Abstract
Monitoring of stem cell differentiation and pluripotency is an important step for the practical use of stem cells in the field of regenerative medicine. Hence, a new non-destructive detection tool capable of in situ monitoring of stem cell differentiation is highly needed. In this study, we report a 3D graphene oxide-encapsulated gold nanoparticle that is very effective for the detection of the differentiation potential of neural stem cells (NSCs) based on surface-enhanced Raman spectroscopy (SERS). A new material, 3D GO-encapsulated gold nanoparticle, is developed to induce the double enhancement effect of graphene oxide and gold nanoparticle on SERS signals which is only effective for undifferentiated NSCs. The Raman peaks achieved from undifferentiated NSCs on the graphene oxide (GO)-encapsulated gold nanoparticles were 3.5 times higher than peaks obtained from normal metal structures and were clearly distinguishable from those of differentiated cells. The number of C=C bonds and the raman instensity at 1656cm−1 was found to show a positive correlation, which matches the differentiation state of the NSCs. Moreover, the substrate composed of 3D GO-encapsulated gold nanoparticles was also effective at distinguishing the differentiation state of single NSC by using electrochemical and electrical techniques. Hence, the proposed technique can be used as a powerful non-destructive in situ monitoring tool for the identification of the differentiation potential of various kinds of stem cells (mesenchymal, hematopoietic, and neural stem cells).
Keywords: Graphene-encapsulated nanomaterials, SERS, electrochemical method, Stem cells, Differentiation, in situ monitoring
1. Introduction
The identification of state of stem cell differentiation is an essential step for the practical use of stem cells in the field of regenerative medicine. The monitoring of cell differentiation is incredibly important especially for the application of neural stem cells (NSCs) to treat devastating diseases accurately such as Alzheimer’s disease [1], Parkinson’s disease [1–3], and spinal cord injuries [4,5]. Many conventional tools have been used to detect the differentiation potential of NSCs, as well as to distinguish undifferentiated NSCs from differentiated neuronal and glial cells [6–10]. However, most of these techniques such as immunostaining and fluorescence-activated cell sorting (FACS) methods necessitate the conjugation of fluorescence dyes onto cells, which are expensive, laborious, time-consuming and even potentially toxic making them unsuitable for clinical use. Moreover, cellular components such as RNAs, DNAs and proteins which are essential for accurate biological analysis are difficult to obtain while maintaining cell viability and are therefore, generally collected after cell lysis. For this reason, there has been an urgent need to develop a highly sensitive and non-invasive tool that enables in situ monitoring of differentiation potential of NSCs and distinguishing between their undifferentiated and differentiated states.
Meanwhile, surface-enhanced Raman spectroscopy (SERS) has been developed as a powerful chemical/biological analysis tool that is reagent-free, as well as rapid and non-destructive [11–13]. A variety of SERS-based techniques have been reported for the detection of disease-related molecules [14], biomolecules [15–17], cancer markers [18] and even for in vivo tumor detection [19,20]. We also have reported a SERS-based cell chip that was capable of analyzing the chemical characteristics of cancer cells, the efficiency of various kinds of anticancer drugs, and their time-dependent effects on human liver cancer (HepG2) cells [21]. The research was then further extended to the fabrication of an anodic aluminum oxide (AAO)-assisted homogeneous nanodot-array that allowed for the discrimination of live/dead cells, the identification of normal/cancer cells and the determination of cell-cycle stage with a high sensitivity and low variability in the SERS signals [22,23]. From these endeavors, we opened the possibility of using SERS techniques as a powerful cell analysis tool that could eliminate the need for any additional probes and/or dyes that would be essential for conventional in vitro analytical tools.
Interestingly, this superior characteristics of SERS as an efficient cell analysis tool can potentially be utilized to distinguish the differentiation status of stem cells. Since SERS is a noninvasive and non-destructive tool, the chemical/biological characteristics of stem cells can be analyzed using SERS-based tools while maintaining stem cell viability and making it suitable for in vivo studies [24]. The major issue faced by SERS-based cell analysis, however, is that the complex structure of living cells composed of many heavy molecules including chromosomes, proteins and lipids [25]. This leads to complex Raman spectra that are difficult to translate into actual chemical/biological information. Fortunately, it was found that undifferentiated stem cells generally have polyunsaturated membranes and unsaturated molecules that are rich in C=C bonds due to aromatic structures which would make it easier to distinguish undifferentiated from differentiated stem cell membrane/components [26,27]. Since Raman spectroscopy is very powerful in its ability to detect the chemical structure of the target analytes, the differentiation potential of stem cells can be efficiently determined using SERS analysis tools. More interestingly, recent studies have reported that graphene can adhere to molecules that contain aromatic structures (high numbers of C=C bonds) through π-π stacking and also functions as an SERS-enhancer itself without the need for conventional metal structures, indicating that graphene can be useful for the detection of specific molecules and/or structures using SERS methods [28,29].
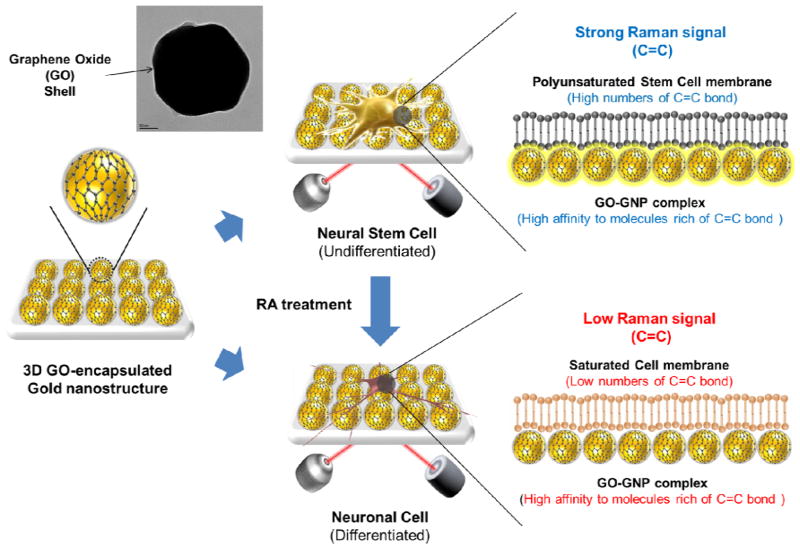
Therefore, in this study, we report a spectroelectrochemical method utilizing 3D graphene oxide-encapsulated gold nanoparticles for the effective detection of the differentiation potential of mouse neural stem cells (mNSCs) (Figure 1). A new material, 3D GO-encapsulated gold nanoparticle, is developed to induce the combination of the chemical/electromagnetic enhancement of Raman signals that is only effective for the quantified stem cell marker (i.e. high degree of C=C saturation). We focused on the absolute and relative values of the Raman signals (Raman intensity of C=C / Raman intensity of C-H) of mNSCs to clearly distinguish its undifferentiated and differentiated states. Due to the high affinity of GO molecules to stem cell markers, the Raman signal can be significantly enhanced for the undifferentiated mNSCs which can eliminate the hard process for the analysis of complex Raman signals generated from stem cells that is normally used in previous studies. Cidofovir and hydroxybenzoate derivatives abundant in undifferentiated stem cells were selected in order to prove the Raman-enhancing ability of 3D GNP-GO complex specific for C=C bond rich molecules, which is important for monitoring the differentiation potential of mNSCs based on SERS tools. Additionally, since GO and its derivatives were found to be excellent for the detection of electrical and electrochemical signals of target molecules, a new electrochemical method utilizing microgap modified with GO-encapsulated nanoparticles was proposed to identify the single undifferentiated and differentiated mNSC by analyzing its own electrochemical features.
