Abstract
The SLC4A7 gene encodes the electroneutral sodium/HCO3 cotransporter NBCn1, which plays important physiological and pathophysiological roles in many cell types. Previous work identified six NBCn1 variants differing in the sequence of the extreme N terminus – MEAD in rat only, MERF in human only – as well as in the optional inclusion of cassettes I, II, and III. Earlier work also left open the question of whether optional structural elements (OSEs) affect surface abundance or intrinsic (per-molecule) transport activity. Here, we demonstrate for the first time that SLC4A7 from one species can express both MEAD- and MERF-NBCn1. We also identify a novel cassette IV of 20 aa, and extend by 10 the number of full-length NBCn1 variants. The alternative N termini and four cassettes could theoretically produce 32 major variants. Moreover, we identify a group of cDNAs predicted to encode just the cytosolic N-terminal domain (Nt) of NBCn1. A combination of electrophysiology and biotinylation shows that the OSEs can affect surface abundance and intrinsic HCO3− transport activity of NBCn1, as expressed in Xenopus oocytes. Specifically, MEAD tends to increase whereas novel cassette IV reduces surface abundance. Cassettes II, III and novel cassette IV all appear to increase the intrinsic activity of NBCn1.
Key points
The human SLC4A7 gene and the mouse Slc4a7 gene each have alternative promoters that can yield two groups of NBCn1 variants, one in which the extreme N terminus begins with MEAD (representing the first four residues of the N-terminal domain (Nt)) and the other in which it begins with MERF.
The mouse Slc4a7 gene contains, and the human SLC4A7 gene is predicted to contain, a novel exon that encodes an alternatively spliced cassette IV of 20 aa in the cytoplasmic Nt domain of NBCn1. This new cassette IV is in a position homologous to that of a previously described cassette in the Nt of NBCn2.
From combinations of known optional structural elements (OSEs), SLC4A7 is theoretically able to produce 32 major variants, of which 16 have now been identified, 10 for the first time in the present study.
With heterologous expression in Xenopus oocytes, the OSEs have strong effects on surface abundance and intrinsic HCO3− transport activity. Cassettes II, III and the novel cassette IV have stimulatory effects on the intrinsic HCO3− transport activity of NBCn1.
Introduction
The electroneutral sodium/bicarbonate cotransporter NBCn1, a member of the solute carrier 4 (SLC4) family that includes nine other genes, is encoded by the SLC4A7 gene, which maps to 3p24 in humans. NBCn1 cDNA was first cloned from skeletal muscle (Pushkin et al. 1999) and then blood vessels (Choi et al. 2000). Functional characterization (Choi et al. 2000) indicates that NBCn1 mediates the largely 4,4-diisothiocyanato-stilbene-2,2-disulfonic acid (DIDS)-insensitive apparent uptake of one Na+ and one HCO3− across the plasma membrane. NBCn1 also exhibits an associated DIDS-stimulated conductance for Na+ and perhaps anions. In a broad range of cell types, the cotransporter plays a critical role in the regulation of intracellular pH (pHi), which, in turn, is important for almost every cell function (Roos & Boron, 1981). Northern blotting studies show that SLC4A7 is expressed in diverse organs, including heart, spleen, skeletal muscle, lung, liver, kidney and testis (Pushkin et al. 1999; Choi et al. 2000).
In the CNS, NBCn1 is highly expressed in multiple brain regions (Cooper et al. 2005; Chen et al. 2007a; Park et al. 2010), including retina (Bok et al. 2003; Lopez et al. 2005). It is also expressed in the cochlea (Bok et al. 2003; Lopez et al. 2005). In both photoreceptor cells and inner-ear hair cells, NBCn1 is present in the synaptic region and, in heterologous expression systems (Reiners et al. 2005), interacts with harmonin (USH1C), very large G-protein coupled receptor VLGR1 (USH2C) and usherin (USH2A). NBCn1 is also present near synapses in cultured hippocampal neurons, where it interacts with the postsynaptic density protein PSD-95 (Park et al. 2010). In Xenopus oocytes, this interaction with PSD-95 enhances the channel conductance, but has no effect on the HCO3− transport activity of NBCn1 (Lee et al. 2012a). In the CNS, expression of NBCn1 – as well as of NBCn2 and the Na+-driven Cl−/HCO3− exchanger NDCBE – is substantially decreased under chronic hypoxia (Chen et al. 2007a, 2008a). In mice, one example of genetic disruption of NBCn1 causes blindness and deafness, associated with sensory neuron degeneration (Bok et al. 2003; Lopez et al. 2005). Interestingly, allelic variations in SLC4A7 may be associated with vulnerability to drug addictions (Ishiguro et al. 2007).
In osteoclasts, NBCn1 mediates a colony-stimuting-factor-1-induced anti-apoptotic increase in pHi (Bouyer et al. 2007), and plays an essential role in the degradation of hydroxyapatite (Riihonen et al. 2010). In the renal medullary thick ascending limb (mTAL), the upregulation of NBCn1 during metabolic acidosis appears to play an important adaptive role in the excretion of NH+4 (Kwon et al. 2002). Moreover, in the mTAL, hypokalaemia substantially upregulates NBCn1 (Jakobsen et al. 2004).
In the cardiovascular system, NBCn1 (and also NBCe1) expression and activity are up-regulated in hypertrophied cardiac myocytes (Yamamoto et al. 2007). Consistent with the aforementioned osteoclast data, knockdown of NBCn1 increases ischaemia-induced apoptosis of coronary endothelial cells (Kumar et al. 2011). It has been shown that the NBCn1-null mice exhibit reduced NO-mediated vasorelaxation and also are mildly hypertensive (Boedtkjer et al. 2011; Schulz & Munzel, 2011). Moreover, a genome-wide association study implicates SLC4A7 polymorphisms in the development of hypertension and increased cardiovascular disease risk in humans (Ehret et al. 2011).
Finally, a single-nucleotide polymorphism in SLC4A7 (SNP# rs4973768 which contains a G→A mutation in the 3′ untranslated region) is strongly associated with increased susceptibility to breast cancer (Ahmed et al. 2009; Antoniou et al. 2010; Long et al. 2010; Sueta et al. 2012). Moreover, in a human breast cancer cell line, the expression of NBCn1 is greatly upregulated by an N-terminal truncation of receptor tyrosine kinase ErbB2 (Lauritzen et al. 2010) that is common in breast cancers, especially at the metastasized stage, and that renders the ErbB2 constitutively active (Christianson et al. 1998). On the other hand, NBCn1 expression decreases in a model (MCF10AT cell line) of breast cancer development (Chen et al. 2007b). Moreover, in this same study, NBCn1 was downregulated in 14 of 22 (and upregulated in only 3 of 22) human clinical samples of invasive ductal carcinomas, compared to matched normal tissues (Chen et al. 2007b).
Like other SLC4 proteins, NBCn1 has a cytosolic amino terminus (Nt), a larger transmembrane domain (TMD) and a short cytoplasmic carboxyl terminus (Ct). Chimeras made between NBCn1 and the electrogenic NBCe1 indicate that elements within both the front and the back halves of the TMD are important for electrogenicity (Choi et al. 2007; Chen et al. 2011) and that, within the back half, the key element is the fourth extracellular loop (Chen et al. 2011).
So far, six NBCn1 variants have been reported from human or rodent (Pushkin et al. 1999; Choi et al. 2000; Cooper et al. 2005). These differ in: (1) the extreme Nt, which is represented (thus far in different species) by sequences starting with MEAD or MERF; and (2–4) the presence/absence of cassettes I (13 aa), II (124 aa in human, 123 aa in rat) and III (36 aa) arising from alternative splicing (for reviews, see Boron et al. 2009; Parker & Boron, 2013). In addition to these three cassettes, a fourth optional cassette (20 aa in the Nt) has been reported, although without a sequence (Yang et al. 2009). Moreover, no published full-length clones contain this cassette. Curiously, the reported alternative Nt sequences beginning with MERF are all from human, whereas those beginning with MEAD are all from rat. It has not been clear whether the MEAD/MERF difference represents species differences or whether the MERF had just not been found in rodents and the MEAD not in humans. If MERF and MEAD exist in the same species, then at least five optional structural elements (OSEs) – i.e. MEAD vs. MERF plus 4 cassettes – could produce as many as 32 NBCn1 variants.
We designed the present study to address two major issues about the molecular physiology of NBCn1: (1) The diversity of NBCn1 products. Specifically, can SLC4A7 from a single species produce NBCn1 variants starting with either MEAD or MERF? Also, what is the molecular identity of the putative fourth cassette, and is this cassette present in full-length clones? (2) The physiological relevance of the structural diversity of NBCn1 products. Specifically, do the five OSEs – i.e. the alternative Nt (MEAD vs. MERF) and four cassettes – influence the plasma membrane abundance and the intrinsic (i.e. per-molecule) HCO3− transport activity of NBCn1. Here we find that the SLC4A7 gene from a single species indeed has alternative promoters that produce variants starting with ‘MEAD’ or ‘MERF’. We definitively identify a new exon in SLC4A7– encoding the novel 20 aa cassette IV in the Nt of NBCn1 – and identify 10 new full-length NBCn1 variants. Finally, our data provide the first evidence that the OSEs of NBCn1 have functional consequences both for surface abundance and for intrinsic activity.
Methods
5′-RACE
5′ Rapid amplification of cDNA ends (5′-RACE) was performed to obtain the 5′ untranslated region (UTR) of human NBCn1 transcripts with Marathon-Ready cDNA libraries prepared from human brain, heart, kidney, liver and skeletal muscle (Clontech, Palo Alto, CA, USA). The adaptor-specific primers AP1 (unnested) and AP2 (nested) supplied with the cDNA libraries were used in PCRs. The SLC4A7-specific unnested antisense primer A7RACR2 (5′-GCATGACTGTTCCATTGAGGATGCAACTCC-3′) and the nested antisense primer A7RACR1 (5′-CGGTGGTGATGTTTGTGTCCGCGATGCC-3′) were used for 5′-RACE. One microlitre of unpurified unnested RACE reaction product was used as template in the nested RACE reaction.
The 5′-RACE products were subcloned into the pCR 2.1 TOPO vector (Invitrogen, Carlsbad, CA, USA), following the manufacturer's protocol. Plasmid DNA was isolated from single colonies and sequenced to obtain the 5′-UTR sequence.
Cloning of full-length NBCn1 cDNAs from human and mouse
Human MEAD-NBCn1 cDNA was amplified by nested PCR from the cDNA libraries of human whole brain, kidney, liver (QUICK-Clone, Clontech), heart and skeletal muscle (Marathon-Ready, Clontech). Primers for the nested PCR were as follows: the sense primer hMEAD-N1-F1 (5′-GGTTCGCTCAGTTCTAGCTTCAGGTTCC-3′) plus antisense primer hN1-R1 (5′-CACAGCACTGGTATAGACTCCCTATTCTTCCC-3′) for unnested PCR and the sense primer hMEAD-F2 (5′-CCTACTAAAGCCAGCCCAGCAGTCG-3′) plus antisense nested primer hN1-R2 (5′-AGTCTCCACGGTGCTCATTACAAACTCCAG-3′) for nested PCR.
The PCR products were subcloned into pCR 2.1 TOPO vector (Invitrogen) and transformed into bacteria. Plasmid DNA was isolated from single colonies for sequencing analysis.
Mouse NBCn1 cDNA was amplified by nested RT-PCR with total RNA preparations of mouse tissue. Transcripts encoding mouse MEAD-NBCn1 were amplified with the sense primer mMEAD-F1 (5′-CCTCTGCCCGTCTCAGTCCTCGC-3′) plus antisense primer mN1-R1 (5′-TGAAGAAAGCCCACAGAGAAGCCAGG-3′) for unnested PCR and the sense primer mMEAD-F2 (5′-actactcccgggACGCCGTTGCCTCTCTCTCCCG-3′) and antisense primer mN1-R2 (5′-actactgcggccgcTCTATGGTGTCCACAACAAATATCTGACGC-3′) for nested PCR. The upper-case letters represent genome sequences; the non-italicized lower-case letters represent random sequences. Italicized letters represent restriction sites that we introduced into the nested primers for subcloning the PCR products into the vector. Transcripts encoding mouse MERF-NBCn1 were amplified with the sense primer mMERF-F1 (5′-GCACTGCCAGAAACAAGACCTACCCTG-3′) plus antisense primer mN1-R3 (5′-ACAGTTACATGAAGAAAGCCCACAGAGAAGCC-3′) for unnested PCR and the sense primer mMERF-F2 (5′-actactcccgggGCCAGAAACAAGACCTACCCTGTCAGTATTAC-3′) plus anti-sense primer mN1-R4 (5′-actactgcggccgCACCACATGGGCAGACTCCTTATTCTACC-3′) for nested PCR. The sense primers for mouse MERF-NBCn1 were designed based upon the predicted exon 2 of mouse Slc4a7, assuming that it is homologous to exon 2 of human SLC4A7. The PCR products were digested with XmaI and NotI, and subcloned into pGH19. Plasmid DNA was isolated from single colonies for identification of NBCn1 variants.
Artificial generation of human NBCn1-A, -B, -D and mouse NBCn1-F
Human NBCn1-A
The full-length sequence of human NBCn1-A (AF047033.1) was previously reported (1999), and here we created it artificially as follows. We amplified by PCR a fragment encoding the Nt from a cDNA clone that contained a partial sequence of human NBCn1-B (which contains MEAD). The forward primer contained the sequence encoding MERF-Nt to replace the unique MEAD-Nt of the human MEAD-NBCn1 clone. We then amplified a second fragment encoding the TMD and Ct from a human NBCn1 clone similar to human NBCn1-A. The overlapping regions of these two fragments contain an EcoRI site that was introduced into the primers used for PCR. The two fragments were digested and ligated to obtain full-length human NBCn1-A.
Human NBCn1-B
NBCn1-B (MEAD), which is identical to NBCn1-A (MERF) except for MEAD/MERF, was previously cloned from rat (Choi et al. 2000). Here we artificially created the human version, starting from the above human NBCn1-A (MERF) cDNA, and replacing the sequence encoding the MERF-Nt with that encoding the MEAD-Nt.
