether oncogenic Ras-expressing cells engage in macropinocytosis in vivo, we employed both a heterotopic xenograft mouse model and an autochthonous mouse model. For the heterotopic model, tumors were injected with FITC-conjugated dextran (FITC-dextran) and intracellular uptake was assessed by fluorescent microscopy of tissue sections. The number of macropinosomes identified as FITC-positive puncta was markedly higher in tumors derived from MIA PaCa-2 cells relative to BxPC-3-derived tumors (Fig. 1e). To confirm that the macropinosomes are a feature of the transplanted rather than the host cells, the tumor sections were stained with human specific anti-CK8 antibody, which selectively labels the transplanted human epithelial cells10 (Fig. 1e). To analyze macropinocytosis in mouse pancreatic tumors, we utilized an autochthonous mouse model of pancreatic cancer11. In this model, animals of the genotype p48-Cre;LSL-KRasG12D;p53−/+ (KPC) develop pancreatic intraepithelial neoplasias (PanINs) within 4 weeks of birth and progress to invasive pancreatic ductal adenocarcinoma (PDAC) between 9 and 13 weeks12. To assess macropinocytic uptake, KPC mice were injected with FITC-dextran at 12 weeks of age and pancreata were subsequently harvested. FITC-positive macropinosomes were detected in CK19-labeled acinoductal cells within mid- to late-stage PanIN lesions of KPC pancreata but not in pancreata from wild-type mice (Supplementary Fig. 3). Altogether, these data indicate that an elevated level of macropinocytosis is an attribute of cancer cells expressing oncogenic Ras both in vitro and in vivo.
In mammals, approximately 70% of the soluble substances found in extracellular fluid can be accounted for by proteins, with serum albumin being the most abundant13. Therefore, we sought to determine whether protein uptake via macropinocytosis can be utilized by oncogenic Ras-expressing cells to meet their metabolic needs for proliferation. As an experimental system, we have employed oncogenic K-Ras-transformed NIH 3T3 (NIH 3T3 [K-RasV12]) cells because of their documented dependence on glutamine14. The expression of oncogenic K-RasG12V in these cells was sufficient to stimulate a robust EIPA-sensitive macropinocytic response as measured by TMR-dextran uptake (Fig. 2a, b). To monitor the internalization of albumin, NIH 3T3 [K-RasV12] cells were incubated with a FITC-labeled form of bovine serum albumin (FITC-BSA). As shown in Figure 2c, FITC-BSA was incorporated into discrete intracellular structures that co-localized with TMR-dextran. Similar uptake was also observed in MIA PaCa-2 and T24 cells (Supplementary Fig. 4), indicating that albumin internalization can occur via macropinocytosis in cancer cells harboring endogenous oncogenic Ras mutations. Inhibition of FITC-BSA uptake by treatment with EIPA confirmed that the uptake mechanism was macropinocytosis (Fig. 2d).
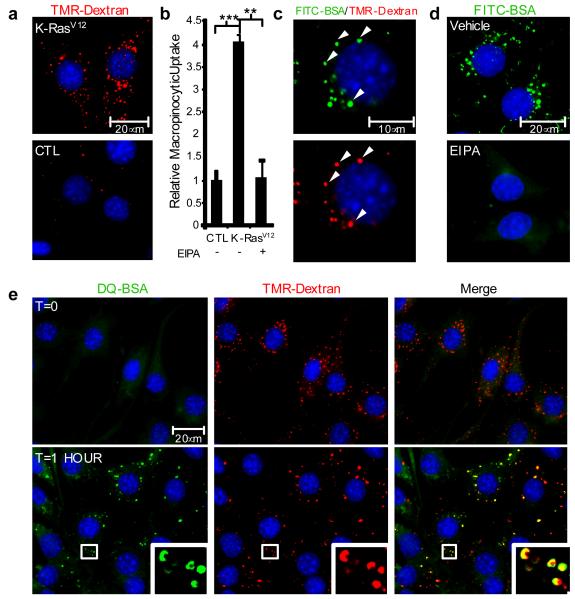
Figure 2. Oncogenic K-Ras-induced macropinocytosis in NIH 3T3 cells mediates the internalization of extracellular albumin that is subsequently targeted for proteolytic degradation.
a, TMR-dextran (red) is internalized at higher levels in NIH 3T3 [K-RasV12] cells (K-RasV12, top panel) compared to untransformed control cells (CTL, bottom panel). b, Quantification of macropinocytic uptake in control cells and NIH 3T3 [K-RasV12] cells incubated with vehicle (DMSO) or with 75 μM EIPA. Data are presented relative to the values obtained for the untransformed control cells. Error bars indicate mean +/− SEM for n=3 independent experiments with at least 300 cells scored per experiment. Statistical significance was determined via t-test; **p<0.01, ***p<0.001. c, FITC-BSA (green) is internalized into discrete puncta that co-localize (white arrowheads) with TMR-dextran (red). d, FITC-BSA uptake is abrogated by treatment with 75 μM EIPA. e, Analysis of DQ-BSA fluorescence in NIH 3T3 [K-RasV12] cells that were co-incubated with DQ-BSA (green) and TMR-dextran (red) and fixed either immediately (T=0) or following a 1 hour chase. The fluorescent signal emanating from DQ-BSA (T=1 HOUR) is an indication of albumin degradation. Insets represent a higher magnification of the boxed areas. Images shown in c, d and e are representative of at least 3 independent experiments.
