Abstract
Currently, all three cytochrome P450 1 (CYP1) monooxygenases are believed to participate in lipid mediator biosynthesis and/or their local inactivation; however, distinct metabolic steps are current unknown. We used multiple-reaction monitoring and LC-UV-MS/MS-based lipid-mediator metabololipidomics to identify and quantify three different lipid-mediator metabolomes in basal peritoneal and zymosan-stimulated inflammatory exudates, comparing Cyp1a1/1a2/1b1(–/–) C57BL/6J-background triple-knockout with C57BL/6J wild-type mice. Significant differences between untreated triple-knockout and wild-type mice were not found for peritoneal cell number or type, or basal CYP1 activities involving 11 identified metabolic steps. Following zymosan-initiated inflammation, 18 lipid mediators were identified including members of the eicosanoids and specialized pro-resolving mediators, i.e. resolvins and protectins. Compared with wild-type mice, Cyp1 triple-knockout mice exhibited increased neutrophil recruitment in zymosan-treated peritoneal exudates. Zymosan stimulation was associated with 8 statistically significantly altered metabolic steps: increased arachidonic acid-derived leukotriene B4 (LTB4) and decreased 5S-hydroxyeicosatetraenoic acid (5S-HETE); decreased docosahexaenoic acid-derived neuroprotectin D1/protectin D1 (NPD1/PD1), 17S-hydroxydocosahexaenoic acid (17S-HDHA), and 14S-HDHA; and decreased eicosapentaenoic acid-derived 18R-hydroxyeicosapentaenoic acid (18R-HEPE), 15S-HEPE, and 12S-HEPE. In neutrophils analyzed ex vivo, elevated LTB4 levels were shown to parallel increased neutrophil numbers, and 20-hydroxy-LTB4 formation was found to be deficient in Cyp1 triple-knockout mice. Together, these results demonstrate novel contributions of CYP1 enzymes to the local metabolite profile of lipid mediators that regulate neutrophilic inflammation.
Introduction
Lipid mediators (LMs) derived from polyunsaturated fatty acids (Fig. 1A) include more than 150 chemicals, many of which have potent bioactivity (45). The ω–6 fatty acid, arachidonic acid (AA), is converted to twelve known classes of LMs: prostaglandins, prostacyclins, thromboxanes, leukotrienes, epoxyeicosatrienoic acids (EETs), hydroxyeicosatetraenoic acids (HETEs), dihydroxyeicosatrienoic acids (DHETEs), hydroperoxyeicosatetraenoic acids (HpETEs), ω– and ω–1 alcohols, lipoxins, hepoxilins, and eoxins (5,18,37). AA is well known to be converted by cytochrome P450 to EETs (44), some of which are active in inflammation (43). On the other hand, the ω–3 fatty acids—docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA)—are converted to resolvins (E-series & D-series), neuroprotectins, and maresins (5,18,21,29,50). Many ω–6 and ω–3 LMs participate in inflammation. Some are more involved in the initiation phase; others participate more in the resolution phase.
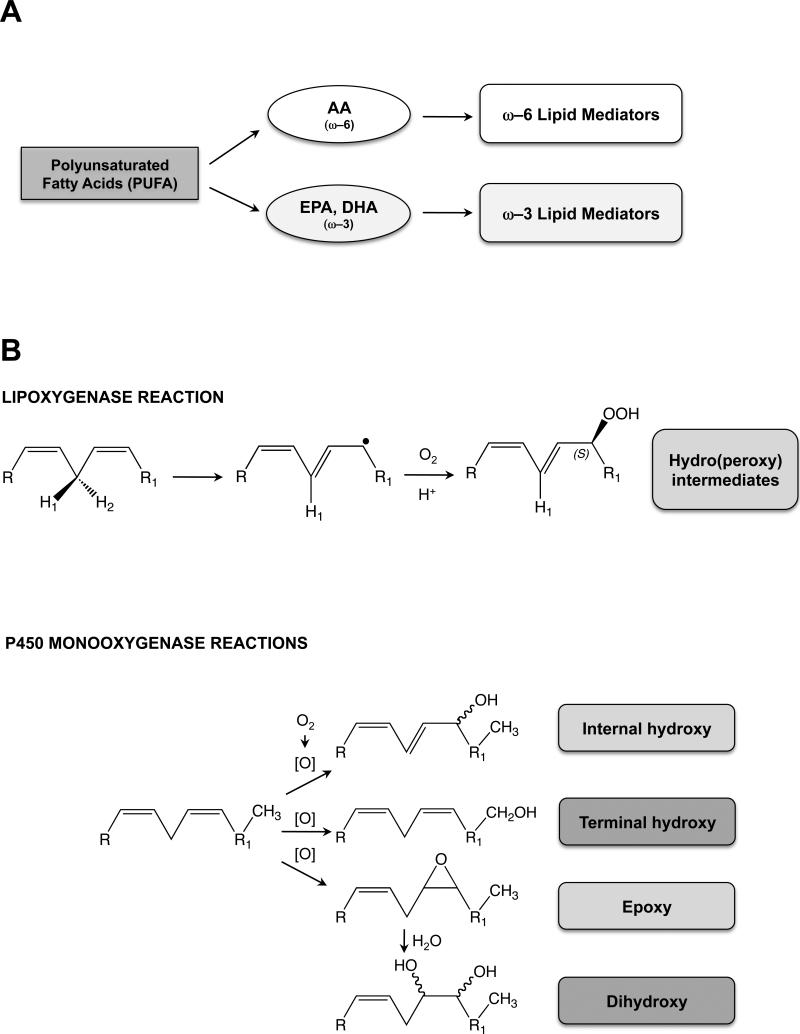
FIGURE 1.
Introduction to polyunsaturated fatty acids, lipoxygenase vs monooxygenase reactions, and LM biosynthesis pathways. A, Scheme depicting the origin of ω–6 and ω–3 LMs. B, Diagram showing the mechanisms for lipoxygenase vs P450-monooxygenase reactions.
Oxidative biosynthesis of LMs is carried out in multiple steps by specific arachidonate lipoxygenases (encoded by six human genes: ALOX5, ALOX12, ALOX12B, ALOX15, ALOX15B, and ALOXE3, and seven mouse genes: Alox5, Alox12, Alox12b, Alox12e, Alox15, Alox8 and Aloxe3). In addition to the cyclooxygenases (COX1, COX2; official names PTGS1, PTGS2) and arachidonate lipoxygenases, there is considerable evidence (5,18,37) suggesting that cytochrome P450 (CYP) monooxygenases also participate in the oxidative biosynthesis and inactivation of LMs. Members of the CYP1, CYP2, CYP3 and CYP4 gene families—as well as CYP5A1 and CYP8A1—are involved in biosynthesis and further inactivation of LMs (38). Although some P450-mediated specific reactions of bioactive LMs have been described [reviewed in Ref (37)], the vast majority remains to be determined; this has been largely due to the technical challenges involved in identifying LMs, particularly with regard to stereochemistry.
The CYP1 gene family encodes three enzymes (CYP1A1, CYP1A2 & CYP1B1) in both human and mouse that are evolutionarily highly conserved—suggesting that mouse CYP1 data are likely able to be extrapolated to human CYP1 functions. On the other hand, the CYP2, CYP3 and CYP4 families are far more complex due to multiple gene-duplication events followed by “genetic drift” during the past 65 million years since human and mouse had a common ancestor. This has resulted in the human genome having 16 functional CYP2, 4 CYP3, and 12 CYP4 protein-coding genes—compared with the mouse genome having 50 functional protein-coding Cyp2, 9 Cyp3, and 20 Cyp4 genes (39).
Lipoxygenases insert both atoms (25), whereas P450 monooxygenases insert one atom (20,30,31), of diatomic oxygen into substrates to form the products (Fig. 1B). Another important distinction between lipoxygenase and P450 monooxygenase reactions is that, although occasionally lipoxygenases can produce epoxides (e.g. leukotriene A4 formation by ALOX5), the major product is a fixed-chirality hydroperoxide; on the other hand, P450 monooxygenases can generate racemic mixtures of internal-monohydroxy products, terminal-monohydroxy products, and epoxides which (following hydrolysis) often proceed to form racemic mixtures of dihydroxy products (Fig. 1B).
Ultimately, among many other functions [detailed in Refs. (37,38)], AA-derived LMs are somewhat more likely to be involved in the pro-inflammatory phase, whereas the DHA-derived bioactive metabolome and EPA-derived bioactive metabolome are LMs that orchestrate the resolution phase of self-limited inflammatory responses. In addition, the location of many of these metabolic reactions is usually highly tissue- and/or cell type-specific (5,18,37).
One approach to resolving the challenging problem of identifying which CYP enzyme participates in what step(s) of the LM cascade involved in acute inflammation would be to use Cyp knockout mouse lines, in combination with the most advanced metabololipidomics (13) for separating and identifying as many unique LM metabolites as possible. The present study capitalizes on this approach. The Cyp1a1/1a2/1b1(–/–) triple-knockout (TKO) mouse (15,36,37) has all 3 highly conserved members of the mammalian Cyp1 gene family genetically deleted. We compared TKO with wild-type (WT) mice during zymosan-induced peritonitis. For this present report, we did not investigate the classical EET products produced by P450 from AA in the LC-MS-MS profiles; rather, we focused instead on their roles in lipoxygenase and cyclooxygenase pathways. Results from the present study indicate that CYP1 monooxygenases play a highly significant role in regulation of at least 8 key steps during LM biosynthesis and their further metabolism; these findings thus provide potentially useful new therapeutic targets for treating inflammation and its natural resolution.
Materials and Methods
Chemicals
Zymosan A was obtained from Sigma-Aldrich Chemical Co. (St. Louis, MO). Liquid-chromatography grade solvents were purchased from Fisher Scientific. Agilent Eclipse Plus C18 columns (50 mm × 4.6 mm × 1.8 μm) and C18 SPE columns were bought from Waters. Synthetic standards for LC-MS/MS quantification and deuterated internal standards were procured from Cayman Chemicals. All chemicals and reagents represented the highest available grades.