Human NBCn1-D
In the present study, we were able to clone human NBCn1-D.1, but not NBCn1-D. We created human NBCn1-D as follows. We utilized a natural BamHI site in the cDNA near the end of the Nt of human NBCn1. We excised human NBCn1-B with XmaI and BamHI to obtain the front half of NBCn1-D, and excised human NBCn1-G with BamHI and HindIII to obtain the back half of NBCn1-D. We ligated these two fragments to obtain the full-length cDNA encoding human NBCn1-D.
Mouse NBCn1-F
The full-length sequence of human NBCn1-F (MERF) has been deposited in GenBank (accession no. AAG16773), but the mouse NBCn1-F has not yet been identified. The clone used in the present study was generated from mouse NBCn1-E (MEAD) by using PCR to replace the cDNA encoding the MEAD-Nt with that encoding the MERF-Nt.
Subcloning of cDNAs encoding NBCn1 variants into pGH19
The cDNA encoding human and mouse NBCn1 variants was subcloned into pGH19, an expression vector for Xenopus oocytes (Trudeau et al. 1995). The human NBCn1 variants were enhanced green fluorescent protein (EGFP)-tagged at the Ct, as described previously, starting from a pGH19 expression vector containing cDNA encoding EGFP (Chen et al. 2008b). Briefly, an XmaI site was introduced before the start codon at the 5′ end of the NBCn1 cDNA. In addition, an AgeI site was introduced in-frame right before the stop codon of the NBCn1 open reading frame. The NBCn1 cDNA fragment was excised by restrictive digestion with XmaI and AgeI, and ligated with the vector fragment. The resulting construct was transformed into ABLE C Competent Cells (Cat. No. 200171, Agilent Technologies, Santa Clara, CA, USA). In total, 250 ml of bacterial culture was grown from one single colony and plasmids containing the cDNA of human NBCn1 were isolated with a QIAGEN Plasmid Midi Kit (Cat. No. 12143, QIAGEN, Germantown, MD, USA) according to the manufacturer's instructions.
The mouse NBCn1 variants were EGFP-tagged at the Nt, starting from pGH19-EGFP-hNBCe1-A, which expresses human NBCe1-A with an EGFP tag at the Nt (Lu et al. 2006). Briefly, an XmaI site was generated by site-directed mutagenesis right before the ‘ATG’ encoding the Met in the motif ‘MSTEN’ in construct pGH19-EGFP-hNBCe1-A. Based upon this new construct, an XhoI site was introduced after the stop codon of NBCe1-A in pGH19. The XhoI site at the 3′ end of the 3′ UTR in pGH19 was then removed by site-directed mutagenesis.
The mouse NBCn1 cDNA was amplified by PCR with sense primer ‘aagcatcccgggATGGAGGCAGACGGGGCC’ (for MEAD-NBCn1, upper case represents the coding region of NBCn1) or ‘atcgcccgggATGGAAAGATTTCAGCTGGCG’ (for MERF-NBCn1) containing an XmaI site and antisense primer ‘actgctcgagACTTTACAATGAAGTTTCAGCATC’ containing an XhoI site. The PCR product was restricted by XmaI and XhoI, and ligated with pGH19-EGFP, which was restricted by XmaI and XhoI. The resulting construct expresses a fusion protein containing EGFP and NBCn1, linked by a 21 aa peptide ‘GQLWQINSPSAEFGLGGLAPG’. The resulting construct was transformed into One Shot TOP10 Chemically Competent Cells (Cat. No. C4040-03, Life Technologies Corporation, Carlsbad, CA, USA). In total, 5 ml of bacterial culture was grown from one single colony and plasmids containing the cDNA of mouse NBCn1 were isolated with a QIAprep Spin Miniprep Kit (Cat. No. 27104, QIAGEN) according to the manufacturer's instructions.
cRNA preparation and oocyte injection
An ovary lobe was excised into small pieces in Ca2+-free NRS (in mm: 82 NaCl, 2 KCl, 20 MgCl2, 5 Hepes; pH 7.50; 200 mOsm). The ovary was digested for 60–70 min with 2 mg ml−1 of Type 1A collagenase (Sigma-Aldrich, St Louis, MO, USA) in Ca2+-free NRS. The dissociated oocytes were rinsed five times with Ca2+-free NRS and five times with standard ND96 (96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2, 5 Hepes; pH 7.50; 200 mOsm). The oocytes were sorted to select stage V–VI cells which were then incubated at 18°C in OR3 medium (pH 7.50 and 200 mOsm) by dissolving one package of Leibovitz's L-15 powder (Gibco, Life Technologies) in 5 mm Hepes and supplementing with 50 ml Pen/Strep (5000 units penicillin and 5000 μg streptomycin).
For the preparation of cRNA, plasmid DNA containing NBCn1 cDNA was linearized by digestion with NotI, and then incubated with the T7 RNA polymerase provided with the T7 mMessage mMachine kit (Ambion, Austin, TX, USA), according to the manufacturer's instructions. The reaction mixture was extracted with phenol and chloroform. The cRNA was precipitated with isopropanol, collected by centrifugation, dissolved in H2O, and stored in aliquots at −80°C. In total, 25 ng cRNA was injected into each oocyte. The oocytes were incubated at 18°C for 4–5 days in OR3 medium for protein expression, and then used for electrophysiological measurements or biotinylation assays.
Measurement of pHi and membrane potential
Our approach for the simultaneous monitoring of membrane potential (Vm) and pHi in an oocyte has recently been reviewed in detail (Musa-Aziz et al. 2010). Briefly, we place an oocyte in a plastic oocyte recording chamber, the channel of which is filled with ND96 (Warner Instruments Corp., Hamden, CT, USA). We impale with two microelectrodes. One is an H+-sensitive microelectrode (the ‘pHi electrode’) that is sensitive to Vm plus the intracellular activity of H+. The other is a KCl-filled electrode (the ‘Vm electrode’). The tip of a third electrode, filled with KCl (the ‘bath electrode’), is placed in the chamber close to the oocyte. The tip of the pHi electrode is filled with an H+-sensitive ionophore (H+-sensitive ionophore I cocktail B; Sigma-Aldrich), a backfill solution making the electrical contact between the ionophore and the microelectrode circuitry. Oocyte pHi is a linear function of the differential outputs of the pHi and Vm electrodes. Vm is given by the differential outputs of the Vm and bath electrodes. Electrode outputs are recorded using an FD223 dual-channel high-impedence electrometer (World Precision Instruments, Inc., Sarasota, FL, USA) and an OC-275 oocyte clamp (Warner Instrument Corp., Hamden, CT, USA). Data are sampled every 500 ms. The differential outputs are provided by a custom made subtraction amplifier and data are acquired using custom made software. Perfusion solutions are delivered to the recording chamber using infusion syringe pumps (catalogue no. 55-2226, Harvard Apparatus, Holliston, MA, USA).
Biotinylation analysis of surface-expressed NBCn1 in Xenopus oocytes
NBCn1 constructs with an EGFP tag at the Nt were heterologously expressed in Xenopus oocytes. The Pinpoint Cell Surface Protein Isolation Kit (catalogue no. 89881, Pierce, Rockford, IL, USA) was employed for the detection of NBCn1 proteins expressed at the plasma membrane of oocytes, as previously described (Chen et al. 2012). Briefly, we incubated oocytes with 240 μg ml−1 Sulfo-NHS-SS-Biotin in phosphate-buffered saline (PBS, diluted to 200 mOsm) for 1 h at 4°C, and stopped the reaction with ‘Quenching Solution’ (250 μl). The oocytes were washed with Tris-buffered saline (TBS, diluted to 200 mOsm), homogenized in 10 ml TBS containing 1% Triton X-100 and EDTA-free proteinase inhibitors (lysis buffer), and then centrifuged for 10 min at 1000 g at 4°C. An aliquot of the resulting supernatant, representing the ‘total protein’, was saved. The rest was incubated at room temperature for 1 h with Immobilized Neutravidin (500 μl). After five washes with 500 μl lysis buffer, the bound proteins were eluted with 1× sample buffer containing 50 mm dithiothreitol (DTT), representing the ‘surface proteins’.
The protein preparations were separated by SDS-PAGE and transferred onto a polyvinylidene difluoride (PVDF) membrane (Millipore, Bedford, MA, USA) for Western blotting. The blots were subsequently probed for NBCn1 with mouse anti-EGFP monoclonal antibody (Clontech) and horseradish peroxidase-conjugated goat anti-mouse secondary antibody (MP Biomedicals, Solon, OH, USA). Following chemiluminescence with Amersham ECL Plus (GE Healthcare, Buckinghamshire, UK), the signals were visualized by X-ray exposure.
Data analysis and statistics
The data for pHi recovery as well as surface abundance are presented as mean ± SEM. One-way analysis of variance (ANOVA) was performed with R (R Core Team, 2013; http://www.r-project.org/). Pairwise comparisons were performed using ANOVA contrasts. A linear regression analysis was performed using R (R Core Team, 2013).
We define the intrinsic activity as the ratio of the mean dpHi/dt to the mean surface protein. To draw statistically valid conclusions about intrinsic activity, it is necessary to take into account the experimental variation in both dpHi/dt as well as surface protein abundance. To take these both into account, we used a maximum likelihood approach for estimation and a likelihood ratio approach for testing (Casella & Berger, 2001). To determine confidence intervals and statistical significance, we used a parametric bootstrapping approach (Efron & Tibshirani, 1994). Denote the ith dpHi/dt measurement by ri and denote the jth surface-protein measurement by sj. Denote the population ratio of dpHi/dt to surface protein for each of the full-length transcript variants (v) by ρv and the mean surface protein for each variant as μv. We assume that sj is normally distributed with mean μv and variance σ2s and that ri is normally distributed with mean ρvμv and variance σ2r. Note that σ2s and σ2r are held in common across all groups. Finally, we assumed that ρv=Σlxvlβl, where xvl represents a set of predictor variables and βl is a coefficient representing the effect sizes. For the estimates of the ratio for each of the variants (Table 4), the X matrix (made up of xvl) was simply the identity matrix (similar in principle to an ANOVA). We then performed pairwise comparisons between each of the variants. We combined the P values for sets of comparisons involving the same OSE, using Fisher's method (Won et al. 2009). Finally, we considered a linear model (Table 5B) in which the xvl variables take on the values 1 or 0, representing the presence or absence of a particular OSE (i.e. the Nt of MEAD vs. MERF, and cassettes I−IV). This approach allows us to test the effect of each OSE individually while adjusting for the effects of all the other OSEs. In each case, we maximized the likelihood using a custom function written in the R statistical computing environment (R Core Team, 2013), and we used a parametric bootstrap approach for inference with 10,000 bootstrap samples. For example, to calculate the confidence intervals for intrinsic activity (see Table 4 and Table 5B, below), we simulated 10,000 new data sets from the random distribution that we described above using the parameter estimates from the real data. We then analysed each of these simulated data sets in the same way we analysed the real data, finding an estimate for each simulated data set. The confidence interval is based on the quantiles of the estimates from the simulated data sets.
Table 4.
Comparison of effects of OSEs on HCO3− transport activity as well as surface abundance of NBCn1
| Nt | I–II–III–IV | dpHi/dt (functional expression) | P | Surface abundance | P | dpHi/dt/surface (intrinsic activity) | P | |
|---|---|---|---|---|---|---|---|---|
| (A) MEAD vs. MERF | ||||||||
| E | MEAD | +−−− | 4.6 (1, 8.1)a | 0.66 | 1.2 (1.1, 1.4) | 0.01 | 3.7 (1.5, 6.0) | 0.28 |
| F | MERF | +−−− | 5.7 (2.3, 9) | 0.9 (0.7, 1.1) | 6.3 (3.4, 9.6) | |||
| G | MEAD | +−+− | 12.2 (9.4, 15.1) | 0.76 | 1.4 (1.3, 1.6) | 0.003 | 8.5 (6.8, 10.3) | 0.16 |
| J | MERF | +−+− | 11.6 (8.7, 14.5) | 1.0 (0.8, 1.2) | 11.4 (8.9, 14.3) | |||
| Combined P valueb | 0.85 | 0.0004 | 0.18 | |||||
| (B) Cassette I | ||||||||
| D | MEAD | +++− | 14.6 (11, 18.1) | 0.22 | 1.0 (0.8, 1.2) | 0.17 | 14.6 (11.3, 18.3) | 0.08 |
| C | MEAD | −++− | 11.3 (7.4, 15.1) | 1.2 (1.0, 1.4) | 9.5 (6.8, 12.4) | |||
| G | MEAD | +−+− | 12.2 (9.4, 15.1) | 0.77 | 1.4 (1.3, 1.6) | 0.001 | 8.5 (6.8, 10.3) | 0.11 |
| I | MEAD | −−+− | 11.6 (8.6, 14.6) | 1.0 (0.8, 1.2) | 12.0 (9.3, 15.2) | |||
| Combined P value | 0.47 | 0.0019 | 0.05 | |||||
| (C) Cassette II | ||||||||
| B | MEAD | ++−− | 10.9 (7.0, 14.8) | 0.02 | 1.0 (0.8, 1.2) | 0.08 | 10.9 (7.7, 14.5) | 0.005 |
| E | MEAD | +−−− | 4.6 (1.0, 8.1) | 1.2 (1.1, 1.4) | 3.7 (1.5, 6.0) | |||
| D | MEAD | +++− | 14.6 (11, 18.1) | 0.32 | 1.0 (0.8, 1.2) | 0.002 | 14.6 (11.3, 18.3) | 0.011 |
| G | MEAD | +−+− | 12.2 (9.4, 15.1) | 1.4 (1.3, 1.6) | 8.5 (6.8, 10.3) | |||
| C | MEAD | −++− | 11.3 (7.4, 15.1) | 0.88 | 1.2 (1.0, 1.4) | 0.11 | 9.5 (6.8, 12.4) | 0.36 |
| I | MEAD | −−+− | 11.6 (8.6, 14.6) | 1.0 (0.8, 1.2) | 12.0 (9.3, 15.2) | |||
| Combined P value | 0.12 | 0.0014 | 0.001 | |||||
| (D) Cassette III | ||||||||
| D | MEAD | +++– | 14.6 (11, 18.1) | 0.18 | 1.0 (0.8, 1.2) | 0.98 | 14.6 (11.3, 18.3) | 0.25 |
| B | MEAD | ++–– | 10.9 (7.0, 14.8) | 1 (0.82, 1.19) | 10.9 (7.7, 14.5) | |||
| G | MEAD | +−+− | 12.2 (9.4, 15.1) | 0.002 | 1.4 (1.3, 1.6) | 0.1 | 8.5 (6.8, 10.3) | 0.012 |
| E | MEAD | +−−− | 4.6 (1.0, 8.1) | 1.2 (1.1, 1.4) | 3.7 (1.5, 6.0) | |||
| J | MERF | +−+− | 11.6 (8.7, 14.5) | 0.01 | 1.0 (0.8, 1.2) | 0.3 | 11.4 (8.9, 14.3) | 0.065 |
| F | MERF | +−−− | 5.7 (2.3, 9.0) | 0.9 (0.7, 1.1) | 6.3 (3.4, 9.6) | |||
| Combined P value | 0.0003 | 0.42 | 0.009 | |||||
| (E) Cassette IV | ||||||||
| N | MERF | +−++ | 14.3 (11.1, 17.5) | 0.2 | 0.7 (0.5, 0.8) | 0.01 | 21.6 (16.6, 28.3) | 0.008 |
| J | MERF | +−+− | 11.6 (8.7, 14.5) | 1.0 (0.8, 1.2) | 11.4 (8.9, 14.3) | |||
| O | MEAD | −−++ | 11.9 (8, 15.8) | 0.9 | 0.6 (0.4, 0.8) | 0.007 | 20.1 (14.0, 28.3) | 0.074 |
| I | MEAD | −−+− | 11.6 (8.6, 14.6) | 1.0 (0.8, 1.2) | 12.0 (9.3, 15.2) | |||
| Combined P value | 0.5 | 0.0009 | 0.005 | |||||
The data are presented as mean (lower of 95% CI, upper of 95% CI). CI: confidence interval.