The macropinocytic internalization and subsequent degradation of albumin could in principle lead to the generation of amino acids that sustain tumor cell bioenergetics and macromolecular synthesis15. To determine whether the internalized albumin is intracellularly degraded, we have utilized a highly self-quenched BODIPY dye conjugated form of BSA (DQ-BSA) that emits a bright fluorescent signal only after proteolytic digestion16. Dual labeling of cells with DQ-BSA and TMR-dextran was used to establish the macropinocytic origin of the degradative compartment. In NIH 3T3 [K-RasV12] cells that were immediately fixed following a 30 minute incubation with DQ-BSA and TMR-dextran (T=0), there was no appreciable DQ-BSA fluorescence detected in macropinosomes (Fig. 2e). However, in cells that were incubated for 30 minutes and subsequently chased for 1 hour in media free of both DQ-BSA and TMR-dextran, DQ-BSA fluorescence was detected in TMR-positive macropinosomes (Fig. 2e). DQ-BSA fluorescence was also detected within macropinosomes after a 1 hour chase in MIA PaCa-2 and T24 cells, indicating that these trafficking events were also occurring in cancer cells harboring endogenous oncogenic Ras mutations (Supplementary Fig. 5). The degradation of macropinocytosed albumin was dependent on lysosomal hydrolases because treatment of MIA PaCa-2 and NIH 3T3 [K-RasV12] cells with bafilomycin A1 prevented the degradation of DQ-BSA (Supplementary Fig. 6). These data demonstrate that oncogenic Ras-expressing cells can harness macropinocytosis for the internalization and degradation of extracellular albumin, and raise the possibility that plasma protein degradation may provide an important source of intracellular amino acids.
Glutamine is a major nutrient for many proliferating cells and is metabolized to glutamate and then α-ketoglutarate to enter central carbon metabolism17. Ras-transformed cells exhibit heightened dependency on glutamine for growth and survival14,18. To test whether the degradation of macropinocytosed albumin results in the production of intracellular glutamine, we directly measured the intracellular concentrations of glutamate and α-ketoglutarate in cells grown either in the absence or presence of albumin. NIH 3T3 [K-RasV12] cells were cultured for 24 hours in complete media (CM) supplemented with physiological concentrations of albumin (2 g/100 mL, 2%). As a control, NIH 3T3 [K-RasV12] cells were grown in CM alone. The addition of albumin to the media led to EIPA-sensitive increases in intracellular concentrations of both glutamate and α-ketoglutarate (Supplementary Fig. 7), indicating that macropinocytic uptake of albumin can increase levels of glutamate and α-ketoglutarate in oncogenic Ras-transformed cells.
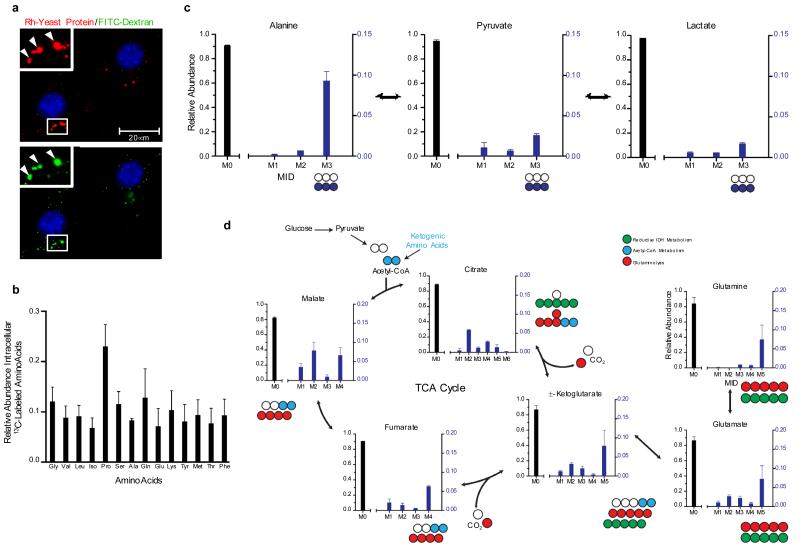
NIH 3T3 [K-RasV12] cells utilize glutamine as a major carbon source for tricarboxylic acid (TCA) cycle anaplerosis19. Therefore, to directly trace the fate of protein-derived amino acids in oncogenic Ras-transformed cells, we cultured NIH 3T3 [K-RasV12] cells in the presence of soluble, heat-inactivated, 13C-labeled yeast protein. Uptake of yeast-derived protein via macropinocytosis was confirmed microscopically by the fluorescent co-localization of rhodamine-labeled protein and FITC-dextran (Fig. 3a). Following 24 hours of culture in low glutamine (0.2 mM) complete medium supplemented with 13C-labeled yeast proteins, intracellular metabolites were extracted and 13C labeling was quantified via gas chromatography/mass spectrometry (GC/MS; Supplementary Table 1)20. Significant labeling of numerous intracellular amino acids, including glutamine, was detected (Fig. 3b), while only low levels of 13C amino acids were detected in media samples incubated for the same period of time without cells (Supplementary Fig. 8). Catabolized yeast protein also entered central metabolism, as evidenced by 13C-labeling of pyruvate, lactate, and various TCA metabolites (Fig. 3c, d). Partially labeled mass isotopomers in numerous metabolites suggested that protein-derived amino acids were potentially being metabolized through several pathways, including glutamine anaplerosis/oxidation, acetyl-CoA metabolism, reductive carboxylation, and serine/glycine cycling (Fig. 3d, Supplementary Fig. 9).