Animals
Generation of the Cyp1a1/1a2/1b1(–/–) triple-knockout mouse line has been detailed (15). TKO animals have been backcrossed for >10 generations onto C57BL/6J to ensure a genetic background similarity of greater than 99.8% (35) to WT C57BL/6J mice. All TKO mice used in experiments were age-matched with WT controls. We found that mice provided with regular lab chow gave highly variable results. Therefore, both WT and TKO mice were maintained—from prior to conception—on flavone-free Laboratory Chow Diet 5001. This diet is purportedly designed to assure minimal inherent biological variation in long-term studies due to its being enriched with ω-3 essential fatty acids (www.labdiet.com); indeed, the 5001 diet provided us with highly reproducible data. Results appeared to be more consistent in males than females; subsequently, for all experiments described herein we chose to study males only, between 50 and 100 days of age. All animal experiments were approved by, and conducted in accordance with, the National Institutes of Health standards for the care and use of experimental animals and the University Cincinnati Medical Center Institutional Animal Care and Use Committee.
Zymosan challenge
An acute inflammatory response was induced with 1 mg of zymosan per mouse (24). Zymosan (insoluble carbohydrate from yeast cell wall) was freshly prepared (2 mg/mL) in sterile 0.9% NaCl, and 0.5 mL was injected intraperitoneally into each mouse; zero-time controls were not treated. At the appropriate time-points (zero, 6 & 9 h), each peritoneal cavity was washed with 5 mL of phosphate-buffered saline (PBS; Cellgro) twice; all baseline peritoneal cells at time zero, and all peritoneal exudates at 6 and 9 h following zymosan challenge, were harvested and centrifuged (560 × g for 6 min). Supernatant fractions were collected and mixed (2:1) ratio with cold methanol (Sigma); samples were then stored at –80°C until metabololipidomics analysis. Cell pellets were resuspended in 1 mL of cold PBS and total cells quantified. In addition, 80 μL of cells was used to prepare histology slides at baseline, while an additional dilution of 1:20 was used to prepare histology slides following zymosan challenge; the amount of lavage exudate recovered per mouse was quantified and used for normalization of total cell counts per mouse. For each group, three to eight mice (WT and TKO) were studied individually, and each experiment was repeated at least twice more.
Cell quantification
Following centrifugation (560 × g for 6 min), total cell counts were quantified from peritoneal lavage at baseline or lavage of peritoneal exudates following zymosan challenge, as well as from cells isolated from peripheral blood or bone marrow. Differential cell counts were quantified following cytospin analysis (on 80 μL of total cell exudate at baseline, an additional 1:20 dilution for conditions involving zymosan challenge) and subsequent Diff-Quik staining (Siemens). We quantified 400 cells/slide, to determine the percentages of neutrophils, monocytes/macrophages, mast cells, eosinophils and lymphocytes.
Sample extraction and LM metabololipidomics
All samples for LC-MS/MS-based analyses were extracted using SPE columns (13). Briefly, columns were equilibrated with 1 column volume of methanol and 2 volumes doubly-distilled water (ddH2O). Prior to extraction, 500 pg of deuterium-labeled internal standards d8-5S-HETE, d4-LTB4 and d4-PGE2 were added to facilitate quantification of sample recovery. Sample supernatant fractions were diluted with 10 volumes of ddH2O, acidified (pH ~3.5), and immediately loaded onto a SPE column. After loading, columns were washed with 1 volume of neutral ddH2O and 1 volume of hexane.
Samples were eluted with 6 mL methylformate and taken to dryness using Speedvac or nitrogen stream. Samples were subsequently suspended in methanol/water for LC-MS/MS. The LC-UV-MS/MS system includes QTrap 3200 equipped with a Shimadzu SIL-20AC auto-injector and LC-20AD binary pump or QTrap 5500, (ABSciex) equipped with an Agilent HP1100 binary pump. An Agilent Eclipse Plus C18 column (50 mm × 4.6 mm × 1.8 μm; or 100 mm × 4.6 mm × 1.8 μm) was used with a gradient of methanol/water/acetic acid of 60:40:0.01 (v/v/v) to 100:0:0.01 at 0.5-mL/min flow rate.
To monitor and quantify the levels of the various LMs, we developed a multiple-reaction-monitoring (MRM) method with signature ion fragments for each molecule. Identification was conducted using published criteria (13) with at least six diagnostic ions. Calibration curves were obtained using synthetic and authentic LM mixtures; these included d8-5S-HETE, d4-LTB4, d4-PGE2, 15-HETE, 12-HETE, 5-HETE, LXA4, LXB4, LTB4, PGE2, PGD2, PGF2α, TXB2, RvE1, RvE2, 18-HEPE, 15-HEPE, 12-HEPE, 5-HEPE, RvD1, RvD2, RvD5, PD1, MaR1, 17-HDHA, 14-HDHA, 7-HDHA, and 4-HDHA at 12.5, 25, 50 and 100 pg (Supplemental Table S1 online). Linear calibration curves for each were obtained with r2 values in the range 0.98-0.99. Quantification was carried out based on peak area of the MRM transition and the linear calibration curve for each compound. Reverse-phase chiral LC-MS-MS was conducted as described (41).
Human and mouse neutrophils
Neutrophils were isolated as described (28), with minor modifications. Specifically, human neutrophils were isolated from peripheral blood; mouse neutrophils were purified from femoral and tibial bone marrow as described (57). Cells were collected in ice-cold Ca2+- and Mg2+-free Hank's balanced salt solution, (HBSS; Life Technologies) supplemented with 0.1% bovine serum albumin (BSA; Sigma); neutrophils were purified using a discontinuous Percoll (GE Healthcare) gradient. Subsequently, neutrophils were layered onto Histopaque 1119 (Sigma), centrifuged at 650 × g for 20 min, and stopped without brake, to separate red blood cells. Purified neutrophils were collected and washed with HBSS, 0.1% BSA, centrifuged for 5 min at 400 × g, and resuspended in PBS with Ca2+ and Mg2+ prior to being stimulated with LTB4. Purity of isolated neutrophils was determined by cytospin analysis followed by Diff-Quik stain (Siemens). Isolation of human neutrophils was performed at CCHMC; all participants provided written informed consent and the study was approved by the CCHMC IRB.
Incubation of neutrophils with LTB4
Cells (1 × 106) were plated in a flat-bottom 96-well plate and stimulated with 500 nM LTB4 at 37°C and 5% CO2. Following a 30-min incubation, the reaction was stopped by addition of 200 μL of cold methanol (Sigma) and cells and cell lysates were snap-frozen. Human neutrophils were used as a LTB4-responsive positive control.
Statistical analysis
All assays were performed in duplicate or triplicate, and average values from each mouse considered as one independent determination. Statistical differences were assessed by Student's pair-wise t-test or chi-square analysis. Data were normally distributed and are presented as means ± S.E.M. All P-values of <0.05 were regarded as statistically significant.
Results
Peritoneal cell numbers and cell types
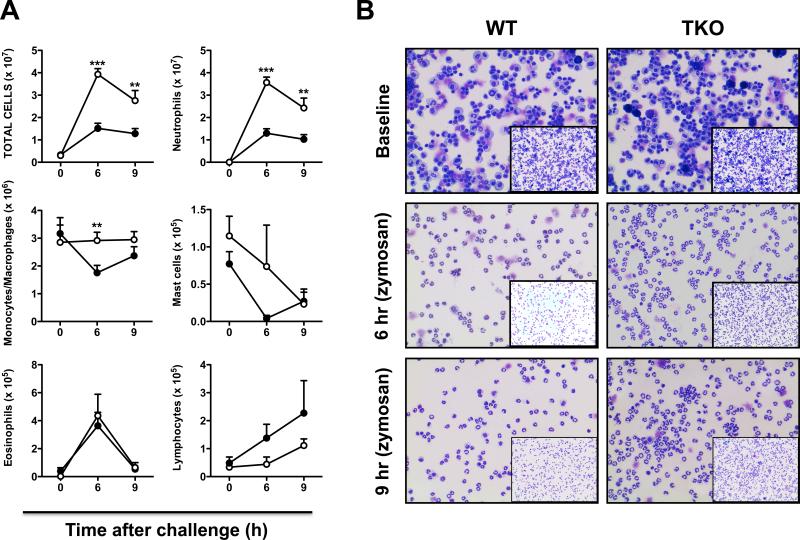
In an earlier study, peritoneal immune cell numbers and types were found to be similar at baseline between WT and TKO mice (15). In contrast, following intraperitoneal zymosan, an exaggerated increase in peritoneal exudate total cell number, neutrophils, and monocyte/macrophages was observed in TKO mice (15). In the present study, zymosan challenge also led to significant increases in total cell numbers in peritoneal exudates of TKO compared with WT mice at 6 h (>14-fold) and 9 h (~10-fold), with significantly increased neutrophil accumulation 6 and 9 h after challenge, and significantly increased monocyte/macrophage accumulation at 6 h after challenge (Fig. 2). Significant differences between WT and TKO mice were not found for eosinophil, mast cell, or lymphocyte numbers.
FIGURE 2.
Peritoneal cell parameters from baseline cells vs acute inflammatory exudates, comparing WT with TKO mice. A, Peritoneal cell numbers and cell types were compared at time zero, 6 h and 9 h following intraperitoneal zymosan administration. In this and subsequent figures, bars and brackets denote means ± S.E.M., using Student's two-tailed t-test (n = 3 mice per genotype). *P <0.05; **P >0.01; ***P<0.001. B, Representative Diff-Quik staining of cells from WT vs TKO at the three time-points. Baseline (neat cells); for 6 h and 9 h following zymosan challenge, a 1:20 dilution of cells was used for quantification.
