Abstract
Polycyclic Aromatic Hydrocarbons (PAHs) are a group of compounds that pose many health threats to human and animal life. They occur in nature as a result of incomplete combustion of organic matter, as well as from many anthropogenic sources including cigarette smoke and automobile exhaust. PAHs have been reported to cause liver damage, red blood cell damage and a variety of cancers. Because of this, methods to reduce the amount of PAHs in the environment are continuously being sought. The purpose of this study was to find soil bacteria capable of degrading high molecular weight PAHs, such as pyrene (Pyr) and benzo[a]pyrene (BaP), which contain more than three benzene rings and so persist in the environment. Bacillus subtilis, identified by fatty acid methyl ester (FAME) analysis, was isolated from PAH contaminated soil. Because it grew in the presence of 33μg/ml each of pyrene, 1-AP and 1-HP, its biodegradation capabilities were assessed. It was found that after a four-day incubation period at 30°C in 20μg/ml pyrene or benzo[a]pyrene, B. subtilis was able to transform approximately 40% and 50% pyrene and benzo[a]pyrene, respectively. This is the first report implicating B. subtilis in PAH degradation. Whether or not the intermediates resulting from the transformation are more toxic than their parent compounds, and whether B. subtilis is capable of mineralizing pyrene or benzo[a]pyrene to carbon dioxide and water, remains to be evaluated.
Keywords: Polycyclic Aromatic Hydrocarbons, bioremediation, Bacillus subtilis
Introduction
Polycyclic Aromatic Hydrocarbons (PAHs) are a class of compounds composed of two or more fused benzene rings [1, 2]. They occur naturally, resulting from the incomplete combustion of organic matter, volcanic eruptions and forest fires. They are also distributed into the environment due to such human activities as cigarette smoking, automobile exhaust, and the processing, production and spillage of petroleum [3, 4]. Many of these compounds have been found to have toxic, mutagenic and/or carcinogenic properties [5–7]. PAHs are photocytotoxic to both plants and animals [8, 9]. They are found in soil and water, and therefore are able to enter vegetation and aquatic organisms that humans will ultimately consume. Their fused benzene ring structure makes PAHs very hydrophobic and therefore, highly stable. These properties make them persist in the environment as recalcitrant pollutants for very long periods of time. For these reasons, the removal of PAHs from the environment is important and necessary.
Biological systems have been widely used in various aspects of the remediation of the environment. Plants, fungi and microorganisms have been used as very effective tools in the removal of toxic metals and harmful organic xenobiotics from the environment. Microbial activity is the most significant cause of PAH removal from the environment, and the ability of bacteria to degrade PAHs has been well documented [10]. This documentation, however, is mainly limited to low molecular weight (LMW) PAHs, which contain fewer than four benzene rings. High-molecular weight (HMW) PAHs, containing four or more benzene rings, are more difficult to degrade because they are extremely hydrophobic [11, 12]. Few bacteria strains have been reported with the ability to degrade HMW-PAHs. The focus of this research is finding soil bacteria that are capable of degrading HMW-PAHs, such as Pyrene (Pyr) and Benzo[a]pyrene (BaP). In this study, we hypothesize that soil bacteria capable of degrading HMW-PAH will be isolated from PAH-contaminated soil.
Materials and Methods
Chemicals and Reagents
Tryptone, bacto-agar, sodium chloride (NaCl), yeast extract, K2HPO4, KH2PO4, (NH4)2SO4, sodium citrate•2H2O, MgSO4•7H2O, Ca(NO3)2, MnCl2, FeSO4, glucose, pyrene (Pyr), 1-aminopyrene (1-AP), 1-hydroxypyrene (1-HP), benzo[a]pyrene (BaP), N, N-dimethylformamide, acetonitrile and HPLC grade hexane were purchased from Sigma-Aldrich Chemical Co., (St. Louis, Missouri). Contaminated soil sample taken from a site in Southern Maryland was provided by Dr. Huey-Min Hwang.