Computed using Fisher's method.
Table 5.
Linear effects of OSEs on relative surface abundance of NBCn1 and intrinsic HCO3− transport activity
| 95% CI | |||||
|---|---|---|---|---|---|
| Coefficient | Estimated effect | Standard error | Lower | Upper | P |
| (A) Surface abundance | |||||
| (Intercept) | 0.73 | 0.15 | 0.45 | 1.02 | <0.0005 |
| MEAD/MERF | 0.25 | 0.09 | 0.07 | 0.43 | 0.01 |
| Cassette I | 0.19 | 0.09 | 0.01 | 0.38 | 0.05 |
| Cassette II | −0.13 | 0.08 | −0.30 | 0.03 | 0.13 |
| Cassette III | 0.13 | 0.08 | −0.04 | 0.29 | 0.14 |
| Cassette IV | −0.45 | 0.10 | −0.64 | −0.26 | <0.0005 |
| (B) Intrinsic (i.e. per molecule) activity (×10−5) | |||||
| (Intercept) | 6.90 | 2.41 | 3.04 | 10.90 | 0.005 |
| MEAD/MERF | −2.01 | 1.52 | −4.58 | 0.46 | 0.21 |
| Cassette I | −0.17 | 1.59 | −2.82 | 2.37 | 0.92 |
| Cassette II | 3.81 | 1.43 | 1.47 | 6.16 | 0.011 |
| Cassette III | 4.39 | 1.34 | 2.18 | 6.56 | 0.003 |
| Cassette IV | 10.62 | 3.01 | 6.09 | 15.95 | <0.0005 |
In this paper, we perform multiple hypothesis tests, so it is necessary to adjust for the number of tests. We consider our investigation of surface protein abundance and intrinsic transporter activity to be separate issues, leading us to argue that the type I error should be controlled separately for the two. For both surface protein abundance and intrinsic activity, our main results consist of only five statistical tests (one for each OSE); other P values and statistical results should be considered more exploratory in nature. Thus, using the Bonferoni correction procedure, P values less than α= 0.05/5 = 0.01 are considered statistically significant.
Results
Detection of 5′-UTR of human SLC4A7 transcripts by 5′-RACE
Figure 1 shows the updated structures of the human SLC4A7 gene (Fig. 1A) and mouse Slc4a7 gene (Fig. 1B). The SLC4A7/Slc4a7 genes contain 28 exons, four of which – exons 8, 10, 15 and 27 – are cassette exons, the entirety of which can be alternatively spliced in or out. A portion of the 3′-end of exon 7 and the entirety of exons 8 and 27 encode the previously described optional cassettes I, II and III, respectively (Choi et al. 2000). Exons 10 and 15 represent two new cassette exons identified in the present study [see details in ‘Identification of cassette IV – a novel optional cassette in mouse Slc4a7’ and ‘Identification of exon 15 deletions (producing truncated Nt products)’ below].
Figure 1. Gene structures of human (A) and mouse (B) SLC4A7.

SLC4A7 contains 28 exons. The grey box, which includes parts of exons 15 and 25, indicates the region encoding the transmembrane domain. The underlined exon numbers refer to exons that give rise to the major NBCn1 variants. Exon 1 encodes ‘MEAD’ and exon 2 encodes ‘MERF’, the two alternative extreme Nts of NBCn1. Exon 7 can be alternatively spliced at its 3′ end to produce cassette I. Exons 8 and 10 can be alternatively spliced in to produce cassettes II and IV, respectively. Exon 15 can be alternatively spliced out to produce the truncated Nt of NBCn1. Exon 27 can be alternatively spliced in to produce cassette III. The asterisk in exon 16 indicates a cryptic stop codon that can lead to truncated Nt proteins [see ‘Identification of exon-15 deletions (producing truncated Nt products)’ in Results]; the other asterisk is the normal stop codon. The arrows indicate the approximate locations of the two anti-sense primers A7RACR1 and A7RACR2 used for 5′-RACE (see ‘5′-RACE’ in Methods). Human SLC4A7 appears to contain an exon 10 that maps from 27404272 to 27404331 in genomic DNA contig NT_022517.18. However, in the present study, we did not obtain any full-length cDNA clones containing sequences corresponding to human exon 10. The diagrams are drawn to scale.
Two different extreme N termini exist in the six published NBCn1 variants. The first 11 aa (starting with the four amino-acid residues ‘MERF’) of the human NBCn1 variants (NBCn1-A and -F) differ from the first 16 aa (starting with ‘MEAD’) of all published rodent NBCn1 variants (NBCn1-B, -C, -D and -E). However, our in silico analysis of genomic DNA reveals that human SLC4A7 (Fig. 1A) and mouse Slc4a7 (Fig. 1B) each contain a predicted exon 1 encoding ‘MEAD’ and predicted exon 2 encoding ‘MERF’. We also localized the exon encoding the ‘MERF’ Nt of NBCn1 in rat genome (not shown). Although MEAD-NBCn1 variants from rat were reported as cDNAs (Choi et al. 2000), we could not localize the ‘MEAD’ exon in rat genome, presumably due to the incomplete nature of the genome. Prior to the present study, no MEAD-NBCn1 variants had been published for humans, and no MERF-NBCn1 variants had been published for rodents.
In a preliminary study, Gill et al. (2006) obtained two full-length as well as several partial cDNA clones corresponding to the NBCn1 Nt starting with ‘MEAD’ by nested PCR from diverse commercial human cDNA libraries, including heart, kidney, liver and skeletal muscle, suggesting that MEAD-NBCn1 variants are expressed in these human tissues. They also obtained two full-length NBCn1 clones starting with ‘MERF’, confirming the expression of MERF-NBCn1 in human reported by Pushkin et al. (1999). However, the study by Gill et al. (2006) was focused on the variations in the Nt of NBCn1. In the present study, we performed 5′-RACE to obtain the 5′-UTR sequences of SLC4A7 transcripts from human cDNA libraries of brain, heart, kidney, liver and skeletal muscle. As shown in Fig. 2, we amplified fragments of ∼0.3 kb in all cases. Sequencing analysis demonstrates that this fragment represents a mixture of two 5′-RACE products: ‘MEAD’ (including exons 1, 3 and 4) and ‘MERF’ (including exons 2, 3 and 4). These results confirm the expression of MEAD-NBCn1 transcripts in human tissues. The ∼1.3 kb product from liver in Fig. 2 consists of a 3′ portion of intron 2–3 as well as exons 3 and 4. This product could represent part of a novel SLC4A7 transcript initiated from a place distinct from the initiation sites of either MERF-NBCn1 or MEAD-NBCn1.
Figure 2. 5′-RACE analysis of human SLC4A7 transcripts.

5′-RACE was performed with cDNA libraries (Marathon-Ready cDNA, Clontech) of human brain, heart, kidney, liver and skeletal muscle. The PCR products were analysed by agarose gel electrophoresis. A major product of ∼0.3 kb (lower arrow) was obtained for all cases. The entire PCR product was cloned into TOPO vector. Multiple random clones (12 for brain, 8 for kidney, 4 for heart, 6 for skeletal muscle and 9 for liver) were sequenced for each organ. Three different types of transcripts were identified. The first includes exons 1 + 3 + 4 encoding ‘MEAD’ (represented by lower arrow). The second includes exons 2 + 3 + 4 encoding ‘MERF’ (also represented by lower arrow). The third type (from liver) initiates from intron 2–3 and may represent a novel transcript (the ∼1.3 kb band represented by the higher arrow) that we did not further pursue in the present study.
Cloning and expression patterns of NBCn1 in human and mouse tissues
Performing nested RT-PCR with primer sets specific for the cDNA encoding human MEAD-NBCn1, we obtained ∼3.5 kb products from human brain, heart and skeletal muscle (Fig. 3A) as well as liver but not kidney (Fig. 3B). Employing a similar approach, but with primer sets specific for mouse MERF-NBCn1 or mouse MEAD-NBCn1, we obtained products of ∼3.5 kb from eye (Fig. 3C) and selected other mouse tissues: heart, skeletal muscle and reproductive tract (data not shown).
Figure 3. PCR cloning of NBCn1 transcripts from human (A and B) and mouse (C) tissues.

Nested PCR yielded ∼3.5 kb bands that correspond to the full-length cDNA of MEAD-NBCn1 variants from human brain, heart, skeletal muscle, liver and kidney. In the skeletal muscle, the ∼30 kb band could represent an immature RNA product. Nested PCR also yielded ∼3.5 kb bands that correspond to the full-length cDNA of MEAD- and MERF-NBCn1 variants from mouse cDNA libraries of eye.
We recovered the 3.5 kb bands from the gel, subcloned the cDNAs into vectors, transformed the constructs into bacteria and then isolated plasmid DNA from single colonies for identification of NBCn1 variants. The result is that we have expanded the number of NBCn1 variants from the previously known six that are predicted to encode full-length proteins (NBCn1-A to -F) to a present total of 21 that are predicted to encode full-length proteins, including:
Sixteen full-length ‘major’ variants (NBCn1-A to -P, Fig. 4). These variants differ from one another based on the selection of MEAD vs. MERF at the extreme N terminus, as well as the optional inclusion of cassettes I–IV. Ten of the 16 major variants are novel, i.e. we identify them for the first time in the present study. The cloning of these 10 new variants confirms ‘MEAD’ and ‘MERF’ as alternative initial sequences in both human and mouse (see section above). Moreover, we describe splicing cassette IV (exon 10) for the first time in the present study (see details below in ‘Identification of cassette IV – a novel optional cassette in mouse Slc4a7’).
Five full-length ‘minor’ variants. Four of these variants differ from major variants by an optional extension to exon 1 in humans. The fifth variant differs from a major variant by an optional extension to exon 26 in mice (see details below in ‘Extensions to exons’).
Figure 4. Summary of known major NBCn1 variants.
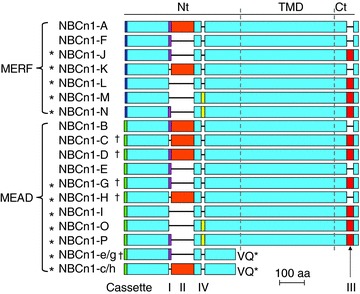
The diagram is based on a sequence alignment for human NBCn1-A (AAD38322.1), mouse NBCn1-B (AFR46591.1), mouse NBCn1-C (AFR46592.1), mouse NBCn1-D (AFR46593.1), human NBCn1-E (ACH61961.1), human NBCn1-F (AAG16773), human NBCn1-G (ACB47400.1), human NBCn1-H (ACH61958.1), mouse NBCn1-I (ADC92004.1), mouse NBCn1-J (ADO51787.1), mouse NBCn1-K (ADO51788.1), mouse NBCn1-L (AFB82586.1), mouse NBCn1-M (AFI43934.1), mouse NBCn1-N (AFB82538.1), mouse NBCn1-O (AFI43933.1), mouse NBCn1-P (AFI43932.1), and human truncated e/g (ACI24741.1) and truncated c/h (ACI24740.1). The light blue boxes represent the regions that are identical in all NBCn1 variants. The green box at the extreme Nt represents the first 16 aa of NBCn1 variants starting with MEAD, whereas the dark-blue box at the extreme Nt represents the first 11 aa of NBCn1 variants starting with MERF. Cassette I (purple) contains 13 aa, cassette II (orange) contains 124 aa in human and 123 aa in mouse and rat, cassette IV (yellow) contains 20 aa, and cassette III (red) contains 36 aa. The truncated product NBCn1-e/g is predicted from truncated transcript types 1 and 2, whereas truncated c/h is predicted from truncated transcript types 3 and 4 (see Table 1). The last two residues ‘VQ’ of the truncated products are unique compared to the full-length NBCn1. †Variants that are also represented by 4 aa extensions to the MEAD module (see Table 1 and Table 2). The length of the predicted amino acid sequences of known NBCn1 variants ranges from 1093 to 1253 aa. The stars indicate the novel variants reported for the first time in the present study.
In addition, we have identified:
Six mRNA variants (Table 1) from human that are predicted to encode three unique truncated protein products containing nearly the entire Nt but lacking the TMD and Ct [see details below in ‘Identification of exon 15 deletions (producing truncated Nt products)’].
Table 1.