Figure 1.
Detection strategy to monitor differentiation of mNSCs. Cells were allowed to attach on the fabricated substrate and detection time for Raman spectra was 1s for all experiments.
2. Materials and Methods
2.1. Materials
GNPs 60 nm in diameter was purchased from BBInternational (Newyork, UK). Aminopropyltriethoxysilane (APTES) and cysteamine hydrochloride were obtained from Sigma-Aldrich (Germany). Single layer graphene oxide (275mg/L) dispersed in water was purchased from Graphene Supermarket (U.S.A.). Dulbecco’s Phosphate-buffered saline (D-PBS) was purchased from STEMCELL Technologies (U.S.A.). GIBCO Minimum Essential Medium (MEM) Alpha and antibiotics were purchased from Invitrogen (U.S.A). L-glutamine, poly-L-lysine (PLL), retinoic acid and 3.7% formaldehyde solution were obtained from Sigma-Aldrich (Germany). Monoclonal anti-mouse/rat nestin antibody, neuron-specific β-III tubulin antibody, Northernlights conjugated anti-mouse IgG emitting 514nm and 574nm were purchased from R&D SYSTEMS(Minneapolis, MN, U.S.A). A 4 well plastic chamber (Lab-Tek(R)) suitable for cell culture was obtained from Thermo fisher scientific (USA). Other chemicals used in this study were obtained commercially and were of reagent grade.
2.2. Cell culture and induction of stem cell differentiation
NE-4C neuroectodermal stem cells were purchased from ATCC (Rockville, MD). Cells were cultured in MEM Alpha supplemented with 10% FBS, 1% antibiotics and 4mM L-glutamine. Cell culture plates were pre-coated with PLL solution (4°C, 12h) prior to the cell seeding on its surface. Cells were maintained under the common cell culture conditions at 37°C in an atmosphere of 5% CO2. To induce stem differentiation, culture media containing 10−6 M retinoic acid (RA) was added and maintained for 3 days. Culture media was changed every 2 days. After confirmation of neurosphere formation and axon growth of mNSCs at day 10, cells were re-dispersed in culture media and seeded again on PLL-coated cell culture plate to achieve differentiated mNSCs.
2.3. Immunostaining of mNSCs
mNSCs were fixed with D-PBS solution containing 3.7% formaldehyde for 20 minutes at 25°C and blocked with D-PBS solution containing 10% normal horse serum, 1% BSA and 0.1% Triton X-100 for 30 minutes at 25°C. After the blocking step, primary antibodies diluted with D-PBS (10 μg/ml) that specifically bind to nestin and β-III tubulin were added to undifferentiated and differentiated NE-4C cells, respectively, and incubated overnight at 4°C. Northernlights conjugated anti-mouse IgG emitting 574nm and 514nm (10μg/ml) were then incubated with cells treated with nestin-specific and β-III tubulin-specific antibody for one hour in the dark, respectively. Finally, 4′,6-diamidino-2-phenylindole (DAPI) was applied and kept for 10 minutes to stain the nuclei of NE-4C cells. A D-PBS solution containing 0.1% BSA was used to wash cells between each step. Confocal laser microscopy (TCS SP2, Germany) was used to obtain the fluorescence images presented in this study.
2.4. MTT viability assay
Approximately 2.1×104 cells were seeded on the different substrates to investigate the mitochondrial activity of the cells using the MTT viability assay. After 48h of incubation, 20 μL of stock MTT (5 mg/mL) solution was added to each chamber, followed by incubation for 3h at 37°C and 5% CO2. Media were removed and formazan that existed in the cells was dissolved with dimethyl sulfoxide (DMSO). Absorbance was measured at 540nm using a Benchmark microplate reader (Bio-Rad, Mississauga, ON, Canada). All measurements were carried out in triplicate in three or more independent experiments.
2.5. Fabrication of 3D GO-encapsulated GNPs on ITO surface and cell chip
ITO-deposited glass substrates were sonicated for 20min using 0.1% Triton™ X-100, deionized water (DIW) and ethanol sequentially for perfect cleaning. The cleaned ITO surface was then sonicated again in a basic piranha solution (1:1:5, H2O2:NH4OH:H2O) for 3 hours. After washing with DIW and drying under a N2 stream, 5% APTES in 95% ethanol (95:5, C2H5OH: H2O) was added to the ITO electrode and incubated for 3h. After rinsing with 95% ethanol, the APTES absorbed ITO surfaces were heated at 100°C for 15min to cure organosilane molecules [30]. Next, a 1cm × 2cm × 0.5cm (length × width × height) chamber was attached on the APTES-functionalized ITO surface to fabricate cell chip chamber using polydimethylsiloxane (PDMS) [31]. GNPs with 60nm in diameter were then added to the cell chip and incubated for 24h at 4°C, followed by washing with DIW and drying under a N2 stream to remove unbound GNPs. GO was further encapsulated on the surface of GNP through electrostatic interaction between the positively charged amine surface of GNP and the negative charged GO as previously reported [32–34]. 10mM Cysteamine hydrochloride was then incubated for 24h at 25°C to obtain positively charged surface, followed by 3 washes with DIW. GO was ultrasoniated for 1h and centrifuged for 20min at 13200rpm to achieve single-layered GO with small size prior to use. GO was then applied to GNP immobilized on the ITO surface and incubated for 6h, followed by another 3 washes with DIW. The fabricated cell chip chamber was sterilized by 70% ethanol and UV for 12h. Finally, 1×105 undifferentiated or differentiated mNSCs were seeded on fabricated cell chip 24h prior to experiments using Raman spectroscopy.
2.6. Characterization of structures and cells based on AFM and SERS
AFM images and Raman spectra were obtained using NTEGRA spectra (AFM-Raman Spectrometer, NT-MDT, Russia) equipped with a liquid nitrogen-cooled CCD camera and an inverted microscope (Olympus IX71). Semi-contact mode was used to detect the topological characteristics of the bare ITO, ITO-60GNP and ITO-60GNP-GO 3D complex. The cantilever (NSG01) used in this study had a typical resonant frequency in the range of 115 to 190 kHz and a force constant of 2.5 to 10N/m. In the case of Raman spectroscopy (SERS), the resolution of the spectrometer in the XY plane was 200 nm and along the Z axis was 500 nm. Raman spectra were recorded using a NIR laser emitting light at a wavelength of 785 nm. The irradiation laser power was 3mW in the sample plane. The Raman spectrum obtained from the same substrate without cells was considered as a background which was the averaged signal of five scans of 1s time intervals from the same range of spectra obtained for cells. All of the Raman signals achieved from cells were subtracted by background spectra. The same method was used to detect the Raman signals of cidofovir (Cido) and methyl 4-hydroxybenzoate (M4H).
2.7. Fabrication of GO-GNP nanoparticles-modified microgap
The photoresist was first patterned on the substrate and Cr/Au (10/100 nm) was deposited on the same substrate. After the removal of remaining photoresist using acetone, photoresist was coated again and exposed to UV through 2nd photo mask for making selectively exposed area between micro-gap. After washing with DIW, PR-removed areas were exposed to O2 plasma for changing the surface to hydrophilic and then, 5% APTES in 95% ethanol (95:5, C2H5OH: H2O) was added and incubated for 2h. After rinsing with 95% ethanol, the APTES modified ITO were heated at 100°C for 15min to cure organosilane molecules. GNPs with 60nm in diameter were then added to the APTES modified area and incubated for 24h at 4°C, followed by washing with DIW and drying under a N2 stream to remove unbound GNPs. 10mM Cysteamine hydrochloride was then incubated for 24h at 25°C to obtain positively charged surface, followed by 3 washes with DIW. The single-layered GO was then finally added to encapsulate GNP on microgap.