Figure 3. Macropinocytic uptake of extracellular protein drives the accumulation of catabolic intermediates and entry of protein-derived amino acids into central carbon metabolism.
a, Rhodamine-labeled (Rh) yeast protein (red) is internalized into puncta (arrowheads) that co-localize with FITC-dextran (green). Insets represent a higher magnification of the boxed areas. b, Uniformly 13C-labeled intracellular amino acid pools were detected in NIH 3T3 [K-RasV12] cells after culture in low glutamine-containing medium (0.2 mM) supplemented with 2% 13C-labeled yeast protein. c, Protein-derived alanine enters central carbon metabolism upon transamination to pyruvate and pyruvate can be directly converted to lactate. M3 reflects fully labeled alanine, pyruvate and lactate, while M0 abundances reflect metabolites with no 13C label. M1 and M2 represent partially labeled species that are not present in significant amounts. d, Atom transition map depicting a model for the entry of amino acid-derived carbons into the TCA cycle and isotopic labeling of various metabolites. Open circles represent unlabeled carbon, and different colored circles highlight labeling patterns that correspond to specific pathways. For all graphs, error bars indicate mean +/− SD for three independent experiments. IDH, isocitrate dehydrogenase; MID, mass isotopomer distribution
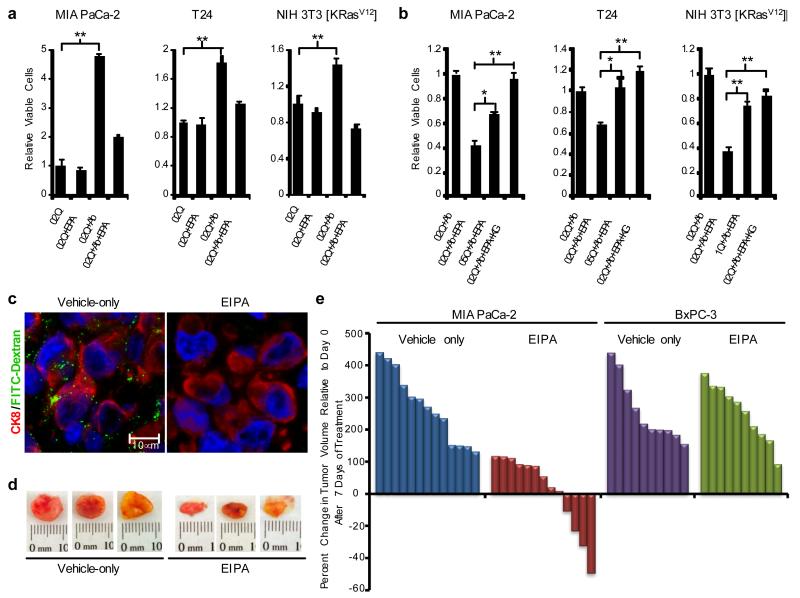
Heightened sensitivity to glutamine deprivation is a hallmark of cancer cells that express oncogenic mutants of Ras14,18. Consistent with this phenotype, we found that MIA PaCa-2, T24 and NIH 3T3 [K-RasV12] cells exhibited decreased proliferation at subphysiological levels of glutamine as measured via the MTT assay or by Syto 60 staining (Supplementary Figs. 10 and 11). We next examined whether cell growth impairment due to glutamine deprivation could be reversed by culturing cells in the presence of physiological levels of albumin. Albumin supplementation of media containing subphysiological glutamine concentrations enhanced proliferation, and this effect was abrogated by EIPA treatment (Fig. 4a, Supplementary Fig. 11). The anti-proliferative response to EIPA observed in these settings was rescued by the addition of either extracellular glutamine or α-ketoglutarate (Fig. 4b). The ability of extracellular albumin to suppress the detrimental effects of glutamine starvation in MIA PaCa-2 cells was dependent on KRAS expression since K-Ras knockdown in these cells diminished the rescuing capacity of albumin (Supplementary Fig. 12). Together, these data indicate that the macropinocytic uptake of albumin could serve to sustain proliferation of oncogenic Ras-transformed cells by constituting a source of glutamine, and potentially other amino acids.