Summary of structural features of truncated human NBCn1 variants
| Cassettes | ||||||||
|---|---|---|---|---|---|---|---|---|
| Truncated types | Nt initial sequence | Accession no. | I | II | IV | III | Exon 15 | Predicted protein product |
| 1 | MEAD | FJ178575 | + | − | − | + | − | NBCn1-e/g |
| 2 | MEAD | FJ178576 | + | − | − | − | − | NBCn1-e/g |
| 3 | MEAD | FJ178574 | − | + | − | − | − | NBCn1-c/h |
| 4 | MEAD | GU354307 | − | + | − | + | − | NBCn1-c/h |
| 5 | MEADa | GU354309 | + | − | − | + | − | NBCn1-e.1/g.1 |
| 6 | MEADa | GU354310 | + | − | − | − | − | NBCn1-e.1/g.1 |
The first exon of these transcripts encoding the MEAD module contains the ‘VTSR’ insert due to the 12 nt extension at the 3′-end of exon 1.
Table 2 summarizes the tissue distributions of the NBCn1 variants in human. Table 3 does likewise for the NBCn1 variants in mouse.
Table 2.
Tissue distribution of human MEAD-NBCn1 variants
| Tissue | NBCn1 variants | Accession no. | Colony frequencya |
|---|---|---|---|
| Brain | NBCn1-G | EU499349 | 8/11 |
| NBCn1-G.1 | GU354308 | 1/11 | |
| Truncated#1 | FJ178575 | 1/11 | |
| Truncated#5 | GU354309 | 1/11 | |
| Heart | Truncated#3 | FJ178574 | 10/10 |
| Skeletal muscle | NBCn1-C.1 | EU934250 | 1/22 |
| NBCn1-E | EU934249 | 3/22 | |
| NBCn1-G | EU499349 | 4/22 | |
| NBCn1-H | EU934246 | 4/22 | |
| NBCn1-H.1 | EU934247 | 3/22 | |
| Truncated#1 | FJ178575 | 2/22 | |
| Truncated#2 | FJ178576 | 2/22 | |
| Truncated#3 | FJ178574 | 1/22 | |
| Truncated#4 | GU354307 | 1/22 | |
| Truncated#6 | GU354310 | 1/22 | |
| Liver | NBCn1-D.1 | EU934248 | 8/8 |
Colony frequency indicates the number of colonies of each variant in the total colonies screened for the specified tissue.
Table 3.
Expression and distribution of mouse NBCn1 variantsa
| Nt initial sequence | Tissue | Variants | Accession no. | Colony frequency |
|---|---|---|---|---|
| MEAD | RT♀b | NBCn1-E | GU386353 | 3/7 |
| NBCn1-G | JQ073566 | 2/7 | ||
| NBCn1-I | GU386352 | 2/7 | ||
| Eye | NBCn1-G | JQ073566 | 8/15 | |
| NBCn1-I | GU386352 | 4/15 | ||
| NBCn1-O | JQ349032 | 2/15 | ||
| NBCn1-P | JQ349031 | 1/15 | ||
| Heart | NBCn1-B | JX254908 | 5/19 | |
| NBCn1-C | JX254909 | 1/19 | ||
| NBCn1-C.2 | JX310364 | 1/19 | ||
| NBCn1-D | JX254910 | 2/19 | ||
| NBCn1-E | GU386353 | 3/19 | ||
| NBCn1-H | JX254911 | 2/19 | ||
| NBCn1-G | JQ073566 | 5/19 | ||
| MERF | Skeletal muscle | NBCn1-A | JX254912 | 15/20 |
| NBCn1-K | HM624051 | 5/20 | ||
| Ovary | NBCn1-J | HM624050 | 12/12 | |
| Testis | NBCn1-J | HM624050 | 2/2c | |
| Eye | NBCn1-J | HM624050 | 11/24 | |
| NBCn1-L | JQ344322 | 1/24 | ||
| NBCn1-M | JQ349033 | 1/24 | ||
| NBCn1-N | JQ347261 | 11/24 |
All mouse variants were first cloned in the present study.
RT♀: female reproductive tract tissues represent the mixture of ovary, uterus and vagina. cIn addition to these two clones, we identified 12 others from testis that lack 114 bp (corresponding to the entire exon 21), leading to deletion of TM7 and part of extracellular loop EL4 in the putative TMD. All 14 clones from testis lack cassette II.
Identification of cassette IV – a novel optional cassette in mouse Slc4a7 (producing major variants)
Compared to the other NBCn1 variants, four clones (NBCn1-M, -N, -O and -P) from mouse eye contain the new splicing cassette IV, which consists of 60 bp. The identification of this new cassette reveals a new alternatively spliced exon (exon 10, see Fig. 1) in Slc4a7 that encodes a peptide of 20 aa (ESASWHCSCGTLGVGLKKPA). In addition to the four full-length variants containing cassette IV (NBCn1-M, -N, -O and -P, Fig. 4), we identified from GenBank a partial IMAGE cDNA (accession no. BC038373.1) as well as an expressed sequence tag (accession no. BF460695.1) – both from mouse retina – that also contain cassette IV.
Although we did not obtain any human clones containing cassette IV, the human SLC4A7 gene is predicted to encode a cassette IV.
Extensions to exons (producing minor variants)
Exon 1 in human
Some human ‘MEAD’ variants include a 12 nt extension (‘GTAACGAGCAGG’) at the 3′-end of exon 1, due to a shift in the splicing-donor site. Thus, these variants contain a 4 aa (‘VTSR’) insert, extending the coding region of the first exon from 16 to 20 aa. We propose to designate these minor variants as NBCn1-C.1, -D.1, -G.1 and -H.1, which are correspondingly identical to NBCn1-C, -D, -G and -H except for the inclusion of the 4 aa insert.
We did not observe any mouse clones analogous to NBCn1-C.1, -D.1, -G.1 and -H.1 in human. Although the homologous region of the mouse Slc4a7 gene does not have a canonical splicing site that would produce a 4 aa extension, it is possible that mice use a non-canonical site. If so, the first four amino acids would be ‘VRSG’.
Exon 26 in mouse
We found one mouse ‘MEAD’ variant that contains a 15 nt extension (‘GTCTGTTCTTTCCAG’) at the 5′-end of exon 26 due to a shift in the splicing-acceptor site. Thus, this variant contains a 5 aa (‘VCSFQ’) insert, extending exon 26 from 33 to 38 aa. We do not yet propose to assign a unique name for this variant, as we have observed it in only one clone.
Identification of exon 15 deletions (producing truncated Nt products)
Table 1 summarizes the major structural features of the six truncation transcripts from human. All six transcripts begin with exon 1 (encoding MEAD) and lack exon 15. They differ from one another by the optional inclusion of the minor extension of exon 1 as well as cassettes I, II and IV. Lacking exon 15 causes a frameshift that leads to a stop codon in exon 16, and thus an early termination near the predicted junction of the Nt and TMD. These six truncation transcripts are predicted to express two major distinct truncated protein products as well as one minor truncated protein product, all starting with the ‘MEAD’ Nt. One major product is the truncated version of NBCn1-E/G; we propose to designate it as NBCn1-e/g. The other major product is a truncated version of NBCn1-C/H, thus designated as NBCn1-c/h (Fig. 4). The minor truncated product NBCn1-e.1/g.1 contains a 4 aa extension in the MEAD exon. None of these predicted truncated soluble proteins has been identified. The tissue distributions of the human truncation variants are summarized in Table 2.
HCO3− transport activity of different NBCn1 variants in Xenopus oocytes
We heterologously expressed NBCn1 variants in Xenopus oocytes. We then superfused individual oocytes with nominally ‘HCO3−-free’ ND96, switched to a solution containing 1.5% CO2/10 mm HCO3− and finally replaced the extracellular Na+ with N-methyl-d-glucamine (NMDG+) in the continuous presence of CO2/HCO3−. Using microelectrodes, we continuously monitored oocyte Vm and pHi.
Human clones
Figure 5A shows the typical recordings from an oocyte expressing human NBCn1-E. Introducing 1.5% CO2/10 mm HCO3− leads to the influx of CO2, which imposes an intracellular acid load, from which pHi begins to recover. The rate of pHi recovery (dpHi/dt) is indicated by the slope of the dashed line. This pHi recovery is much faster in an oocyte expressing NBCn1-G (Fig. 5B) but extremely slow in a H2O-injected control oocyte (Fig. 5C), where the acid extrusion may reflect the activity of an endogenous Na–H exchanger. Thus, nearly all of the pHi recovery in Fig. 5A and B is due to the influx of HCO3− (or a related species such as CO2−3 or the NaCO3− ion pair) via NBCn1 expressed in the plasma membrane. The dpHi/dt in the NBCn1-expressing oocytes, when corrected for the background dpHi/dt in H2O-injected controls, represents the functional expression of NBCn1 – defined as the product of intrinsic (or per-molecule) activity and surface expression of NBCn1 in oocytes. The removal of extracellular Na+ halts the pHi recovery and leads to a slower fall in pHi that is similar in oocytes expressing NBCn1-E and NBCn1-G or injected with H2O. These acidifications could in part reflect (1) blockade of NBCn1 (in oocytes expressing NBCn1) and endogenous Na–H exchangers with unmasking of background acid loading (Boron et al. 1979; Boyarsky et al. 1990), (2) reversal of some combination of these transporters with no background acid loading, or (3) a combination of 1 and 2. Any contribution from endogenous Na–H exchange (Fig. 5C) would presumably have been the same in the oocytes expressing NBCn1 and thus would not affect our estimate of the functional activity of NBCn1.
Figure 5. Representative recordings of intracellular pH (pHi) and membrane potential (Vm) from oocytes expressing human NBCn1-E-EGFP (A), NBCn1-G-EGFP (B) or H2O-injected control oocytes (C).

The oocytes were initially superfused with Hepes buffer (standard ND96), then with 1.5% CO2/10 mm HCO3− for 15 min, followed by the removal of extracellular Na+ (replacing Na+ with NMDG+) for 10 min in the continuous presence of CO2/HCO3−, and then followed by a second exposure to 1.5% CO2/10 mm HCO3−. The slopes of the traces indicated by dashed lines represent pHi recovery rates during the first exposure of CO2/HCO3−.
Note that, in the oocytes expressing NBCn1, we observed no abrupt changes in Vm upon switching from ND96 to CO2/HCO3− (arrow, lower panels in Fig. 5A and B), consistent with the electroneutral cotransport of Na+ and HCO3− by NBCn1. The removal of extracellular Na+, however, causes a large hyperpolarization in oocytes expressing NBCn1. This hyperpolarization, which reflects the associated cation conductance of NBCn1 (Choi et al. 2000), is not present in H2O-injected oocytes (arrow, lower panel in Fig. 5C).
To compare the effect of structural variations on the activity of NBCn1, we expressed a series of human NBCn1 variants in Xenopus oocytes and performed electrophysiology measurements, following the protocol used in Fig. 5. Among the NBCn1 variants whose dpHi/dt data are summarized in Fig. 6, we cloned full-length human NBCn1-D.1, -G and -H for the first time from any species, and NBCn1-E for the first time from human. We constructed full-length human NBCn1-B and -D cDNA based on rodent sequences. A general observation is that we found it difficult to obtain satisfactory yields of plasmids containing human NBCn1 cDNA and thus employed midi- rather than mini-preps. It is possible that the cDNAs of human NBCn1 variants are not stable due to some endogenous recombination mechanism in bacteria. Nevertheless, once we obtained plasmids containing human cDNA, we obtained good functional expression in oocytes.
Figure 6. Summary of pHi recovery rates (dpHi/dt) of oocytes expressing human NBCn1 EGFP-tagged constructs (A), untagged constructs (B) or various NBCn1-G constructs (C).

The full-length cDNAs encoding human NBCn1-B and -D have yet to be cloned. Thus, we artificially generated them based on the known rat and mouse full-length clones. D.1 represents the minor variant of NBCn1-D that contains the ‘VTSR’ extension in the MEAD module. All human NBCn1 variants in A were tagged with EGFP at the Ct. NBCn1-B and -H in B and NBCn1-G in C contain no EGFP tag. The NBCn1-G-ΔIII in C was created by introducing a stop codon immediately after the ‘VKALK’ motif before cassette III, on the background of human NBCn1-G. Oocytes were superfused in a protocol like that in Fig. 5. dpHi/dt represents the pHi recovery rate during the first CO2/HCO3− exposure, analogous to the slope of the dashed line in Fig. 5. For all groups of NBCn1 variants or the truncated NBCn1 construct, the dpHi/dt values are significantly different from that of the control H2O-injected oocytes, based on a one-way ANOVA followed by Dunnett's multiple comparison. Bars for control H2O in B and C are reproduced from A. An asterisk indicates groups significantly different by one-way ANOVA followed by post hoc Tukey's comparison. Numbers in parenthesis: N of oocytes.
The NBCn1 clones summarized in Fig. 6A were all tagged at the Ct with EGFP. The salient observation was that the dpHi/dt of human NBCn1-G is ∼40% higher than that of the other human NBCn1 variants (significant by one-way ANOVA), which were indistinguishable from one another. A comparison of NBCn1-E versus NBCn1-G suggests that the presence of cassette III enhances functional expression.
The two clones summarized in Fig. 6B were not EGFP tagged. This was the only approach by which we could obtain plasmid for NBCn1-H. We chose to compare this variant with NBCn1-B, which differs from NBCn1-H in having cassette I. We observed no difference between the two in functional expression.
In Fig. 6C, the first two constructs differ by the presence of the EGFP tag in NBCn1-G. We observed no significant difference in functional expression. The third construct NBCn1-G-ΔIII – a version of NBCn1-G that we truncated in the Ct, right before cassette III – exhibits a substantially reduced functional expression, confirming that cassette III is stimulatory.
Because of the practical problems in generating plasmids of the human NBCn1 clones, it was not practical to perform biotinylation studies to determine surface expression, which would be necessary to assess the intrinsic activities of the variants. Therefore, we turned to mouse clones.
Mouse clones
To begin the process of evaluating the functional importance of each known OSE of NBCn1, we examined both dpHi/dt (following the protocol in Fig. 5) and NBCn1 surface expression for a series of 10 mouse NBCn1 variants. All variants were tagged with EGFP at the Nt. Figure 7 summarizes the dpHi/dt data and the legend summarizes the statistical analysis, which shows that NBCn1-E and -F are different from each of the other NBCn1 variants.
Figure 7. Summary of pHi recovery rates (dpHi/dt) of oocytes expressing different mouse NBCn1 variants.