2.8. Electrical and electrochemical measurement of single mNSC
Cyclic voltammetry (CV) were performed using a potentiostat (CHI-600, CHInstrments, Austin, TX, USA). The three-electrode system composed of a cell-attached microgap electrode, a platinum (Pt) auxiliary electrode, and an Ag/AgCl/1M KCl reference electrode were used as previously described [35]. For electrochemical measurement, cells were washed with PBS (0.01M, pH 7.4). The scan rate for the all electrochemical measurements was 30mV/s. I/V curve was obtained using a semiconductor parameter analyzer (B1500A, Agilent technologies, USA) equipped with probe station. The range of voltage was −500mV to 500mV and the step voltage was 2mV. PBS (0.01M) was used for the electrical study.
3. Results
3.1. Confirmation of 3D graphene oxide-encapsulated gold nanoparticles fabricated on ITO surface
Figure 2a shows a schematic diagram that represents four differently-fabricated substrates, which were modified with graphene (Substrate B), 60nm GNP (Substrate C) and GO-encapsulated 60nm GNP (Substrate D) on ITO surface. Topological characteristics of GO-encapsulated GNP immobilized on an ITO surface were confirmed by atomic force microscopy (AFM) and scanning electron microscopy (SEM) as shown in Figure 2b-d. Specifically, a sheet-like structure of GO bound on nanoparticles was found surrounding the GNPs (Figure 2c), which was clearly different from the normal GNP modified ITO surface, as well as bare APTES-functionalized ITO surface (SFigure 1a, b). The average peak-to-peak distance was 100 nm and 142 nm, respectively, indicating that the density of GNPs and GNP-GO seeded on the ITO surfaces were similar to each other and would not affect the enhancement capability of the fabricated surfaces. The average height and the surface roughness were 48.6 nm and 11.5 nm, respectively, for the ITO-60GNP and 67.7 nm and 14.4 nm, respectively, for the ITO-60GNP-GO substrate. The increase in height may be due to the presence of GO on the surface of the GNPs, which also led to the increase in surface roughness. The high-resolution AFM image of the 60GNP-GO complexes immobilized on the ITO electrode was shown in Figure 2c and its structural characteristics were also confirmed by Raman spectroscopy, which clearly shows the clear D and G bands of GO (Figure 2e). The large area-scanned AFM images also indicate that both Substrate C and Substrate D are proper SERS study with low signal variations (SFigure 2). Finally, GO-GNP nanoparticles not immobilized on the ITO surface were characterized by transmission electron microscopy (TEM) and UV/vis spectroscopy to demonstrate the successful encapsulation of GO with GNP nanoparticles (SFigure 3a-d, SFigure 4a, b). Based on these results, we can confirm the successful encapsulation of GO with GNPs, which was fabricated as either nanoparticles or nanopatterned arrays.
Figure 2.
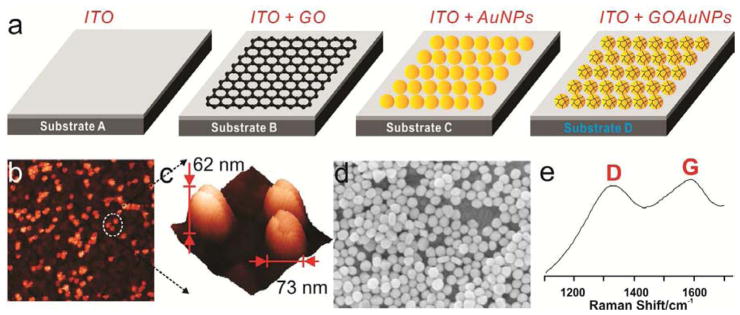
(a) Schematic diagram representing four different substrates modified with graphene (Substrate B), gold nanoparticles (Substrate C) and GO-encapsulated GNP (Substrate D). (b) AFM images of Substrate D and (c) high resolution AFM image of (b) for the confirmation of GO-encapsulated GNP nanoparticles. The X and Y axis were 3um × 3um and 200nm × 200nm for image (b) and (c), respectively, and the scan rate was 0.5Hz and 0.1Hz for image (b) and (c), respectively. (D) SEM image of Substrate D with a high concentrations of GO-encapsulated GNP. (e) Raman spectra of Substrate D to verify the encapsulation of GO on the surface of the GNPs.
3.2. SERS of mNSCs on 3D graphene oxide-encapsulated gold nanoparticles
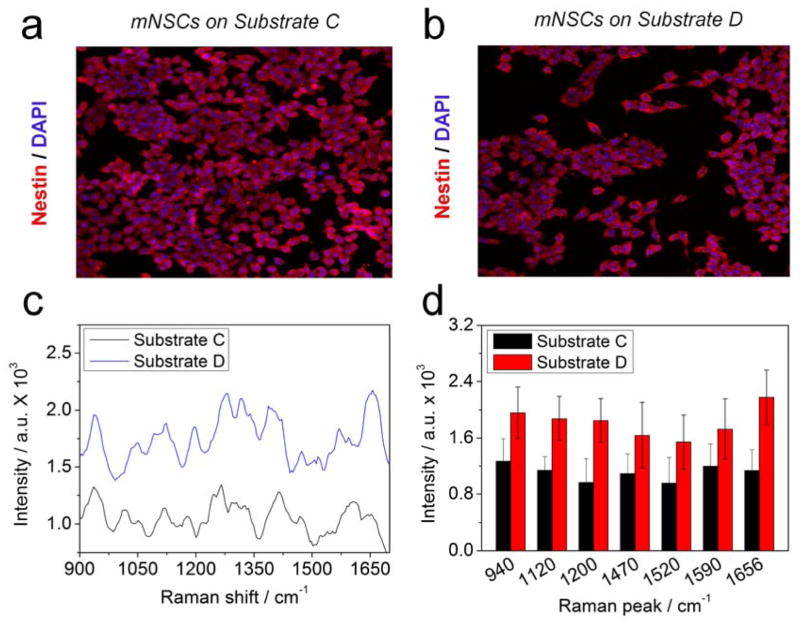
Undifferentiated mNSCs were stained with nestin, a representative marker of undifferentiated neural stem cells, to confirm the undifferentiated state of our mNSCs. Confocal fluorescence images showed mNSCs on Substrate A to D, respectively, indicating that the NSCs maintained an undifferentiated state regardless of the substrates used (Figure 3a, b and SFigure 5a, b). Both Substrate A and Substrate B failed to achieve a clear peak even though the Substrate B showed a higher intensity than Substrate A (SFigure 5c, d). However, the Raman peaks became very clear due to the electromagnetic enhancement of the Raman signals in the presence of 60 nm GNPs or GO-encapsulated GNP on the ITO surface (Figure 3c). Raman peaks appeared at 940 cm−1 (α-helix, C-C backbone str. and C-O-C str. of carbohydrate/proteins), 1051 cm−1 (C-C trans and gauche str. of lipids), 1092 cm−1 (C-O and C-C str. of carbohydrates), 1120 cm−1 (C-O-H def., C-O str. and C-C str. of carbohydrates), 1200–1350 cm-1 (Amide III), 1266 cm−1 (alkyl=C-H cis str. of lipids), 1340 cm−1 (C-H2 def., C-O-H bending of carbohydrates), 1470 cm−1 (C-H def. of lipids/carbohydrates and C-H2 bending of proteins), 1520 cm−1 (C=C str. of carotene of carotenoid), 1590 cm−1 (Amide I) and 1656 cm−1 (C=C str. of lipids and Amide I of proteins) [21,22,36]. As hypothesized, NSCs on Substrate D showed a higher intensity than Substrate C for all peaks (Figure 3d). The enhancement of the intensity of Raman peak was especially evident at 1656 cm−1, which is due to the C=C bond, indicating that polyunsaturated fatty acids such as arachidonic acid, eicosa pentaenoic acid and hexaenoic acid exist in the membrane of NSCs and have a strong affinity to GO modified GNP surfaces (SFigure 6). Since GO strongly attaches to molecules containing aromatic structures rich in C=C bonds through π-π stacking and C=C saturation is generally higher in undifferentiated stem cells than differentiated cells, we hypothesized that these characteristics of GO combined with GNPs can be suitable for the detection of the differentiation potential of NSCs.