Figure 4. Macropinocytosis is required for albumin-dependent cancer cell proliferation in vitro and for tumor growth in vivo.
a, The compromised proliferation of oncogenic Ras-expressing cells resulting from growth in media containing subphysiological concentrations of glutamine (0.2 mM, 0.2Q) is reversed by supplementation with 2% albumin (0.2Q+Alb) and this effect is inhibited by treatment with 25 μM EIPA (0.2Q+Alb+EIPA). Total viable cell counts were measured via MTT assay after 6 days of growth. Data are presented relative to the values obtained for the 0.2Q condition. b, The effects of EIPA treatment (25 μM) are suppressed by increasing the glutamine levels in the growth media to the indicated concentrations (i.e. 0.5Q indicates 0.5 mM glutamine) or the addition to the medium of 7 mM dimethyl α-ketoglutarate (KG). Data are presented relative to the values obtained for the 0.2Q+Alb condition. For both (a) and (b), error bars indicate mean +/− SEM for n=3 independent experiments. Statistical significance was determined via t-test; *p<0.05, **p<0.01. c-e, EIPA inhibits macropinocytosis in vivo and reduces tumor growth in a subcutaneous heterotopic xenograft model of pancreatic cancer. c, Representative images from sections of FITC-dextran (green) injected MIA PaCa-2 tumor xenografts from mice treated with EIPA or vehicle only controls after 7 days of treatment. The human pancreatic cancer cells are marked by anti-CK8 staining (red). d, Representative digital photographs of dissected tumors from mice treated with EIPA or vehicle only controls. e, Waterfall plots indicating the percent change in tumor volume after seven days of treatment relative to baseline (Day 0 of treatment) for tumors derived from MIA PaCa-2 cells or BxPC-3 cells.
To investigate whether the uptake and degradation of albumin is a unique feature of oncogenic Ras-induced macropinocytosis, we have utilized NIH 3T3 cells expressing SrcY527F (NIH 3T3 [SrcY527F]), a constitutively active form of Src21. Consistent with previous reports,22-24 NIH 3T3 [SrcY527F] cells displayed increased levels of macropinocytosis relative to untransformed control cells (Supplementary Fig. 13a). Moreover, Src-induced macropinosomes displayed the capacity to internalize and degrade extracellular albumin (Supplementary Fig. 13b). Importantly, the decrease in proliferation of Src-transformed cells following glutamine deprivation (Supplementary Fig. 10) could be rescued by extracellular albumin (Supplementary Fig. 13c), suggesting that the utilization of albumin by cancer cells to augment amino acid supply can be mediated by inducers of macropinocytosis other than Ras.
The above observations implicate macropinocytosis in providing nutrients to sustain cancer cell proliferation; therefore we sought to directly evaluate the role of macropinocytosis in tumor growth. Mice bearing MIA PaCa-2-derived heterotopic tumors were treated with EIPA or vehicle via osmotic pump. EIPA administration commenced when tumors attained an average volume of 50-100 mm3. After seven days of treatment, FITC-dextran was delivered intratumorally and the tumors were excised. Tumor tissue harvested from EIPA-treated animals displayed a reduction in macropinocytic uptake of FITC-dextran compared to vehicle only controls (Fig. 4c). Moreover, relative to control tumors, those from EIPA-treated animals displayed an attenuation in growth and, in some cases regression (Fig. 4d, e). In contrast, EIPA administration had no effect on the growth rate of tumors derived from BxPC-3 cells, which display low levels of macropinocytosis (Fig. 4e). These results suggest that a reduction in macropinocytic capacity may compromise tumor growth.
