Abstract
Germ cells and adult stem cells maintain tissue homeostasis through a finely tuned program of responses to both physiological and stress-related signals. PLZF (Promyelocytic Leukemia Zinc Finger protein), a member of the POK family of transcription factors, acts as an epigenetic regulator of stem cell maintenance in germ cells and haematopoietic stem cells. We identified L1 retrotransposons as the primary targets of PLZF. PLZF-mediated DNA methylation induces silencing of the full-length L1 gene and inhibits L1 retrotransposition. Furthermore, PLZF causes the formation of barrier-type boundaries by acting on inserted truncated L1 sequences in protein coding genes. Cell stress releases PLZF-mediated repression, resulting in L1 activation/retrotransposition and impaired spermatogenesis and myelopoiesis. These results reveal a novel mechanism of action by which, PLZF represses retrotransposons, safeguarding normal progenitor homeostasis.
Keywords: epigenetic, repression, transposon
Introduction
Epigenetic modifications regulate gene expression and contribute to silencing mechanisms specifying lineage and developmental stages of cells (Azuara et al, 2006). Methylation of cytosine-5 at CpG dinucleotides directly influences gene expression and patterns of DNA methylation are tightly regulated in mammals (Stadler et al, 2011). It is unclear precisely how these are determined and to what extent this process is involved in cell fate determination. The majority of methylated cytosines reside in transposable elements (TEs) that must be silenced in mammals and it is well accepted that DNA methylation represents the primary mechanism of transposition suppression in the host genome (Hancks and Kazazian, 2012). The most commonly found TEs are non-LTR transposons. These are subdivided into long and short interspersed nuclear elements (LINE 1 or L1, and SINE, respectively). L1 elements constitute ∼17% of the human genome, including full-length, truncated and mutated copies (Lander et al, 2001; Goodier and Kazazian, 2008). L1 elements propagate in the genome by retrotransposition and contain two open reading frames: ORF1 encodes a protein with RNA binding activity (Martin et al, 2005) and ORF2 encodes a protein with endonuclease/reverse transcriptase activities necessary for retrotransposition (Mathias et al, 1991; Feng et al, 1996). L1s play a significant role in shaping the mammalian genome and have been implicated in human genome evolution and instability (Kazazian, 2004) and oncogenesis by generating insertion mutations (Cordaux and Batzer, 2009; Hancks and Kazazian, 2012). While L1 retrotransposition has been extensively studied in the male germ line (for review, Bao and Yan, 2012), it now appears that L1 activity can be detected in somatic cells (Coufal et al, 2009; Kano et al, 2009; Muotri et al, 2010; Faulkner, 2011; Lee et al, 2012; Solyom et al, 2012). Interest in L1 is growing because they have been implicated in regulating gene expression on a global scale (Slotkin and Martienssen, 2007; Faulkner et al, 2009). L1s also facilitate the transposition of two other subtypes of non-autonomous non-LTR retrotransposons, SINEs (of which a typical representative is Alu) and SVAs (named after Sine, VNTR variable number of tandem repeats, and Alu) (Dewannieux et al, 2003; Hancks et al, 2011; Raiz et al, 2012).
Safeguarding the genome from retrotransposition is essential for the integrity of the genome. TEs are silenced via the mechanism of chromatin inactivation and this process has not been fully elucidated. Sequence features and epigenetic marks buried within these repetitive elements such as specific binding sites for DNA-interacting proteins need to be unveiled in order to understand how these TEs might guide the formation of heterochromatin.
Members of the POK (POZ and Kruppel zinc finger (ZF)) family of proteins induce epigenetic changes, including histone modifications and DNA methylation, thus regulating the chromatin state. Accordingly, many findings within the past few decades have underscored the importance of the POK family in development, stem cell biology and oncogenesis (Kelly and Daniel, 2006). PLZF (Promyelocytic Leukemia Zinc Finger protein, also known as zbtb16) is a member of the POK family (Costoya, 2007) and was identified when it was found to be a component of a rare chromosomal translocation in acute promyelocytic leukemia (APL) (Chen et al, 1993). We and others have sought to define its function in normal and malignant cells. PLZF plays an important role in the maintenance of haematopoietic stem cells (HSCs) (Doulatov et al, 2009) and germ cells in adult tissues (Buaas et al, 2004) and during embryonic development (Barna et al, 2000). PLZF is recognized as an important regulator of cell growth, self-renewal and differentiation through its sequence-specific negative transcriptional activity (Guidez and Zelent, 2001; Barna et al, 2002; Guidez et al, 2007). Studies examining PLZF alterations have revealed the mechanism of APL leukemogenesis, and have defined the repressive functions of PLZF (Chen et al, 1994; Guidez et al, 1998; He et al, 1998). Through the recruitment of histone deacetylases, DNA methyltransferases and nuclear corepressors and the propagation of a repressive chromatin environment following sequence-specific binding, PLZF exerts local chromatin remodelling activity leading to gene silencing (Barna et al, 2002; Guidez et al, 2005, 2007). PLZF is characterized by two important domains: the BTB/POZ domain, an evolutionarily conserved protein–protein interaction domain that promotes dimerization of the POK members (Bardwell and Treisman, 1994), and the Kruppel-like ZF that participate in various protein–protein interactions and mediate sequence-specific binding to DNA through a consensus genomic motif (A-T/G-G/C-T-A/C-A/C-A-G-T, Li et al, 1997). PLZF DNA binding activity is modulated by a specific acetylation site (aa 632–652) within its last ZF, which is a target of the coactivator and Histone Acetyl Transferase (HAT) protein, p300 (Figure 1A, Guidez et al, 2005).
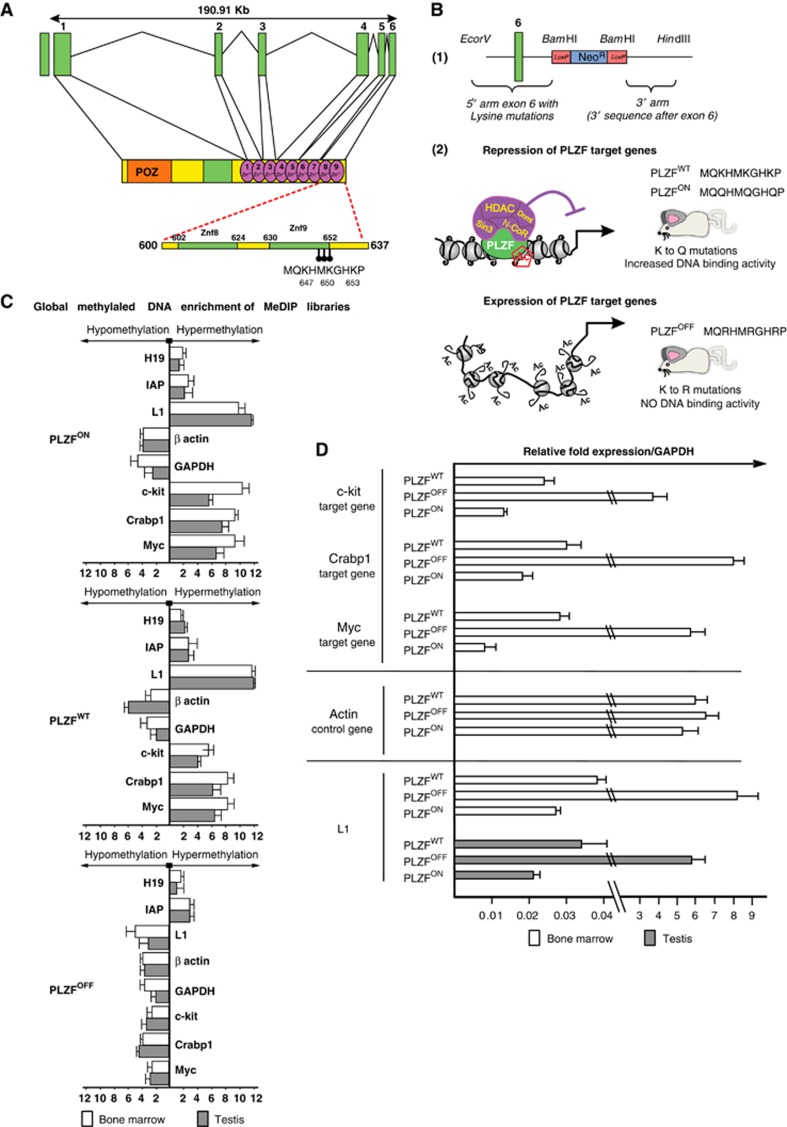
Figure 1.
