Abstract
Expression of claudin-1, a tight junction protein, is highly upregulated in colon cancer. We have reported that claudin-1 expression in colon cancer cells is epigenetically regulated as histone deacetylase (HDAC) inhibitors decrease claudin-1 messenger RNA (mRNA) stability and thus expression. In this regard, our data suggested a role of the 3′-untranslated region (UTR) in the regulation of HDAC-dependent regulation of claudin-1 mRNA stability. In the current study, we demonstrate, based on our continued investigation, that the ELAV-like RNA-binding proteins (RBPs), human antigen R (HuR) and tristetraprolin (TTP) associate with the 3′-UTR of claudin-1 mRNA to modulate the latter’s stability. Ribonomic and site-directed mutagenesis approaches were used to confirm the binding of HuR and TTP to the 3′-UTR of claudin-1. We further confirmed their roles in the stabilization of claudin-1 mRNA, under conditions of HDAC inhibition. In summary, we report that HuR and TTP are the critical regulators of the posttranscriptional regulation of claudin-1 expression in colon cancer cells. We also demonstrate that inhibition of HDACs by trichostatin treatment decreased the binding of HuR while increasing the binding of TTP to the 3′-UTR of claudin-1. Additionally, we provide data showing transcriptional regulation of claudin-1 expression, through the regulation of transcription factor Sp1. Taken together, we demonstrate epigenetic regulation of claudin-1 expression in colon cancer cells at the transcriptional and posttranscriptional levels.
Introduction
Chromatin remodeling plays an important role in the regulation of gene transcription. Evidence suggests that acetylation and deacetylation of the histone or non-histone proteins play a significant role in chromatin remodeling and transcription regulation (1). Trichostatin-A (TSA) and sodium butyrate are two classical and widely used inhibitors of histone deacetylases (HDACs) (2). TSA and sodium butyrate are suggested to increase gene transcription through the promotion of the acetylation level of histones and chromatin relaxation. However, in addition to transcriptional activators, HDAC inhibitors are also involved in the posttranscriptional regulation (3) and acetylation of cytoplasmic proteins.
Posttranscriptional regulation is tightly controlled by the interaction of specific mRNA sequences (cis-acting elements) with specific trans-acting factors such as RNA-binding proteins (RBPs; (4)). Of interest, the AU-rich cis-acting elements (ARE) are the critical cis-acting elements in the 3′-untranslated regions (3′-UTRs) of many short-lived oncogenes (4). A number of ARE-binding proteins have been identified, including ELAV (embryonic lethal abnormal vision in Drosophila melanogaster) protein family members such as human antigen R (HuR), tristetraprolin (TTP), AU-rich-binding/degradation factor (AUF1) and so on (5).
HuR is implicated in a variety of pathologies in which destabilization of many labile key mRNAs is causally linked with the onset and the course of disease (5). HuR stabilizes ARE-containing mRNAs of several inducible genes, such as c-fos, c-myc, cyclooxygenase-2, tumor necrosis factor-α and so on (4,6). In addition, HuR associates with TTP in a homeostatic complex to regulate the cytoplasmic levels of relevant RNAs (7). TTP is the prototype of a group of CCCH tandem zinc finger proteins, characterized by a CCCH zinc finger motif (8). In contrast with the effects of HuR, TTP promotes the rapid decay of ARE-containing mRNAs (7). Dysregulation of claudin-1 expression is reported in colon, skin and ovarian cancers (9,10). Claudin-1 3′-UTR (2.3 kb) contains putative AREs. In previous studies, we have demonstrated that HDAC inhibition inhibits claudin-1 expression (3) in colon cancer cells. Based on our continued investigation, we demonstrate here that HuR and TTP play key roles in HDAC inhibitor-dependent posttranscriptional regulation of claudin-1 expression. We also provide data that transcriptional regulation through the modulation of Sp1 contributes to the HDAC inhibitor-dependent regulation of claudin-1 expression.
Materials and methods
Plasmids and reagents
The antibodies against claudin-1, claudin-4 and HuR were purchased from Invitrogen (San Francisco, CA), and the anti-TTP antibody was from Santa Cruz Biotechnology (Santa Cruz, CA). The anti-β-actin antibody, trichostatin-A, actinomycin D, and the control- and HuR-specific short hairpin RNAs (shRNAs) were purchased from Sigma–Aldrich (St Louis, MO).