The importance of albumin catabolism in human Ras-driven tumors depends on the extent to which the tumors require certain amino acids in excess of the amounts readily attainable in free form from their environment. In human cancers, oncogenic Ras mutations are most common in adenocarcinomas of the pancreas25,26. In preliminary metabolomic experiments, we have measured the abundances of 123 known water-soluble metabolites in human pancreatic cancer tumor specimens compared to benign adjacent pancreatic tissue. The most depleted metabolite in pancreatic tumor tissue in comparison to adjacent normal tissue was glutamine, with serine and glycine also decreased (data not shown). A common feature of these amino acids is their consumption in nucleotide synthesis. These preliminary data suggest that human pancreatic tumor tissue is depleted of these free amino acids and raise the possibility that in these depleted conditions, the acquisition of amino acids through macropinocytosis and protein degradation could contribute to tumor growth. In previous experiments investigating human tumor metabolism, Holm et al. found that the production of nitrogen waste from some colorectal tumors could be in 10-fold excess relative to their uptake of free amino acids27. The Ras-induced use of plasma proteins as a source of precursors for macromolecular synthesis and anaplerosis could explain both of these observations, a possibility that remains to be explored in future studies.
Recent years have witnessed a renewed appreciation of the altered metabolic behavior of tumor cells and the critical role that such metabolic reprogramming plays in conferring growth and survival advantages to tumor cells. Here we provide evidence that macropinocytosis-mediated internalization of extracellular protein and its subsequent intracellular degradation may define a mechanism for amino acid supply in Ras-transformed cancer cells. Moreover, these findings raise the question of whether the inhibition of macropinocytosis can be utilized for therapeutic targeting in a subset of cancers.
Methods
Cell Culture
All cells were maintained under 5% CO2 at 37°C in medium supplemented with 10% FBS (Gibco). MIA PaCa-2 and T24 cells were maintained in DMEM (Invitrogen); BxPC-3 and 5637 cells were maintained in RPMI (Gibco) supplemented with 1mM sodium pyruvate (Cellgro) and NIH 3T3 cells were maintained in DMEM supplemented with 1X MEM nonessential amino acids (Sigma).
Macropinosome Visualization and Quantification
Cells were seeded onto glass coverslips. 24-48 hours after cell seeding, cells were serum starved for 18 hours. Macropinosomes were marked utilizing a high molecular weight TMR-dextran (D1818, Invitrogen) uptake assay wherein TMR-dextran was added to serum-free medium at a final concentration of 1 mg/mL for 30 minutes at 37°C. At the end of the incubation period, cells were rinsed five times in cold PBS and immediately fixed in 3.7% formaldehyde. Cells were DAPI-treated to stain nuclei and coverslips mounted onto slides using DAKO Mounting Media (DAKO). Images were captured using an Axiovert 200 inverted fluorescent microscope (Zeiss) and analyzed using the ‘Analyze Particles’ feature in ImageJ (NIH). The total particle area per cell was determined from at least 5 fields that were randomly selected from different regions across the entirety of each sample.
RNAi-mediated Knockdown of K-Ras
K-Ras knockdown was achieved through the pTRIPZ doxycycline inducible shRNA expression lentivirus system (Open Biosystems). shRNA sequences for Scramble sh (5′-GGAAGTGCAATTATTCTATTA-3′) and K-Ras sh#1 (5′-TAGTTGGAGCTTGTGGCGTAG-3′) were cloned into pTRIPZ according to the manufacturer’s protocol. The K-Ras sh#2 construct was purchased from Open Biosystems (Clone ID: V2THS_275818). MIA PaCa-2 cells were transduced with lentiviral particles containing the indicated pTripz scramble shRNA or K-Ras shRNAs and selected with 2 μg/ml puromycin (Calbiochem) for 3 days. ShRNA expression was induced with 1 μg/mL doxycycline and efficient knockdown was confirmed by immunoblotting with K-Ras-specific antibodies (sc-30, Santa Cruz Biotechnology).
Mice
For the heterotopic xenografts, female homozygous NCr nude mice (Taconic) were injected subcutaneously in both flanks at 8 weeks of age with 1 × 106 cells mixed at a 1:1 dilution with BD Matrigel (BD Biosciences) in a total volume of 100μL. When tumors reached an average volume of 500 mm3, 1mg of fixable FITC-Dextran (D1820, Invitrogen) diluted in PBS to a volume of 100 μL was injected intratumorally. At two hours post-injection, tumors were removed and rapidly frozen in tissue freezing medium. Experimental cohorts of at least three mice per pancreatic cancer cell line, each mouse implanted in both flanks, were used in triplicate experiments. To quantify FITC-dextran uptake in the tumors, the total particle area per cell was determined from at least 5 sections per tumor with 5 fields analyzed per section, totaling at least 15 randomly selected fields per tumor.
For the autochthonous model, pancreata from p48-Cre;LSL-KRasG12D;p53−/+ (KPC) mice, at 12 weeks of age, were injected with 2 mg of FITC-dextran and subsequently harvested. As a control, we analyzed pancreatic tissue from wild-type mice. Frozen sections from sectioned pancreata were analyzed using standard microscopic techniques and macropinocytosis-positive cells were identified by the visualization of FITC-positive puncta.