Preparation of Media
Luria Bertani (LB) broth contained 5.0g tryptone, 5.0g NaCl and 2.5g yeast extract per liter of distilled water. Minimum Medium (MM) was prepared by dissolving 13.9g K2HPO4, 6.0g KH2PO4, 2.0g (NH4)SO4, 1.9 g Sodium Citrate • 2H2O, 0.2g MgSO4 • 7H2O, 0.1ml 1M Ca(NO3)2, 1.0ml of 0.1M MnCl2 and 0.1ml of 0.01M FeSO4 in 1L of distilled water. Glucose stock solutions (10%) were prepared. All solid media contained 1.8% agar along with the other appropriate constituents. Stock solutions (1.0mg/ml) of pyrene (Pyr), 1-aminopyrene (1-AP) and 1-hydroxypyrene (1-HP) and benzo[a]pyrene (BaP) were prepared in N, N-dimethylformamide (DMF).
Isolation of Bacteria from PAH-Contaminated Soil
One gram of PAH-contaminated soil was re-suspended in 100ml of sterile dH2O with vigorous stirring for 10 minutes. This suspension was then allowed to settle at room temperature for 30 minutes. A 100μl sample of the supernatant was spread onto a LB agar plate and incubated at 30°C. Colonies from the LB plate were then spread onto Minimum Medium plus 1% glucose (MG) plates and incubated at 30°C. These spread and transfer processes were repeated several times on MG, MM, and finally, MM+PAH (1μg/ml each Pyr, 1-AP, 1-HP and BaP). Eleven (11) distinct isolates were obtained. One colony of each of the eleven isolates was used to inoculate 5ml of MG and incubated at 30°C in an orbital shaker at a speed of 150 rpm. The cultures were then stored in 10% glycerol at 4°C and labeled Tgr1-11. A 200μl aliquot of each isolate from frozen stock was used to inoculate 10 ml of MM+PAH and incubated at 30°C in an orbital shaker at a speed of 150 rpm for 24hr. Over a four week period, the PAH concentration was increased from 10, 15, 20, 25 to ~33μg/ml. The isolates that survived were subsequently sent to Dr. Veronica Acosta at the USDA in Lubbock, TX for identification by Fatty Acid Methyl Ester (FAME) analysis.
Growth of Tgr3 in Various Media
Five milliliters of MM were inoculated with 100μl of Tgr3. Cultures were incubated in an orbital shaker whose cover was wrapped with aluminium foil to exclude light. Temperature was set for 30°C and shaker speed was 150 rpm for 24 hours. From the 24 hour culture, a subculture was grown in MM until it reached midlog phase (12–14 hours). A 500μl sample of the midlog culture was used to inoculate 25ml of MM, MG and MM supplemented with 20μg/ml pyrene or benzo[a]pyrene. Turbidity measurements were taken using a Klett Colorimeter from the time of inoculation (to) until measurements remained constant.
Tgr3 Viability in the Presence of Different PAH
Five milliliters of MM were inoculated with 100μl of Tgr3. Cultures were incubated in an orbital shaker whose cover was wrapped with aluminum foil to exclude light. Temperature was set for 30°C and shaker speed was 150 rpm for 24 hours. From the 24 hour culture, a subculture was grown in MM until it reached midlog phase. A 100μl sample of the midlog culture was used to inoculate 5ml of MM, MG and MM supplemented with Pyr or BaP, to a final concentration of 20μg/ml. The cultures were incubated at 30°C for 4 days, with shaking. Every 24 hours, serial dilutions of the cultures were made and spread onto a MM plate. Plates were incubated until colonies appeared (~48 hours) and then counted.
Assessment of Degradation Capacity of Tgr3
Five milliliters of MM were inoculated with 100μl of Tgr3. Cultures were incubated in an orbital shaker whose cover was wrapped with aluminum foil to exclude light. Temperature was set for 30°C and shaker speed was 150 rpm for 24 hours. From the 24 hour culture, a subculture was grown in MM until it reached midlog phase. Half of the midlog culture was autoclaved at 121°C/15psi for 15 minutes. A 100μl sample of the midlog culture or heat-killed culture was used to inoculate 5ml of MM supplemented with pyrene or benzo[a]pyrene, to a final concentration of 20μg/ml. The cultures were incubated at 30°C for 4 days, with shaking. Every 24 hours, the PAH was extracted with 2.0 ml of HPLC grade hexane, 1.0ml at a time, vortexed at maximum speed for 20 seconds, and incubated at −20°C to aid with layer separation.