Cell culture
Human colon cancer cell lines (SW480 and SW620) were cultured in RPMI (Invitrogen), breast cancer cells (MDA-MB-468) and Madin Darby canine kidney (MDCK-II) cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen), which was supplemented with 10% fetal bovine serum and antibiotics. Cells were maintained at 37°C in a humidified atmosphere with 5% CO2.
Rapid amplification of complementary DNA ends
The 3′-UTR of human claudin-1 transcript was amplified using the SMART™ (switching mechanism at 5′ end of RNA transcript) rapid amplification of complementary DNA (cDNA) ends (RACE) cDNA amplification kit (BD Biosciences, Clontech, Palo Alto, CA), as per manufacturer’s instructions, and the cDNA generated from the RNA was isolated from SW480 cells. PCR was carried out using a forward primer (5′-CACAGAGGCAAAAGGAGAAAATC-3′) from human claudin-1 codons and the universal primer provided in the kit. The PCR product of 2.5kb was gel purified and cloned into the pCRII-TOPO vector (Invitrogen). Positive clones were identified by restriction digestion with EcoRI, and the nucleotide sequences of the two ends of the insert were determined by sequencing.
PCR amplification of the 3′-UTR elements
For the amplification of the 3′-UTR fragments of claudin-1, the upper XbaI linker-primer 5′ -CACAGAGGCAAAAGGAGAAAATC-3′ of the 3′-UTR of claudin-1 was used in combination with the following lower XbaI linker-primer to establish fragments of different lengths: UTR1(2.5 kb): 5′-ACCACACCTGGAAACAG ACC-3′; UTR2 (1.9 kb): 5′-AATGGGTTTCTTGCCTTAACC-3′; UTR3 (1.5 kb): 5′-TTTCCATTCTTTCAGCTGTG-3′; UTR4 (964 bp): 5′-ACCT TTTTGTTCCCCATTCC-3′; UTR5 (525 bp): 5′-TTTGCCACAAGACCTA GCCT-3′; UTR6 (384 bp): 5′ TGGTATATTTTCTTTTTCG-3′; UTR7 (274 bp): 5′-TACTCAATTGGGGGAA GGG-3′.
Construction of plasmid constructs
The chimeric firefly luciferase reporter construct of claudin-1 3′-UTR was generated using standard protocol. The full-length 2.5 kb 3′-UTR of claudin-1 and its different fragments (UTR1–UTR7) were amplified and subcloned into the pGL3-Luc plasmid (Promega) at the XbaI site to generate various chimeric pGL3-Luc-claudin-1 3′-UTR reporter constructs.
Reporter assays
Reporter assays were performed as described previously (3). Briefly, cells were seeded on 12-well plates in triplicate and allowed to grow overnight to reach 50–70% confluence. Then cells were cotransfected with a firefly luciferase reporter construct and a reference construct that contained Renilla reniformis luciferase, phRL-TK (Promega), using Lipofectamine Plus reagents (Invitrogen). Luciferase activity was measured 48h later using the Dual Luciferase Reporter Assay System kit (Promega) in an Optocomp II luminometer (MGM Instruments, Hamden, CT). Firefly luciferase activity was normalized to R. reniformis luciferase activity and plotted as mean ± SD from three independent experiments.
Western blot and immunofluorescence analysis
Standard protocols were used as described previously (11). Secondary antibodies were either donkey anti-rabbit Rhodamine Red-X or goat anti-mouse fluorescein isothiocyanate conjugates.
Analysis of mRNA stability
This procedure was performed as described previously (3). Briefly, actinomycin D (10 μg/ml; Sigma–Aldrich) was used to inhibit nascent RNA synthesis. SW620 or SW480 cells were pretreated with TSA (200ng/ml) for 4h and then with actinomycin D (10 μg/ml) for 0, 4, 8, 16, 24, 36 and 48 h, after which the samples were collected for RNA isolation.
RNA immunoprecipitations
Lysates were prepared from confluent SW480 cells. Equal amounts of protein lysates were used (100–300 μg). HuR monoclonal antibody or isotype control IgG1 were precoated onto Protein-A Sepharose beads and extensively washed. Lysates from each cell line were initially preabsorbed with 30 μg of IgG1. Individual pulldowns were performed at 4°C and 2 µg of recovered RNA per immunoprecipitation was amplified to generate cDNA. Claudin-1 3′-UTR elements were validated by PCR, as described previously (12).