To evaluate the effects of EIPA on macropinocytosis in vivo, mice heterotopically transplanted with MIA PaCa-2 cells, as described above, were treated via osmotic pump (Alzet, Model 1004) when tumors attained an average volume of 50-100 mm3 (approximately 2 weeks after transplantation). EIPA (20 mg/mL in a 1:4 dilution of DMSO and PBS) was administered at a pump rate of 0.11 μL/hr. As a control, we employed animals treated with vehicle only (DMSO in PBS). Animals were intratumorally injected with FITC-dextran at various time points (4-7 days) during the course of treatment. Following a subsequent one hour time interval, tumors were harvested, sectioned and analyzed using standard microscopic techniques. Each experimental group consisted of 5 animals and at least 5 sections per pancreas were analyzed.
For tumor growth assays, the animals were randomized into control and treated groups. Treatment, as described above, was initiated two weeks post-transplantation when the tumors had attained an average volume of 50-100 mm3. Volumes of the subcutaneous tumors were calculated based on measurements obtained in a blinded fashion via digital calipers after seven days of treatment.
Metabolite Quantification
NIH 3T3 [K-RasV12] cells were grown in DMEM with/without the supplementation of 2% BSA for 24 hours prior to harvesting. Cells were lysed in a buffered 1% Triton solution and lysates were deproteinized using PCA-precipitation (K808-200, BioVision). Relative metabolite concentrations were then determined utilizing colorimetric assay kits: glutamate/glutamine (EGLN-100, Bioassay Systems) and α-ketoglutarate (K677-100, BioVision).
Generation of 13C-Labeled Proteins
A prototrophic haploid SK1 strain of Saccharomyces cereviseae was grown to OD 4.0 in synthetic complete media lacking amino acids (DIFCO) and containing 2% glucose that was either normal isotopic, or uniformly 13C-labeled (Cambridge Isotopes). Whole cell extracts were prepared according to Tsakraklides et al.28. In brief, cells were harvested, resuspended in 100 mM HEPES (pH 7.6); 0.8 mM sorbitol; 10 mM magnesium acetate; 2 mM EDTA; 300 mM potassium glutamate and lysed using a Sample Prep 6870 Freezer/Mill (Spex Certi Prep Group). Lysate was treated with DNase (Ambion) and RNAseH (Invitrogen) and dialyzed against phosphate buffered saline. Endogenous protease and glutaminase activity were eliminated by adding the protein to FBS and incubating for 30 minutes at 56°C. The resulting labeled and unlabeled protein was either 1) tagged with NHS-Rhodamine (Pierce) according to manufacturer’s instructions for visualization in confocal microscopy experiments or 2) used to supplement cell culture media for subsequent tracing experiments.
Metabolite extraction
For extracellular metabolite analysis, media was collected and metabolites extracted using 0.3 mL ice cold acetone containing 1μg norvaline. Extracts clarified by centrifugation and the supernatant evaporated under nitrogen. For intracellular metabolite analysis, cells were rinsed three times with cold saline and quenched with cold methanol. One half volume of cold water containing 1μg norvaline was added, cell lysate was collected, and one volume of chloroform was added to each sample. Following extraction the aqueous phase was collected and evaporated under nitrogen.
Gas chromatography/mass spectrometry (GC/MS) analysis
Dried polar metabolites were dissolved in 20 μl of 2% methoxyamine hydrochloride in pyridine (Thermo) and held at 37°C for 1.5 hours. After dissolution and reaction, tert-butyldimethylsilyl (TBDMS) derivatization was initiated by adding 30 μl N-methyl-N-(tert-butyldimethylsilyl)trifluoroacetamide (MBTSTFA) + 1 % tert-butyldimethylchlorosilane (TBDMCS; Regis) and incubating at 37°C for 1 hour. GC/MS analysis was performed using an Agilent 6890 GC equipped with a 30m DB-35MS capillary column connected to an Agilent 5975B MS operating under electron impact (EI) ionization at 70 eV. One μl of sample was injected in splitless mode at 270°C, using helium as the carrier gas at a flow rate of 1 ml min−1. For measurement of organic and amino acids, the GC oven temperature was held at 100°C for 3 min and increased to 300°C at 3.5° min−1. The MS source and quadrupole were held at 230°C and 150°C, respectively, and the detector was run in scanning mode, recording ion abundance in the range of 100 – 605 m/z. Mass isotopomer distributions (MIDs) were determined by integrating the appropriate ion fragments30 listed in Supplementary Table 1 and corrected for natural isotope abundance using in house algorithms adapted from Fernandez et al.31.