Comparison of WT vs TKO levels of LMs at baseline
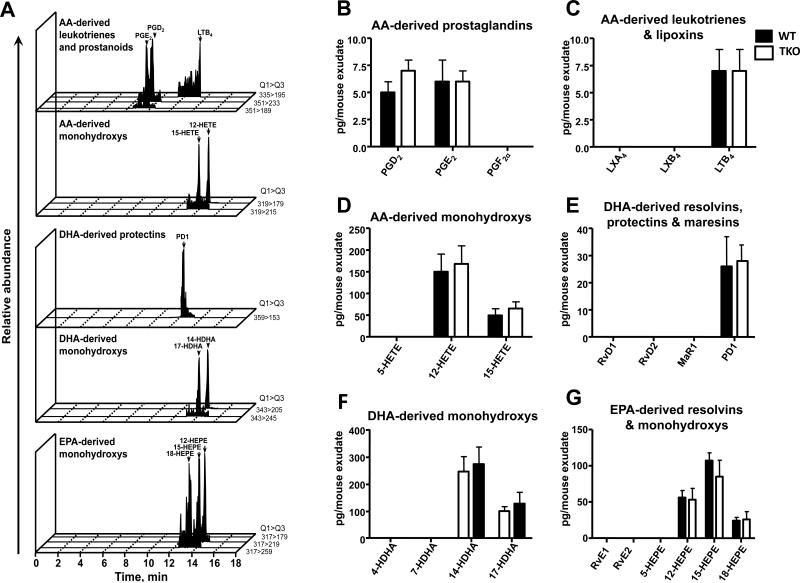
To determine the contributions of CYP1 monooxygenases in LM biosynthetic pathways and LM profiles in vivo, we initially quantified LM levels in untreated mice (Fig. 3). Illustrations of representative chromatography (Fig. 3A) unequivocally demonstrate that each of the bioactive LMs was identified using strict reported criteria (cf. “Materials and Methods”). Of the 24 LMs screened, 11 were detected in baseline peritoneal cells of WT vs TKO mice: prostaglandin D2 (PGD2) and PGE2, LTB4, and 12-HETE and 15-HETE derived from AA (Figs. 3B, 3C & 3D); PD1 and 14-HDHA and 17-HDHA derived from DHA (Figs. 3E & 3F); and 12-HEPE, 15-HEPE and 18-HEPE derived from EPA (Fig. 3G). Statistically significant differences were not found between TKO and WT peritoneal cell LMs without zymosan challenge.
FIGURE 3.
Baseline peritoneal LM levels are not significantly different between untreated WT and TKO mice. Peritoneal lavage samples were obtained from untreated WT or TKO mice, and LM levels were assessed by LM-metabololipidomics. A, Representative MRM chromatograms of LMs identified in peritoneal exudates. LM levels for B, AA-derived prostaglandins, C, leukotrienes and lipoxins, D, monohydroxy-ETE pathway markers. LM levels for E, DHA-derived resolvins, protectins and maresins, and F, monohydroxy-DHA pathway markers. LM levels for G, EPA-derived resolvins and monohydroxy-EPA pathway markers. Results are expressed as means ± S.E.M. (n=3 mice per group).
Comparison of WT vs TKO levels of LMs in exudate 6 h into inflammation
Of the LMs screened via targeted LC-MS-MS-based lipidomics, 17 were identified in peritoneal exudates from WT vs TKO mice treated with zymosan for 6 h (data not shown). Hence, besides those 11 identified in baseline peritoneal cells (Fig. 3), 6 additional LMs and pathway markers were found following 6 h of inflammation. These included: PGF2α, LXA4, 5-HETE, 4-HDHA, RvE2, and 5-HEPE. Moreover, compared with WT mice, zymosan-treated TKO mice revealed a trend towards increased AA-derived prostaglandins and LTB4 levels—as well as decreased 12-HETE, DHA-derived PD1 and 14-HDHA, and EPA-derived 15-HEPE levels (data not shown).
Comparison of WT vs TKO levels of LMs in exudate 9 h into inflammation
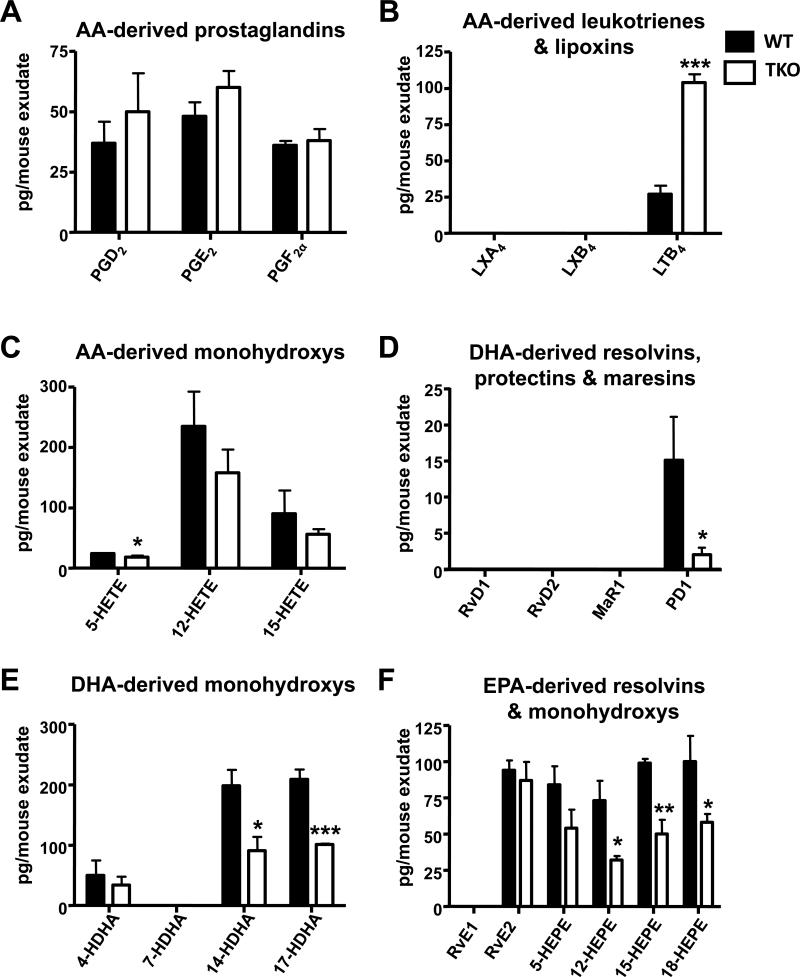
Of the LMs and pathways profiled, 16 were identified in the peritoneal exudate of WT vs TKO mice treated with zymosan for 9 h (Fig. 4). LXA4, which had been found at low levels at 6 h into inflammation, was no longer detectable at 9 h. Whereas LTB4 levels rose dramatically between 6 and 9 h of zymosan treatment in TKO mice, 7 other LMs were statistically significantly lowered in TKO—compared with that in WT mice (Fig. 4 & Table S1).
FIGURE 4.
LM levels from peritoneal exudates are elevated in TKO mice, 9 h following zymosan treatment. The same two dozen LM levels are listed in the same order as in Figs. 3B-3G. Results are expressed as means ± S.E.M., using Student's two-tailed t-test (n = 3 mice per genotype). *P<0.05; **P<0.01; ***P<0.001 TKO differences vs WT.
Therefore, of the 3 functionally distinct LM metabolomes screened (i.e. AA-, DHA- and EPA-bioactive metabolomes), statistically significantly alterations were obtained in 8 LMs and pathway markers during inflammation in TKO when compared with WT mice: TKO mice exhibited a ~3.9-fold increase in LTB4 levels, a ~7.5-fold decrease in PD1, and between 1.7- and 2.3-fold decrease in 5-HETE, PD1, 14-HDHA, 17-HDHA, 12-HEPE, 15-HEPE and 18-HEPE levels, and a 1.3-fold decrease in 5-HETE (Fig. 4).
LTB4 metabolism ex vivo
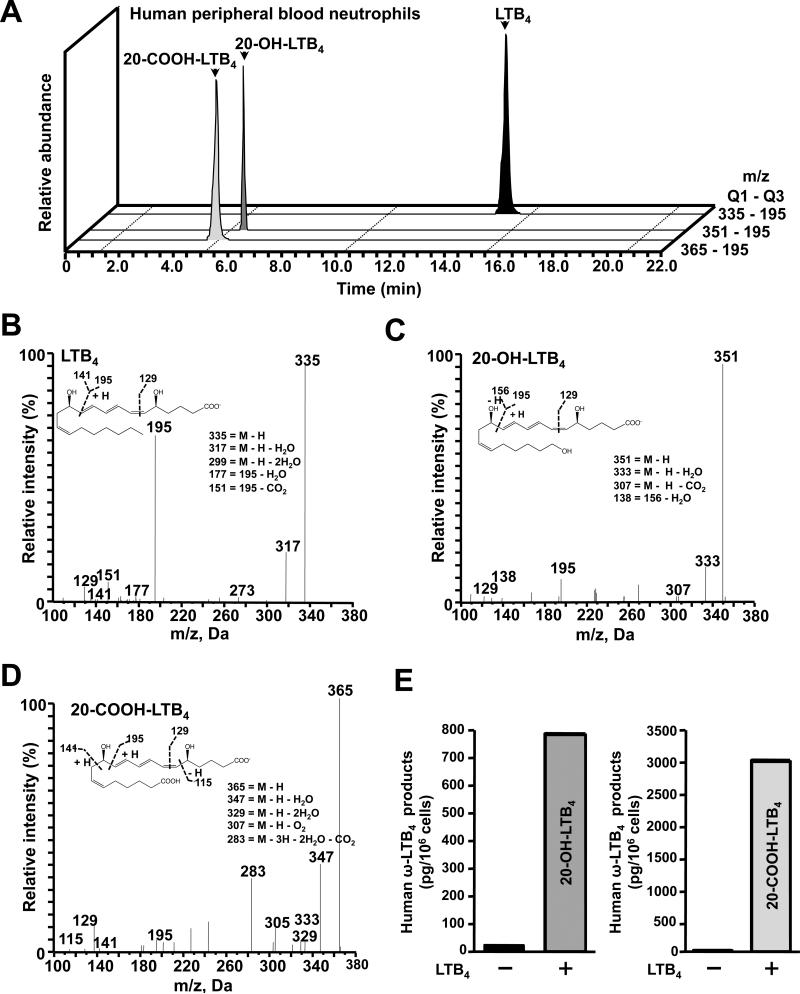
We chose to explore LTB4 metabolism further, in peritoneal and bone marrow neutrophils ex vivo. A proof-of-principle experiment was first carried out with human neutrophils. When the substrate LTB4 was added to human peripheral blood neutrophils, ~4-fold more 20-COOH-LTB4 than 20-OH-LTB4 (Figs. 5A & 5E) was produced. The confirmed mass-to-charge ratio of the parent compound LTB4 was m/z 335 (Fig. 5B), of 20-OH-LTB4 m/z 351 (Fig. 5C), and of 20-COOH-LTB4 m/z 365 (Fig. 5D). This ex vivo experiment with human neutrophils thus validates identification of LTB4 and two further metabolites that were reported earlier to be produced by a member of the human CYP4 family (56).