Figure 3.
Confocal fluorescence images of undifferentiated mNSCs on (A) Substrate C and (B) Substrate D. Nestin, a general protein marker for undifferentiated neural stem cells, is a red stain, whereas the nuclei were stained blue using DAPI. (C) Raman spectra of mNSCs on Substrate C and D. (D) Intensities of the Raman peaks of obtained from (C). All the Raman spectra of mNSCs were subtracted by the Raman spectra of same substrates without cells to eliminate the background signals.
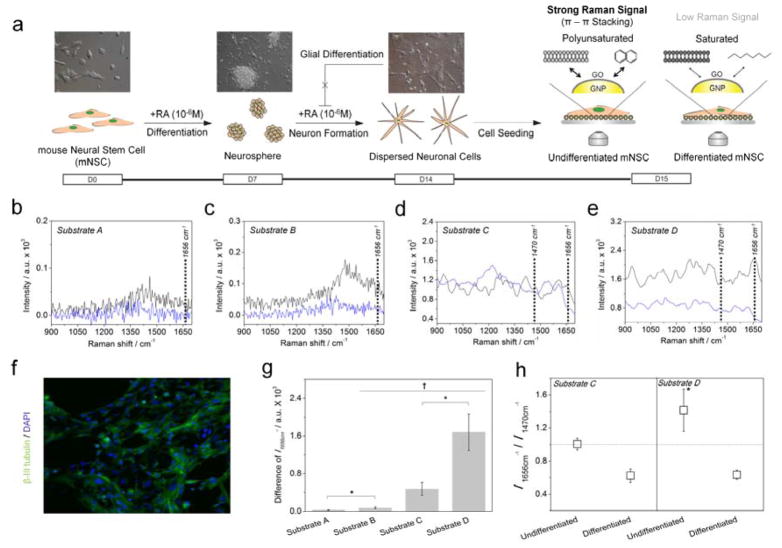
3.3. 3D GNP-GO composites to detect the differentiation potential of mNSC
Mouse NSCs (mNSCs) were treated with retinoic acid (RA) to induce the neural differentiation of NSCs (Figure 4a) [37,38]. This was followed by immunostaining with a β-III tubulin-specific antibody. Figure 4f shows β-III tubulin/DAPI double stained mNSCs proving their successful differentiation to neuronal cells. The undifferentiated and differentiated NSCs, seeded on Substrate A-D for 24 hours, were then exposed to near infrared laser (785 nm) for 1 s to obtain Raman spectra from target cells. As shown in Figure 4b, no difference was observed from cells on Substrate A due to the low intensity of Raman signals. Cells on Substrate B gave a slight intensity difference in the Raman signals when comparing undifferentiated and differentiated cells. However, the Raman peaks were not clear enough to distinguish it from the background signal and additionally, the intensity difference was not large enough to detect the differentiation potential of the mNSCs (Figure 4c). Unlike cells on Substrate A and Substrate B, Figure 4d and 4e shows clear Raman peaks, which were obtained from mNSCs on Substrate C and Substrate D, respectively. In the case of Substrate D, peaks at 1047 cm−1 (C-H in plane Phe structures of protein) and 1220 cm−1 (Amide III) were increased in differentiated mNSCs when compared with that of undifferentiated cells, while 1266 cm−1 (alkyl=C-H cis str. of lipids), 1606 cm−1 (C=C of proteins) and 1668 cm−1 (C=C str. of lipids) were decreased in differentiated cells. However, in the case of Substrate D, intensities of all Raman peaks obtained from undifferentiated mNSCs were much higher than that of differentiated cells, which enabled easy and clear distinguishment of the undifferentiated/differentiated state of NSCs.
Figure 4.
(a) Schematic diagram representing the method to detect the undifferentiated and differentiated state of mNSCs using 3D GO-encapsulated gold nanoparticles. Raman spectra of (—) undifferentiated or (
 )differentiated mNSCs on (b) Substrate A, (c) Substrate B, (d) Substrate C and (e) Substrate D. (f) Confocal fluorescence images of differentiated mNSCs on the Substrate D showing the successful differentiation of mNSCs to neuronal cells. (g) Intensity difference of Raman peak at 1656 cm−1 (C=C bond) achieved from undifferentiated mNSCs subtracted by differentiated cells. (h) Relative values of the Raman intensity at 1656 cm−1 divided by the intensity at 1470 cm−1. All the Raman spectra of mNSCs were subtracted by the Raman spectra of same substrates without cells to eliminate the background signals. Results are medians of Raman signals obtained from ten different spots (†p <0.05, N=3, ANOVA test and *p <0.05, Student’s t-test).
)differentiated mNSCs on (b) Substrate A, (c) Substrate B, (d) Substrate C and (e) Substrate D. (f) Confocal fluorescence images of differentiated mNSCs on the Substrate D showing the successful differentiation of mNSCs to neuronal cells. (g) Intensity difference of Raman peak at 1656 cm−1 (C=C bond) achieved from undifferentiated mNSCs subtracted by differentiated cells. (h) Relative values of the Raman intensity at 1656 cm−1 divided by the intensity at 1470 cm−1. All the Raman spectra of mNSCs were subtracted by the Raman spectra of same substrates without cells to eliminate the background signals. Results are medians of Raman signals obtained from ten different spots (†p <0.05, N=3, ANOVA test and *p <0.05, Student’s t-test).
Since undifferentiated stem cells have polyunsaturated fatty acid rich in C=C bond than normal/differentiated cells, we specifically focused on the Raman peak at 1656–1668 cm−1 assigned to C=C str. of lipids which is an indicator of the C=C saturation of cells. As shown in Figure 4g, the intensity difference of the Raman peak at 1656 cm−1 (C=C bond of lipids) between the undifferentiated and differentiated mNSCs on Substrate D was 3.5 times higher than that of Substrate C. The SERS mapping images that scanned whole cells (32 X 32 points, total: 1024 points) also proves GO-GNP-induced Raman enhancement only specific for undifferentiated mNSCs (SFigure 6). Moreover, the relative values of the Raman spectra (Raman intensity at 1656 cm−1 divided by Raman intensity at 1470 cm−1) achieved from undifferentiated NSCs on Substrate D was higher than 1 (I1656 cm−1/ I1470 cm−1 = 1.41), indicating that Substrate D were more effective at enhancing the molecules rich in C=C bond than the ITO-60GNP surface (I1656 cm−1/I1470 cm−1 = 1.01) as shown in Figure 4h. The relative ratio from the Raman spectra achieved from differentiated NSCs were 0.62 and 0.63 for Substrate C and Substrate D, respectively, indicating that both substrates showed similar performances in regard to the signal enhancement of cells containing molecules not rich in C=C bonds. The GO-modified 3D structure was also found to be proper for long-term culture of NSCs cell that was confirmed by MTT viability assay (SFigure 7). Hence, the differentiation potential of neural stem cells can be more simply and sensitively monitored by using the ITO-60GNP-GO 3D complex as a SERS-active surface than normal noble metal modified surfaces.