Glutamine Deprivation Assays
Cells were plated in 96-well plate format at a density of 1000-1500 cells/well. 32 hours after seeding, cells were rinsed briefly in PBS and incubated in the indicated glutamine deprivation media. Glutamine-free DMEM media was supplemented to the indicated concentration of glutamine in the presence of 10% dialyzed FBS and 25 mM Hepes. For some conditions, media was supplemented with BSA (Fraction V, fatty acid-free, nuclease- and protease-free, #126609, Calbiochem). The nominal 0.2% albumin concentration inherently present in complete media was supplemented such that the final concentration of BSA in the media was 2%. For rescue experiments, dimethyl α-ketoglutarate (Sigma) was used at 7 mM29. For all experiments, media was replaced every 24 hours. Viable cell counts were obtained utilizing the MTT assay and relative cell number was determined by Syto 60 staining. 5-(N-Ethyl-N-isopropyl) amiloride (EIPA, Invitrogen) was diluted in DMSO and used at the indicated concentrations with vehicle only controls consisting of DMSO alone. Images depicting cell number in 96-well format were obtained by staining with Syto60 (S11342, Molecular Probes), a red fluorescent nucleic acid stain. Plates were then scanned with an Odyssey Imager (Li-cor) at 700 nm.
Supplementary Material
Acknowledgments
We are grateful to members of the Bar-Sagi laboratory for their helpful comments and discussions; and N. Fehrenbacher and M. Philips for sharing cell lines. This work was supported by NIH grant R01CA055360 to D.B.-S. C.C. was supported by a Canadian Institutes of Health Research postdoctoral fellowship and an AACR postdoctoral fellowship provided by the Pancreatic Cancer Action Network. M.V.H. acknowledges support from the Burroughs Wellcome Fund, the Damon Runyon Cancer Research Foundation, the Smith Family, the Stern family, the Broad Institute and the NCI (P01-CA117969 and P30-CA14051-39). J.J.K. was supported by a Hope Funds for Cancer Research Fellowship (HFCR-11-03-01). C.B.T., J.A.D. and J.D.R. acknowledge support by the Stand Up To Cancer (SU2C) Pancreatic Cancer Dream Team Award. All animal care and procedures were approved by the Institutional Animal Care and Use Committee at NYU School of Medicine. The Histopathology Core of NYU School of Medicine is partially supported by the National Institutes of Health (grant 5 P30CA016087-32). Troma I, an antibody that recognizes CK8, was contributed by P. Brulet and R. Kemler and made available by the Developmental Studies Hybridoma Bank under the auspices of the NICHD.
Footnotes
Author Contributions
C.C. and D.B-S. conceived the cell biological and cell growth experiments. C.C. carried out the macropinocytic assays, microscopy, and proliferation assays. C.C. and R.G.S. carried out the xenograft experiments. C.C. and E.G. carried out the K-Ras knockdown experiments. S.M.D., S.J.P., C.M.M. and M.G.V.H. conceived and carried out the 13C-labelling experiments. J.J.K., S.H., M.N., J.A.D., C.B.T. and J.D.R. conceived and carried out the human metabolomics analysis.
References
- 1.Bar-Sagi D, Feramisco JR. Induction of membrane ruffling and fluid-phase pinocytosis in quiescent fibroblasts by ras proteins. Science. 1986;233:1061–1068. doi: 10.1126/science.3090687. [DOI] [PubMed] [Google Scholar]
- 2.Porat-Shliom N, Kloog Y, Donaldson JG. A unique platform for H-Ras signaling involving clathrin-independent endocytosis. Mol Biol Cell. 2008;19:765–775. doi: 10.1091/mbc.E07-08-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh AB, Bar-Sagi D. Differential activation of the Rac pathway by Ha-Ras and K-Ras. J Biol Chem. 2001;276:15609–15615. doi: 10.1074/jbc.M010573200. [DOI] [PubMed] [Google Scholar]
- 4.Lopez-Crapez E, Chypre C, Saavedra J, Marchand J, Grenier J. Rapid and large-scale method to detect K-ras gene mutations in tumor samples. Clinical chemistry. 1997;43:936–942. [PubMed] [Google Scholar]
- 5.Aoki K, Yoshida T, Sugimura T, Terada M. Liposome-mediated in vivo gene transfer of antisense K-ras construct inhibits pancreatic tumor dissemination in the murine peritoneal cavity. Cancer Res. 1995;55:3810–3816. [PubMed] [Google Scholar]
- 6.Ivanov AI. Pharmacological inhibition of endocytic pathways: is it specific enough to be useful? Methods Mol Biol. 2008;440:15–33. doi: 10.1007/978-1-59745-178-9_2. [DOI] [PubMed] [Google Scholar]
- 7.West MA, Bretscher MS, Watts C. Distinct endocytotic pathways in epidermal growth factor-stimulated human carcinoma A431 cells. J Cell Biol. 1989;109:2731–2739. doi: 10.1083/jcb.109.6.2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capon DJ, Chen EY, Levinson AD, Seeburg PH, Goeddel DV. Complete nucleotide sequences of the T24 human bladder carcinoma oncogene and its normal homologue. Nature. 1983;302:33–37. doi: 10.1038/302033a0. [DOI] [PubMed] [Google Scholar]