FIGURE 5.
Human peripheral blood neutrophils preferentially convert LTB4 to 20-COOH-LTB4 more than 20-OH-LTB4. A, Representative MRM traces for LTB4, 20-OH-LTB4 and 20-COOH-LTB4. Accompanying MS/MS spectra employed for the rigorous identification of B, LTB4. C, 20-OH-LTB4 and D, 20-COOH-LTB4. E, 20-OH-LTB4 and 20-COOH-LTB4 levels in human peripheral blood neutrophils following ex vivo incubation without, vs with, the substrate LTB4.
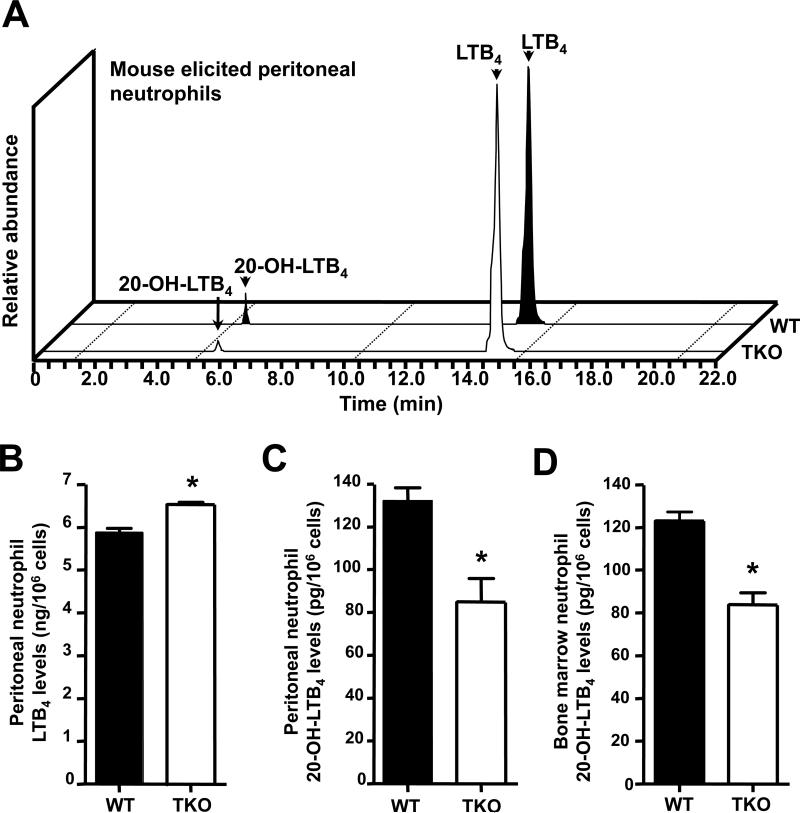
Peritoneal and bone marrow neutrophils were then isolated from TKO and WT mice that had received 4 h of zymosan treatment. Before incubation with the substrate LTB4, peritoneal neutrophils from TKO mice displayed significant increases in LTB4 levels, compared with those from WT neutrophils (Figs. 6A & 6B). After incubation with the substrate LTB4, relative to the WT, TKO mice showed about two-thirds as much 20-OH-LTB4 in both peritoneal neutrophils (Fig. 6C) and bone marrow neutrophils (Fig. 6D).
FIGURE 6.
Impairment of further metabolism of LTB4 in TKO neutrophils. Neutrophils from peritoneal exudates and bone marrow were obtained from WT and TKO mice following 4 h of zymosan treatment. Cells were counted and incubated with LTB4. A, Representative MRM traces for LTB4 and 20-OH-LTB4 obtained from WT and TKO leukocyte incubations. B, LTB4 in peritoneal neutrophils before incubation with substrate. C, 20-OH-LTB4 levels in peritoneal neutrophils and D, 20-OH-LTB4 levels in bone marrow neutrophils, following incubation with the substrate LTB4. Results for B through D are expressed as means ± S.E.M. (n=3 mice per group). *P <0.05. Note the differences in values on the Y-axis between B vs the C and D panels.
Curiously, whereas a significant difference in 20-OH-LTB4 levels was found between TKO and WT elicited peritoneal neutrophils, the 20-COOH-LTB4 (downstream oxidized metabolite) was not detected in either TKO or WT (Fig. 6A). In contrast, high levels of 20-COOH-LTB4 were clearly demonstrable with human neutrophils (Figs. 5A, 5D & 5E); these data indicate that mice apparently do not have the CYP4 enzyme that is equivalent in function to the human CYP4 enzyme responsible for 20-COOH-LTB4 formation (56). In fact, the amount of 20-COOH-LTB4 in human neutrophils was ~4 times greater than the amount of 20-OH-LTB4 (Fig. 5E).
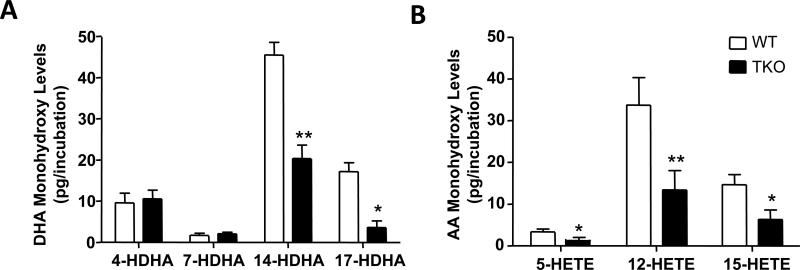
In order to determine the contribution of CYP1 enzymes to LM biosynthesis, we next assessed the production of LM precursors by peritoneal exudate leukocytes. Here we found that 14-HDHA, 17-HDHA, 12-HETE and 15-HETE levels were significantly lower in TKO leukocyte incubations, when compared with those of WT leukocyte incubations (Fig. 7). Finally, using reverse-phase chiral LC-MS-MS (41), we investigated the levels of R/S-enantiomers for each of the monohydroxy acids that had been found to be significantly decreased in TKO peritoneal leukocyte incubations. This analysis demonstrated a ~59 ± 9% fall in the levels of 14-HDHA, 17-HDHA, 12-HETE and 15-HETE; these four metabolites in TKO carry the hydroxyl group in the R-enantiomeric position when compared with that in WT leukocyte incubations. For these four metabolites in TKO that carry the hydroxyl group in the S-enantiomeric position, we found a ~62 ± 14% diminution in the 14-HDHA, 17-HDHA, 12-HETE and 15-HETE levels. These results provide further evidence for the contribution of CYP1 enzymes to LM precursor biosynthesis.
FIGURE 7.
Contribution of CYP1 enzymes to LM biosynthesis, following zymosan challenge. The four most significant changes in the generation of LM precursors by peritoneal exudate leukocytes included 14-HDHA, 17-HDHA, 12-HETE and 15-HETE levels; these metabolites were significantly lower in TKO, compared with those in WT mice.
Discussion
In the present study, we have examined the combined contributions of the 3 CYP1 enzymes in LM pathways involved during acute inflammation (13). We started with the hypothesis that absence of all 3 CYP1 enzymes would alter the LM metabololipidomics profile at precise steps during inflammation and its resolution. Specifically, if a LM was increased in TKO relative to WT, this would suggest that one or more of the CYP1 enzymes might be critical in a downstream metabolic step in this pathway, i.e. absence of CYP1 would lead to a build-up in levels of the upstream LM. In contrast, if a specific LM was decreased in TKO relative to WT, this would suggest that one or more of the CYP1 enzymes is critical in an upstream event involved in formation of the metabolite in this pathway, i.e. absence of CYP1 would lead to a diminution of the downstream LM. Elucidation of P450 enzyme specificity for any particular step in the initiation phase or resolution phase of inflammation should aid in developing novel drugs for treatment of various forms of acute inflammation—as well as chronic inflammatory disorders such as cardiovascular disease, rheumatoid arthritis, periodontitis, and Alzheimer disease (45,52).
Leukocyte trafficking within the site of inflammation is a dynamic process regulated by chemical cues produced at the site of injury and/or infection. In the present report, we focused on the initiation phase of the inflammatory response, employing metrics that map leukocyte recruitment dynamics in response to a self-limited challenge (2). Our present results demonstrate that TKO mice display altered leukocyte recruitment dynamics in response to a self-limited challenge with elevated neutrophil recruitment following zymosan challenge and significantly higher exudate monocytes/macrophages levels at the peak of inflammation, when compared with their WT littermates. These results, together with the finding that exudate pro-inflammatory LM levels (vide infra) were elevated in TKO mice, indicate that TKO mice display a pro-inflammatory phenotype. Our present findings are concurrent with those made in models of non-resolving (8) and delayed-resolving (16) inflammation models; these data underscore the contribution of CYP1 enzymes to inflammation and its timely resolution.