Mouse NBCn1 variants are tagged with EGFP at the Nt. Oocytes expressing mouse NBCn1 were superfused using a protocol like that shown in Fig. 5. The data set for H2O-injected control oocytes is independent of that summarized in Fig. 6. A one-way ANOVA shows that the omnibus overall P value is 4.5 × 10−11, indicating statistically significant differences among different variants and H2O. Moreover, a post hoc Tukey's analysis following the ANOVA shows that NBCn1-E and -F are significantly different from the other NBCn1 variants, but not H2O. The insignificance of NBCn1-E and -F vs. H2O-injected oocytes is simply a result of the small sample size and lack of power. An asterisk indicates the groups significantly different by one-way ANOVA followed by post hoc Tukey's comparison. Numbers in parenthesis: N of oocytes.
Surface expression of NBCn1 variants in Xenopus oocytes
To examine whether the OSEs affect the expression of EGFP-NBCn1 at the plasma membrane of Xenopus oocytes, we performed biotinylation assays. In Fig. 8A, which shows a Western blot for total NBCn1 protein, each variant is represented by three bands. The lowest molecular weight bands – ranging from ∼140 to ∼170 kDa – presumably represent the non-glycosylated or core-glycosylated NBCn1 monomer (predicted molecular weight ranging from 152.0 to 169.4 kDa, depending on the size of the OSEs included in the variant). The next bands – ranging from ∼160 to ∼190 kDa – presumably represent the fully glycosylated EGFP-NBCn1 monomer. Finally, the bands in the range ∼290 to ∼400 kDa presumably represent EGFP-NBCn1 dimers. Compared to other variants, NBCn1-N and -O – and to a lesser extent NBCn1-B and -D – have a greater fraction of total NBCn1 protein that run as dimers by SDS-PAGE.
Figure 8. Western blots of total (A) and surface (B) NBCn1, and summary of relative surface abundance (C) of mouse NBCn1 variants in Xenopus oocytes.
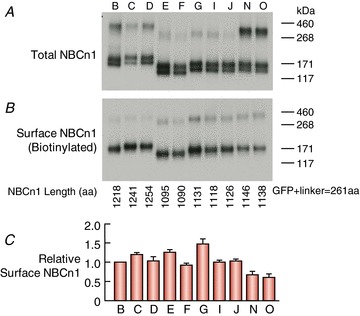
For total protein abundance, we loaded the equivalent of 0.25 oocyte per lane. For surface protein abundance using a biotinylation approach, we pooled 20 oocytes for each experiment and loaded the gel with material from the equivalent of only 1 oocyte. In each lane in B, we compute the sum of densities of the lower molecular weight band (monomer) and the higher molecular weight band (dimer), and then normalize this sum to the sum of densities for lane 1 (i.e. NBCn1-B). C, summary of the relative abundance of surface NBCn1 proteins from 3–4 independent experiments, like that shown in B. A one-way ANOVA (omnibus overall P value = 1.2 × 10−5) followed by a post hoc Tukey's comparison shows that NBCn1-N and -O are both significantly different from NBCn1-C, -E and -G. In addition, NBCn1-G is significantly different from NBCn1-F and -I.
Figure 8B shows a Western blot for biotinylated surface proteins. Note that, for the surface proteins, the blot reveals only the fully glycosylated EGFP-NBCn1 monomers as well as dimers. We did not detect bands presumably representing non-glycosylated or core-glycosylated monomers, consistent with the idea that these are immature forms of the transporters and are not delivered to the plasma membrane. Note that, whereas the Western blots of total protein (Fig. 8A) reveal certain constructs that are enriched as dimers (NBCn1-N ≅ -O > -B ≅ -D > others), the blot of surface protein (Fig. 8B) does not reveal such a difference. The NBCn1 protein that runs at the higher molecular weight presumably reflects dimers that are resistant to denaturation, a property that – among NBCn1 proteins that are not at the cell surface – appears to depend on the combination of OSEs.
Figure 8C summarizes the relative protein levels of NBCn1 variants expressed at the plasma membrane of Xenopus oocytes, and the legend summarizes the statistical analysis.
Discussion
The electroneutral Na/HCO3− cotransporter NBCn1, first cloned from human skeletal muscle (Pushkin et al. 1999) and rat blood vessels (Choi et al. 2000), is widely expressed in diverse tissues, playing critically important physiological as well as pathological roles. In the present study, we extensively examined the diversity of NBCn1 products expressed in human and mouse tissues. We identified two new OSEs (MEAD vs. MERF in a single species, and cassette IV) and 10 new full-length NBCn1 variants. In addition, we used a combination of electrophysiology and biotinylation to examine surface abundance and HCO3− transport activity of 10 mouse NBCn1 variants as expressed in Xenopus oocytes.
Sources of diversity in SLC4A7 products
Elements of diversity
The ability of cells to generate alternative transcription products – the result of using different promoters and alternative splicing – is an important mechanism for regulating gene expression. Most genes from mammals contain alternative promoters and alternatively spliced exons (Pajares et al. 2007; Davuluri et al. 2008), enabling the production of multiple products from a single gene. This multiplicity is probably important for adjusting the phenotype of the protein as a function of developmental programme, cell type, and responses to signals and stresses. Among the SLC4 gene products, those of SLC4A7 are by far the richest in terms of transcript diversity (Parker & Boron, 2013). The known NBCn1 structural variations include two distinct Nts and five major alternative splicing exons:
The MEAD module (16 aa) and the MERF module (11 aa), which probably arise from alternative promoters (discussed below) that use exons 1 and 2, respectively. In addition, the alternative splicing of the 3′ end of exon 1 gives rise to a longer MEAD module (16 aa + 4 aa = 20 aa).
Cassette I (13 aa) in the Nt, which arises from alternative splicing within the 3′ end of exon 7.
Cassette II (124 aa in human, 123 aa in rodent) in the Nt, which is encoded by exon 8.
Cassette IV (20 aa) in the Nt, which is encoded by exon 10.
The Nt truncations, which arise from the splicing-out of exon 15.
Cassette III (36 aa) in the Ct, which is encoded by exon 27.
In the present study, we describe for the first time that MEAD and MERF variants, which we now appreciate are encoded by exons 1 and 2, occur in the same species. Moreover, we describe variants involving exons 10 and 15 for the first time. Our new findings greatly expand the potential coding capacity of SLC4A7. The combination of MEAD(16)/MEAD(20)/MERF + Cassette I + Cassette II + Cassette IV + Cassette III could give rise to as many as 48 full-length protein variants.
Proposed nomenclature
Of these 48 variants, we propose to consider as ‘major’ the 32 variants that involve:
An alternative promoter, such as MEAD(16) vs. MERF.
The splicing in/out of an entire exon, such as cassettes II, IV or III.
The alternative splicing of an exon that changes (add/deletes) amino acids equivalent to no less than 0.5% of total protein length. Thus, cassette I qualifies under this rule.
We propose to continue naming these major variants as NBCn1-α, where the α represents an upper case English letter in the order of discovery. When the number of major variants exceeds 26 – as is soon likely to be the case – we propose naming them NBCn1-AA, -AB, -AC and so on.
In addition, we propose to consider as ‘minor’ the alternative splicing of an exon that modifies protein length by less than 0.5% of total protein length. Thus, we would consider as minor the 4 aa extension of exon 1, which produces the 20 aa version of MEAD. We propose that a minor variant have a base name that is the same as that of the associated major variant, appended by a period and a numeral. The numerals would increase in the order of discovery. Thus, the 4 aa extensions NBCn1-C, -D, -G and -H would be known as NBCn1-C.1, -D.1,-G.1 and -H.1 (Table 2). We have not discussed the 5 aa extension to exon 26 because we observed it in only one mouse clone; however, it would be named NBCn1-C.2. Note that our definitions of ‘major’versus‘minor’ are simply structural, with no implications in the effects of the OSEs on the function of the transporter.
Note that the excision of exon 15 could give rise to as many as 48 additional transcripts (± cassette III) encoding 24 truncated protein products involving combinations of all of the above OSEs except for cassette III. We propose that truncated variants have a base name that is the same as that of the associated two full-length major or minor variants, but with lower case letters separated by a solidus. Thus, our two newly described truncations would become NBCn1-e/g and NBCn1-c/h. Transcripts predicted to encode truncated Nts containing the 4 aa MEAD extension would be named NBCn1-e.1/g.1, and so on.
Physiological relevance of diversity
A particular organism may not make all of the possible 32 full-length major variants and 16 full-length minor variants, even at the level of mRNA. Nevertheless, the list of full-length NBCn1 variants – verified at the level of cDNA cloning – now reaches 16 major variants (10 of which are reported for the first time in the present study) as well as four minor variants that are the result of 4 aa extensions to the MEAD module (all of which are reported for the first time here). Many issues remain to be addressed. For each variant, it will be important to know: (1) transcript abundance, (2) protein abundance, (3) plasma membrane protein abundance (or, in the case of epithelial cells, abundance in apical vs. basolateral membranes), (4) intrinsic (or per-molecule) HCO3− transport activity, (5) intrinsic Na+ conductance, (6) interaction of specific OSEs with protein binding partners and (7) other post-translational regulation. Most of these items are likely to vary markedly among cell types.
Presumably the organism derives some benefit from the large number of OSEs for NBCn1. For example, as discussed below, different combinations of OSEs establish different intrinsic NBCn1 basal HCO3− transport rates. For example, Na+-coupled HCO3− transporters of the SLC4 family have auto-stimulatory and auto-inhibitory domains (McAlear et al. 2006; Parker et al. 2008; Lee et al. 2012b) that may correspond to portions of OSEs. In the present paper, we show that, at least in oocytes, OSEs can modulate surface abundance (as summarized in Table 4 and Table 5A, and discussed below in ‘Effects of optional structural elements on surface abundance and intrinsic activity of NBCn1’). Different combinations of OSEs could also produce different basal intrinsic conductances (Choi et al. 2000), alter the response of NBCn1 to a diverse array of signalling cascades, and permit interactions with specific binding partners such as IRBIT (Shirakabe et al. 2006; Lee et al. 2012b) or calcineurin (Danielsen et al. 2013). Perhaps such functional diversity is beneficial because NBCn1 plays key – and distinct – roles in different cell types, both during development and in the mature animal. For example, in the renal thick ascending limb, basolateral NBCn1 may maintain a sufficiently high pHi to permit NaCl and NH+4 absorption (Aalkjaer et al. 2004). In vascular smooth muscle, NBCn1 has been implicated in the regulation of vascular tone and blood pressure (Boedtkjer et al. 2011). And in the CNS, NBCn1 is widely expressed in neurons in diverse brain regions (Bok et al. 2003; Cooper et al. 2005; Chen et al. 2007a; Park et al. 2010). In any of these cells, NBCn1 could have substantial effects on pHi (by virtue of HCO3− transport) and membrane potential (by virtue of its intrinsic conductance).
Finally, the OSEs of NBCn1 could play a role in pathophysiology. The molecular weight of NBCn1 expressed in human breast cancer is consistently higher than that expressed in normal tissues (Boedtkjer et al. 2012). This larger size of NBCn1 in breast cancer could reflect a change in alternative splicing of SLC4A7 products, although it could also be the result of a change in protein modification in the tumour tissues.
New optional structural elements
In the present study, we describe for the first time the alternative use of exon 1 vs. exon 2, the splicing in/out of exon 10 and the splicing in/out of exon 15.
Exon 1/MEAD vs. Exon 2/MERF
The transcripts encoding the MEAD-NBCn1 variants and the MERF-NBCn1 variants have distinct 5′-UTRs, indicating that SLC4A7 contains two alternative promoters. The present study is the first demonstration that SLC4A7 from the same species is able to express both MEAD- and MERF-type Nts. In addition, the present study for the first time reveals the 4 aa extension to exon 1 that yields four novel minor variants: NBCn1-C.1, -D.1,-G.1 and -H.1.
Exon 10/cassette IV
The 20 aa cassette IV in the Nt of NBCn1 is the fourth major alternative splicing cassette to be identified in SLC4A7 products. Yang et al. (2009) have also reported the expression of a 20 aa cassette in the Nt domain of NBCn1 from rat tissues. However, neither nucleotide nor amino acid sequence was reported. A personal communication (I. Choi, Emory University, Atlanta, GA, USA) allowed us to confirm that our new 20 aa cassette (identified in NBCn1 from mouse eye) is homologous to that present in rat NBCn1 products. Genomic sequence analysis shows that this 60 bp optional cassette – encoding a sequence ‘ESASWHCSCGTLGVGLKRPA’– is present in SLC4A7 from both rat and human. The amino acid sequence of the mouse cassette IV is identical to that of human and rat counterparts except that, in mouse, the third residue from the end is a lysine rather than an arginine (underlined above).
We analysed the potential functional sites in mouse cassette IV using the online tool Eukaryotic Linear Motif (http://elm.eu.org/search/; Dinkel et al. 2012). ‘SCGTLGVG’ contains a potential phosphorylation site for casein kinase CKI. This site also constitutes a potential binding motif for the FHA (forkhead-associated) domain, which is a phosphopeptide–peptide binding domain identified in protein kinases and transcription factors (Hofmann & Bucher, 1995). The ‘ESASWHCS’ motif contains a potential phosphorylation site for glycogen synthase kinase GSK3 (Hur & Zhou, 2010). The ‘KKPAVDMNF’ motif (first four residues derived from cassette IV) provides a potential docking site for mitogen-activated protein kinase MAPK (Tanoue & Nishida, 2002).
We find it interesting that the position of the novel 20 aa cassette IV (encoded by exon 10) within the Nt of NBCn1 is homologous to the position of the 30 aa optional cassette (insert A; encoded by exon 8) in the Nt of NBCn2 (Giffard et al. 2003; Liu et al. 2011). However, the sequences of these two OSEs from NBCn1 and NBCn2 are not conserved at all, consistent with the hypothesis that they originated from different sources during the evolution of SLC4A7 and SLCA10.