PLZF-induced DNA methylation throughout the mouse genome. (A) Schematic representation of the structure of the mouse PLZF gene. Green boxes represent exons and connected black straight lines represent intronic regions. The PLZF protein structure is shown under the intronic-exonic organization of the PLZF gene. The orange box represents the POZ domain; the green box represents the proline-rich region; and the green circles represent individual zinc fingers. The acetylation site is situated at the C-terminal end of the most C-terminal zinc finger (9 Zn++), encoded by exon 6, and consists of three lysine residues (K 647, 650 and 653; described in Guidez et al, 2005). These amino acids and their position are shown in the acetylation site sequence. (B) PLZF-targeting vectors and animal models. (1) Schematic representation of the targeting vector pKo scrambler used for the generation of the knock-in embryonic cells. The green box represents exon 6 of the murine PLZF gene, which encodes zinc finger 9 of the PLZF protein. The neomycin cassette is shown in blue and is flanked by two LoxP sites (red boxes), which are used to remove the resistance cassette after recombination. (2) Schematic representation of the impact of the mutant PLZF on chromatin structure in knock-in mice. The PLZFOFF knock-in model expresses a PLZF protein that is unable to bind DNA (due to the mutation of the key lysine residues described above to arginine) and do not recruit co-repressor complex to target sequences; thus, the PLZF-associated genes are constitutively active. The PLZFON knock-in model expresses a PLZF protein that constitutively binds to DNA (due to mutation of the lysine residues in the PLZF acetylation site to glutamine) and recruits like the PLZFWT a co-repressor complex; thus, the PLZF-associated genes are constitutively repressed. (C) Global methylated DNA enrichment of the MeDIP libraries. DNA libraries were prepared from the BM and testis from PLZFOFF, PLZFON and PLZFWT mice. MeDIP (immunoprecipitation of methylated DNA using an antibody raised against 5-methylcytosine (5mC)) was performed to purify genomic methylated DNA regions. The enriched DNA was monitored using quantitative PCR for sequences known to be methylated in mice (the H19 gene and repeated sequences (IAP and L1 elements)) in comparison to input DNAs, as shown in the PLZFWT diagram. Housekeeping genes (GAPDH and beta actin) were used as negative controls. To validate the effect of the PLZF mutants in mice, we performed quantitative PCR of previously described PLZF target genes, including c-kit, Crabp1 and Myc, from the methylation-enriched DNA to determine their levels of DNA methylation (hypomethylation in PLZFOFF samples compared to hypermethylation in PLZFON and PLZFWT samples). (D) Gene expression associated with the presence of mutant or wild-type PLZF proteins. RNAs from mouse tissues were isolated and gene expression was assessed by quantitative PCR using GAPDH expression as a control. Known PLZF targets (c-kit, Myc and Crabp1 genes, A.1) were used to monitor PLZF repressive function while actin served as an internal control (A.2). L1 RNA expression levels were monitored in the bone marrow and testis to correlate the DNA methylation pattern and gene expression of these PLZF-regulated repeats.
Here, we ask whether the PLZF-induced epigenetic programming might contribute to gene regulation in the haematopoietic tissue through the interaction of PLZF with specific TEs.
Results
PLZF binds to methylated L1 sequences throughout the mouse genome
To understand PLZF’s involvement in epigenetic regulation at the molecular level, we established two knock-in PLZF mouse lines either bearing a PLZF loss-of-function mutant that lacked DNA binding activity (PLZFOFF) or a PLZF gain-of-function mutant that constitutively bound to its genomic DNA targets (PLZFON) (described in Guidez et al, 2005 and Figure 1B). The PLZFOFF model recapitulates both the testicular phenotype associated with the PLZF knockout mouse model (Costoya et al, 2004), with an increase in spontaneous apoptosis of purified testicular cells (Supplementary Figure 1) and the biallelic loss phenotype (PLZF−/−) in humans (Fischer et al, 2008), leading to male infertility due to the deregulation of germ cell maintenance. In line with PLZF expression in HSC, the maintenance of haematopoietic progenitor cells is altered in the PLZFON and PLZFOFF mutants with a subsequent increase or loss of these cells, respectively (Supplementary Figure S1).
Cells from the bone marrow (BM) and testis of PLZFOFF and PLZFON mice were analysed using an immunocapture approach to identify differentially methylated DNA sequences. First, we looked at the PLZF target genes. In PLZFOFF mice, the constitutive loss of PLZF DNA binding activity induced the hypomethylation of CpG dinucleotides situated in CpG islands in the c-kit, CrabpI and Myc genes in both BM and testis (Figure 1C), associated with a correlated increase in the mRNA expression levels of these genes (Figure 1D). In the PLZFON model, the constitutive interaction of PLZF with its genomic DNA targets induced hypermethylation and gene expression levels similar to those observed in the wild-type PLZF mouse (PLZFWT) (Figures 1C and D). Control genes such as the housekeeping gene actin and known mouse methylated control genes (IAP and H19) were not affected (Figures 1C and E). Thus, the identification of different activities of the PLZF mutants, validated on PLZF target genes, confirmed the epigenetic function of PLZF in our mouse models. We then subjected MeDIP-purified DNA to next-generation sequencing and identified 188 differentially methylated regions (DMRs) genome-wide. In silico analysis of these DMRs identified genomic repeat elements; >50% of these were long and small interspersed nuclear elements such as long (LINE 1s or L1s) and short interspersed elements (SINEs) (Figure 2A; Supplementary Table SI). Considering the role of L1 in gene expression, these novel results prompted us to further investigate PLZF’s binding to methylated L1 DNA sequences and to determine the purpose of such a PLZF-induced epigenetic regulation.
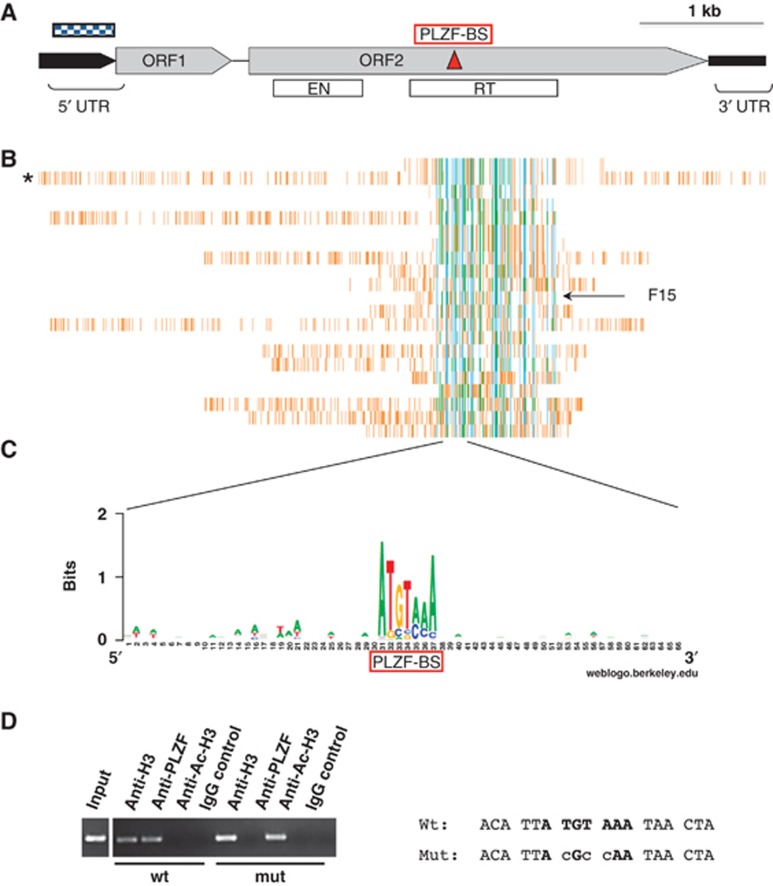
Figure 2.
PLZF binds to genomic DNA L1 retrotransposon sequences. (A) Schematic representation of the human L1 element. Black boxes represent the 5′ and 3′ untranslated regions (5′ UTR and 3′ UTR); grey boxes represent the two open reading frames (ORF1 and ORF2); and the dotted box represents the CpG island found in the human L1 5′ UTR. The white boxes represent the coding sequences of the endonuclease (ENT) and the reverse transcriptase (RT). (B) Comparative PLZF-associated chromatin fragment alignment. PLZF-associated chromatin fragments were aligned with the human full-length L1 sequence (indicated by *) using http//:ebi.ac.uk/clustalW. A common domain encompassing parts of the ORF2 is found in all purified L1 sequences and has perfect homology with purified fragment 15 (F15). Sequence information is summarized in Supplementary Table SII. (C) Alignment of the L1 central region. The deduced consensus sequence (determined http//:weblogo.berkeley.edu/logo.cgi) for the PLZF-BS (red box) (ATGTAAA) nucleotides is conserved throughout the L1 PLZF-purified genomic sequences. (D) ChIP assay. ChIP was performed in 293T cells transfected with wild-type (wt) or mutant (mut) PLZF-BS F15_tk-luc to assess the acetylation level (anti-AcH3) and the PLZF binding activity (anti-PLZF) at these sequences. The level of histone H3 protein in the vicinity of this site (anti-H3) was used as a positive control for the ChIP procedure and to normalize the quantity of input DNA for each sample. An irrelevant polyclonal antibody was used as a negative control (IgG control). When the PLZF-BS is mutated, PLZF does not bind the L1 sequence and histone acetylation is detected (anti-AcH3).