3.4. 3D GNP-GO composites to detect stem cell markers based on SERS
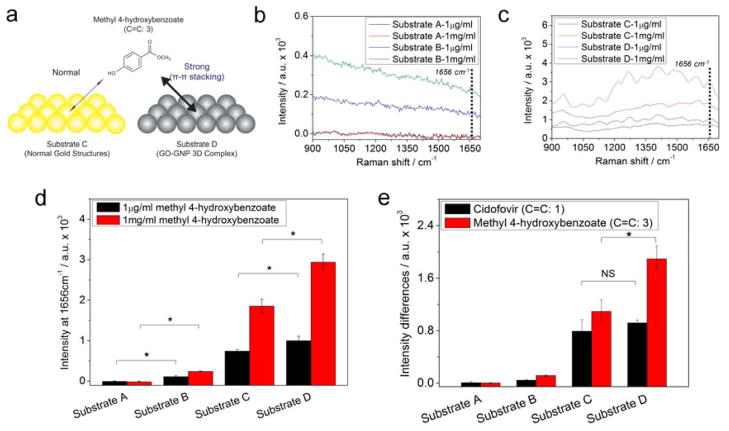
To prove the hypothesis that the enhancement of Raman signals came from the high affinity of polyunsaturated fatty acids that exist in stem cell membrane to GO surface, cidofovir (Cido) and methyl 4-hyroxybenzoate (M4H) were selected for additional SERS study. Since cidofovir and hydroxy benzoate derivatives are molecules that exist in stem cells [26], they are proper candidates to study and understand the mechanism of signal enhancement. Specifically, M4H which possess three C=C bond in its aromatic structure may have stronger interaction with Substrate D than Substrate C through π-π stacking (Figure 5a). As shown in Figure 5b, the Raman signals of M4H achieved from Substrate B showed clear differences that correspond to the different concentrations of M4H (1μg/mL and 1mg/mL, respectively), while Substrate A gave no difference in Raman spectra with the same concentration of M4H. The SERS mapping images obtained from 16 X 16 points of Raman signals at 1656cm−1 (total: 256 points) also showed similar results that of Figure 5a (SFigure 8). Substrate B was also effective for the detection of different concentrations of Cido. However, the intensity difference of the Raman spectra between different concentrations of Cido (1μg/mL and 1mg/mL, respectively) was too weak and was not easily distinguishable from the background noise (SFigure 9a). The GO-induced difference of the Raman peak intensity was found to be clearer in Substrate C than the Substrate B. As shown in Figure 5c, the clear Raman peak of M4H was observed from Substrate D while Substrate C showed weak Raman peaks for the same molecule. However, in the case of Cido, Substrate C gave higher Raman signals than Substrate D for both low (1μg/mL) and high (1mg/mL) concentrations. Since GO has a high affinity for molecules containing aromatic rings through π-π stacking, GO acts as weak barrier for the enhancement of Raman signals of molecules that are not preferred by GO leading to weak SERS signals compared to that obtained from normal metal substrate (SFigure 9b). Hence, we focused on the Raman peak at 1656 cm−1 assigned to C=C bond to compare the intensity achieved from Cido and M4H, respectively. The Raman peak assigned to 1656 cm−1 was significantly higher in Substrate D for M4H (Figure 5d), while the same peak of Cido was higher in Substrate C for both low and high concentrations of analytes, respectively (SFigure 9c). Interestingly, the intensity of the Raman peaks assigned to C=C bond (1656cm−1) of M4H achieved from Substrate B were also remarkable compared to that of Substrate A, indicating that GO was effective for enhancing the Raman signal of M4H. Finally, the difference of the Raman intensities between high and low concentrations of Cido and M4H at 1656 cm−1 obtained from Substrate A to D were calculated to confirm the structure-dependent SERS effect of the GNP-GO 3D complex. As shown in Figure 5E, the enhancement of the Raman signal (1656cm−1) of Cido, which contains one C=C bond in its molecular structure, was similar for Substrate C and Substrate D. However, in the case of M4H, which contains three C=C bond, the enhancement of the Raman signals (1656cm−1) was evident for Substrate D when compared with that of Substrate C. These results suggest that the GO modified SERS-active surface is effective for molecules containing high numbers of C=C bonds in its aromatic hydrocarbons, which is very powerful for the detection of the undifferentiated state of NSCs.
Figure 5.
(a) Schematic illustration representing the differences of Raman intensity induced by the π-π interaction between the GO-GNP and molecules containing three C=C bond. Raman spectra of 1μg/ml and 1mg/ml of M4H obtained from (b) Substrate A and Substrate B, (c) Substrate C and Substrate D. (d) Intensities of Raman signals at 1656cm−1 (C=C bond) of M4H using Substrate A to D. (e) Intensity difference of Raman signals between Raman intensity of 1mg/ml Cido or M4H subtracted by 1μg/ml of Cido or M4H. Background signals were removed by subtracting Raman peaks of substrates from Raman peaks of chemicals. Results are medians of Raman signals obtained from five different spots (*p <0.05, Student’s t-test. ‘NS’ indicating that the two groups were not significantly different at p=0.05).