Analysis of PAH Extracted from Transformation Experiment
The PAHs extracted from completed transformation experiments were analyzed using Waters HPLC system consisting of dual λ absorbance detector and a Supercosil LC-PAH (5μm, 25.0cm × 4.6mm ID) column. Analysis was performed in gradient mode from 60% water (solvent A) to 100% acetonitrile (solvent B) at a flow rate of 1.5ml/min. After isocratic run for 5 minutes in 40% solvent B, the mobile phase was changed by a linear gradient over 25 minutes to solvent B and elution continued for a further 10 minutes. The UV light absorption was monitored at 2 nm wavelength intervals from 210 to 600nm. The wavelength used to integrate peaks was 254 nm.
Results
Isolation of Bacteria from PAH-Contaminated Soil
We spread 100μl aliquots of the supernatant from suspension of PAH-contaminated soil in water on LB plates. Successive spread and transfer of bacteria to different growth media supplemented with selected PAH, allowed us to isolate about a dozen bacteria. Isolates were grown in MM for 24 hours prior to initial characterization using Gram staining according to the Becton-Dickinson Gram stain protocol. As shown in Table 1, 11 distinct isolates were obtained. These isolates were further characterized using fatty acid methyl ester (FAME) analysis. Fatty acids were extracted from pure bacteria isolates using the procedure described for pure culture isolates by the Microbial Identification System (MIS, Microbial ID, Inc., Newark, DE). The fatty acid methyl esters (FAME) were extracted and sent to the USDA Plant Stress and Water Conservation Laboratory in Lubbock, TX where they were analyzed in a 6890 GC Series II (Agilent Technologies). FAMEs were identified, and their relative peak areas were determined by the MIS Aerobe method of the MIDI system (Microbial ID, Inc., Newark, DE). The aerobic library for the TSBA 40 method (Version 4.10) was used to identify the bacteria. Isolates 3, 4, (Tgr3 and Tgr4) were identified as Bacillus subtilis and Burkholderia cepacia, respectively. Both isolates 7 and 9 (Tgr7 and Tgr9) were identified as Pseudomonas cepacia.
Table 1.
Characteristics of soil bacterial isolates: Isolates were grown in MM for 24 hours and Gram stained according to the Becton-Dickinson Gram Stain Protocol Growth and Viability of Tgr3
| Bacterium | Characteristics | ||
|---|---|---|---|
|
| |||
| Morphology | Gram Stain | Other Features | |
| Tgr1 | Rod Shaped | − | Cloudy w/yellow flocculation |
| Tgr 2 | Rod Shaped | − | Cloudy white |
| Tgr 3 | Rod Shaped | + | Cloudy white |
| Tgr 4 | Rod Shaped | − | Cloudy white |
| Tgr 5 | Rod Shaped | − | Cloudy white |
| Tgr 7 | Rod Shaped | + | Cloudy white |