Source data for this figure is available on the online supplementary information page.
PLZF induces epigenetic modifications to full-length L1 retrotransposon at the 5′ UTR region and ORF2 site via a PLZF-binding site
The canonical full-length L1 retrotransposon consists of a 5′ UTR containing an internal RNA polymerase promoter, two open reading frames (ORF1 and ORF2) and a 3′ UTR containing a polyadenylation signal (reviewed in Hancks and Kazazian, 2012 and Figure 2B). Thus, L1 retrotransposons encode for proteins required for their mobilization. Because PLZF specifically induces DNA methylation alteration in regions containing L1 sequences, we investigated whether PLZF could interact directly with genomic L1 retrotransposons in human haematopoietic cells. PLZF chromatin immunoprecipitation (ChIP) was performed in PLZF expressing human myeloid progenitor cells (KG1a) and the PLZF-bound DNA sequences were purified and sequenced. Sequence analysis of the purified genomic DNA fragments revealed that 57.5% (19/33) of the PLZF-bound ChIP fragments lay within L1 elements including the full-length L1 retrotransposon, confirming the finding of the genome-wide MeDIP-seq data in mice and further indicating that L1 elements are bona fide PLZF target sequences (Figure 2C; Supplementary Table SII). Of note, the remaining PLZF-bound sequences contain non-LTR retrotransposons such as SINE/Alu, but no SVA elements. Alignment of the ChIP sequences revealed an overlapping critical region of 450 bp in the centre of the full-length L1 retrotransposon, within the ORF2 but not in the promoter region (Figure 2C). An in silico search for putative PLZF-BS corroborated the presence of only one PLZF-BS (a 7-bp Hoxd11-like PLZF-BS motif, ATGTAAA; Barna et al, 2002), located within the ORF2 (nt 2634–2640) and not in the promoter region (Figure 2B). Interestingly, a comparative alignment of the PLZF-L1-interacting DNA sequences revealed a high degree of conservation of the PLZF-BS within the 19 ChIP-purified L1 sequences (Figures 2C and D). Notably, the human and mouse PLZF-BS are 100% identical, and an alignment of mammalian reference L1 DNA sequences containing the PLZF-BS reveals a high conservation between species with little sequence variation (Supplementary Figure S2A). T/A to C mutations within the PLZF-BS were able to abolish PLZF recruitment to L1 DNA sequences in vivo (Figure 2D). Because the L1 PLZF-BS is situated 2 kb downstream from the 5′ CpG island of the full-length L1 retrotransposon promoter, we questioned whether the PLZF-DNA binding at this site could be involved in the epigenetic modifications at the L1 promoter. Histone acetylation and protein binding activities were evaluated in an endogenous PLZF non-expressing cell line, 293T, expressing ectopic PLZF. PLZF ChIP was followed by semi-quantitative PCR spanning the full-length L1 sequence. The results of PLZF over expression show that PLZF is first recruited at the L1 PLZF-BS in ORF2 (Supplementary Figure S4), followed by the 3′–5′ propagation of a repressive chromatin environment towards the 5′ CpG UTR site (Figure 3A). In a sequential ChIP assay, the recruitment of DNA methylase (DNMT1) and histone deacetylase (HDAC1) proteins at the L1/PLZF-BS region (Figure 3C; Supplementary Figure S3B) confirmed local histone deacetylation and DNA methylation (Figure 3B PCR1; Supplementary Figure S4). Notably, HDAC3 is known to deacetylate PLZF and is thus not associated with the repressor complex recruited by PLZF (Figure 3C). The propagation signal terminates with the histone deacetylation of the L1 promoter at the 5′ UTR, followed by the recruitment of the methyl DNA binding protein (MeCP2, Figure 3B; MBD1, Supplementary Figure S3) and methylation of the CpG island located in the L1 promoter (Figure 3D). Thus, the induction of a non-permissive chromatin state at the L1 promoter correlates with PLZF-specific binding to a single distal 3′ L1 binding site.
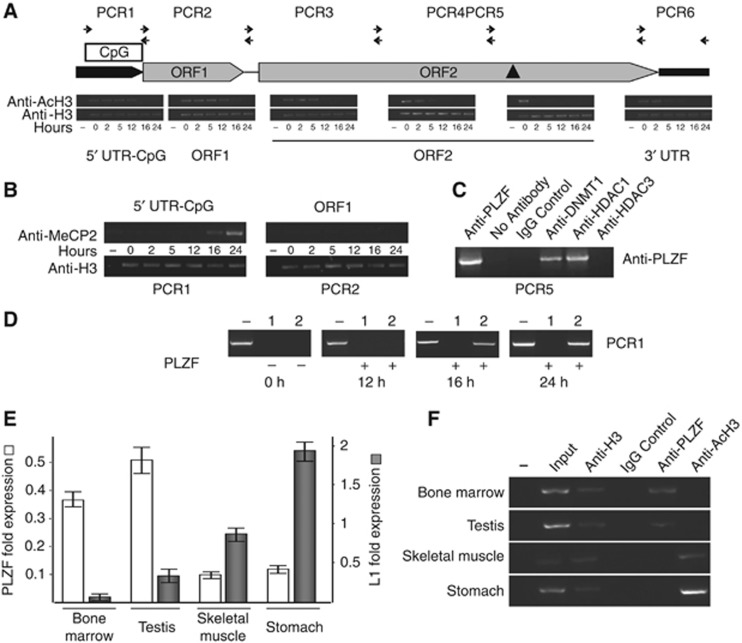
Figure 3.
PLZF induces epigenetic modifications at L1 retrotransposon loci. (A) Assessment of the histone H3 acetylation levels within the full-length L1 element after PLZF expression. Throughout the ChIP experiment, the ChIP efficiency was assessed by using an anti-histone H3 antibody (panel anti-H3) as a positive control. Histone acetylation levels were determined using an antibody raised against the acetylated form of histone H3 (panel anti-AcH3). All ChIP experiments were performed from a single chromatin preparation for each time point (0, 2, 5, 12, 16 and 24 h). Various segments of genomic DNA situated at the ORF2 (PCR3, PCR 4 and PCR5), as well as the ORF1 (PCR2) and the 5′ (PCR1) and 3′ (PCR6) UTR regions were tested at various time points after PLZF transfection in 293T cells. The results are shown as images of the PCR amplification products on agarose gels. The expression and DNA binding of the PLZF protein are shown in Supplementary Figure S3A. (B) PLZF-induced DNA methylation and methyl DNA binding protein recruitment. An antibody against MeCP2 (anti-MeCP2) was used to assess the abundance of MeCP2 proteins bound to L1 genomic DNA, particularly within the 5′ UTR region containing a CpG island (PCR1, 5′ UTR-CpG). (C) PLZF recruits a histone deacetylase and DNA methylase complex to L1 genomic sequences. Chromatin prepared from PLZF-expressing haematopoietic KG1 cells was subjected to sequential ChIP. An anti-PLZF antibody (anti-PLZF, left of the gel picture) was used during the first precipitation round, and purified complexes were again precipitated using the specific antibodies shown at the top of the image. Positive (anti-PLZF) and negative (no antibody and ChiP-grade IgG antibody (IgG control)) controls were used to validate the immunoprecipitation efficiency. Anti-HDAC (histone deacetylase 1 (HDAC1) and 3 (HDAC3)) and anti-Dnmt1 (DNA methyl transferase 1) were used to test the association of these proteins with the PLZF-bound L1 genomic sequences. PCR amplification was used to visualize purification of the genomic region of interest (PCR 5, genomic sequence containing the PLZF-BS, black triangle in schematic (A)). Other ChIP-L1 interacting fragments show the same PLZF-induced recruitment of proteins to DNA targets (Supplementary Figure S3). (D) The effect of PLZF expression in 293T cells on the methylation status of the L1 5′ UTR. The ectopic expression of the PLZF in these cells induced methylation in the 5′ UTR region, as indicated by the appearance of a specific band in lane 2 at 16 and 24 h post PLZF transfection, indicating the presence of DNA methylation by blocking the methylation sensitive-endonuclease activity. Thus, the presence of an amplicon revealed that this specific DNA sequence is methylated. Lane −, no enzyme input control; lane 1, HaeIII digest (as a control for digestion); lane 2, HpaI digest (methylation-sensitive endonuclease). (E) PLZF and L1 gene expressions in adult mouse tissues measured by real-time PCR. Low or high-PLZF expressing level tissues were collected from adult wild-type mice, RNAs were extracted and the levels of PLZF and L1 transcripts were assessed by quantitative PCR. (F) Histone acetylation and PLZF recruitment. ChIP was performed in adult mouse tissues to assess the acetylation level (anti-AcH3) and the PLZF binding activity (anti-PLZF) at L1 element. The level of histone H3 protein in the vicinity of the mouse PLZF-BS (anti-H3) was used as a positive control for the ChIP procedure and to normalize the quantity of input DNA for each tissue. An irrelevant ChIP-grade polyclonal antibody was used as a negative control (IgG control).