Interestingly, statistically significant differences in LM levels between TKO and WT mice were not found in peritoneal cells at baseline, but only after zymosan stimulation conditions. This observation is consistent with the fact that it is well known CYP1 levels in immune tissues are very low or negligible under basal conditions (55). After zymosan stimulation, an endogenous inducer of CYP1, believed to be one or more LMs, or after treatment of the animal with a foreign chemical such as a PAH or dioxin, then CYP1 levels in immune cells become dramatically increased.
After 6 h of zymosan challenge, compared with WT, TKO peritoneal exudates showed a trend of increased prostaglandins D2, E2 and F2α, along with a 2.3-fold increase in LTB4 levels; on the other hand, relative to WT mice, TKO exudates showed a ~6-fold decrease in PD1 levels and noticeable decreases in 12-HETE, 14-HDHA and 15-HEPE. Following 9 h of zymosan-initiated inflammation, TKO demonstrated statistically significant differences from WT exudates for eight chemicals: strikingly increased LTB4 and decreased PD1 levels, and decreased amounts of 5-HETE, 14-HDHA, 17-HDHA, 12-HEPE, 15-HEPE and 18-HEPE levels. The reason for the diminished levels in AA-derived 5-HETE in TKO inflammatory exudates after 9 h of zymosan (Fig. 4) is not presently known; the small decrease in 5-HETE could be caused by various factors. One possibility is perhaps lowered overall ALOX5 activity is coupled with more efficient conversion of 5-HpETE to LTB4; another possibility is that at least a portion of 5-HETE formation represents the direct metabolism of AA by CYP1 (10,26).
These results indicate that, prior to challenge, contribution of the three CYP1 enzymes to LM biosynthesis and/or further metabolism is not significant, indicating that resident peritoneal cells do not express functionally appreciable amounts of these enzymes at levels that would regulate LM levels in the absence of challenge. Upon challenge, leukocytes are then recruited to the peritoneal cavity; in particular polymorphonuclear leukocytes, which possess the CYP1 enzymes, and which lead to alterations in the exudate LM profiles in WT and TKO mice. Future studies will need to address the contribution of the CYP1 enzymes and the role(s) of dynamic cellular traffic at the site of inflammation and their contribution to pro-resolving mediator biosynthesis during inflammation resolution.
We conclude that one or more CYP1 enzymes participate highly significantly in at least 8 specific metabolic steps of LM pathways in the acute inflammatory response. Furthermore, our data indicate that CYP1 monooxygenases contribute to the overall balance of local autacoids, i.e. deletion of an inactivation pathway can give rise to accumulation of a specific LM during the time course of both the initiation and resolution phases of inflammation in the intact animal.
AA-derived LTB4 is known to be a potent chemo-attractant—generated from activated innate immune cells such as neutrophils, macrophages and mast cells (42) with mice lacking either 5-LOX or LTA4 hydrolase; these two enzymes are responsible for biosynthesis of this potent mediator, displaying decreased leukocyte recruitment following zymosan challenge (4). Thus, our finding of zymosan-elicited increases in the number of these three cell types in TKO compared with WT exudates is consistent with the subsequent observation of increased LTB4 levels in TKO mice following zymosan challenge.
TKO peritoneal and bone marrow neutrophils ex vivo show a diminished capacity to metabolize LTB4, but this effect appears small, relative to the ~4-fold increase in LTB4 observed in peritoneal exudates from TKO mice. It is not known with certainty whether this increase reflects a requirement of CYP1 for LTB4 oxidation in vivo in these mice, or whether it reflects some compensatory decreased level of a human CYP4 enzyme that is known to metabolize LTB4 in neutrophils (56). Curiously, CYP4A14 and CYP4A10 mRNA expression was found to be 2- to 3-fold lower in liver of Cyp1a2(–/–) single-knockout mice as compared with WT mice (54); however, when TKO was compared with WT liver under more rigorous microarray conditions in a later study (15), no mouse CYP4 mRNA was found to be significantly up- or down-regulated.
Alternatively, increased LTB4 levels in TKO mice might be explained by the increase in neutrophils present in TKO exudates rather than by LTB4 catabolism. Nevertheless, there appears to be an intriguing CYP1-mediated effect on TKO cellular dynamics in the peritoneal cavity as a result of inflammation; the present study does not determine with certainty the mechanism of this effect. In any event, CYP1 participation in the step from LTB4 to 20-OH-LTB4 appears to be very highly likely.
It is not clear that differences in LTB4 metabolism in neutrophils ex vivo (Fig. 6) can account for the increased LTB4 observed in the intact mouse; this is only one of several possible explanations. For example, LTB4 could be higher because the higher number of neutrophils present in the TKO exudate might produce greater amounts of LTB4. Another possibility is that LTB4 levels could be greater due to compensatory up-regulation of lipoxygenases, or effects secondary to the clearly altered state of inflammation that exists in the TKO peritoneal cavity. Even in isolated neutrophils ex vivo, it remains to be determined with absolute certainty that any of the CYP1 enzymes is directly responsible for altered LM metabolism.
Conversion of 20-OH-LTB4 to 20-COOH-LTB4 in human neutrophils is the main route of LTB4 inactivation (56); this finding was also corroborated in Fig. 5, whereas this enzymatic reaction appears not to occur in mouse neutrophils (Fig. 6). It has been known for more than two decades that even one altered amino-acid residue can dramatically modify P450 substrate specificity (27); this is the major reason why substrate specificities of various P450 enzymes differ between mouse and human—especially among members of the CYP2, CYP3 and CYP4 families (39) in which orthologs between the two species cannot be determined conclusively. In addition to ω-oxidation pathways, an alternative dehydrogenation pathway has been described for the inactivation of LTB4 which involves the dehydrogenation of the hydroxyl group on C-12 by 12-hydroxydehydrogenase (PGR/LTB4DH) to produce 12-oxo-LTB4 (11,61).
The role of CYP1 enzymes in the inactivation of LTB4 in mouse neutrophils is of interest and the main point of the experiments described herein. For this present report, we first investigated the pathway known for LTB4 inactivation in human neutrophils that express the human orthologs of these enzymes (56). Here we showed that human neutrophils converted LTB4 to the P450-mediated metabolites 20-OH-LTB4 and 20-COOH-LTB4, as reported elsewhere (56). Formation of these two metabolites are described first, in order to follow sequential metabolic steps regulated by distinct enzymes. The formation of 20-OH-LTB4 occurs first and is primarily regulated by CYP1 enzymes, which in humans is then converted by CYP4 enzymes to 20-COOH-LTB4 (56). For a direct comparison, we isolated mouse neutrophil populations obtained from two distinct sites: peritoneum (collected in response to zymosan challenge) and bone marrow polymorphonuclear leukocytes; each converted LTB4 to 20-OH-LTB4. Of note, 20-COOH-LTB4 was not produced in these incubations from two mouse neutrophil populations—unlike human neutrophils. Quantification of both LTB4 and 20-OH-LTB4 levels in these incubations demonstrated that, in the absence of the CYP1 enzymes (in TKO mice) there was an accumulation of LTB4 with a higher recovery. These results indicate that CYP1 enzymes are important in the inactivation of this initiating signal, which is also pro-inflammatory via ω-hydroxylation in mice. This is in line with the elevated LTB4 levels found within inflammatory exudates from TKO mice, in which we also found elevated polymorphonuclear leukocyte recruitment to the peritoneum upon zymosan challenge.
Although LTB4 biosynthesis in self-limited inflammatory exudates is not restricted to neutrophils, in this model LTB4 production coincides with increased exudate neutrophil levels (14), and in the absence of CYP1 enzymes LTB4 levels accumulated at the site of inflammation that led to the exaggerated inflammatory response (Fig. 2). Together, these findings suggest that CYP1 enzymes in phagocytes exert a pivotal role in inflammation-resolution.
Absence of CYP1 in TKO mice results in lowered formation of the DHA-derived 17S-HDHA It is noteworthy that, in human polymorphonuclear leukocytes (51), double dioxygenation via a second lipoxygenase produces 10,17-diHpDHA, which is then reduced by peroxidase activity to form 10S,17S-dihydroxydocosa-4Z,7Z,11E,13E,15Z,19Z-hexaenoic acid (10S,17S-diHDHA), an isomer of neuroprotectin NPD1/PD1 (32). It would also be possible to produce this PD1 isomer via a second sequential P450 step. e.g. P450-mediated 17S-HDHA conversion to the 10,17-diHDHA isomer of PD1. The double geometry of this isomer is different from an enzymatic epoxide-dependent pathway and epoxide-hydrolase reaction (32,52), which can then produce PD1 and other specialized pro-resolving mediators as their main route in human leukocytes. Also during the resolution phase of inflammation, absence of CYP1 in TKO mice leads to lowered formation of DHA-derived PD1.
Finally, the present study shows that CYP1 contributes to the levels of EPA-derived 15-HEPE and 12-HEPE, as well as involvement in the oxidative step from EPA-derived 18-HpEPE to 18R-HEPE. This latter step is a bottleneck to the generation of resolvins of the E-series. Thus, CYP1 ablation might also affect production of a number of pro-resolving LMs, in addition to the ones mentioned above that include the AA-derived lipoxins, as well as the D-series resolvins. CYP1 ablation appears to supply the required precursors, i.e. 17-HDHA and 15-HETE that are then further converted by lipoxygenases to D-series resolvins and lipoxins, respectively (23,40,49).
Metabololipidomic analysis of the products made ex vivo by exudate leukocytes, comparing WT and TKO mice, demonstrated a significant reduction in TKO LM precursors (i.e. 17-HDHA, 14-HDHA, 15HETE, 12-HETE and 5-HETE) (Fig. 7). These results are in line with the hypothesis that deletion of CYP1 enzymes is, at least in part, responsible for the differences found in exudate LM levels between WT and TKO mice.