3.5. 3D GNP-GO composites to detect electrical & electrochemical characteristics of single mNSC
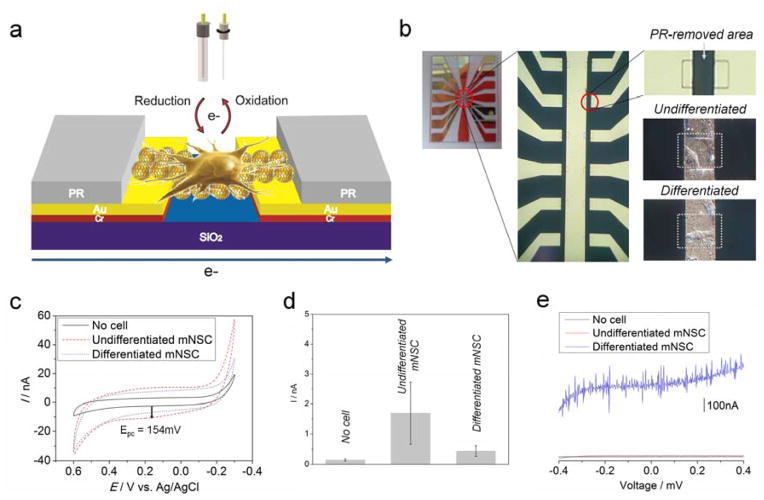
Since the fabricated GO-encapsulated GNP nanoparticles are conductive and more adhesive to unsaturated stem cell membrane than saturated ones, the electrical and electrochemical characteristics of mNSC were analyzed to study of the difference in the features that are induced by differentiation. As shown in Figure 6a, single undifferentiated or differentiated mNSCs were immobilized on the microgap modified with GO-GNP nanoparticles that bridge two separated Au electrodes, by covering other areas with photoresist (PR). This was done to monitor the electrical and electrochemical responses generated from single mNSC. Figure 6b shows the fabricated microgap and the small PR-removed area where single undifferentiated or differentiated mNSC attached to its surface. We have previously reported that cells have its own redox characteristics which can be detected by cyclic voltammetry [39,40]. Since cell differentiation normally induce huge changes of intracellular and/or extracellular environment, the redox characteristics of single mNSC in two different states (undifferentiated/differentiated) also can be verified by voltammetric method using fabricated GO-GNP-modified microgap substrate. A weak oxidation peak appeared at 154mV (Epc) achieved from undifferentiated single mNSC while bare GO-GNP nanoparticles-bridged microgap showed no redox peak at the same condition (Figure 6c). However, single differentiated neuronal cells showed no oxidation peak around 154mV which was clearly different from undifferentiated mNSC, indicating that GO-GNP nanoparticles can be effective for identifying the differentiation level of single cells through analysis of the electrochemical response (Figure 6d). Significantly, the oxidation peak only appeared from undifferentiated mNSCs, also may be due to the high affinity of the GO surface for the unsaturated molecules that contain aromatic structure (high numbers of C=C bonds) through π-π stacking, the same mechanism explained in the SERS study. We also compared the electrochemical signals achieved from undifferentiated mNSCs on GO and GO-encapsulated GNP surface to confirm the superiority of GO-GNP composites in terms of electrochemical enhancement. To this end, we cultured undifferentiated mNSCs on the cell chip composed of GO and GO-GNP surface, followed by the detection of redox signals using cyclic voltammetry. Large populations of cells were used to check the surface-dependent characteristics of electrochemical signals achieved from mNSCs. As a result, the voltammogram obtained from cells cultured on GO-GNP structure showed very similar voltammetric signals with that of Figure 6 (SFigure 10). However, in case of signals achieved from mNSCs on GO, no oxidation or reduction peak was observed. Hence, it can be concluded that GO-encapsulated gold nanoparticles possess an ability to enhance electrochemical signals from cells via double enhancement from GO and gold particles, which is not possible using GO as an electrode for electrochemical study. Finally, I–V curves were obtained from a single mNSC bridging two Au microgap to compare the current responses between the undifferentiated and differentiated state. As shown in Figure 6e, differentiated single neuronal cells gave higher current intensities than undifferentiated mNSC within the full voltage range (−400mV to 400mV). This may be due to a large number of neurites existing in neuronal cells, which accelerate electron transfer between two metal electrode. Since GO is not a highly-conductive material due to the many branches that contain hydroxyl groups, the high affinity of GO surface to stem cell membrane didn’t affects the conductance induced by cell immobilization. These results suggest that the GO-GNP nanoparticles are also useful for monitoring the differentiation level of mNSC by using electrical and/or electrochemical tools which can be combined with SERS techniques.
Figure 6.
(a) Schematic illustration describing the detection strategy of electrochemical and electrical characteristics of undifferentiated/differentiated single mNSC. (b) Picture of microelectrodes representing the microgap where PR was selectively removed by the 2nd mask to detect the electrochemical and electrical signals generated or transferred from single cells. White squares indicate undifferentiated/differentiated mNSC adhered on PR-removed area where GOencapsulated GNPs were immobilized between two metal electrodes prior to cell seeding. (c) Electrochemical responses from undifferentiated/differentiated mNSC attached on microgap which were achieved by cyclic voltammetry. The voltammogram was averaged from three different experiments. (d) Intensities of cathodic peaks at 154mV obtained from GO-GNP modified microgap without cell, electrodes with undifferentiated and differentiated mNSC. Error bars are the mean ± standard deviation of three different experiments. (e) I/V curve achieved from undifferentiated/differentiated mNSC showing the differences of current intensities and patterns induced by the electrons that passed through cells on GO-GNP nanoparticles bridging microgap.
4. Discussion
SERS has been considered a powerful ‘fingerprint technology’ which can overcome the weak signal intensities of normal Raman methods [16,41,42]. However, it is also true that SERS and Raman show a limited ability to analyze cellular components such as proteins, ligands, enzymes and nucleic acids due to the complexity of cells [25]. Since Raman spectroscopy is very efficient for the investigation of the molecular structures of target materials, SERS could be more useful than any other method to probe the specific chemical structures of biomarkers, which are very complex and have a high variety of molecular weights, in in vitro or in vivo studies. Previously, Wu et al. have reported the potential of this kind of new application of Raman tools by profiling oil-producing microalgae to determine the degree of unsaturation of lipids based on single-cell laser-trapping Raman spectroscopy (LTRS) [36]. Li et al. also have recently reported the cell imaging method using GO as a Raman reporter which was found to be efficient to investigate the uptake of Au/Go hybrids and its mechanism based on SERS technique [43].
Fortunately, undifferentiated stem cells were found to have many molecules that contain a high number of C=C bonds (high degree of C=C saturation), which are also highly adhesive to the surface of GO [26]. The high number of C=C bonds allows for the maintenance of its ‘chemical plasticity’ and for mediating differentiation through the control of the redox status of oxidative pathways, which has been reported to be an important factor for determining the stemness of target cells [27]. Since Raman spectroscopy is a very effective tool for profiling/quantifying the chemical structures of target cells, GO-encapsulated GNP structure was very effective for enhancing the Raman signals of undifferentiated NSCs selectively and was far better than normal GNP modified surface (Figure 3c). In contrast, differentiated NSCs on an ITO-60GNP surface showed higher Raman signals than an ITO-60GNP-GO 3D complex which was distinct from that of undifferentiated NSCs (Figure 4d, e). These distinct characteristics of the ITO-60GNP-GO 3D complex that selectively enhance the Raman signal from undifferentiated NSCs can enable rapid, easy and non-destructive in situ monitoring of the differentiation potential of NSCs by simply comparing the absolute and relative values of C=C bonds obtained from the Raman signal intensity assigned to C=C bonds (1656cm−1) divided by that of C-H bonds (1470cm−1). Since the ratio of C=C/C-H bonds represent the relative amount of C=C in the cellular components of the target cell, this value can be used as an indicator of C=C saturation of NSCs by means of Raman techniques without the use of any fluorescence dyes or pretreatment steps. Interestingly, a weak enhancement of Raman signal was observed from the undifferentiated NSCs on 2D ITO-GO surface, which was different from the bare ITO surface, indicating that GO has some ability to enhance the signal from polyunsaturated fatty acids that exist in the membrane of undifferentiated NSCs. To understand the mechanism underlying signal enhancement, Cido and M4H were studied. The GO-induced Raman enhancement was clearer in M4H, which contains three C=C bonds, than Cido, which contains only one C=C bond due to the stronger π-π stacking between the molecules and surface of GO. The enhancement of Raman signals induced by GO was effective regardless of the presence of GNPs, even though the enhancement was much higher in the ITO-GNP-GO complex than in the ITO-GO substrate due to the electromagnetic enhancement phenomenon. Since C=C bond-rich molecules are much more abundant in undifferentiated NSCs than differentiated NSCs, we can conclude that these C=C bond-rich molecules contributed to the difference of Raman signals that originated from the undifferentiated and differentiated NSCs on the ITO-60GNP-GO 3D complex. As GO was reported as an excellent enhancer of electrochemical signals such as specific molecules containing aromatic rings [44], single undifferentiated mNSC containing highly polyunsaturated membrane components showed a clear oxidation peak at 154mV absent in differentiated single neuronal cells. Hence, this electrochemical technique can be useful for the detection of the differentiation level and/or for the identification of undifferentiated and differentiated state of single stem cells when combined with the SERS technique.