Source data for this figure is available on the online supplementary information page.
A change in the acetylation and methylation of L1 chromatin induced by PLZF is correlated with the regulation of L1 expression. In PLZFWT mice, L1 mRNA expression levels are negatively correlated with PLZF expression in BM, testis, skeletal muscle and stomach tissues (Figure 3E). The differential expression levels in these tissues correlate with the 5′ UTR methylation status of the L1 retrotransposon. The histone H3 acetylation, as determined by ChIP, shows that the chromatin in the 5′ UTR regulatory regions of these target genes is in a closed state in high PLZF-expressing tissues (BM and testis) and in an open state in low PLZF-expressing tissues (muscle and stomach) (Figure 3F). Interestingly, L1 expression levels are increased in PLZFOFF tissues compared to wild-type and PLZFON tissues, similar to the patterns observed for known PLZF target genes (c-myc, c-kit and CRABP1), underscoring that the L1 retrotransposon is a novel PLZF target (Figure 1D). Furthermore, these differential L1 expression levels are associated with the DNA methylation status determined by the MeDIP analysis (Figure 1C; Supplementary Figure S5). These results indicate that PLZF regulates the epigenetic state of full-length L1 elements, including the 5′ UTR regulatory region, in PLZF-expressing BM and testis, both in human and in mouse cells.
We then show that a member of the DNMT protein family, DNMT1, is recruited in the presence of PLZF to the L1 retrotransposons to induce specific DNA methylation of L1 sequences. Other DNMT members do interact with PLZF in vitro (data not shown), but we were unable to confirm their recruitment to L1 sequences in vivo. Furthermore, the methyl DNA binding protein MeCP2 is recruited to the L1 5′ UTR following PLZF-induced DNA methylation indicating that L1 repression could be directly mediated by MeCP2 binding at the L1 promoter (Figure 3B). Our findings indicate that the binding of PLZF to the ORF2-L1 DNA-BS leads to epigenetic regulation at the 5′ UTR regulatory region of the full-length L1 retrotransposon.
PLZF induces transcriptional repression by binding to L1 truncated elements inserted into coding genes
The results of the MeDIP experiment performed in this study in mouse and human cells reveal that the L1 sequences interacting with PLZF are located near or within the 3′ UTR of coding genes and in intronic regions (75% of the human PLZF-L1 ChIP purified sequences and 44.6% of mouse PLZF-associated DMRs; Supplementary Tables SI and SII).
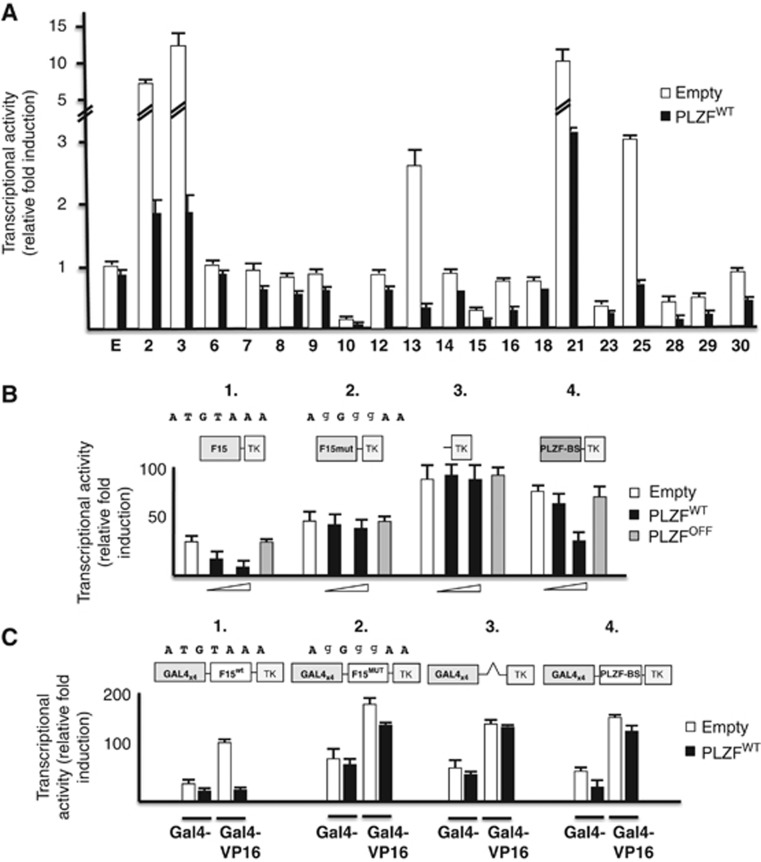
To assess whether PLZF binding to truncated L1 elements altered gene transcription, the 19 ChIP-purified genomic fragments (Supplementary Table SII) were cloned into two different luciferase plasmid reporters (pt109-tk-luc and pt109-GAL4BS-tk-luc) and transfected into non-PLZF expressing 293T cells in the absence or presence of a PLZF expressing plasmid. The analysis of these DNA sequences demonstrates PLZF-dependent repression of transcription by L1 inserted elements (Figure 4A) and that this occurs only in the presence of an intact PLZF-BS (Figure 4B). Inserted L1 elements are active modifiers of the human genome and may act as chromatin barrier to regulate gene expression. Here, L1-related gene repression was challenged with GAL4-VP16 activating transcriptional fusion proteins to assess the potential barrier functions of L1 fragments in the presence of PLZF (Figure 4C). While the expected transcriptional activation induced by the GAL4-VP16 fusion protein was observed in the presence of wild-type L1 genomic sequences alone, the co-expression of PLZF triggered a repressive state that persisted even in the presence of strong GAL4 activators such as VP16 (Figure 4C, panels 1 and 2). These results indicate that the PLZF-L1 interaction generates a non-permissive heterochromatin block, inhibiting the propagation of the VP-16-induced open chromatin state towards the promoter and block the reporter gene expression. Interestingly, this barrier function was not noted in reporter constructs that contained only synthetic PLZF-BS or L1 sequences with mutated PLZF-BS (Figure 4C, panels 2 and 4). Hence, PLZF uses L1 loci scattered throughout the genome as barrier boundaries.
Figure 4.
Transcriptional activities of PLZF-interacting L1 sequences. (A) Transcriptional repression activity of PLZF measured in purified L1 element genomic fragments. The fragments containing L1 sequences were cloned upstream of the thymidine kinase minimal promoter driving the expression of the luciferase gene (L1-tk-luc) and transiently transfected into 293T cells in the presence of 100 ng of various expression plasmids containing no insert or wild-type PLZF. The expression of the L1-tk-luc constructs was compared to the empty reporter vector. All L1 fragments show an inhibitory transcriptional activity in the presence of PLZF protein (black bar). (B) Transcriptional repression activity of PLZF via the L1 element binding site. The fragment containing the binding site was cloned upstream of the thymidine kinase minimal promoter driving the expression of the Luciferase gene F15-tk-luc (F15-TK) and F15mutant-tk-luc (containing the mutated PLZF BS (F15mut-TK), see sequence above) and transiently transfected into 293T cells in the presence of expression plasmids containing no insert, wild-type PLZF (100 and 200 ng) or a DNA binding-deficient point mutant (PLZFOFF) (200 ng). The expression of the F15-tk-Luc was compared to that of the empty reporter (tk-luc, negative control) and a 3-repeat copy of a previously validated PLZF BS (PLZF-BS-tk-luc) reporter vector (positive control for PLZF transcriptional repression activity). As observed in panel 1, PLZF (black histogram) reduces the transcriptional activity of the F15-TK reporter in a dose-dependent manner, while the mutant PLZF (grey histogram), which is unable to bind L1 DNA, does not affect its activity, as observed for the responsive PLZF reporter (PLZF-BS-TK, panel 4). The transcriptional activities of the empty reporter (TK, panel 3) and the L1 reporter containing the PLZF-BS mutant (F15mut-TK, panel 2) are not regulated by PLZF expression. (C) Barrier-like function of L1 PLZF-interacting sequences. L1 PLZF-interacting sequences containing the PLZF-binding site (BS) were cloned between four copies of the GAL4 BS and the thymidine kinase minimal promoter (TK) driving the expression of the Luciferase gene and transiently transfected into 293T cells in the presence of expression plasmids containing no insert, wild-type PLZF (200 ng) and/or Gal4-VP16 activating fusion protein (50 ng). Empty (GAL4 × 4-TK), F15 PLZF BS mutant (GAL4 × 4-F15mut-TK) and PLZF BS only (GAL4 × 4-PLZF-BS-TK) reporters were strongly activated by the expression of a GAL4-VP16 fusion activator (GAL4-VP16) but not by VP16 alone (panels 2, 3 and 4). In contrast, wild-type F15 PLZF BS (GAL4 × 4-F15wt-TK, panel 1) is refractory to GAL4-VP16 activation in the presence of PLZF expression (black histogram).