Of note in the present analysis, although we found a significant decrease in 18-HEPE production in TKO mice, this did not correlate with a parallel drop in RvE2 levels, an observation that may reflect a difference in the further distal conversion of RvE2 in TKO mice, which leads to an apparent accumulation of this mediator in the inflammatory exudates. Although in the present study we see no detectable differences between TKO and WT resolvin levels, this may reflect issues of detection limits for these mediators.
TKO mice exhibited ~2-fold lower levels of LXA4 than WT mice at 6 h, but not at 9 h (Fig. 4), of zymosan. This finding suggests that LXA4 formation from 15S-hydroxy-5(6)-epoxy-ETE, at least initially, might involve contributions from CYP1 enzyme(s), but then other oxidative enzymes perhaps might compensate to continue producing sufficient LXA4 levels after 9 h of zymosan-initiated inflammation. Of possible relevance, LXA4 itself (46) as well as two downstream metabolites of LTA4—5S,6R-DHETE and 5S,6S-DHETE—(9) have been suggested to act as endogenous ligands for aryl hydrocarbon receptor (AHR), a transcription factor involved in regulating all three CYP1 genes (34,37). AHR-dependent activity is highly likely to be required during the acute inflammatory response in these peritoneal exudate cells.
In addition to the TKO mouse described herein, numerous other P450 knockout mouse lines have been generated—including to date Cyp1b1(–/–) (3), Cyp2e1(–/–) (59), Cyp2j5(–/–) (1) and Cyp4a10(–/–) (33) single-knockouts, Cyp1a1/1a2(–/–), and Cyp1a1/1b1(–/–) and Cyp1a2/1b1(–/–) double-knockouts (58) and, more recently, ablation of the entire Cyp2c (47), Cyp2d (48) and Cyp3a (19,60) gene subfamily clusters. Experiments similar to the present study should now be possible to identify participation of the specific CYP1A1, CYP1A2, CYP1B1, CYP2, CYP3 and CYP4 enzymes in distinct steps of the LM metabolic pathways during inflammation. Most likely, as we have found with the CYP1 enzymes, numerous LM steps will be altered in the absence of the CYP2, CYP3 and CYP4 monooxygenases. Moreover, we predict that redundancy of CYP enzymes for many of these LM biosynthesis steps will likely be found during the acute inflammatory response.
Whereas mouse vs human CYP1 enzymes display quite similar substrate specificities and inducers that up-regulate CYP1 expression (34), substrate specificities vary to a much greater degree in the CYP2, CYP3 and CYP4 families, because many gene-duplication events have occurred such that one cannot assign orthologs between the mouse and human genomes (39). Hence, we predict that “humanized” P450 mouse lines—in which the “knocked-in” human specific CYP2, CYP3 or CYP4 gene is expressed in place of the mouse ortholog—will provide the ultimate tool for further understanding the role of each human CYP2, CYP3 and CYP4 monooxygenase during LM biosynthesis/inactivation. Furthermore, use of such tools would directly confirm the clinical importance of each P450 enzyme in a specific LM biosynthetic step. Identification of such steps should be useful in the future for drug targeting and, as such, promise a strong therapeutic potential.
Humanized P450 mouse lines so far reported include hCYP1A1/1A2 (6,22,53), hCYP1B1 (Frank Gonzalez, personal communication), hCYP2C9 (47), hCYP2D6 (12), hCYP2E1 (7), and hCYP3A4 (17). Of note, the first humanized hCYP2D6 (12) and hCYP3A4 (17) mouse lines had the human gene inserted into the mouse genome that included all of the mouse Cyp2d and Cyp3a genes.
It should be mentioned that, in the present study, we screened for changes in the profiles and levels of several novel LMs and pathway markers from the AA metabolome (prostaglandins, leukotrienes, lipoxins), and the DHA and EPA metabolomes (resolvins, protectins and maresins), as well as their pathway markers relevant during the acute inflammatory response and its resolution. Whether CYP1 monooxygenases participate in metabolic steps involving any of the more than 125 other LMs during inflammation have not yet been assessed in these TKO mice. Moreover, the present study monitored the levels of basal peritoneal cells vs zymosan-initiated inflammatory exudates. Dozens of other cell types—plus numerous LM-mediated physiological as well as pathological stimuli (37,38,43), in addition to zymosan-initiated inflammation—also remain to be scrutinized via this approach in future studies.
In summary, our hypothesis was that global ablation of all three CYP1 enzymes combined, and comparison of genetically-modified TKO mice with WT mice, would uncover a number of disruptions in the signature profiles of LM pathways during an acute inflammatory response. Hence, to this end, we compared basal CYP1 activities as well as zymosan-stimulated CYP1 activities. Whereas no statistically significant differences were found in untreated baseline TKO vs WT peritoneal cells, 8 statistically significant alterations were uncovered between TKO and WT peritoneal exudates during inflammation. Specifically, compared with WT, TKO mice revealed statistically significant large increases in LTB4 and decreases in PD1 levels; TKO also showed significant decreased amounts of 5-HETE, 14-HDHA, 17-HDHA, 12-HEPE, 15-HEPE and 18R-HEPE. Because of CYP1 participation in these metabolic steps and absence of CYP1 in TKO mice, the present study shows that one or more of the CYP1 enzymes play(s) a role in polyunsaturated fatty acid metabolism, in addition to further LM metabolism by cyclooxygenases and ALOXs; the possibility exists of direct CYP1 involvement. Further experiments will be required to determine the precise step at which each CYP1 monooxygenase participates in order to effect the 8 changes in LM levels observed in the present study.
Compared with WT, TKO mice also show substantial differences in cell population in response to zymosan. It is conceivable that—at one or more of these metabolic steps—CYP1 enzymes might clear metabolites related to the zymosan challenge, rather than metabolizing LMs directly; failure to clear those metabolites in the absence of CYP1 might lead to an alteration in local cellular response which, in turn, might produce a change in LM mediator production by cyclooxygenases and ALOXs.
Therefore, future experiments—in zymosan-challenged mice genetically lacking only one of the Cyp1a1, Cyp1a2 or Cyp1b1 genes—will be necessary to further dissect which specific CYP1 monooxygenase participates in each particular metabolic step. Because of the high degree of conserved functions and substrate specificities known to exist between the three mouse and three human CYP1 enzymes, such a reductionist approach should lead to elucidation of clinically important novel drug targets.
Supplementary Material
Acknowledgments
We thank our colleagues, especially Larry Marnett (Vanderbilt) for valuable discussions and/or careful reading of the manuscript. We are indebted to Dr. Frank J Gonzalez for sharing his Cyp1b1(–/–) knockout line more than a decade ago, which has helped immensely in our combined-knockout mouse breeding studies.
Grant Support: This project was funded by Cystic Fibrosis Foundation RDP Center Component II Grant (C.L.K.), National Institutes of General Medical Sciences [Grant P01 GM095467] (C.N.S.), and National Institute of Environmental Health Sciences [Grant T32 ES016646] (M.G.-P.), and [Grants R01 ES008147, R01 ES014403 and P30 ES06096] (D.W.N.).
Abbreviations used
- AA
arachidonic acid
- Cyp1
cytochrome P450 1 gene family encoding three enzymes
- CYP1A1/CYP1A2/CYP1B1
cytochrome P450 1A1/1A2/1B1 mRNA & protein
- Cyp1a1, Cyp1a2, Cyp1b1
cytochrome P450 (family 1) genes in mouse
- DHA
docosahexaenoic acid
- EPA
eicosapentaenoic acid
- EPI
enhanced product ion
- ETEs
eicosatetraenoic acids
- GC-MS/MS
gas chromatography coupled with tandem mass spectrometry
- HpDHAs
hydroperoxydocosahexenoic acids
- HpETEs
hydroperoxyeicosatetraenoic acids
- HDHA
hydroxydocosahexenoic acids
- HEPEs
hydroxyeicosapentaenoic acids
- HETEs
hydroxyeicosatetraenoic acids
- HpEPEs
hydroperoxyeicosapentaenoic acids
- LC/HPLC
high-performance liquid chromatography
- LC-UV
liquid chromatography-UV spectrometry
- LC-UV-MS/MS
liquid chromatography-UV coupled with tandem mass spectrometry
- LTA4, LTB4
leukotrienes A4, B4
- LXA4, LXB4
lipoxins A4, B4
- MaR1
Maresin (macrophage mediators in resolving inflammation, dihydroxydocosahexaenoic acid)
- MRM
multiple-reaction monitoring
- PD1 or [N]PD1
neuroprotectin/protectin D1
- PGH2, PGD2, PGE2, PGF2α
prostaglandins H2, D2, E2, F2α
- PTGS2
prostaglandin G synthase-2 (inducible cyclooxygenase-2; COX2)
- RvE1, RvE2, RvD1, RvD2, RvD3, RvD4, RvD5, RvD6
resolvins E1, E2. D1, D2, D3, D4, D5, D6
- TxA2, TxB2
thromboxanes A2, B2
- TKO
Cyp1a1/1a2/1b1(–/–) triple-knockout on >99.8% C57BL/6J background
- TOF
time-of-flight, used to determine mass-to-charge (m/z) ratio in mass spectrometer
- WT
C57BL/6J wild-type mouse
Footnotes
Present address of C. L. Karp: The Bill & Melinda Gates Foundation, Seattle, WA 98102
Conflict of interest: C.N.S. is an inventor on patents [resolvins] assigned to the Brigham and Women's Hospital and licensed to Resolvyx Pharmaceuticals. C.N.S. is scientific founder of Resolvyx Pharmaceuticals and owns equity in the company. The interests of C.N.S. were reviewed and are managed by the Brigham and Women's Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. The other coauthors have declared that no conflict of interest exists.