Even more remarkably, the technique introduced in this study can potentially be applied to other types of stem cells (embryonic, mesenchymal and hematopoietic stem cells) and/or progenitor cells which are undergoing differentiation or are maintained in an undifferentiated state, because the unsaturated state of the cell membrane is a common characteristic of undifferentiated stem cells [26]. Cancer stem cells, cancer cell that have the ability to self-renew and differentiate, have recently become a hot issue in the field of cancer diagnosis/treatment due to their high resistivity to chemotherapy and their ability to form new tumors [45,46]. Since cancer stem cells are also thought to maintain ‘chemical plasticity’ in a similar manner as stem cells, our developed method can potentially be applied to the discrimination of cancer stem cells from normal cancer cells. Hence, the proposed spectroelectrochemical tool utilizing 3D GO-encapsualted gold nanoparticles can be used for various kinds of stem cell-based research and therapy and also as a rapid, sensitive and in situ monitoring tool for determining both the undifferentiated and differentiated state of stem cells, leading to advancements in the practical use of stem cells as a new regeneration medicine.
5. Conclusion
In this report, we have fabricated 3D graphene oxide-encapsulated gold nanoparticles to determine the differentiation potential of NSCs based on spectroelectrochemical tool, which was highly effective for monitoring of stem cell differentiation. The fabricated structures exhibit excellent performance in enhancing the Raman signals from undifferentiated NSCs, especially at the 1656 cm−1 Raman peak that is assigned to C=C bonds. Both the absolute value of the Raman peak at 1656cm−1 and the relative value of the Raman intensity at 1656 cm−1 (C=C bond) divided by the Raman intensity at 1470 cm−1 (C-H bond) from undifferentiated NSCs was found to be higher in undifferentiated NSCs than differentiated NSCs. Since undifferentiated stem cells normally have a large number of molecules that are rich in C=C bonds (high degree of C=C saturation), our new material combined with SERS and electrochemical technique will be very effective for in situ monitoring of the undifferentiated and differentiated state or differentiation level of various kinds of stem cells.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (2009-0080860) and by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science, ICT & Future Planning (2005-2001333). K.-B.L would like to acknowledge the support from the NIH Director’s Innovator Award [(1DP20D006462–01). He also thanks Perry Yin, Birju Shah, and KBLEE group for their scientific input and valuable suggestions for the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Steindler DA, Okun MS, Scheffler B. Stem cell pathologies and neurological disease. Mod Pathol. 2012;25:157–62. doi: 10.1038/modpathol.2011.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Daadi MM, Grueter BA, Malenka RC, Redmond DE, Jr, Steinberg GK. Dopaminergic neurons from midbrain-specified human embryonic stem cell-derived neural stem cells engrafted in a monkey model of Parkinson’s disease. PLoS One. 2012;7:e41120. doi: 10.1371/journal.pone.0041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kriks S, Shim J, Piao J, Ganat YM, Wakeman DR, Xie Z, et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. 2011;480:547–U177. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X, Katsanevakis E, Liu X, Zhang N, Wen X. Engineering neural stem cell fates with hydrogel design for central nervous system regeneration. Prog Polym Sci. 2012;37:1105–29. [Google Scholar]
- 5.Donnelly EM, Lamanna J, Boulis NM. Stem cell therapy for the spinal cord. Stem Cell Res Ther. 2012;3:24. doi: 10.1186/scrt115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danova-Alt R, Heider A, Egger D, Cross M, Alt R. Very small embryonic-like stem cells purified from umbilical cord blood lack stem cell characteristics. PLoS One. 2012;7:e34899. doi: 10.1371/journal.pone.0034899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ganat YM, Calder EL, Kriks S, Nelander J, Tu EY, Jia F, et al. Identification of embryonic stem cell-derived midbrain dopaminergic neurons for engraftment. J Clin Invest. 2012;122:2928–39. doi: 10.1172/JCI58767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piao S, Kim IG, Lee JY, Hong SH, Kim SW, Hwang T, et al. Therapeutic effect of adipose-derived stem cells and BDNF-immobilized PLGA membrane in a rat model of cavernous nerve injury. J Sex Med. 2012;9:1968–79. doi: 10.1111/j.1743-6109.2012.02760.x. [DOI] [PubMed] [Google Scholar]
- 9.Bujan J, Pascual G, Corrales C, Gomez-Gil V, Rodriguez M, Bellon J. Muscle-derived stem cells in tissue engineering: defining cell properties suitable for construct design. Histol Histopathol. 2005;20:891–9. doi: 10.14670/HH-20.891. [DOI] [PubMed] [Google Scholar]
- 10.Xu Y, Huang S, Ma K, Fu X, Han W, Sheng Z. Promising new potential for mesenchymal stem cells derived from human umbilical cord Wharton’s jelly: sweat gland cell-like differentiative capacity. J Tissue Eng Regen Med. 2012;6:645–54. doi: 10.1002/term.468. [DOI] [PubMed] [Google Scholar]
- 11.Notingher I, Green C, Dyer C, Perkins E, Hopkins N, Lindsay C, et al. Discrimination between ricin and sulphur mustard toxicity in vitro using Raman spectroscopy. J R Soc Interface. 2004;1:79–90. doi: 10.1098/rsif.2004.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Votteler M, Berrio DAC, Pudlas M, Walles H, Stock UA, Schenke-Layland K. Raman spectroscopy for the non-contact and non-destructive monitoring of collagen damage within tissues. J Biophotonics. 2012;5:47–56. doi: 10.1002/jbio.201100068. [DOI] [PubMed] [Google Scholar]
- 13.Thuy NT, Tam PD, Tuan MA, Le AT, Tam LT, Thu VV, et al. Detection of pathogenic microorganisms using biosensor based on multi-walled carbon nanotubes dispersed in DNA solution. Curr Appl Phys. 2012;12:1553–60. [Google Scholar]
- 14.Wang Y, Irudayaraj J. A SERS DNAzyme biosensor for lead ion detection. Chem Commun. 2011;47:4394–6. doi: 10.1039/c0cc04140h. [DOI] [PubMed] [Google Scholar]
- 15.Das G, Patra N, Gopalakrishnan A, Zaccaria RP, Toma A, Thorat S, et al. Fabrication of large-area ordered and reproducible nanostructures for SERS biosensor application. Analyst. 2012;137:1785–92. doi: 10.1039/c2an16022f. [DOI] [PubMed] [Google Scholar]
- 16.Li M, Zhang J, Suri S, Sooter LJ, Ma D, Wu N. Detection of adenosine triphosphate with an aptamer biosensor based on surface-enhanced Raman scattering. Anal Chem. 2012;84:2837–42. doi: 10.1021/ac203325z. [DOI] [PubMed] [Google Scholar]
- 17.Alivisatos P. The use of nanocrystals in biological detection. Nat Biotechnol. 2004;22:47–52. doi: 10.1038/nbt927. [DOI] [PubMed] [Google Scholar]