PLZF binds to L1 retrotransposon mRNA sequences
Taking into consideration that the L1-PLZF BS was present within the L1 ORF2 at an RNA stem loop secondary structure (Supplementary Figure S6A) and that members of the ZF protein family of trans-acting factors, such as PLZF, are also known to interact with mRNA and regulate their stability (Burdach et al, 2012), we hypothesized that PLZF could also be involved in L1 RNA regulation via direct interaction of PLZF to L1 mRNAs. In vitro RNA EMSA shows that PLZF binds ORF2 mRNA probes, an interaction not observed with PLZF-BS mutant probes (Supplementary Figure S6B). These mutations destabilize the stem loop secondary RNA structure associated with the wild-type L1 PLZF-BS sequence (Supplementary Figure S6A). PLZF/L1 mRNA complexes were also detected in vivo in human and mouse tissues (Figure 5A; Supplementary Figure S6C). We performed in vitro translation assays using wild-type and mutant L1 containing PLZF-BS mRNA templates and show that increasing amounts of PLZF protein could significantly decrease the ORF2 peptide level produced from wild-type template. These data correlate the integrity of the interaction between PLZF and the stem loop with poor translation (Figure 5B). To further understand the dual interaction of PLZF with DNA and RNA L1 sequences, we challenged PLZF function under cellular stress.
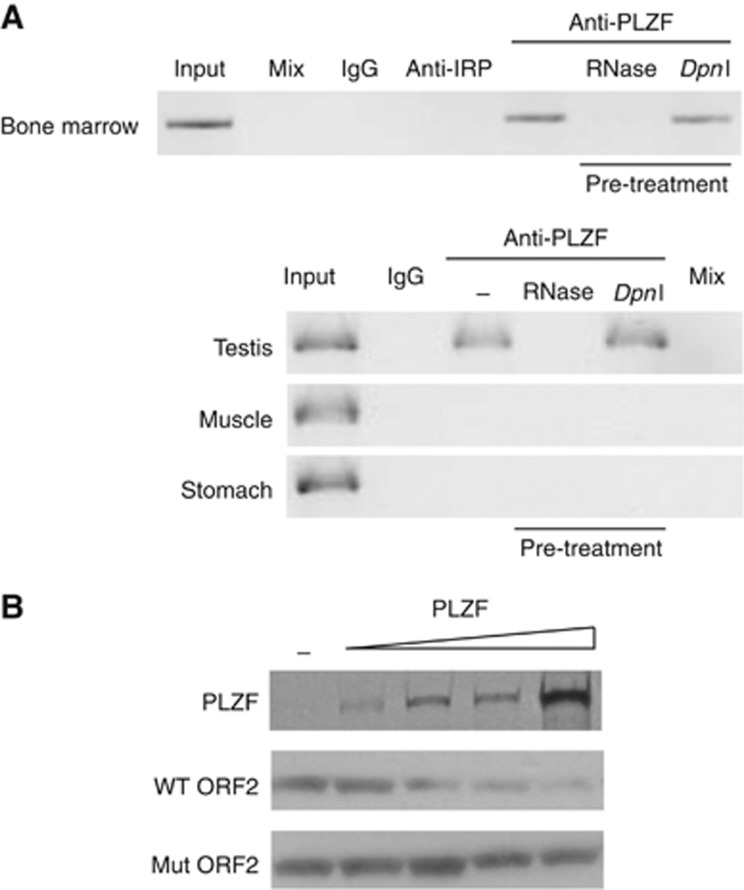
Figure 5.
PLZF interacts with L1 mRNAs and regulates their translation. (A) RNA-IP in mouse tissues. Immunoprecipitation with IgG, anti-IRP (control antibodies) and anti-PLZF was realized on mouse cell extracts from bone marrow, testis, muscle and stomach. Specific L1 PCR was performed and the presence of amplicon revealed an interaction between PLZF and its L1 target RNA. Pre-treatments by RNase or DpnI enzymes were used to validate the specificity of the RNA/PLZF interaction to degrade specific templates of RNA or possible genomic DNA contamination. (B) In vitro translation assay. ORF2 RNA templates were used to translate biotinylated ORF2 peptides in the presence or absence of PLZF protein. Incubation of the RNA templates with increasing amount of PLZF protein (PLZF panel) decreases the production of Wt ORF2 peptide (panel Wt ORF2), while ORF2 RNA template exhibiting mutations in the PLZF-BS are not affected by PLZF presence (Mut ORF2).
Source data for this figure is available on the online supplementary information page.
Cell stress induces PLZF delocalization and antagonizes the PLZF-L1 interaction leading to increased L1 retrotransposition
During induced cell stress in haematopoietic human KG1a cells, PLZF relocates from the nucleus speckles to the cytoplasm (Doulatov et al, 2009; Figure 6A1, panels 0 to 3 h and Figure 6A2), antagonizing the PLZF-L1 interaction leading to the release of L1 sequences from their PLZF-bound state (Figure 6B). The expression of L1 sequences is increased along with histone H3 acetylation (Figure 6B, lane 3; Figure 6C1), in line with the reported data showing mobilization and activation of L1 elements in the genome under cellular stress (Farkash et al, 2006; Goodier and Kazazian, 2008). The remaining low PLZF/L1 mRNA interaction observed, underscores the required presence of PLZF under stress conditions and its function in stem cell maintenance (Figure 6C2). These results in KG1a cells reinforce the observed loss of HSC self-renewal in PLZFOFF mutant mice that can be now correlated to hypomethylated L1 sequences (Figure 1C; Supplementary Figure S1).
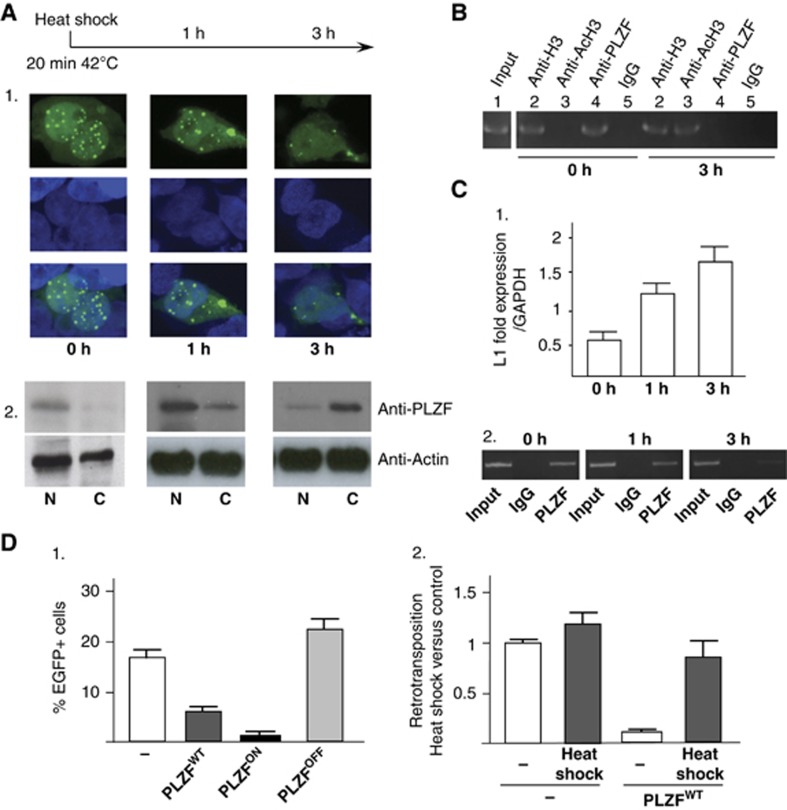
Figure 6.