References
- 1.Athirakul K, Bradbury JA, Graves JP, DeGraff LM, Ma J, Zhao Y, Couse JF, Quigley R, Harder DR, Zhao X, Imig JD, Pedersen TL, Newman JW, Hammock BD, Conley AJ, Korach KS, Coffman TM, Zeldin DC. Increased blood pressure in mice lacking cytochrome P450 2J5. FASEB J. 2008;22:4096–4108. doi: 10.1096/fj.08-114413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bannenberg GL, Chiang N, Ariel A, Arita M, Tjonahen E, Gotlinger KH, Hong S, Serhan CN. Molecular circuits of resolution: formation and actions of resolvins and protectins. J Immunol. 2005;174:4345–4355. doi: 10.4049/jimmunol.174.7.4345. [DOI] [PubMed] [Google Scholar]
- 3.Buters JT, Sakai S, Richter T, Pineau T, Alexander DL, Savas U, Doehmer J, Ward JM, Jefcoate CR, Gonzalez FJ. Cytochrome P450 CYP1B1 determines susceptibility to 7,12-dimethylbenzo[a]anthracene-induced lymphomas. Proc Natl Acad Sci U. S. A. 1999;96:1977–1982. doi: 10.1073/pnas.96.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrum RS, Goulet JL, Snouwaert JN, Griffiths RJ, Koller BH. Determination of the contribution of cysteinyl leukotrienes and leukotriene B4 in acute inflammatory responses using 5-lipoxygenase- and leukotriene A4 hydrolase-deficient mice. J Immunol. 1999;163:6810–6819. [PubMed] [Google Scholar]
- 5.Campbell EL, Serhan CN, Colgan SP. Antimicrobial aspects of inflammatory resolution in the mucosa: a role for proresolving mediators. J Immunol. 2011;187:3475–3481. doi: 10.4049/jimmunol.1100150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung C, Ma X, Krausz KW, Kimura S, Feigenbaum L, Dalton TP, Nebert DW, Idle JR, Gonzalez FJ. Differential metabolism of 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) in mice humanized for CYP1A1 and CYP1A2. Chem Res Toxicol. 2005;18:1471–1478. doi: 10.1021/tx050136g. [DOI] [PubMed] [Google Scholar]
- 7.Cheung C, Yu AM, Ward JM, Krausz KW, Akiyama TE, Feigenbaum L, Gonzalez FJ. The CYP2E1-humanized transgenic mouse: role of CYP2E1 in acetaminophen hepatotoxicity. Drug Metab Dispos. 2005;33:449–457. doi: 10.1124/dmd.104.002402. [DOI] [PubMed] [Google Scholar]
- 8.Chiang N, Fredman G, Backhed F, Oh SF, Vickery T, Schmidt BA, Serhan CN. Infection regulates pro-resolving mediators that lower antibiotic requirements. Nature. 2012;484:524–528. doi: 10.1038/nature11042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiaro CR, Morales JL, Prabhu KS, Perdew GH. Leukotriene A4 metabolites are endogenous ligands for the AH receptor. Biochemistry. 2008;47:8445–8455. doi: 10.1021/bi800712f. [DOI] [PubMed] [Google Scholar]
- 10.Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. Metabolism of retinoids and arachidonic acid by human and mouse cytochrome P450 1B1. Drug Metab Dispos. 2004;32:840–847. doi: 10.1124/dmd.32.8.840. [DOI] [PubMed] [Google Scholar]
- 11.Clish CB, Levy BD, Chiang N, Tai HH, Serhan CN. Oxidoreductases in lipoxin A4 metabolic inactivation: a novel role for 15-ono-prostaglandin 13-reductase/leukotriene B4 12-hydroxydehydrogenase in inflammation. J Biol Chem. 2000;275:25372–25380. doi: 10.1074/jbc.M002863200. [DOI] [PubMed] [Google Scholar]
- 12.Corchero J, Granvil CP, Akiyama TE, Hayhurst GP, Pimprale S, Feigenbaum L, Idle JR, Gonzalez FJ. The CYP2D6 humanized mouse: effect of the human CYP2D6 transgene and HNF4α on the disposition of debrisoquine in the mouse. Mol Pharmacol. 2001;60:1260–1267. doi: 10.1124/mol.60.6.1260. [DOI] [PubMed] [Google Scholar]
- 13.Dalli J, Serhan CN. Specific lipid mediator signatures of human phagocytes: microparticles stimulate macrophage efferocytosis and pro-resolving mediators. Blood. 2012;120:e60–e72. doi: 10.1182/blood-2012-04-423525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dalli J, Winkler JW, Colas RA, Arnardottir H, Cheng CY, Chiang N, Petasis NA, Serhan CN. Resolvin D3 and aspirin-triggered resolvin D3 are potent immunoresolvents. Chem Biol. 2013;20:188–201. doi: 10.1016/j.chembiol.2012.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dragin N, Shi Z, Madan R, Karp CL, Sartor MA, Chen C, Gonzalez FJ, Nebert DW. Phenotype of the Cyp1a1/1a2/1b1(−/−) triple-knockout mouse. Mol Pharmacol. 2008;73:1844–1856. doi: 10.1124/mol.108.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fredman G, Li Y, Dalli J, Chiang N, Serhan CN. Self-limited versus delayed resolution of acute inflammation: temporal regulation of pro-resolving mediators and microRNA. Sci Rep. 2012;2:639. doi: 10.1038/srep00639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Granvil CP, Yu AM, Elizondo G, Akiyama TE, Cheung C, Feigenbaum L, Krausz KW, Gonzalez FJ. Expression of the human CYP3A4 gene in the small intestine of transgenic mice: in vitro metabolism and pharmacokinetics of midazolam. Drug Metab Dispos. 2003;31:548–558. doi: 10.1124/dmd.31.5.548. [DOI] [PubMed] [Google Scholar]
- 18.Greene ER, Huang S, Serhan CN, Panigrahy D. Regulation of inflammation in cancer by eicosanoids. Prostaglandins Other Lipid Mediat. 2011;96:27–36. doi: 10.1016/j.prostaglandins.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hasegawa M, Kapelyukh Y, Tahara H, Seibler J, Rode A, Krueger S, Lee DN, Wolf CR, Scheer N. Quantitative prediction of human pregnane X receptor and cytochrome P450 3A4-mediated drug-drug interaction in a novel multiple humanized mouse line. Mol Pharmacol. 2011;80:518–528. doi: 10.1124/mol.111.071845. [DOI] [PubMed] [Google Scholar]
- 20.Hayaishi O, Katagiri M, Rothberg S. Mechanism of the pyrocatechase reaction. J Am Chem Soc. 1955;77:5450–5451. [Google Scholar]
- 21.Ji RR, Xu ZZ, Strichartz G, Serhan CN. Emerging roles of resolvins in the resolution of inflammation and pain. Trends Neurosci. 2011;34:599–609. doi: 10.1016/j.tins.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang Z, Dalton TP, Jin L, Wang B, Tsuneoka Y, Shertzer HG, Deka R, Nebert DW. Toward the evaluation of function in genetic variability: characterizing human SNP frequencies and establishing BAC-transgenic mice carrying the human CYP1A1_CYP1A2 locus. Hum Mutat. 2005;25:196–206. doi: 10.1002/humu.20134. [DOI] [PubMed] [Google Scholar]
- 23.Kim TH, Kim GD, Jin YH, Park YS, Park CS. Omega-3 fatty acid-derived mediator, resolvin E1, ameliorates 2,4-dinitrofluorobenzene-induced atopic dermatitis in NC/Nga mice. Int Immunopharmacol. 2012;14:384–391. doi: 10.1016/j.intimp.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 24.Kolaczkowska E, Chadzinska M, Scislowska-Czarnecka A, Plytycz B, Opdenakker G, Arnold B. Gelatinase B/matrix metalloproteinase-9 contributes to cellular infiltration in a murine model of zymosan peritonitis. Immunobiology. 2006;211:137–148. doi: 10.1016/j.imbio.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Kuhn H, Wiesner R, Alder L, Fitzsimmons BJ, Rokach J, Brash AR. Formation of lipoxin B by the pure reticulocyte lipoxygenase via sequential oxygenation of the substrate. Eur J Biochem. 1987;169:593–601. doi: 10.1111/j.1432-1033.1987.tb13650.x. [DOI] [PubMed] [Google Scholar]
- 26.Lee CA, Lawrence BP, Kerkvliet NI, Rifkind AB. 2,3,7,8-Tetrachlorodibenzo-p-dioxin induction of cytochrome P450-dependent arachidonic acid metabolism in mouse liver microsomes: evidence for species-specific differences in responses. Toxicol Appl. Pharmacol. 1998;153:1–11. doi: 10.1006/taap.1998.8468. [DOI] [PubMed] [Google Scholar]
- 27.Lindberg RL, Negishi M. Alteration of mouse cytochrome P450coh substrate specificity by mutation of a single amino-acid residue. Nature. 1989;339:632–634. doi: 10.1038/339632a0. [DOI] [PubMed] [Google Scholar]
- 28.Lowell CA, Fumagalli L, Berton G. Deficiency of SRC family kinases p59/61HCK and p58c-FGR results in defective adhesion-dependent neutrophil functions. J Cell Biol. 1996;133:895–910. doi: 10.1083/jcb.133.4.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mas E, Croft KD, Zahra P, Barden A, Mori TA. Resolvins D1, D2, and other mediators of self-limited resolution of inflammation in human blood following n-3 fatty acid supplementation. Clin Chem. 2012;58:1476–1484. doi: 10.1373/clinchem.2012.190199. [DOI] [PubMed] [Google Scholar]
- 30.Mason HS. Mechanisms of oxygen metabolism. Science. 1957;125:1185–1188. doi: 10.1126/science.125.3259.1185. [DOI] [PubMed] [Google Scholar]
- 31.Mason HS, Fowlkes WL, Peterson E. Oxygen transfer and electron transport by the phenolase complex. J Am Chem Soc. 1955;77:2914–2915. [Google Scholar]