| Tgr 8 | Rod Shaped | + | Cloudy white |
| Tgr 9 | Rod Shaped | − | Cloudy white |
| Tgr 10 | Rod Shaped | + | Slightly cloudy |
| Tgr 11 | Rod Shaped | − | Cloudy white |
| Tgr 12 | Rod Shaped | − | Slightly cloudy |
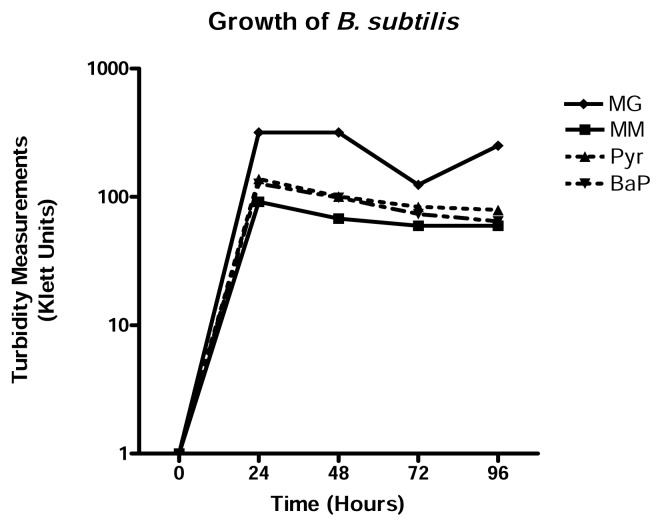
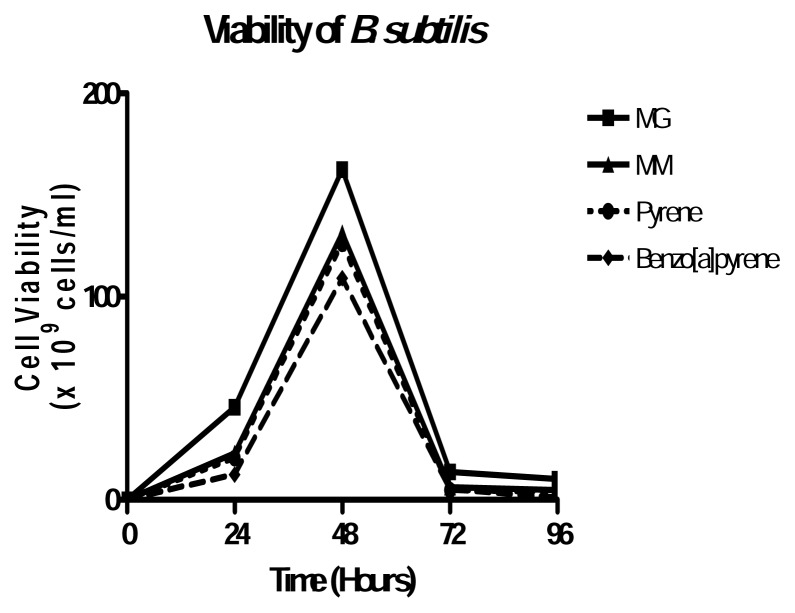
To assess the potential of B. subtilis as a PAH degrader, several investigations were done. B. subtilis is able to grow in 20μg/ml pyrene and benzo[a]pyrene. Growth physiology results suggest that the growth of B. subtilis was not affected by 20μg/ml pyrene or benzo[a]pyrene because its growth in either PAH was almost identical to that in minimum medium alone (Figure 1). Turbidity measurements taken after every 24 hours of growth also indicated that B. subtilis reached exponential phase within the first 24 hours of growth and began to decline slowly thereafter in each growth medium. Figure 2 shows that B. subtilis achieves maximum viability at 48 hours in all media used. As expected, MG is still the best growth medium, with virtually no difference between MM, MM+Pyr and MM+BaP.
Figure 1.
Growth physiology of B. subtilis in MM, MG, and MM+PAH. B. subtilis was grown in each medium for 4 consecutive days. Turbidity measurements were taken every 24 hours using a Klett colorimeter. Results represent the mean +/−SD values of experiment performed in triplicate.
Figure 2.
Viability of B. subtilis in MM, MG and MM+PAH. B. subtilis grown in each medium was spread onto MM plates every 24 hours. Plates were incubated at 30°C for 48 hours and colonies were counted. Results represent the mean +/− SD values of experiment performed in triplicate.
Transformation Experiments
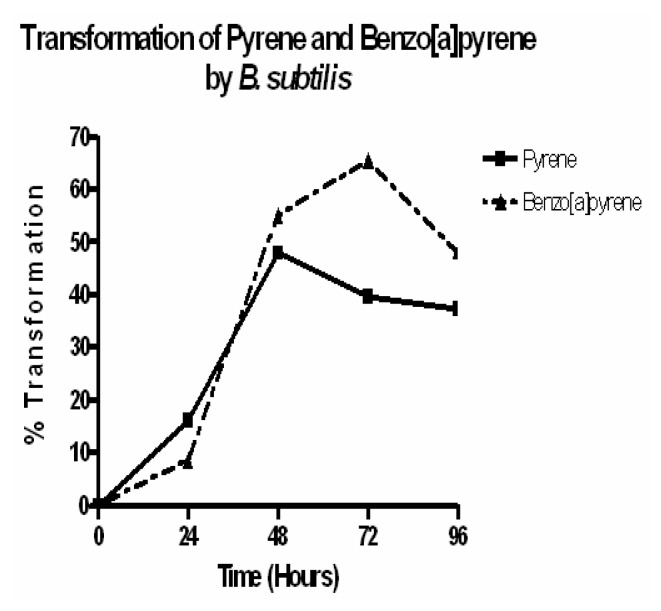
PAH transformation experiments revealed that B. subtilis has the ability to transform pyrene and benzo[a]pyrene in its growth medium. PAH transformation occurred within the first 24 hours of growth in 20μg/ml pyrene and benzo[a]pyrene, with approximately 15% and 8% of the pyrene and benzo[a]pyrene being transformed, respectively (Figure 3). B. subtilis reached its peak growth (~ 1.25 × 10 11cells/ml) after 48 hours in pyrene, where it transformed about 55% of the pyrene in the growth medium (Figure 3). After 48 hours of growth, B. subtilis transformed about 50% of the benzo[a]pyrene in its growth medium, where it was also at its peak growth (~ 1.09 × 1011 cells/ml). Even though the number of viable cells significantly decreased after 72 hours of growth in benzo[a]pyrene, B. subtilis continued to transform up to about 65% of the benzo[a]pyrene present. As far as we know, this is the first report suggesting a potential role for B. subtilis in the degradation of PAH.