- 18.Lin D, Feng S, Pan J, Chen Y, Lin J, Chen G, et al. Colorectal cancer detection by gold nanoparticle based surface-enhanced Raman spectroscopy of blood serum and statistical analysis. Opt Express. 2011;19:13565–77. doi: 10.1364/OE.19.013565. [DOI] [PubMed] [Google Scholar]
- 19.Qian X, Peng X, Ansari DO, Yin-Goen Q, Chen GZ, Shin DM, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nat Biotechnol. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 20.Zavaleta C, de la Zerda A, Liu Z, Keren S, Cheng Z, Schipper M, et al. Noninvasive Raman spectroscopy in living mice for evaluation of tumor targeting with carbon nanotubes. Nano Lett. 2008;8:2800–5. doi: 10.1021/nl801362a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El-Said WA, Kim T, Kim H, Choi J. Detection of effect of chemotherapeutic agents to cancer cells on gold nanoflower patterned substrate using surface-enhanced Raman scattering and cyclic voltammetry. Biosens Bioelectron. 2010;26:1486–92. doi: 10.1016/j.bios.2010.07.089. [DOI] [PubMed] [Google Scholar]
- 22.El-Said WA, Kim T, Kim H, Choi J. Analysis of intracellular state based on controlled 3D nanostructures mediated surface enhanced Raman scattering. PLoS One. 2011;6:e15836. doi: 10.1371/journal.pone.0015836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung M, El-Said WA, Choi J. Fabrication of gold nanodot arrays on a transparent substrate as a nanobioplatform for label-free visualization of living cells. Nanotechnology. 2011;22:235304. doi: 10.1088/0957-4484/22/23/235304. [DOI] [PubMed] [Google Scholar]
- 24.Schulze HG, Konorov SO, Caron NJ, Piret JM, Blades MW, Turner RFB. Assessing differentiation status of human embryonic stem cells noninvasively using Raman microspectroscopy. Anal Chem. 2010;82:5020–7. doi: 10.1021/ac902697q. [DOI] [PubMed] [Google Scholar]
- 25.Willets KA. Surface-enhanced Raman scattering (SERS) for probing internal cellular structure and dynamics. Anal Bioanal Chem. 2009;394:85–94. doi: 10.1007/s00216-009-2682-3. [DOI] [PubMed] [Google Scholar]
- 26.McMurray RJ, Gadegaard N, Tsimbouri PM, Burgess KV, McNamara LE, Tare R, et al. Nanoscale surfaces for the long-term maintenance of mesenchymal stem cell phenotype and multipotency. Nat Mater. 2011;10:637–44. doi: 10.1038/nmat3058. [DOI] [PubMed] [Google Scholar]
- 27.Yanes O, Clark J, Wong DM, Patti GJ, Sanchez-Ruiz A, Benton HP, et al. Metabolic oxidation regulates embryonic stem cell differentiation. Nat Chem Biol. 2010;6:411–7. doi: 10.1038/nchembio.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee YH, Polavarapu L, Gao N, Yuan P, Xu Q. Enhanced optical properties of graphene oxide-Au nanocrystal composites. Langmuir. 2012;28:321–6. doi: 10.1021/la204047a. [DOI] [PubMed] [Google Scholar]
- 29.Ling X, Xie L, Fang Y, Xu H, Zhang H, Kong J, et al. Can graphene be used as a substrate for Raman enhancement? Nano Lett. 2010;10:553–61. doi: 10.1021/nl903414x. [DOI] [PubMed] [Google Scholar]
- 30.Ballarin B, Cassani MC, Scavetta E, Tonelli D. Self-assembled gold nanoparticles modified ITO electrodes: The monolayer binder molecule effect. Electrochim Acta. 2008;53:8034–44. [Google Scholar]
- 31.Kim T, El-Said WA, Choi J. Highly sensitive electrochemical detection of potential cytotoxicity of CdSe/ZnS quantum dots using neural cell chip. Biosens Bioelectron. 2012;32:266–72. doi: 10.1016/j.bios.2011.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Myung S, Solanki A, Kim C, Park J, Kim KS, Lee K. Graphene-encapsulated nanoparticle-based biosensor for the selective detection of cancer biomarkers. Adv Mater. 2011;23:2221–5. doi: 10.1002/adma.201100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang S, Feng X, Ivanovici S, Muellen K. Fabrication of graphene-encapsulated oxide nanoparticles: Towards high-performance anode materials for lithium storage. Angew Chem -Int Edit. 2010;49:8408–11. doi: 10.1002/anie.201003485. [DOI] [PubMed] [Google Scholar]
- 34.Myung S, Yin PT, Kim C, Park J, Solanki A, Reyes PI, et al. Label-free polypeptide-based enzyme detection using a graphene-nanoparticle hybrid sensor. Adv Mater. 2012;24:6081–87. doi: 10.1002/adma.201202961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kafi MA, Kim T, An JH, Choi J. Electrochemical cell-based chip for the detection of toxic effects of bisphenol-A on neuroblastoma cells. Biosens Bioelectron. 2011;26:3371–5. doi: 10.1016/j.bios.2010.12.030. [DOI] [PubMed] [Google Scholar]
- 36.Wu H, Volponi JV, Oliver AE, Parikh AN, Simmons BA, Singh S. In vivo lipidomics using single-cell Raman spectroscopy. Proc Natl Acad Sci U S A. 2011;108:3809–14. doi: 10.1073/pnas.1009043108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlett K, Madarasz E. Retinoic acid induced neural differentiation in a neuroectodermal cell line immortalized by p53 deficiency. J Neurosci Res. 1997;47:405–15. [PubMed] [Google Scholar]
- 38.Gudas LJ, Wagner JA. Retinoids regulate stem cell differentiation. J Cell Physiol. 2011;226:322–30. doi: 10.1002/jcp.22417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kafi MA, Kim T, An JH, Choi J. Fabrication of cell chip for detection of cell cycle progression based on electrochemical method. Anal Chem. 2011;83:2104–11. doi: 10.1021/ac102895b. [DOI] [PubMed] [Google Scholar]
- 40.Kim T, El-Said WA, An JH, Choi J. ITO/gold nanoparticle/RGD peptide composites to enhance electrochemical signals and proliferation of human neural stem cells. Nanomedicine: NBM. 2013;9:336–44. doi: 10.1016/j.nano.2012.08.006. [DOI] [PubMed] [Google Scholar]
- 41.Chen K, Han H, Luo Z, Wang Y, Wang X. A practicable detection system for genetically modified rice by SERS-barcoded nanosensors. Biosens Bioelectron. 2012;34:118–24. doi: 10.1016/j.bios.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 42.Fang J, Liu S, Li Z. Polyhedral silver mesocages for single particle surface-enhanced Raman scattering-based biosensor. Biomaterials. 2011;32:4877–84. doi: 10.1016/j.biomaterials.2011.03.029. [DOI] [PubMed] [Google Scholar]
- 43.Liu Q, Wei L, Wang J, Peng F, Luo D, Cui R, et al. Cell imaging by graphene oxide based on surface enhanced Raman scattering. Nanoscale. 2012;4:7084–9. doi: 10.1039/c2nr32525j. [DOI] [PubMed] [Google Scholar]
- 44.Kim Y, Bong S, Kang Y, Yang Y, Mahajan RK, Kim JS, et al. Electrochemical detection of dopamine in the presence of ascorbic acid using graphene modified electrodes. Biosens Bioelectron. 2010;25:2366–9. doi: 10.1016/j.bios.2010.02.031. [DOI] [PubMed] [Google Scholar]
- 45.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–9. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 46.Soltysova A, Altanerova V, Altaner C. Cancer stem cells. Neoplasma. 2005;52:435–40. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.