Cellular stress and PLZF-induced L1 regulation. (A) Localization of the PLZF protein by immunofluorescence and western blotting during heat-shock stress in KG1 haematopoietic cells. KG1 cells were submitted to heat shock (20 min at 42°C, 0 h) and then allowed to recover for 1 or 3 h (1 and 3 h). PLZF immunofluorescence was performed as described in Guidez et al (2005) at these time points. PLZF, which is found in nuclear foci (0 h) under normal conditions, is detected in the cytoplasm of stressed cells (1 and 3 h) by both immunofluorescence (1) and western blotting analysis (2). Actin was used as negative control for translocation from the nucleus to the cytoplasm. (B) PLZF interaction with L1 sequences. PLZF DNA association with L1 sequences was evaluated by ChIP following heat-shock treatment. Negative (IgG, lane 5) and positive (anti-histone H3, lane 2) control antibodies were used to assess the specificity of the immunoprecipitation, while acetylation levels (anti-Ac histone H3, lane 4) and PLZF DNA binding (anti-PLZF, lane 3) indicated an increase in histone acetylation and a loss of PLZF DNA binding to L1 sequences under stress conditions. Lane 1 represents 10% of the input DNA used for PCR detection. (C) L1 expression during cellular stress. L1 expression was determined by qPCR relative to GAPDH expression. (D) (1) PLZF expression modulates the L1 retrotransposition frequency. The 293T cells transfected with the L1-EGFP were co-transfected with empty (−) or PLZFWT, PLZFON and PLZFOFF mutant expression vectors. The percentage of cells expressing EGFP was measured by flow cytometry. Percentages of EGFP-positive cells (% of EGFP+ cells) were scored on day 8 in the absence or presence of PLZF expression followed by heat shock-induced cellular stress (dotted boxes). (2) Effect of L1 retrotransposition during cellular stress. 294T cells, transfected with the L1-EGFP reporter plasmid in the presence of an empty (−) or PLZF expression (PLZFWT) vectors, were submitted to heat shock (20 min at 42°C) and returned to culture. Percentage of EGFP-positive cells was scored by flow cytometry and retrotransposition frequencies were compared to the control cells (transfected with the empty expression vector under normal cell culture conditions).
Source data for this figure is available on the online supplementary information page.
To further assess the negative effect of cell stress on PLZF function on L1 mobility, we tested L1 retrotransposition in the presence of wild-type PLZFWT, and the PLZFON and PLZFOFF mutants in a cultured cell assay upon heat shock-induced cellular stress. L1 retrotransposition frequency was measured by transfecting an active human element (containing the described PLZF DNA binding site), L1RP, tagged with an EGFP reporter as described previously (Ostertag et al, 2001; Farkash et al, 2006) in the presence or absence of PLZF. L1 insertion of sufficient length into a transcriptionally permissive location in the genome will express EGFP. As shown in Figure 6D, L1-EGFP cells were detected in 293T cells transfected with an empty or with the PLZFOFF expression vectors. Co-transfection with PLZF or PLZFON expression vectors leads to a decrease in EGFP-positive cells indicating a reduction in L1 retrotransposition frequency. However, when the transfected cells were submitted to heat shock-associated cellular stress the percentage of L1-EGFP in the presence of PLZFWT is increased indicating cell stress abolishes PLZFWT function and induces an increase in L1 retrotransposition is correlated with an increase in L1 retrotransposition (Figure 6D2).
Discussion
Together, these data suggest that PLZF binding to DNA and RNA L1 sequences is crucial for the inhibition of L1 expression at transcriptional levels in PLZF-expressing tissues. DNA binding activity of PLZF alters the local chromatin structure repressing the transcription of L1 retrotransposons. These two mechanisms are important for maintaining the tight repressive state, which are considered as the major players involved in direct (Beck et al, 2010; Ewing and Kazazian, 2011) and indirect retrotransposition in mammals (Dewannieux et al, 2003; Hancks et al, 2011; Raiz et al, 2012).
DNA interacting proteins, besides PLZF, have been reported to bind to the L1 retrotransposon including Runx3, p53 and SRY associated with the activation of L1 transcription (Tchénio et al, 2000; Yang et al, 2003; Harris et al, 2009). Until now, only one other transcription factor, the retinoblastoma protein (Rb), has been reported to induce L1 transcriptional repression by binding to the L1 promoter (Montoya-Durango et al, 2009) and we have previously shown that Rb and PLZF factors could cooperate to repress specific target promoters (Petrie et al, 2008). In this study, we show that PLZF is able to bind a specific DNA binding site, located outside the L1 promoter, to recruit proteins with epigenetic enzymatic activities (HDAC1 and DNMT1) inducing specific histone deacetylation and DNA methylation. Thus, distal PLZF-DNA binding induces deacetylation and DNA methylation at the L1 5′ UTR and the specific recruitment of methyl DNA-binding protein. Interestingly, L1 repression mechanisms have been associated with such epigenetic regulators as Dnmt3L. The loss of the DNA Methyl transferase Dnmt3L in mouse prevents L1 methylation in the testis leading to meiotic catastrophe, illustrating a crucial role for the DNMT protein in L1-CpG methylation (Bourc'his and Bestor, 2004; Schaefer et al, 2007). However, the mechanism by which DNMTs are specifically targeted and recruited to L1 sequences is not fully understood. DNA methylation is a crucial mechanism leading to L1 repression, the recruitment of methyl DNA binding proteins (MDBs) is also critical to implement full epigenetic repression of these sequences. Knock-out experiments of the MBD protein, MeCP2, are crucial to demonstrate the control of L1 repression in brain (Muotri et al, 2010). In the haematopoietic tissue, following PLZF-induced DNA methylation, MeCP2 also binds to the L1 promoter offering a molecular mechanism by which this MBD protein could be specifically targeted to the L1 promoter by the presence of PLZF. While PLZF is able to induce specific repression of full-length L1 retrotransposon in testicular and haematopoietic tissues, it appears that PLZF also uses truncated L1 sequences scattered throughout the genome, to establish putative chromatin boundaries and may thus play a direct role in the epigenetic regulation induced by L1 sequences of the genome (Slotkin and Martienssen, 2007). To note, human genome-wide studies have shown that one-quarter of expressed reference sequences contains an L1 retrotransposon in their 3′ UTR or intronic regions, associated with reduced gene expression (Faulkner et al, 2009). Here, we show that PLZF targets L1 sequences located exclusively within these genomic regions indicating that PLZF could be involved in maintaining the tissue-specific pattern of gene expression induced by L1 sequences. Of note, only a fraction of truncated L1s will contain the PLZF-BS sequence since most of the 5′ truncated L1s are <2 kb in length. The L1-PLZF interaction could underlie a novel regulatory function of PLZF through the contribution of L1 elements in the transcriptome of testicular and haematopoietic somatic cells.
A puzzling feature of L1 expression is its poor translation, this is not accounted by transcript instability, but rather by poor L1 transcript elongation suggesting a mammalian-specific mechanism for negatively regulating L1 expression (Han et al, 2004). Additionally, introduction of a thermostable stem loop in the inter-ORF spacer can reduce ORF2 protein translation (Alisch et al, 2006). Here, we proposed that PLZF interaction with a specific L1 RNA stem loop, located in the ORF2 region, might explained the poor translation of the L1 transcript translation by stabilization of the secondary RNA structure. Other than a secondary mechanism by which PLZF could regulate L1 expression in adult-PLZF expressing tissues, it appears that the specific L1 RNA-PLZF interaction might be of importance in keeping under control L1 activation during cellular stress. As PLZF epigenetic functions are abrogated during cellular stress, we have shown that PLZF/L1 RNA interaction still persists in order to maintain low L1 mRNA levels indicating a possible safeguard mechanism regulated by PLZF.
We have shown for the first time that the repressor PLZF is crucial for inducing DNA methylation of L1 sequences, the primary event of transposition suppression, and that PLZF-induced repression of L1 expression is also supplemented by a second degree of regulation at the RNA levels. This regulation is associated with the inhibition of L1 mobilization, indicating that PLZF is involved in the maintenance of L1 DNA dormant-state in somatic cells (Supplementary Figure S7).