Figure 3.
Transformation of benzo[a]pyrene and pyrene by B. subtilis. B. subtilis was grown in MM plus 20μg/ml BaP or Pyr for 4 consecutive days. Every 24 hours, the PAH was extracted with HPLC grade hexane and analyzed by HPLC. Results represent the mean values of experiment performed in triplicate.
Discussion
Isolation of Potential HMW-PAH Degrading Bacteria
Soil bacteria are frequently examined for their abilities to degrade PAHs because of the prevalence of the compounds in soils. In previous research, bacteria have been isolated from contaminated soils in areas such as coal gasification sites, creosote-contaminated sites and oil fields [1, 13–14]. These sites are usually assumed to be contaminated with PAHs because of these anthropogenic activities. Since bacteria are present virtually everywhere in nature, it is reasonable to expect them to be found in PAH-contaminated sites. Those bacteria that survive in such sites are likely to be able to degrade or metabolize the contaminant. Few bacteria are known that can degrade HMW PAHs. White rot fungi are more often associated with HMW PAH degradation than bacteria. They are believed to produce such enzymes as lignin peroxidase and P450 monooxygenases for use in PAH degradation [15]. As the search for HMW PAH degrading bacterium goes on, more genera are being identified as degraders. The Mycobacterium genus is the family that is most often associated with degradation of HMW PAHs [14, 16, 17]. Mycobacterium austroafricanum GTI-23 can utilize phenanthrene, fluoranthene and pyrene as a sole source of carbon and energy, and mineralize 300μg/ml pyrene in liquid culture [18]. It can also partially degrade 300μg/ml fluorene and benzo[a]pyrene. After 17 days of growth, M. austroafricanum completely degraded [4, 5, 9, 10 14C]Pyrene to 14CO2.
Another Mycobacterium, strain RJGII-135, isolated from a coal gasification site, was able to degrade more than 45% [14C ]Pyrene, with metabolites such as methylated -4-phenanthtrene-carboxylic acid, methylated-4,5-Phenanthrene dicarboxylic acid and 4,5-pyrene-dihydrodiol being produced [14]. Although significantly slower, RJGII-135 also degraded [14C]benzo[a]pyrene, producing metabolites cis-4-(7-hydroxypyrene-8-yl)-2-oxobut-3-enoic acid, methylated 4,5-chrysene-dicarboxylic acid, cis-7,8-dihydrodiol-BaP, 7,8-dihydro-pyrene-8-carboxylic acid. This organism was also able to degrade benzo[a]anthracene at a rate similar to that of pyrene. Mycobacterium sp. strain AP1 grew in mineral medium with pyrene as the sole source of carbon [16]. After 6 days of growth, the bacterium decreased the amount of pyrene in the growth medium from 180 g/ml to 50 g/ml while producing, for the first time in literature, metabolite 6,6-dihydroxy-2,2-biphenyl dicarboxylic acid.
Pseudomonas stutzeri strain P16, Pseudomonas saccharophila strain P15, Bacillus cereus strain P21 and Sphingomonas yanoikuyae strain R1 all play an active role in pyrene metabolism when grown in minimum salts buffer [19]. Strains P16 and P21 are able to transform pyrene into the first intermediate in degradation by aerobic organisms, cis-4,5-dihydro-4,5-dihydroxypyrene. Strains P15 and R1 were not able to transform pyrene into any intermediates, but they were able to further metabolize cis-4,5-dihydro-4,5-dihydroxypyrene into pyrene-4,5-dione.