Exon 15/truncated NBCn1 products
Omission of exon 15 in SLC4A7 transcripts causes early termination in the open reading frame of NBCn1 around the junction of the Nt and TMD (Fig. 4). We identified six such transcripts in human, and these correspond to two major and one minor variant – NBCn1-e/g, -c/h and -e.1/g.1, respectively – predicted to encode only the Nt domain of NBCn1 (Fig. 1). We also identified in mouse NBCn1 transcripts lacking exon 15 (data not shown). Although Yang et al. (2009) also reported rat NBCn1 clones that are truncated near the end of the Nt domain, no sequence information is available for the truncated rat transcripts. Whether the truncated Nt protein product is present in cells – and if so, where – remains to be addressed. As to the potential physiological relevance of the truncated products of SLC4A7, one might speculate that the truncated Nts might serve as a docking site for cytosolic proteins, or might interfere with the interaction between the Nt and TMD of a full-length NBCn1.
Two truncated protein products, corresponding to only the cytosolic Nt domain of AE3 (anion exchanger SLC4A3), have been identified by Western blot from brain and cardiac tissue (Morgans & Kopito, 1993).
Effects of OSEs on surface abundance and intrinsic activity of NBCn1
Of the 32 potential major full-length variants of NBCn1 (based on five major OSEs: MEAD/MERF and cassettes I–IV), we have now examined 10 from mouse. For each OSE, we have at least two variant pairs for which the only difference between pair members is the presence or absence of a particular OSE. Because the roles of these OSEs may depend upon their context within NBCn1 (i.e. the combination of other OSEs), a full accounting will eventually require the examination of all 32 potential full-length major variants, not all of which may occur in nature. Nevertheless, we are now in a position in which pairwise comparisons among the variants begin to provide information on the functional contribution of each OSE.
To assess the functional effects of OSEs, we estimated the intrinsic activity of our 10 mouse clones, for which we have both dpHi/dt data (Fig. 7) and surface biotinylation data (Fig. 8C). In the first step, we subtract the mean dpHi/dt of control H2O-injected oocytes from individual oocytes expressing NBCn1 to obtain the mean NBCn1-dependent dpHi/dt (Fig. 9A). These values reflect the functional expression, which depends on the product of surface abundance and the intrinsic activity of the transporter. In the second step, using a likelihood-based approach as described in the Methods, we normalize the NBCn1-dependent dpHi/dt data of individual oocytes to the mean relative surface biotinylation signal – a measure of surface abundance – of the corresponding NBCn1 variant. Here, we implicitly assume that the density of NBCn1 proteins in the plasma membrane does not influence the apparent activity of individual NBCn1 molecules. Our statistical approach involves some complexity because we must account for the variation in both the numerator and the denominator of the fraction. Furthermore, because we have not carried out a balanced, full factorial experiment examining the activity of all possible variants, it is difficult to dissect the functional contribution of each OSE. Figure 9B displays the estimated intrinsic activity for each of the variants.
Figure 9. NBCn1-dependent functional expression (A) and intrinsic HCO3− transport activity (B).
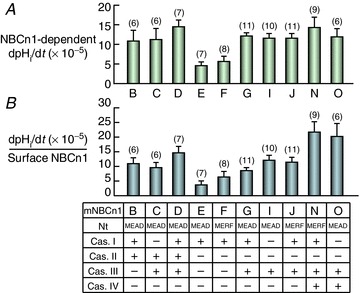
In A, we subtract from the dpHi/dt values of individual NBCn1-expressing oocytes the mean dpHi/dt value of all H2O-injected control oocytes (studied contemporaneously with NBCn1-expression oocytes) to obtain the NBCn1-dependent dpHi/dt (a measure of functional expression). A one-way ANOVA (overall P value = 0.0013) followed by a post hoc Tukey's comparison shows that the differences between the following pairs are significant: D/E, D/F, E/G, E/N and F/N. In B, we normalize the NBCn1-dependent dpHi/dt values from individual oocytes (contributing to the bars in A) to the mean surface NBCn1 levels (Fig. 8C) using a likelihood-based approach. The standard errors were produced using a bootstrap procedure. These normalized dpHi/dt values are indices of the intrinsic HCO3− transport rate of NBCn1 (i.e. the per-molecule transport activity). A bootstrap test, followed by a very conservative Bonferroni correction, shows that the following pairs are significantly different: E/D, E/I, E/J, E/N, E/O, F/N, F/O and G/N. Numbers in parenthesis: N of oocytes.
Table 4 shows the results of pairwise comparisons of variants that differ by only one OSE. For each pair, we obtained unadjusted raw P values (not adjusted with Bonferoni correction) for functional expression, surface abundance and intrinsic activity using ANOVA contrasts. Finally, using a Fisher's method for each set of comparisons, we obtained combined P values that are suitable for judging the overall statistical significance of each OSE. The combined P values represent tests of the main hypotheses in this paper. Because we evaluate each parameter for five different OSEs (Table 4A–E), combined P values of less than 0.05/5 = 0.01 are considered significant. Note that the P values for each individual comparison, while interesting, do not represent our main results, and should not be over-interpreted because that would entail the adjusting for more than five tests.
Table 5 shows the results of two linear regression analyses, one for surface abundance (Table 5A) and one for intrinsic activity (Table 5B). In this approach, surface abundance or intrinsic activity is conceptualized as a linear function of the effects of each OSE, allowing us to estimate the effect of each OSE while adjusting for the other OSEs. We carried out this approach using standard multiple linear regressions for the relative abundance (Table 5A) and using the likelihood/bootstrap approach described in the Methods for intrinsic activity (Table 5B). The estimated effects in Table 5B are the βl coefficient values for l= 1, …, 5 described in the Methods. We can reject the null hypothesis that all linear coefficients are zero because the overall omnibus P values are <0.0005 for both surface abundance and intrinsic activity. As we will see below, this linear regression approach leads to the conclusion that at least some of the OSEs influence the relative abundance of NBCn1 and some influence the intrinsic activity.
Although not shown in Table 5, an additional test for interaction terms yields P= 0.015 for surface abundance and P= 0.090 for intrinsic activity. Thus, it is likely that the OSEs influence the relative abundance in a complex, non-linear manner. Although our current data do not provide strong evidence that intrinsic activity is related to the OSEs in a complex manner, and a linear model appears to predict intrinsic activity adequately, this is probably the result of low statistical power. Indeed, there is at least some suggestive statistical evidence that interactions do in fact exist. In the presence of interactions, the linear effects reported in Table 5 may be thought of as the average linear effect over the various transcripts present in the data, and are thus still informative.
Note that a fundamental difference between the analyses in Table 4 and Table 5 is that each panel of Table 4 considers data from 2 or 3 pairs of variants (a total of 4–6 variants). Each row of Table 5A or B considers data from all 10 variants.
Effect of MEAD vs. MERF
We can make two pairs of comparisons in which the only difference between mouse NBCn1 variants is the presence of MEAD vs. MERF: (1) NBCn1-E vs. NBCn1-F and (2) NBCn1-G vs. NBCn1-J. As shown in Table 4A, the combined P values show that MEAD vs. MERF has a significant effect on the surface abundance, but not on either functional expression or intrinsic activity of NBCn1 in Xenopus oocytes. It appears that MEAD tends to increase the surface abundance compared to MERF as seen in pairs E/F and G/J. These results are corroborated by Table 5, which shows that MEAD has a positive influence on surface abundance (Table 5A: estimated effect = 0.25, P= 0.01), but no significant effect on the intrinsic activity (Table 5B: estimated effect =−2.01, P= 0.21).
Effect of cassette I
We can make two pairs of comparisons to isolate the effect of cassette I on the function of NBCn1: (1) NBCn1-D vs. NBCn1-C and (2) NBCn1-G vs. NBCn1-I. As summarized in Table 4B, the combined P values show that cassette I has a significant effect on surface abundance, but not on either functional expression or intrinsic activity. In Table 5, the P values for both surface abundance (Table 5A) and intrinsic activity (Table 5B) exceed the critical value of 0.05/5 = 0.01. Thus, Table 4B and Table 5A provide conflicting insight into the effect of cassette I on surface abundance. This conflict may be the result of non-linear interactions between the OSEs.
Effect of cassette II
We can make three pairs of comparisons to isolate the effect of cassette II. As summarized in Table 4C, the combined P values show that cassette II has significant effects on both surface abundance and intrinsic activity, but not the functional expression of NBCn1 variants. For surface abundance, the direction of the effect is not consistent, suggesting a complex interaction (although it is consistent for the pairs with small P values). For intrinsic activity, the predominant effect seems to be stimulation by cassette II. In Table 5A, the lack of statistical significance for surface expression is probably the result of complex interactions. In Table 5B, the P value of 0.011 is virtually at the level of statistical significance, indicating that cassette II enhances intrinsic activity (estimated effect = 3.81), consistent with the trend in Table 4C.
Effect of cassette III
As summarized in Table 4D, we can make three pairs of comparisons to isolate the effects of cassette III. The combined P values show that cassette III has a significant effect on the functional expression and intrinsic activity, but not on surface abundance. The presence of cassette III tends to increase both functional expression and intrinsic activity. As shown in Table 5A, cassette III does not appear to have an overall linear influence on the surface abundance. However, as shown in Table 5B, cassette II does have a strong stimulatory effect (estimated effect = 4.39, P= 0.003) on intrinsic activity.
Effect of cassette IV
As summarized in Table 4E, we can make two pairs of comparisons to isolate the effects of cassette IV. The combined P values show that cassette IV has significant effects on surface abundance and intrinsic activities, but not functional expression. The presence of cassette IV tends to lower the surface abundance, but substantially increases the intrinsic activities of NBCn1. Table 5 corroborates these conclusions. The presence of cassette IV has an inhibitory effect on surface abundance (Table 5A: estimated effect =−0.45, P < 0.0005) but a substantial stimulatory effect on intrinsic activity (Table 5B: estimated effect = 10.62, P < 0.0005). Note that, for both surface abundance and intrinsic activity, the estimated effects of cassette IV have magnitudes far larger than for all other OSEs examined.
Concluding remarks
In the present study, we have made the following major observations.
We demonstrate for the first time that, under the control of distinct promoters, the SLC4A7 gene from a single species is able to produce two types of transcripts encoding two alternative Nts, one starting with ‘MEAD’ and the other with ‘MERF’.
We identify two new major alternative splicing units: exon 10 (encoding cassette IV) and exon 15 (lack of which causes truncation of NBCn1 near the boundary between the Nt and TMD).
Based upon the above findings, we identify 10 new major and four new minor full-length NBCn1 variants, plus two major and one minor truncated variants encoded by six SLC4A7 transcripts.
We show that the OSEs can affect both the surface abundance and the intrinsic activity of the transporter. MEAD tends to increase surface abundance, whereas cassette IV reduces it. Cassettes II, III and IV all appear to enhance intrinsic HCO3− transport activity.
The five identified OSEs are capable of generating an unusual diversity of NBCn1 variants, the expression of which is likely to be cell type-specific in vivo. Although we characterized these OSEs in the context of Xenopus oocytes, the structure–function relationships that we have begun developing for the five OSEs of NBCn1 are likely to apply in varying degrees to different subgroups of mammalian cell types that natively express NBCn1. The reason for caution is two types of diversity. First, each cell type presumably has its own set of rules (Muth & Caplan, 2003) governing the delivery and retrieval of proteins – the two processes that determine abundance on the plasma membrane. Thus, how these rules apply to the effect of the five OSEs on the surface abundance of NBCn1 probably varies from one cell type (including oocytes) to the next, and during the life of an individual cell. Second, the ability of a particular OSE to influence intrinsic activity may depend on the roster, concentration and status (e.g. phosphorylation) of binding partners that interact with the OSE. These binding partner properties certainly vary from one cell type (including oocytes) to the next, and throughout the life of an individual cell. In other words, no universal cell type exists – certainly not for cells in culture – for evaluating the effects of OSEs on NBCn1 variants in any truly native system. Nevertheless, our present oocyte work on 10 of the 32 possible NBCn1 variants has already revealed that all OSEs except cassette I can have a major impact on surface abundance or intrinsic activity.
Future work on NBCn1 OSEs could expand in three directions: (1) examination of the associated Na+ conductance of NBCn1, which we did not measure in the present study; (2) consideration of the other 22 theoretical NBCn1 variants; and (3) assessment of the roles of alternative transcription and alternative splicing of NBCn1 in the long list of human diseases associated with NBCn1. Note that an expansion of the number of analysed variants could reveal a role for cassette I, and would carry with it the increased statistical power that would allow one to evaluate how the impact of an OSE depends on its context (i.e. the other OSEs present in the variant).
Acknowledgments
We thank Duncan Wong and Dale Huffman for computer support.
Glossary
- CnAβ
calcineurin A beta
- Ct
cytosolic carboxy-terminal domain
- DIDS
4,4-diisothio-cyanatostilbene-2,2-disulfonic acid
- EGFP
enhanced green fluorescent protein
- IRBIT
inositol trisphosphate (IP3)-receptor (IP3R) binding protein released with IP3
- mTAL
renal medullary thick ascending limb
- NBC
Sodium/bicarbonate cotransporter
- NDCBE
sodium-driven Cl-HCO3− exchanger
- NMDG
N-methyl-d-glucamine
- Nt
cytosolic amino-terminal domain
- OSE
optional structural element
- PSD-95
postsynaptic density protein 95
- RACE
rapid amplification of cDNA ends
- SLC4
solute carrier family 4
- TM
transmembrane segment
- TMD
transmembrane domain
- USH
Usher syndrome
- UTR
untranslated region
Additional information
Competing interests
None declared.
Author contributions
Conception and design of the experiments: Y.L., X.Q., H.S.G., M.D.P., L.-M.C. and W.F.B; performing the experiments and data collection: Y.L., X.Q., L.-M.C., M.D.P. and D.-K.W; data analysis and interpretation: Y.L., X.Q., N.M., L.-M.C. and W.F.B.; drafting: L.-M.C., X.Q. and W.F.B.; final approval: Y.L., X.Q., D.-K.W., H.S.G., Y.-M.G., N.M., M.D.P., L.-M.C. and W.F.B.
Funding
This work was supported by NIH grant NS18400 (W.F.B.) and NSFC grants 30900513 (Y.L.), 31271208 (L.-M.C.) and 31000517 (L.-M.C.), and partially supported by grants 2010MS123 (Y.L.) and 2013TS083 (L.-M.C.) from the Fundamental Research Funds for the Central Universities of China. X.Q. was supported by grants 09POST2060873 and 11POST7670014 from the American Heart Association. H.S.G. was supported by NIH grants F32DK075258 and K01DK082646. W.F.B. gratefully acknowledges the support of the Myers/Scarpa endowed chair.