Materials and methods
MeDIP and second-generation sequencing
DNA libraries were prepared from mouse tissues for multiplexed-paired-end sequencing on the Illumina GAIIx platform. Genomic DNA samples were quantified by Qubit DS DNA analysis and 6 μg of DNA was sonicated to obtain an average fragment size of ∼200 bp using the covaris S2 sonicator (Covaris, Massachusetts). Briefly, each sample was sonicated at 10% duty cycle, at intensity 5 with 200 cycles per burst, for 180 s at +4°C. Libraries were then prepared from the sonicated DNA samples using the NEBNEXT DNA library preparation kit (NEB, USA). After each process (sonication, blunt-ending, A-tailing, adapter ligation) sample clean-up was performed using a QIAquick PCR purification kit (QIAGEN, USA). Following library preparation, MeDIP was performed using 4 μg of each library, using a previously described method (Mohn et al, 2009). All samples (including inputs) were then amplified by LM-PCR and DNA libraries were tagged with a unique 6 NT index to allow pooling of 12 libraries from mice with the same background. This enabled the sequencing of multiplexed PLZFWT, PLZFOFF and PLZFON libraries on the same flow cell (indexes 1 and 7 for BM and indexes 2 and 8 for testis).
Samples were loaded on a flow cell and analysed in the Illumina genome analyzer.
The full protocol for the MeDIP-seq approach, that is, the coupling of MeDIP with next generation, short-read sequencing technologies is almost identical to the approach described in Down et al (2008). High-throughput sequencing with the libraries made from BM and testis DNA generates clusters, which are imaged and then converted into paired sequence reads (one read of 36 bp at the 5′end and one read of 36 bp at the 3′ end of each fragment). Reads generated during the Illumina sequencing run were 36 bp in length but represent fragments of DNA that were anywhere between 150 and 350 bp in length based on the size selected when gel purifying the adapter-ligated library. Library size of both single and pooled libraries was determined by Bioanalyzer (Agilent, USA). The samples reads were filtered and aligned to the genome and data were analysed by a binomial model to call regions with significantly different methylation levels between libraries. These examples show 1000, bp genomic regions where differences in methylation have been observed (see Supplementary Figure S5). The validation of DNA methylation enrichment was achieved using qPCR, with region-specific primers, which were designed for two regions known to be methylated and two unmethylated control regions; bactin and GAPDH. To quantify the amount of DNA methylation in these regions, the ratio of ΔCT of the MeDIP and input samples is calculated. This is done by comparing MeDIP samples against an input (sonicated library DNA was set aside before MeDIP was performed for use as input DNA).
ChIP and sequential ChIP assays
293T cells in 10-cm plates were co-transfected with 2 μg of PLZF expression vector and the L1 plasmid EF06R graciously provided by Dr Eline T Luning Prak University of Pennsylvania (described in Nucleic Acids Res 34: 1196, 2006) using the calcium phosphate precipitation method. Immunoprecipitation of plasmid DNA plus associated histones was carried out at various times after transfection according to a previously published protocol (Guidez et al, 2005, 2007), with the following modifications. Histone/DNA complexes were cross-linked by addition of 1% formaldehyde to the medium and incubation at 37°C for 10 min. After lysis, the chromatin was sonicated to 0.2–1.0 kb and diluted 10-fold in IP buffer (0.01% SDS, 1% Triton X-100, 2 mM EDTA, 20 mM Tris–HCl, pH 8.0, 150 mM NaCl, plus protease inhibitors). Protein samples for western blotting were taken prior to dilution; control samples for assaying input DNA were taken after dilution and decross-linked. Various antibodies were used for the immunoprecipitation, and the DNA/histone complexes were collected overnight with protein A/G-Sepharose beads (Santa Cruz Biotechnology). In sequential ChIP experiments (Guidez et al, 2007), complexes were eluted by incubation for 30 min at 37°C in 25 μl 10 mM DTT. After centrifugation, the supernatant was diluted 20 times with Re-ChIP buffer (1% Triton X-100, 2 mM EDTA, 150 mM NaCl, 20 mM Tris–HCl, pH 8.1) and subjected again to the ChIP procedure. After decross-linking of DNA, sequences were detected by semi-quantitative PCR using primers derived from sequences from the different human L1 genomic region (see Supplementary Materials and Methods, primer list). The number of cycles was determined empirically to give results that fall within the linear range of the particular PCR assay.
KG1 cells or dounce-homogenized cells from tissue samples were cross-linked and submitted to the ChIP procedure as described above.
RNA pull-down (RIP-Chip)
RNA pull-down was conducted as described in Keene et al (2006). In short, KG1 cells or murine cells were resuspended in polysome lysis buffer (PLB) supplemented with RNase and proteases inhibitors. A/G beads were precleared in 5% BSA in PLB and antibodies of interest were added and incubate overnight on rotating wheel at 4°C. mRNA lysates were added to the antibody mixture and incubated at 4°C for 4 h. Following washes, the mRNAs from the immunoprecipitated pellets were isolated by adding Trizol reagent. RNAs were reverse-transcribed using Moloney Murine Leukemia reverse transcriptase (M-MLV-RT; Gibco BRL) and random hexamers primers (Amersham), as suggested by manufacturer’s instructions. Sequences were detected by nested PCR using the primers described in Supplementary Materials and Methods. To ensure that RNA samples had no genomic contamination, DpnI endoculease pre-digested samples were assessed by PCR. Furthermore, pre-treatment with RNase A (Roche) of the immunoprecipitated purified RNA particles was carried out to assess that amplified products were only amplified from RNA template.
L1-EGFP transposition assay
Transient transfections using 293T cells were performed using the calcium phosphate precipitation method. On day 0, 1 × 104 293T cells were transfected with 0.350 μg of L1-EGFP (EF06R) with or without expression vectors (0.1 μg) and with carrier DNA to a total of 0.5 μg in total. Heat shock was carried out on day 2 for 20 min at 42oC and cells were returned to culture to recover. On day 8, cells were harvested and the percentage of EFGP+ cells analysed by flow cytometry (BD FACSCalibur), gating on live cells by forward/side scatter and propidium iodide/AnnexinV exclusion (as described in the Annexin V apoptotic detection Kit APC, eBioscience). All transfections were performed at least three times and in duplicates.
Supplementary Material
Acknowledgments
We thank Dr Eline T Luning Prak for the gift of the EF06R plasmid; R Roberts, L Delva and B Cassinat for critical reading of the manuscript. R Nancel and E Savariau for their help in designing the figures; Genoway in establishing the PLZF knock-in lines. Dr R Schultz for his help in the analysis of the MeDIP-seq data. This research was supported by grants from the Infertility Research Trust (IRT trust, Sheffield UK), the Leukaemia and Lymphoma Research (LLR, UK), the Kay Kendall Leukaemia Fund (KKLF, UK) (to FG) and The Wellcome Trust 085448/Z/08/Z (to WP) and 084358/Z/07/Z (to RJO).
Footnotes
The authors declare that they have no conflict of interest.