Conclusions
Isolates 3, 4, (Tgr3 and Tgr4) were identified as Bacillus subtilis and Burkholderia cepacia, respectively. Both isolates 7 and 9 (Tgr7 and Tgr9) were identified as Pseudomonas cepacia by fatty acid methyl ester analysis. Results of transformation experiments clearly suggest that B. subtilis transforms about 40% of pyrene and 50% of benzo[a]pyrene in its growth medium. B. subtilis has previously been used in the bioremediation of selenite, a mutagenic selenium compound that causes base-pair substitution in DNA, because of its ability to convert selenite to elemental selenium [20]. These transformation results are significant because as far as we know, this is the first report suggesting a potential role for B. subtilis in the degradation of PAH. This is even more fascinating considering the fact that Tgr3 was isolated from the same soil sample as Burkholderia cepacia, isolate 4 (Tgr4) and Pseudomonas cepacia, isolates 7 and 9 (Tgr7 and Tgr9), which have previously been reported to degrade HMW-PAH [21–22]. This finding warrants further extensive investigation of Tgr3 isolate. The toxicity of the metabolites produced during transformation must be assessed. Mineralization of pyrene and benzo[a]pyrene by Tgr3 is yet to be determined. If as these results seem to suggest B. subtilis is able to degrade HMW-PAH, its use in the clean-up of PAH-contaminated sites will go a long way in improving the health of the environment for the benefit of humankind.
Acknowledgements
This research was supported in part by a grant from the Army Research Office (Grant# DAAD 19-01-1-0733), awarded to Jackson State University, and in part by a grant from the National Institutes of Health Research Centers in Minority Institutions, NIH-RCMI (Grant #1G12RR13459) awarded to Jackson State University. Part of this work was used by Rochelle Hunter to satisfy the Masters Degree thesis requirement at Jackson State University. I thank Dr. Veronica Acosta for her technical assistance in identifying the isolates by Fatty Acid Methyl Ester (FAME) analysis.
References
- 1.Geiselbrecht A., Hedlund B., Tichi M., Staley J. Isolation of marine polycyclic aromatic hydrocarbon (PAH)-degrading Cycloclasticus strains from the Gulf of Mexico and comparison of their PAH degradation ability with that of Puget Sound Cycloclasticus strains. Appl Environ Micro. 1998;64(12):4703–4710. doi: 10.1128/aem.64.12.4703-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cerniglia C. Microbial metabolisms of polycyclic aromatic hydrocarbons. Adv Appl Micro. 1984;30:31–71. doi: 10.1016/s0065-2164(08)70052-2. [DOI] [PubMed] [Google Scholar]
- 3.Samanta S., Singh O., Jain R. Polycyclic aromatic hydrocarbons: environmental pollution and bioremediation. Trends Biotech. 2002;20(6):243–248. doi: 10.1016/s0167-7799(02)01943-1. [DOI] [PubMed] [Google Scholar]
- 4.Kastner M., Breuer-Jammali M., Mahro B. Impact of inoculation protocols, salinity and pH on the degradation of polycyclic aromatic hydrocarbons (PAHs) and survival of PAH-degrading bacteria introduced into soil. Appl Environ Micro. 1998;64(1):359–362. doi: 10.1128/aem.64.1.359-362.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Villemur R., Deziel E., Benachenhou A., Marcoux J., Gauthier E., Lepine F., Beaudet R., Commeau Y. Two-liquid-phase slurry bioreactors to enhance the degradation of high-molecular-weight polycyclic aromatic hydrocarbons in soil. Biotecnol Prog. 2000;16:966–972. doi: 10.1021/bp000118j. [DOI] [PubMed] [Google Scholar]
- 6.Guerin T. Bioremediation of phenols and polycyclic aromatic hydrocarbons in creosote contaminated soil using ex-situ land treatment. J Haz Mat. 1999;B65:305–315. doi: 10.1016/s0304-3894(99)00002-3. [DOI] [PubMed] [Google Scholar]
- 7.Dagher F., Deziel E., Lirette P., Paquette G., Bisaillon J. G., Villemur R. Comparative study of five polycyclic aromatic hydrocarbon degrading bacterial strains isolated from contaminated soils. Can J Micro. 1997;43:368–377. doi: 10.1139/m97-051. [DOI] [PubMed] [Google Scholar]