- 32.Miyata J, Fukunaga K, Iwamoto R, Isobe Y, Niimi K, Takamiya R, Takihara T, Tomomatsu K, Suzuki Y, Oguma T, Sayama K, Arai H, Betsuyaku T, Arita M, Asano K. Dysregulated synthesis of protectin D1 in eosinophils from patients with severe asthma. J Allergy Clin Immunol. 2012 doi: 10.1016/j.jaci.2012.07.048. in press. [DOI] [PubMed] [Google Scholar]
- 33.Nakagawa K, Holla VR, Wei Y, Wang WH, Gatica A, Wei S, Mei S, Miller CM, Cha DR, Price E, Jr, Zent R, Pozzi A, Breyer MD, Guan Y, Falck JR, Waterman MR, Capdevila JH. Salt-sensitive hypertension is associated with dysfunctional Cyp4a10 gene and kidney epithelial sodium channel. J Clin Invest. 2006;116:1696–1702. doi: 10.1172/JCI27546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. Role of aryl hydrocarbon receptor-mediated induction of the CYP1 enzymes in environmental toxicity and cancer. J Biol Chem. 2004;279:23847–23850. doi: 10.1074/jbc.R400004200. [DOI] [PubMed] [Google Scholar]
- 35.Nebert DW, Dalton TP, Stuart GW, Carvan MJ., III “Gene-swap knock-in” cassette in mice to study allelic differences in human genes. Ann. N Y. Acad Sci. 2000;919:148–170. doi: 10.1111/j.1749-6632.2000.tb06876.x. [DOI] [PubMed] [Google Scholar]
- 36.Nebert DW, Galvez-Peralta M, Shi Z, Dragin N. Inbreeding and epigenetics: beneficial as well as deleterious effects. Nat Rev Genet. 2010;11:662. doi: 10.1038/nrg2664-c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nebert DW, Karp CL. Endogenous functions of the aryl hydrocarbon receptor (AHR): intersection of cytochrome P450 1 (CYP1)-metabolized eicosanoids and AHR biology. J Biol Chem. 2008;283:36061–36065. doi: 10.1074/jbc.R800053200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nebert DW, Wikvall K, Miller WL. Human cytochromes P450 in health and disease. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120431. doi: 10.1098/rstb.2012.0431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes, and alternative-splice variants. Pharmacogenetics. 2004;14:1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 40.Oh SF, Dona M, Fredman G, Krishnamoorthy S, Irimia D, Serhan CN. Resolvin E2 formation and impact on inflammation resolution. J Immunol. 2012;188:4527–4534. doi: 10.4049/jimmunol.1103652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh SF, Pillai PS, Recchiuti A, Yang R, Serhan CN. Pro-resolving actions and stereoselective biosynthesis of 18-S E-series resolvins in human leukocytes and mouse inflammation. J. Clin. Invest. 2011;121:569–581. doi: 10.1172/JCI42545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohnishi H, Miyahara N, Gelfand EW. The role of leukotriene B4 in allergic diseases. Allergol. Int. 2008;57:291–298. doi: 10.2332/allergolint.08-RAI-0019. [DOI] [PubMed] [Google Scholar]
- 43.Panigrahy D, Edin ML, Lee CR, Huang S, Bielenberg DR, Butterfield CE, Barnes CM, Mammoto A, Mammoto T, Luria A, Benny O, Chaponis DM, Dudley AC, Greene ER, Vergilio JA, Pietramaggiori G, Scherer-Pietramaggiori SS, Short SM, Seth M, Lih FB, Tomer KB, Yang J, Schwendener RA, Hammock BD, Falck JR, Manthati VL, Ingber DE, Kaipainen A, D'Amore PA, Kieran MW, Zeldin DC. Epoxyeicosanoids stimulate multiorgan metastasis and tumor dormancy escape in mice. J Clin Invest. 2012;122:178–191. doi: 10.1172/JCI58128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proctor KG, Falck JR, Capdevila J. Intestinal vasodilation by epoxyeicosatrienoic acids: arachidonic acid metabolites produced by a cytochrome P450 monooxygenase. Circ. Res. 1987;60:50–59. doi: 10.1161/01.res.60.1.50. [DOI] [PubMed] [Google Scholar]
- 45.Samuelsson B. Role of basic science in the development of new medicines: examples from the eicosanoid field. J Biol Chem. 2012;287:10070–10080. doi: 10.1074/jbc.X112.351437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaldach CM, Riby J, Bjeldanes LF. Lipoxin A4: a new class of ligand for the AH receptor. Biochemistry. 1999;38:7594–7600. doi: 10.1021/bi982861e. [DOI] [PubMed] [Google Scholar]
- 47.Scheer N, Kapelyukh Y, Chatham L, Rode A, Buechel S, Wolf CR. Generation and characterization of novel cytochrome P450 Cyp2c gene cluster knockout and CYP2C9 humanized mouse lines. Mol Pharmacol. 2012;82:1022–1029. doi: 10.1124/mol.112.080036. [DOI] [PubMed] [Google Scholar]
- 48.Scheer N, Kapelyukh Y, McEwan J, Beuger V, Stanley LA, Rode A, Wolf CR. Modeling human cytochrome P450 2D6 metabolism and drug-drug interaction by a novel panel of knockout and humanized mouse lines. Mol Pharmacol. 2012;81:63–72. doi: 10.1124/mol.111.075192. [DOI] [PubMed] [Google Scholar]
- 49.Serhan CN. Novel lipid mediators and resolution mechanisms in acute inflammation: to resolve or not? Am J Pathol. 2010;177:1576–1591. doi: 10.2353/ajpath.2010.100322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serhan CN, Fredman G, Yang R, Karamnov S, Belayev LS, Bazan NG, Zhu M, Winkler JW, Petasis NA. Novel pro-resolving aspirin-triggered DHA pathway. Chem Biol. 2011;18:976–987. doi: 10.1016/j.chembiol.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Serhan CN, Gotlinger K, Hong S, Lu Y, Siegelman J, Baer T, Yang R, Colgan SP, Petasis NA. Anti-inflammatory actions of neuroprotectin D1/protectin D1 and its natural stereoisomers: assignments of dihydroxy-containing docosatrienes. J Immunol. 2006;176:1848–1859. doi: 10.4049/jimmunol.176.3.1848. [DOI] [PubMed] [Google Scholar]
- 52.Serhan CN, Petasis NA. Resolvins and protectins in inflammation resolution. Chem Rev. 2011;111:5922–5943. doi: 10.1021/cr100396c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shi Z, Chen Y, Dong H, Amos-Kroohs RM, Nebert DW. Generation of a ‘humanized’ hCYP1A1_1A2_Cyp1a1/1a2(−/−)_Ahrd mouse line harboring the poor-affinity aryl hydrocarbon receptor. Biochem Biophys Res Commun. 2008;376:775–780. doi: 10.1016/j.bbrc.2008.09.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith AG, Davies R, Dalton TP, Miller ML, Judah D, Riley J, Gant T, Nebert DW. Intrinsic hepatic phenotype associated with the Cyp1a2 gene as shown by cDNA expression microarray analysis of the knockout mouse. EHP. Toxicogenomics. 2003;111:45–51. [PubMed] [Google Scholar]
- 55.Stejskalova L, Dvorak Z, Pavek P. Endogenous and exogenous ligands of aryl hydrocarbon receptor: current state of art. Curr Drug Metab. 2011;12:198–212. doi: 10.2174/138920011795016818. [DOI] [PubMed] [Google Scholar]
- 56.Sumimoto H, Minakami S. Oxidation of 20-hydroxyleukotriene B4 to 20-carboxyleukotriene B4 by human neutrophil microsomes. Role of aldehyde dehydrogenase and leukotriene B4 ω-hydroxylase (cytochrome P-450LTBω) in leukotriene B4 ω-oxidation. J Biol Chem. 1990;265:4348–4353. [PubMed] [Google Scholar]
- 57.Szczur K, Zheng Y, Filippi MD. The small Rho GTPase Cdc42 regulates neutrophil polarity via CD11b integrin signaling. Blood. 2009;114:4527–4537. doi: 10.1182/blood-2008-12-195164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uno S, Dalton TP, Dragin N, Curran CP, Derkenne S, Miller ML, Shertzer HG, Gonzalez FJ, Nebert DW. Oral benzo[a]pyrene in Cyp1 knockout mouse lines: CYP1A1 important in detoxication, CYP1B1 metabolism required for immune damage independent of total-body burden and clearance rate. Mol Pharmacol. 2006;69:1103–1114. doi: 10.1124/mol.105.021501. [DOI] [PubMed] [Google Scholar]
- 59.Valentine JL, Lee SS, Seaton MJ, Asgharian B, Farris G, Corton JC, Gonzalez FJ, Medinsky MA. Reduction of benzene metabolism and toxicity in mice that lack CYP2E1 expression. Toxicol Appl. Pharmacol. 1996;141:205–213. doi: 10.1006/taap.1996.0277. [DOI] [PubMed] [Google Scholar]
- 60.van Herwaarden AE, Wagenaar E, van der Kruijssen CM, van Waterschoot RA, Smit JW, Song JY, van der Falk MA, van Tellingen O, van der Hoorn JW, Rosing H, Beijnen JH, Schinkel AH. Knockout of cytochrome P450 3A yields new mouse models for understanding xenobiotic metabolism. J Clin Invest. 2007;117:3583–3592. doi: 10.1172/JCI33435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yokomizo T, Izumi T, Takahashi T, Kasama T, Kobayashi Y, Sato F, Taketani Y, Shimizu T. Enzymatic inactivation of leukotriene B4 by a novel enzyme found in the porcine kidney. Purification and properties of leukotriene B4 12-hydroxydehydrogenase. J Biol Chem. 1993;268:18128–18135. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.