- 9.Knowles MA, Williamson M. Mutation of H-ras is infrequent in bladder cancer: confirmation by single-strand conformation polymorphism analysis, designed restriction fragment length polymorphisms, and direct sequencing. Cancer research. 1993;53:133–139. [PubMed] [Google Scholar]
- 10.Wagner M, et al. A murine tumor progression model for pancreatic cancer recapitulating the genetic alterations of the human disease. Genes Dev. 2001;15:286–293. doi: 10.1101/gad.184701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hingorani SR, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 2005;7:469–483. doi: 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 12.Nolan-Stevaux O, et al. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stehle G, et al. Plasma protein (albumin) catabolism by the tumor itself--implications for tumor metabolism and the genesis of cachexia. Crit Rev Oncol Hematol. 1997;26:77–100. doi: 10.1016/s1040-8428(97)00015-2. [DOI] [PubMed] [Google Scholar]
- 14.Gaglio D, Soldati C, Vanoni M, Alberghina L, Chiaradonna F. Glutamine deprivation induces abortive s-phase rescued by deoxyribonucleotides in k-ras transformed fibroblasts. PLoS One. 2009;4:e4715. doi: 10.1371/journal.pone.0004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mates JM, et al. Glutamine homeostasis and mitochondrial dynamics. Int J Biochem Cell Biol. 2009;41:2051–2061. doi: 10.1016/j.biocel.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Reis RC, Sorgine MH, Coelho-Sampaio T. A novel methodology for the investigation of intracellular proteolytic processing in intact cells. Eur J Cell Biol. 1998;75:192–197. doi: 10.1016/S0171-9335(98)80061-7. [DOI] [PubMed] [Google Scholar]
- 17.Newsholme EA, Crabtree B, Ardawi MS. Glutamine metabolism in lymphocytes: its biochemical, physiological and clinical importance. Q J Exp Physiol. 1985;70:473–489. doi: 10.1113/expphysiol.1985.sp002935. [DOI] [PubMed] [Google Scholar]
- 18.Wu MC, Arimura GK, Yunis AA. Mechanism of sensitivity of cultured pancreatic carcinoma to asparaginase. International journal of cancer. Journal international du cancer. 1978;22:728–733. doi: 10.1002/ijc.2910220615. [DOI] [PubMed] [Google Scholar]
- 19.Gaglio D, et al. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol Syst Biol. 2011;7:523. doi: 10.1038/msb.2011.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metallo CM, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature. 2012;481:380–384. doi: 10.1038/nature10602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirai H, Varmus HE. Sh2 Mutants of C-Src That Are Host-Dependent for Transformation Are Transdominant Inhibitors of Mouse-Cell Transformation by Activated C-Src. Genes Dev. 1990;4:2342–2352. doi: 10.1101/gad.4.12b.2342. [DOI] [PubMed] [Google Scholar]
- 22.Kasahara K, et al. Role of Src-family kinases in formation and trafficking of macropinosomes. J Cell Physiol. 2007;211:220–232. doi: 10.1002/jcp.20931. [DOI] [PubMed] [Google Scholar]
- 23.Mettlen M, et al. Src triggers circular ruffling and macropinocytosis at the apical surface of polarized MDCK cells. Traffic. 2006;7:589–603. doi: 10.1111/j.1600-0854.2006.00412.x. [DOI] [PubMed] [Google Scholar]
- 24.Veithen A, Cupers P, Baudhuin P, Courtoy PJ. v-Src induces constitutive macropinocytosis in rat fibroblasts. J Cell Sci. 1996;109(Pt 8):2005–2012. doi: 10.1242/jcs.109.8.2005. [DOI] [PubMed] [Google Scholar]
- 25.Bos JL. ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 26.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holm E, et al. Substrate balances across colonic carcinomas in humans. Cancer research. 1995;55:1373–1378. [PubMed] [Google Scholar]
- 28.Tsakraklides V, Bell SP. Dynamics of pre-replicative complex assembly. The Journal of biological chemistry. 2010;285:9437–9443. doi: 10.1074/jbc.M109.072504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wise DR, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antoniewicz MR, Kelleher JK, Stephanopoulos G. Accurate assessment of amino acid mass isotopomer distributions for metabolic flux analysis. Anal Chem. 2007;79:7554–7559. doi: 10.1021/ac0708893. [DOI] [PubMed] [Google Scholar]
- 31.Fernandez CA, Des Rosiers C, Previs SF, David F, Brunengraber H. Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J Mass Spectrom. 1996;31:255–262. doi: 10.1002/(SICI)1096-9888(199603)31:3<255::AID-JMS290>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.