- 8.Schirmer K., Xhan A. G. J., Greenberg B. M., Dixon D. G., Bols N. C. Ability of 16 priority PAHs to be phototoxic to a cell line from rainbow trout gill. Toxicol. 1998;127:143–155. doi: 10.1016/s0300-483x(98)00031-6. [DOI] [PubMed] [Google Scholar]
- 9.Shyong E., Lu Y., Goldstein A., Lebwohl M., Wei H. Synergistic enhancement of H2O2 production in human epidermoid carcinoma cells by benzo[a]pyrene and ultraviolet A radiation. Toxicol Appl Pharm. 2003;188:104–109. doi: 10.1016/s0041-008x(03)00018-8. [DOI] [PubMed] [Google Scholar]
- 10.Daane L., Harjone I., Zylstra G., Haggblom M. Isolation and characterization of polycyclic aromatic hydrocarbon-degrading bacteria associated with the rhizosphere of salt marsh plants. Appl Environ Micro. 2001;67(6):2683–2691. doi: 10.1128/AEM.67.6.2683-2691.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gauthier E., Deziel E., Villlemur R., Juteau P., Lepine F., Beaudet R. Initial characterization of new bacteria degrading high-molecular weight polycyclic aromatic hydrocarbons isolated from a 2-year enrichment in a two-liquid-phase culture system. J Appl Micro. 2003;94:301–311. doi: 10.1046/j.1365-2672.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- 12.Kanaly R., Bartha R., Watanabe K., Harayama S. Rapid mineralization of benozo[a]pyrene by a microbial consortium growing on diesel fuel. Appl Environ Micro. 2000;66(10):4205–4211. doi: 10.1128/aem.66.10.4205-4211.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samanta S. K., Chakrabort A. K., Jain R. K. Degradation of phenanthrene by different bacteria: evidence for novel transformation sequences involving the formation of 1-naphthol. Appl Micro Biotech. 1999;53:98–107. doi: 10.1007/s002530051621. [DOI] [PubMed] [Google Scholar]
- 14.Schneider J., Grosser R., Jayasimhulu K., Xue W., Warshawsky D. Degradation of pyrene, benzo[a]anthracene and benzo[a]pyrene by Mycobacterium sp. Strain RJGII-135, isolated from a former coal gasification site. Appl Environ Micro. 1996;62:13–19. doi: 10.1128/aem.62.1.13-19.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harayama S. Polycyclic aromatic hydrocarbon bioremediation design. Curr Opin Biotech. 1997;8:268–273. doi: 10.1016/s0958-1669(97)80002-x. [DOI] [PubMed] [Google Scholar]
- 16.Vila J., Lopez Z., Sabate J., Minguillon C., Solanas A. M., Grifoll M. Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp. Strain AP1: actions of the isolate on two and three-ring polycyclic aromatic hydrocarbons. Appl Environ Micro. 2001;67:5497–5505. doi: 10.1128/AEM.67.12.5497-5505.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Churchill S. A., Harper J. P., Churchill P. F. Isolation and characterization of a Mycobacterium species capable of degrading three and four ring aromatic and aliphatic hydrocarbons. Appl Environ Micro. 1999;65:549–552. doi: 10.1128/aem.65.2.549-552.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bogan B. W., Lahner L. M., Sullian W. R., Paterek J. R. Degradation of straight-chain aliphatic and high-molecular-weight polycyclic aromatic hydrocarbons by a strain of Mycobacterium austroafricanum. J. Appl Micro. 2003;94:230–239. doi: 10.1046/j.1365-2672.2003.01824.x. [DOI] [PubMed] [Google Scholar]
- 19.Kazunga C., Aitken M. Products from incomplete metabolism of pyrene by polycyclic aromatic hydrocarbon-degrading bacteria. Appl Environ Micro. 2000;66(5):1917–1922. doi: 10.1128/aem.66.5.1917-1922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Noda M., Takano T., Sakurai H. Mutagenic activity of selenium compounds. Mut Res. 1979;66(2):175–179. doi: 10.1016/0165-1218(79)90063-6. [DOI] [PubMed] [Google Scholar]
- 21.Juhasz A. L., Britz M. L., Stanley G. A. Degradation of high molecular weight polycyclic aromatic hydrocarbons by Pseudomonas cepacia. Biotech Lett. 1996;18:577–582. [Google Scholar]
- 22.Juhasz A. L., Britz M. L., Stanley G. A. Degradation of fluoranthene, pyrene, benz[a]anthracene and dibenz[a,h]anthracene by Burkholderia cepacia. J Appl Micro. 1997;83:189–198. [Google Scholar]