References
- Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV (2006) Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev 20: 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuara V, Perry P, Sauer S, Spivakov M, Jørgensen HF, John RM, Gouti M, Casanova M, Warnes G, Merkenschlager M, Fisher AG (2006) Cromatin signatures of pluripotent cell lines. Nat Cell Biol 8: 532–538 [DOI] [PubMed] [Google Scholar]
- Bao J, Yan W (2012) Male germline control of transposable elements. Biol Reprod 86: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R (1994) The POZ domain: a conserved protein-protein interaction motif. Genes Dev 8: 1664–1677 [DOI] [PubMed] [Google Scholar]
- Barna M, Hawe N, Niswander L, Pandolfi PP (2000) Plzf regulates limb and axial skeletal patterning. Nat Genet 24: 166–172 [DOI] [PubMed] [Google Scholar]
- Barna M, Merghoub T, Costoya JA, Ruggero D, Brandford M, Bergia A, Samori B, Pandolfi PP (2002) Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell 3: 499–510 [DOI] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV (2010) LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc'his D, Bestor TH (2004) Meiotic catastrophe and retrotransposon reactivation in male germ cells. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE (2004) Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 36: 647–652 [DOI] [PubMed] [Google Scholar]
- Burdach J, O'Connell MR, Mackay JP, Crossley M (2012) Two-timing zinc finger transcription factors liaising with RNA. Trends Biochem Sci 37: 199–205 [DOI] [PubMed] [Google Scholar]
- Chen Z, Brand NJ, Chen A, Chen SJ, Tong JH, Wang ZY, Waxman S, Zelent A (1993) Fusion between a novel Krüppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. EMBO J 12: 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Guidez F, Rousselot P, Agadir A, Chen SJ, Wang ZY, Degos L, Zelent A, Waxman S, Chomienne C (1994) PLZF-RAR alpha fusion proteins generated from the variant t(11;17)(q23;q21) translocation in acute promyelocytic leukemia inhibit ligand-dependent transactivation of wild-type retinoic acid receptors. Proc Natl Acad Sci USA 91: 1178–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA (2009) The impact of retrotransposons on human evolution. Nat Rev Genet 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costoya JA (2007) Functional analysis of the role of POK transcriptional repressors. Brief Funct Genomic Proteomic 6: 8–18 [DOI] [PubMed] [Google Scholar]
- Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP (2004) Essential role of PLZF in maintenance of spermatogonial stem cells. Nat Genet 36: 653–659 [DOI] [PubMed] [Google Scholar]
- Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, Morell M, O'Shea KS, Moran JV, Gage FH (2009) L1 retrotransposition in human neural progenitors cells. Nature 460: 1127–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M, Esnault C, Heidman T (2003) LINE-mediated retrotransposition of marked Alu sequences. Nat Genet 35: 41–48 [DOI] [PubMed] [Google Scholar]
- Doulatov S, Notta F, Rice KL, Howell L, Zelent A, Licht JD, Dick JE (2009) PLZF is a regulator of homeostatic and cytokine-induced myeloid development. Genes Dev 23: 2076–2087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Down TA, Rakyan VK, Turner DJ, Flicek P, Li H, Kulesha E, Gräf S, Johnson N, Herrero J, Tomazou EM, Thorne NP, Bäckdahl L, Herberth M, Howe KL, Jackson DK, Miretti MM, Marioni JC, Birney E, Hubbard TJ, Durbin R et al. (2008) A bayesian deconvolution strategy for immunoprecipitation-based DNA methylome analysis. Nat Biotechnol 26: 779–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH Jr (2011) Whole-genome resequensing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res 6: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkash EA, Kao GD, Horman SR, Prak ET (2006) Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res 34: 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner GJ (2011) Retrotransposons: mobile and mutagenic frm conception to death. FEBS Lett 585: 11589–11594 [DOI] [PubMed] [Google Scholar]
- Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Kawai J, Forrest AR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P (2009) The regulated retrotransposon transcriptome of mammalian cells. Nat Genet 41: 563–571 [DOI] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH Jr, Boeke JD (1996) Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposotion. Cell 87: 905–916 [DOI] [PubMed] [Google Scholar]
- Fischer S, Kohhase J, Böhm D, Schweiger B, Hoffmann D, Heitmann M, Horsthemke B, Wieczorek D (2008) Biallelic loss of function of the promyelocityc leukaemia zinc finger (PLZF) gene causes severe skeletal defects and genital hypoplasia. J Med Genet 45: 731–737 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Kazazian HH Jr (2008) Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135: 23–45 [DOI] [PubMed] [Google Scholar]
- Guidez F, Howell L, Isalan M, Cebrat M, Alani RM, Ivins S, Hormaesche I, McConnell MJ, Pierce S, Cole PA, Licht J, Zelent A (2005) Histone acetyltransferase activity of p300 is required for transcriptional repression by the promyelocytic leukemia zinc finger protein. Mol Cell Biol 25: 5552–5566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidez F, Ivins S, Zhu J, Söderström M, Waxman S, Zelent A (1998) Reduced retinoic acid-sensitiviness of nuclear receptor corepressor binding to PML- and PLZF-RARalpha underlie molecular pathogenesis and treatment of acute promyelocytic leukemia. Blood 91: 2634–2642 [PubMed] [Google Scholar]
- Guidez F, Parks S, Wong H, Jovanovic JV, de Thé H, Solomon E, Grimwade D (2007) RARalpha-PLZF overcomes PLZF-mediated repression of CRABPI, contributing to retinoid resistance in t(11;17) acute promyelocytic leukemia. Proc Natl Acad Sci USA 104: 18694–18699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidez F, Zelent A (2001) Role of nuclear receptor corepressors in leukemogenesis. Curr Top Microbiol Immunol 254: 165–185 [DOI] [PubMed] [Google Scholar]
- Han JS, Szask ST, Boeke JD (2004) Transcriptional disruption by L1 transposon and implications for mammalian transcriptiomes. Nature 429: 268–274 [DOI] [PubMed] [Google Scholar]
- Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH Jr (2011) Retrotransposon of marked SVA elements by human L1s in culture cells. Hum Mol Genet 20: 3386–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr (2012) Active human transposons: variation and disease. Curr Opin Genet Dev 22: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CR, Dewan A, Zupnick A, Normart R, Gabriel A, Prives C, Levine AJ, Hoh J (2009) p53 responsive elements in human transposons. Oncogene 28: 3857–3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He LZ, Guidez F, Tribioli C, Peruzzi D, Ruthardt M, Zelent A, Pandolfi PP (1998) Distinct interactions of PML-RARalpha and PLZF-RARalpha with co-repressors determine differential response to RA in APL. Nat Genet 18: 126–135 [DOI] [PubMed] [Google Scholar]
- Kang SI, Choi HW, Kim IY (2011) Redox-mediated modification of PLZF by SUMO-1 and ubiquitin. Biochem Biophys Acta 1809: 285–294 [DOI] [PubMed] [Google Scholar]
- Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, Kazazian HH Jr (2009) L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev 23: 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH Jr (2004) Mobile elements: drivers of genome evolution. Science 303: 1626–1632 [DOI] [PubMed] [Google Scholar]
- Keene JD, Komisarow JM, Friedersdorf MB (2006) RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc 1: 302–307 [DOI] [PubMed] [Google Scholar]
- Kelly KF, Daniel JM (2006) POZ for effect--POZ-ZF transcription factors in cancer and development. Trends Cell Biol 16: 578–587 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K et al. (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ 3rd, Lohr JG, Harris CC, Ding L, Wilson RK, Wheeler DA, Gibbs RA, Kucherlapati R, Lee C, Kharchenko PV, Park PJ, Cancer Genome Atlas Research Network. (2012) Landscape of somatic retrotransposotion in human cancers. Science 337: 967–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JY, English MA, Ball HJ, Yeyati PL, Waxman S, Licht JD (1997) Sequence-specific DNA binding and transcriptional regulation by the promyelocytic leukemia zinc finger protein. J Biol Chem 272: 22447–22455 [DOI] [PubMed] [Google Scholar]
- Martin SL, Cruceanu M, Branciforte D, Wai-Lun LiP, Kwok SC, Hodges RS, Williams MC (2005) LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J Mol Biol 348: 549–561 [DOI] [PubMed] [Google Scholar]
- Mathias SL, Scott AF, Kazazian HH Jr, Boeke JD, Gabriel A (1991) Reverse transcriptase encoded by a human transposable element. Science 254: 1808–1810 [DOI] [PubMed] [Google Scholar]
- Mohn F, Weber M, Schübeler D, Roloff TC (2009) Methylated DNA immunoprecipitation (MeDIP). Methods Mol Biol 507: 55–64 [DOI] [PubMed] [Google Scholar]
- Montoya-Durango DE, Liu Y, Teneng I, Kalbfleisch T, Lacy ME, Steffen MC, Ramos KS (2009) Epigenetic control of mammalian LINE-1 retrotransposon by retinoblastoma proteins. Mutat Res 665: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muotri AR, Marchetto MC, Coufal NG, Oefner R, Yeo G, Nakashima K, Gage FH (2010) L1 retrotransposon in neurons is modulated by MECP2. Nature 468: 443–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag EM, Prak ET, DeBerardinis RJ, Moran JV, Kazazian HH Jr (2001) Determination of L1 retrotransposon kinetics in cultured cells. Nucleic Acids Res 28: 1418–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrie K, Guidez F, Zhu J, Howell L, Owen G, Chew YP, Parks S, Waxman S, Licht J, Mittnacht S, Zelent A (2008) Retinoblastoma protein and the leukemia-associated PLZF transcription factor interact to repress target gene promoters. Oncogene 27: 5260–5266 [DOI] [PubMed] [Google Scholar]
- Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Löwer J, Strätling WH, Löwer R, Schumann GG (2012) The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res 40: 1666–1683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer CB, Ooi SK, Bestor TH, Bourc'his D (2007) Epigenetic decisions in mammalian germ cells. Science 316: 398–399 [DOI] [PubMed] [Google Scholar]
- Slotkin RK, Martienssen R (2007) Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 8: 272–285 [DOI] [PubMed] [Google Scholar]
- Solyom S, Ewing AD, Rahrmann EP, Doucet T, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, Wheelan S, Upton KR, Shukla R, Faulkner GJ, Largaespada DA, Kazazian HH Jr (2012) Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res 22: 2328–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Liebert F, Shöler A, van Nimwegen E, Wirbelauer C, Orkeley EJ, Gaidatzis D, Tiwari WK, Schübeler D (2011) DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480: 490–495 [DOI] [PubMed] [Google Scholar]
- Tchénio T, Casella JF, Heidmann T (2000) Members of the SRY family regulate the human LINE retrotransposons. Nucleic Acids Res 28: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Zhang L, Zhang Y, Kazazian HH Jr (2003) An important role for RUNX3 in human L1 transcription and retrotransposition. Nucleic Acids Res 31: 4929–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.