Author's present addresss
H. S. Gill: The George Washington University & The GW Medical Faculty Associates, Department of Medicine; Division of Renal Diseases & Hypertension, 2300 I Street NW, Ross Hall 436B, Washington, DC 20052, USA.
References
- Aalkjaer C, Frische S, Leipziger J, Nielsen S, Praetorius J. Sodium coupled bicarbonate transporters in the kidney, an update. Acta Physiol Scand. 2004;181:505–512. doi: 10.1111/j.1365-201X.2004.01324.x. [DOI] [PubMed] [Google Scholar]
- Ahmed S, Thomas G, Ghoussaini M, Healey CS, Humphreys MK, Platte R, Morrison J, Maranian M, Pooley KA, Luben R, Eccles D, Evans DG, Fletcher O, Johnson N, dos SS I, Peto J, Stratton MR, Rahman N, Jacobs K, Prentice R, Anderson GL, Rajkovic A, Curb JD, Ziegler RG, Berg CD, Buys SS, McCarty CA, Feigelson HS, Calle EE, Thun MJ, Diver WR, Bojesen S, Nordestgaard BG, Flyger H, Dork T, Schurmann P, Hillemanns P, Karstens JH, Bogdanova NV, Antonenkova NN, Zalutsky IV, Bermisheva M, Fedorova S, Khusnutdinova E, Kang D, Yoo KY, Noh DY, Ahn SH, Devilee P, van Asperen CJ, Tollenaar RA, Seynaeve C, Garcia-Closas M, Lissowska J, Brinton L, Peplonska B, Nevanlinna H, Heikkinen T, Aittomaki K, Blomqvist C, Hopper JL, Southey MC, Smith L, Spurdle AB, Schmidt MK, Broeks A, van Hien RR, Cornelissen S, Milne RL, Ribas G, Gonzalez-Neira A, Benitez J, Schmutzler RK, Burwinkel B, Bartram CR, Meindl A, Brauch H, Justenhoven C, Hamann U, Chang-Claude J, Hein R, Wang-Gohrke S, Lindblom A, Margolin S, Mannermaa A, Kosma VM, Kataja V, Olson JE, Wang X, Fredericksen Z, Giles GG, Severi G, Baglietto L, English DR, Hankinson SE, Cox DG, Kraft P, Vatten LJ, Hveem K, Kumle M, Sigurdson A, Doody M, Bhatti P, Alexander BH, Hooning MJ, van den Ouweland AM, Oldenburg RA, Schutte M, Hall P, Czene K, Liu J, Li Y, Cox A, Elliott G, Brock I, Reed MW, Shen CY, Yu JC, Hsu GC, Chen ST, nton-Culver H, Ziogas A, Andrulis IL, Knight JA, Beesley J, Goode EL, Couch F, Chenevix-Trench G, Hoover RN, Ponder BA, Hunter DJ, Pharoah PD, Dunning AM, Chanock SJ, Easton DF. Newly discovered breast cancer susceptibility loci on 3p24 and 17q23.2. Nat Genet. 2009;41:585–590. doi: 10.1038/ng.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniou AC, Beesley J, McGuffog L, Sinilnikova OM, Healey S, Neuhausen SL, Ding YC, Rebbeck TR, Weitzel JN, Lynch HT, Isaacs C, Ganz PA, Tomlinson G, Olopade OI, Couch FJ, Wang X, Lindor NM, Pankratz VS, Radice P, Manoukian S, Peissel B, Zaffaroni D, Barile M, Viel A, Allavena A, Dall’Olio V, Peterlongo P, Szabo CI, Zikan M, Claes K, Poppe B, Foretova L, Mai PL, Greene MH, Rennert G, Lejbkowicz F, Glendon G, Ozcelik H, Andrulis IL, Thomassen M, Gerdes AM, Sunde L, Cruger D, Birk JU, Caligo M, Friedman E, Kaufman B, Laitman Y, Milgrom R, Dubrovsky M, Cohen S, Borg A, Jernstrom H, Lindblom A, Rantala J, Stenmark-Askmalm M, Melin B, Nathanson K, Domchek S, Jakubowska A, Lubinski J, Huzarski T, Osorio A, Lasa A, Duran M, Tejada MI, Godino J, Benitez J, Hamann U, Kriege M, Hoogerbrugge N, van der Luijt RB, van Asperen CJ, Devilee P, Meijers-Heijboer EJ, Blok MJ, Aalfs CM, Hogervorst F, Rookus M, Cook M, Oliver C, Frost D, Conroy D, Evans DG, Lalloo F, Pichert G, Davidson R, Cole T, Cook J, Paterson J, Hodgson S, Morrison PJ, Porteous ME, Walker L, Kennedy MJ, Dorkins H, Peock S, Godwin AK, Stoppa-Lyonnet D, de PA, Mazoyer S, Bonadona V, Lasset C, Dreyfus H, Leroux D, Hardouin A, Berthet P, Faivre L, Loustalot C, Noguchi T, Sobol H, Rouleau E, Nogues C, Frenay M, Venat-Bouvet L, Hopper JL, Daly MB, Terry MB, John EM, Buys SS, Yassin Y, Miron A, Goldgar D, Singer CF, Dressler AC, Gschwantler-Kaulich D, Pfeiler G, Hansen TV, Jonson L, Agnarsson BA, Kirchhoff T, Offit K, Devlin V, Dutra-Clarke A, Piedmonte M, Rodriguez GC, Wakeley K, Boggess JF, Basil J, Schwartz PE, Blank SV, Toland AE, Montagna M, Casella C, Imyanitov E, Tihomirova L, Blanco I, Lazaro C, Ramus SJ, Sucheston L, Karlan BY, Gross J, Schmutzler R, Wappenschmidt B, Engel C, Meindl A, Lochmann M, Arnold N, Heidemann S, Varon-Mateeva R, Niederacher D, Sutter C, Deissler H, Gadzicki D, Preisler-Adams S, Kast K, Schonbuchner I, Caldes T, de la HM, Aittomaki K, Nevanlinna H, Simard J, Spurdle AB, Holland H, Chen X, Platte R, Chenevix-Trench G, Easton DF. Common breast cancer susceptibility alleles and the risk of breast cancer for BRCA1 and BRCA2 mutation carriers: implications for risk prediction. Cancer Res. 2010;70:9742–9754. doi: 10.1158/0008-5472.CAN-10-1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boedtkjer E, Moreira JM, Mele M, Vahl P, Wielenga VT, Christiansen PM, Jensen VE, Pedersen SF, Aalkjaer C. Contribution of Na+, HCO3−-cotransport to cellular pH control in human breast cancer: a role for the breast cancer susceptibility locus NBCn1 (SLC4A7) Int J Cancer. 2012;132:1288–1299. doi: 10.1002/ijc.27782. [DOI] [PubMed] [Google Scholar]
- Boedtkjer E, Praetorius J, Matchkov VV, Stankevicius E, Mogensen S, Fuchtbauer AC, Simonsen U, Fuchtbauer EM, Aalkjaer C. Disruption of Na+, HCO3− cotransporter NBCn1 (slc4a7) inhibits NO-mediated vasorelaxation, smooth muscle Ca2+ sensitivity, and hypertension development in mice. Circulation. 2011;124:1819–1829. doi: 10.1161/CIRCULATIONAHA.110.015974. [DOI] [PubMed] [Google Scholar]
- Bok D, Galbraith G, Lopez I, Woodruff M, Nusinowitz S, BeltrandelRio H, Huang WH, Zhao SL, Geske R, Montgomery C, Van Sligtenhorst I, Friddle C, Platt K, Sparks MJ, Pushkin A, Abuladze N, Ishiyama A, Dukkipati R, Liu WX, Kurtz I. Blindness and auditory impairment caused by loss of the sodium bicarbonate cotransporter NBC3. Nat Genet. 2003;34:313–319. doi: 10.1038/ng1176. [DOI] [PubMed] [Google Scholar]
- Boron WF, Chen L, Parker MD. Modular structure of sodium-coupled bicarbonate transporters. J Exp Biol. 2009;212:1697–1706. doi: 10.1242/jeb.028563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boron WF, McCormick WC, Roos A. pH regulation in barnacle muscle fibers: dependence on intracellular and extracellular pH. Am J Physiol. 1979;237:C185–193. doi: 10.1152/ajpcell.1979.237.3.C185. [DOI] [PubMed] [Google Scholar]
- Bouyer P, Sakai H, Itokawa T, Kawano T, Fulton CM, Boron WF, Insogna KL. Colony-stimulating Factor-1 increases osteoclast intracellular pH and promotes survival via the electroneutral Na/HCO3 cotransporter NBCn1. Endocrinology. 2007;148:831–840. doi: 10.1210/en.2006-0547. [DOI] [PubMed] [Google Scholar]
- Boyarsky G, Ganz MB, Cragoe EJ, Jr, Boron WF. Intracellular pH dependence of Na–H exchange and acid loading in quiescent and arginine vasopressin-activated mesangial cells. Proc Natl Acad Sci U S A. 1990;87:5921–5924. doi: 10.1073/pnas.87.15.5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casella G, Berger RL. Stastical Inference. 2nd edn. Pacific Grove, CA: Duxbury Press; 2001. [Google Scholar]
- Chen LM, Choi I, Haddad GG, Boron WF. Chronic continuous hypoxia decreases the expression of SLC4A7 (NBCn1) and SLC4A10 (NCBE) in mouse brain. Am J Physiol Regul Integr Comp Physiol. 2007a;293:R2412–2420. doi: 10.1152/ajpregu.00497.2007. [DOI] [PubMed] [Google Scholar]
- Chen LM, Haddad GG, Boron WF. Effects of chronic continuous hypoxia on the expression of SLC4A8 (NDCBE) in neonatal versus adult mouse brain. Brain Res. 2008a;1238:85–92. doi: 10.1016/j.brainres.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Kelly ML, Rojas JD, Parker MD, Gill HS, Davis BA, Boron WF. Use of a new polyclonal antibody to study the distribution and glycosylation of the sodium-coupled bicarbonate transporter NCBE in rodent brain. Neuroscience. 2008b;151:374–385. doi: 10.1016/j.neuroscience.2007.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Liu Y, Boron WF. Role of an extracellular loop in determining the stoichiometry of Na/HCO3 cotransporters. J Physiol. 2011;589:877–890. doi: 10.1113/jphysiol.2010.198713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LM, Qin X, Moss FJ, Liu Y, Boron WF. Effect of simultaneously replacing putative TM6 and TM12 of human NBCe1-A with those from NBCn1 on surface abundance in Xenopus oocytes. J Membr Biol. 2012;245:131–140. doi: 10.1007/s00232-012-9421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Choong LY, Lin Q, Philp R, Wong CH, Ang BK, Tan YL, Loh MC, Hew CL, Shah N, Druker BJ, Chong PK, Lim YP. Differential expression of novel tyrosine kinase substrates during breast cancer development. Mol Cell Proteomics. 2007b;6:2072–2087. doi: 10.1074/mcp.M700395-MCP200. [DOI] [PubMed] [Google Scholar]
- Choi I, Aalkjaer C, Boulpaep EL, Boron WF. An electroneutral sodium/bicarbonate cotransporter NBCn1 and associated sodium channel. Nature. 2000;405:571–575. doi: 10.1038/35014615. [DOI] [PubMed] [Google Scholar]
- Choi I, Yang HS, Boron WF. The electrogenicity of the rat sodium–bicarbonate cotransporter NBCe1 requires interactions among transmembrane segments of the transporter. J Physiol. 2007;578:131–142. doi: 10.1113/jphysiol.2006.114959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TA, Doherty JK, Lin YJ, Ramsey EE, Holmes R, Keenan EJ, Clinton GM. NH2-terminally truncated HER-2/neu protein: relationship with shedding of the extracellular domain and with prognostic factors in breast cancer. Cancer Res. 1998;58:5123–5129. [PubMed] [Google Scholar]
- Cooper DS, Saxena NC, Yang HS, Lee HJ, Moring AG, Lee A, Choi I. Molecular and functional characterization of the electroneutral Na/HCO3 cotransporter NBCn1 in rat hippocampal neurons. J Biol Chem. 2005;280:17823–17830. doi: 10.1074/jbc.M408646200. [DOI] [PubMed] [Google Scholar]
- Danielsen AA, Parker MD, Lee S, Boron WF, Aalkjaer C, Boedtkjer E. Splice cassette II of Na+,HCO3− cotransporter NBCn1 (slc4a7) interacts with calcineurin A: implications for transporter activity and intracellular pH control during rat artery contractions. J Biol Chem. 2013;288:8146–8155. doi: 10.1074/jbc.M113.455386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–177. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- Dinkel H, Michael S, Weatheritt RJ, Davey NE, Van RK, Altenberg B, Toedt G, Uyar B, Seiler M, Budd A, Jodicke L, Dammert MA, Schroeter C, Hammer M, Schmidt T, Jehl P, McGuigan C, Dymecka M, Chica C, Luck K, Via A, Chatr-Aryamontri A, Haslam N, Grebnev G, Edwards RJ, Steinmetz MO, Meiselbach H, Diella F, Gibson TJ. ELM–the database of eukaryotic linear motifs. Nucleic Acids Res. 2012;40:D242–251. doi: 10.1093/nar/gkr1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B, Tibshirani RJ. An Introduction to the Bootstrap. New York: Chapman & Hall/CRC; 1994. [Google Scholar]
- Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ, Pihur V, Vollenweider P, O’Reilly PF, Amin N, Bragg-Gresham JL, Teumer A, Glazer NL, Launer L, Zhao JH, Aulchenko Y, Heath S, Sober S, Parsa A, Luan J, Arora P, Dehghan A, Zhang F, Lucas G, Hicks AA, Jackson AU, Peden JF, Tanaka T, Wild SH, Rudan I, Igl W, Milaneschi Y, Parker AN, Fava C, Chambers JC, Fox ER, Kumari M, Go MJ, van der HP, Kao WH, Sjogren M, Vinay DG, Alexander M, Tabara Y, Shaw-Hawkins S, Whincup PH, Liu Y, Shi G, Kuusisto J, Tayo B, Seielstad M, Sim X, Nguyen KD, Lehtimaki T, Matullo G, Wu Y, Gaunt TR, Onland-Moret NC, Cooper MN, Platou CG, Org E, Hardy R, Dahgam S, Palmen J, Vitart V, Braund PS, Kuznetsova T, Uiterwaal CS, Adeyemo A, Palmas W, Campbell H, Ludwig B, Tomaszewski M, Tzoulaki I, Palmer ND, Aspelund T, Garcia M, Chang YP, O’Connell JR, Steinle NI, Grobbee DE, Arking DE, Kardia SL, Morrison AC, Hernandez D, Najjar S, McArdle WL, Hadley D, Brown MJ, Connell JM, Hingorani AD, Day IN, Lawlor DA, Beilby JP, Lawrence RW, Clarke R, Hopewell JC, Ongen H, Dreisbach AW, Li Y, Young JH, Bis JC, Kahonen M, Viikari J, Adair LS, Lee NR, Chen MH, Olden M, Pattaro C, Bolton JA, Kottgen A, Bergmann S, Mooser V, Chaturvedi N, Frayling TM, Islam M, Jafar TH, Erdmann J, Kulkarni SR, Bornstein SR, Grassler J, Groop L, Voight BF, Kettunen J, Howard P, Taylor A, Guarrera S, Ricceri F, Emilsson V, Plump A, Barroso I, Khaw KT, Weder AB, Hunt SC, Sun YV, Bergman RN, Collins FS, Bonnycastle LL, Scott LJ, Stringham HM, Peltonen L, Perola M, Vartiainen E, Brand SM, Staessen JA, Wang TJ, Burton PR, Artigas MS, Dong Y, Snieder H, Wang X, Zhu H, Lohman KK, Rudock ME, Heckbert SR, Smith NL, Wiggins KL, Doumatey A, Shriner D, Veldre G, Viigimaa M, Kinra S, Prabhakaran D, Tripathy V, Langefeld CD, Rosengren A, Thelle DS, Corsi AM, Singleton A, Forrester T, Hilton G, McKenzie CA, Salako T, Iwai N, Kita Y, Ogihara T, Ohkubo T, Okamura T, Ueshima H, Umemura S, Eyheramendy S, Meitinger T, Wichmann HE, Cho YS, Kim HL, Lee JY, Scott J, Sehmi JS, Zhang W, Hedblad B, Nilsson P, Smith GD, Wong A, Narisu N, Stancakova A, Raffel LJ, Yao J, Kathiresan S, O’Donnell CJ, Schwartz SM, Ikram MA, Longstreth WT, Jr, Mosley TH, Seshadri S, Shrine NR, Wain LV, Morken MA, Swift AJ, Laitinen J, Prokopenko I, Zitting P, Cooper JA, Humphries SE, Danesh J, Rasheed A, Goel A, Hamsten A, Watkins H, Bakker SJ, van Gilst WH, Janipalli CS, Mani KR, Yajnik CS, Hofman A, Mattace-Raso FU, Oostra BA, Demirkan A, Isaacs A, Rivadeneira F, Lakatta EG, Orru M, Scuteri A, la-Korpela M, Kangas AJ, Lyytikainen LP, Soininen P, Tukiainen T, Wurtz P, Ong RT, Dorr M, Kroemer HK, Volker U, Volzke H, Galan P, Hercberg S, Lathrop M, Zelenika D, Deloukas P, Mangino M, Spector TD, Zhai G. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giffard RG, Lee YS, Ouyang YB, Murphy SL, Monyer H. Two variants of the rat brain sodium-driven chloride bicarbonate exchanger (NCBE): developmental expression and addition of a PDZ motif. Eur J Neurosci. 2003;18:2935–2945. doi: 10.1046/j.1460-9568.2003.03053.x. [DOI] [PubMed] [Google Scholar]
- Gill HS, Dutcher LS, Parker MD, Boron WF. Identification and cloning of alternative N termini of human NBCn1. Proc Physiol Soc. 2006;3:C20. [Google Scholar]
- Hofmann K, Bucher P. The FHA domain: a putative nuclear signalling domain found in protein kinases and transcription factors. Trends Biochem Sci. 1995;20:347–349. doi: 10.1016/s0968-0004(00)89072-6. [DOI] [PubMed] [Google Scholar]
- Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat Rev Neurosci. 2010;11:539–551. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro H, Walther D, Arinami T, Uhl GR. Variation in a bicarbonate co-transporter gene family member SLC4A7 is associated with propensity to addictions: a study using fine-mapping and three samples. Addiction. 2007;102:1320–1325. doi: 10.1111/j.1360-0443.2007.01877.x. [DOI] [PubMed] [Google Scholar]
- Jakobsen JK, Odgaard E, Wang W, Elkjaer ML, Nielsen S, Leipziger J, Aalkjaer C. Functional up-regulation of basolateral Na+-dependent HCO3− transporter NBCn1 in medullary thick ascending limb of K+-depleted rats. Pflugers Arch. 2004;448:571–578. doi: 10.1007/s00424-004-1303-4. [DOI] [PubMed] [Google Scholar]
- Kumar S, Flacke JP, Kostin S, Appukuttan A, Reusch HP, Ladilov Y. SLC4A7 sodium bicarbonate co-transporter controls mitochondrial apoptosis in ischaemic coronary endothelial cells. Cardiovasc Res. 2011;89:392–400. doi: 10.1093/cvr/cvq330. [DOI] [PubMed] [Google Scholar]
- Kwon TH, Fulton C, Wang W, Kurtz I, Frokiaer J, Aalkjaer C, Nielsen S. Chronic metabolic acidosis upregulates rat kidney Na-HCO cotransporters NBCn1 and NBC3 but not NBC1. Am J Physiol Renal Physiol. 2002;282:F341–351. doi: 10.1152/ajprenal.00104.2001. [DOI] [PubMed] [Google Scholar]
- Lauritzen G, Jensen MB, Boedtkjer E, Dybboe R, Aalkjaer C, Nylandsted J, Pedersen SF. NBCn1 and NHE1 expression and activity inΔNErbB2 receptor-expressing MCF-7 breast cancer cells: contributions to pHi regulation and chemotherapy resistance. Exp Cell Res. 2010;316:2538–2553. doi: 10.1016/j.yexcr.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Lee S, Yang HS, Kim E, Ju EJ, Kwon MH, Dudley RK, Smith Y, Yun CC, Choi I. PSD-95 interacts with NBCn1 and enhances channel-like activity without affecting Na/HCO3 cotransport. Cell Physiol Biochem. 2012a;30:1444–1455. doi: 10.1159/000343332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, Boron WF, Parker MD. Relief of autoinhibition of the electrogenic Na-HCO3 cotransporter NBCe1-B: role of IRBIT vs. amino-terminal truncation. Am J Physiol Cell Physiol. 2012b;302:C518–526. doi: 10.1152/ajpcell.00352.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Xu JY, Wang DK, Boron WF, Chen LM. Expression and distribution of NBCn2 (Slc4a10) splice variants in mouse brain: cloning of novel variant NBCn2-D. Brain Res. 2011;1390:33–40. doi: 10.1016/j.brainres.2011.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J, Shu XO, Cai Q, Gao YT, Zheng Y, Li G, Li C, Gu K, Wen W, Xiang YB, Lu W, Zheng W. Evaluation of breast cancer susceptibility loci in Chinese women. Cancer Epidemiol Biomarkers Prev. 2010;19:2357–2365. doi: 10.1158/1055-9965.EPI-10-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez IA, Acuna D, Galbraith G, Bok D, Ishiyama A, Liu W, Kurtz I. Time course of auditory impairment in mice lacking the electroneutral sodium bicarbonate cotransporter NBC3 (slc4a7) Brain Res Dev Brain Res. 2005;160:63–77. doi: 10.1016/j.devbrainres.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Lu J, Daly CM, Parker MD, Gill HS, Piermarini PM, Pelletier MF, Boron WF. Effect of human carbonic anhydrase II on the activity of the human electrogenic Na/HCO3 cotransporter NBCe1-A in Xenopus oocytes. J Biol Chem. 2006;281:19241–19250. doi: 10.1074/jbc.M602181200. [DOI] [PubMed] [Google Scholar]
- McAlear SD, Liu X, Williams JB, McNicholas-Bevensee CM, Bevensee MO. Electrogenic Na/HCO3 cotransporter (NBCe1) variants expressed in Xenopus oocytes: functional comparison and roles of the amino and carboxy termini. J Gen Physiol. 2006;127:639–658. doi: 10.1085/jgp.200609520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgans CW, Kopito RR. Generation of truncated brain AE3 isoforms by alternate mRNA processing. J Cell Sci. 1993;106:1275–1282. doi: 10.1242/jcs.106.4.1275. [DOI] [PubMed] [Google Scholar]
- Musa-Aziz R, Boron WF, Parker MD. Using fluorometry and ion-sensitive microelectrodes to study the functional expression of heterologously-expressed ion channels and transporters in Xenopus oocytes. Methods. 2010;51:134–145. doi: 10.1016/j.ymeth.2009.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muth TR, Caplan MJ. Transport protein trafficking in polarized cells. Annu Rev Cell Dev Biol. 2003;19:333–366. doi: 10.1146/annurev.cellbio.19.110701.161425. [DOI] [PubMed] [Google Scholar]
- Pajares MJ, Ezponda T, Catena R, Calvo A, Pio R, Montuenga LM. Alternative splicing: an emerging topic in molecular and clinical oncology. Lancet Oncol. 2007;8:349–357. doi: 10.1016/S1470-2045(07)70104-3. [DOI] [PubMed] [Google Scholar]
- Park HJ, Rajbhandari I, Yang HS, Lee S, Cucoranu D, Cooper DS, Klein JD, Sands JM, Choi I. Neuronal expression of sodium/bicarbonate cotransporter NBCn1 (SLC4A7) and its response to chronic metabolic acidosis. Am J Physiol Cell Physiol. 2010;298:C1018–1028. doi: 10.1152/ajpcell.00492.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD, Boron WF. The divergence, actions, roles, and relatives of sodium coupled bicarbonate transporters. Physiol Rev. 2013;93:803–959. doi: 10.1152/physrev.00023.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker MD, Bouyer P, Daly CM, Boron WF. Cloning and characterization of novel human SLC4A8 gene products encoding Na+-driven Cl−/HCO3− exchanger variants NDCBE-A, -C, and -D. Physiol Genomics. 2008;34:265–276. doi: 10.1152/physiolgenomics.90259.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pushkin A, Abuladze N, Lee I, Newman D, Hwang J, Kurtz I. Cloning, tissue distribution, genomic organization, and functional characterization of NBC3, a new member of the sodium bicarbonate cotransporter family. J Biol Chem. 1999;274:16569–16575. doi: 10.1074/jbc.274.23.16569. [DOI] [PubMed] [Google Scholar]
- Reiners J, van WE, Marker T, Zimmermann U, Jurgens K, te BH, Overlack N, Roepman R, Knipper M, Kremer H, Wolfrum U. Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum Mol Genet. 2005;14:3933–3943. doi: 10.1093/hmg/ddi417. [DOI] [PubMed] [Google Scholar]
- Riihonen R, Nielsen S, Vaananen HK, Laitala-Leinonen T, Kwon TH. Degradation of hydroxyapatite in vivo and in vitro requires osteoclastic sodium-bicarbonate co-transporter NBCn1. Matrix Biol. 2010;29:287–294. doi: 10.1016/j.matbio.2010.01.003. [DOI] [PubMed] [Google Scholar]
- Roos A, Boron WF. Intracellular pH. Physiol Rev. 1981;61:296–434. doi: 10.1152/physrev.1981.61.2.296. [DOI] [PubMed] [Google Scholar]
- Schulz E, Munzel T. Intracellular pH: a fundamental modulator of vascular function. Circulation. 2011;124:1806–1807. doi: 10.1161/CIRCULATIONAHA.111.061226. [DOI] [PubMed] [Google Scholar]
- Shirakabe K, Priori G, Yamada H, Ando H, Horita S, Fujita T, Fujimoto I, Mizutani A, Seki G, Mikoshiba K. IRBIT, an inositol 1,4,5-trisphosphate receptor-binding protein, specifically binds to and activates pancreas-type Na+/HCO3− cotransporter 1 (pNBC1) Proc Natl Acad Sci USA. 2006;103:9542–9547. doi: 10.1073/pnas.0602250103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sueta A, Ito H, Kawase T, Hirose K, Hosono S, Yatabe Y, Tajima K, Tanaka H, Iwata H, Iwase H, Matsuo K. A genetic risk predictor for breast cancer using a combination of low-penetrance polymorphisms in a Japanese population. Breast Cancer Res Treat. 2012;132:711–21. doi: 10.1007/s10549-011-1904-5. [DOI] [PubMed] [Google Scholar]
- Tanoue T, Nishida E. Docking interactions in the mitogen-activated protein kinase cascades. Pharmacol Ther. 2002;93:193–202. doi: 10.1016/s0163-7258(02)00188-2. [DOI] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. URL= http://www.R-project.org. [Google Scholar]
- Trudeau MC, Warmke JW, Ganetzky B, Robertson GA. HERG, a human inward rectifier on the voltage-gated potassium channel family. Science. 1995;269:92–95. doi: 10.1126/science.7604285. [DOI] [PubMed] [Google Scholar]
- Won S, Morris N, Lu Q, Elston RC. Choosing an optimal method to combine P-values. Stat Med. 2009;28:1537–1553. doi: 10.1002/sim.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto T, Shirayama T, Sakatani T, Takahashi T, Tanaka H, Takamatsu T, Spitzer KW, Matsubara H. Enhanced activity of ventricular Na+-HCO3-cotransport in pressure overload hypertrophy. Am J Physiol Heart Circ Physiol. 2007;293:H1254–1264. doi: 10.1152/ajpheart.00964.2006. [DOI] [PubMed] [Google Scholar]
- Yang HS, Cooper DS, Rajbhandari I, Park HJ, Lee S, Choi I. Inhibition of rat Na+HCO3− cotransporter (NBCn1) function and expression by the alternative splice domain. Exp Physiol. 2009;94:1114–1123. doi: 10.1113/expphysiol.2009.048603. [DOI] [PubMed] [Google Scholar]