Site-directed mutagenesis of the claudin-1 3′-UTR
Mutation/s of ARE were induced using the Quikchange II-E Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer’s instructions. For each mutation, the core pentamer AUUUA (ATTTA in the DNA sequence) was mutated to AGGUA (AGGTA in the DNA). The forward primers used are listed. AREs 1 and 2 of the human claudin-1 3′-UTR (NM_021101.4) were mutated using the following forward primers, respectively: mut1: AGCATACTTAAAATATC TCTAAAATAGGTAAATGTAGGTAATTCCATA TTGATG AAGA TGTTTATTGG; mut2 : TATACATATGTAACAGTCAA ATATC A GGTACTCTTCTTCATTAGCTTTGGGTGC.
Chromatin immunoprecipitation assay
This assay was performed with the chromatin immunoprecipitation (ChIP) assay kit (Upstate Biotechnology, Temecula, CA) according to the manufacturer’s instructions, as described previously (11).
Statistical analysis
The mRNA half-life was analyzed by the one-phase exponential decay of non-linear regression. The level of significance was determined using Duncan’s multiple range tests. Quantitative densitometry values were compared by the unpaired Student’s t-test. Different groups were compared using analysis of variance and the exact P values are reported.
Results
The claudin-1 3′-UTR contains AREs that help regulate claudin-1 mRNA stability
We have previously reported that inhibition of HDAC activity decreases claudin-1 expression in colon cancer cells (3). A decrease in mRNA stability underlies this decrease and our data suggested a potential role of claudin-1 3′-UTR in this regulation. To obtain further experimental evidence for a causal role of claudin-1 3′-UTR in HDAC-dependent regulation of claudin-1 mRNA stability, we used expression constructs containing either the claudin-1 coding sequence (pCMVclaudin-1), the coding sequence and the 3′-UTR (pCMVclaudin-1–3′- UTR) or the coding sequence and the 3′-UTR (where the 3′-UTR is flipped in the 3′→5′ orientation; pCMVclaudin-1-FLIP). SW480 colon cancer cells were used for transient transfection of these constructs and the effect on claudin-1 mRNA stability was determined in response to treatment with the HDAC inhibitor, TSA. Results showed a decrease in claudin-1 mRNA stability in SW480vector- or SW480claudin1-3′- UTR-transfected cells but not in SW480claudin-1- or SW480claudin-1FLIP-transfected cells, in response to TSA treatment (Figure 1A and Supplementary Figure 1, available at Carcinogenesis Online). Thus, our studies supported the causal importance of the 3′-UTR in regulation of claudin-1 mRNA stability.
Fig. 1.
The 3′-UTR of claudin-1 is important for its mRNA stability. (A) Immunoblot analysis to determine claudin-1 expression. SW480 cells were transfected with 0.8 µg pCMV-script, pCMVclaudin-1, pCMVclaudin-1–3′-UTR or pCMVclaudin-1-FLIP. Transfected cells were then treated with increasing concentrations of TSA for 36h; (B) Schematic representation of claudin-1 3′-UTR and the AU-rich sequences in it; (C) Schematic representation of UTR1–UTR7 regions that were cloned into the 3′-position of a luciferase reporter gene to create mammalian expression constructs pGL3–claudin-1–UTR1 to pGL3–claudin-1–UTR7; and (D) Effect of TSA treatment upon claudin-1 promoter activity. SW480 and SW620 cells were transiently transfected with pGL3–claudin-1–3′-UTR fragments (UTR1–UTR7) or pGL3 basic along with the reference reporter (phRL-TK). The pGL3 basic (vector alone) construct served as control and a reference reporter plasmid was cotransfected to verify transfection efficiency. Four hours later, cells were treated with methanol (control) or TSA (200ng/ml) for 36 h. Cells were lysed and luciferase activity in the lysates was measured. Each bar represents the mean ± SD of at least three experiments. ***P < 0.001.
Presence of a long 3′-UTR, as in claudin-1 mRNA, implies potential posttranscriptional regulatory mechanisms (13). AREs present in the UTR play an important role in the regulation of mRNA stability through their association with RBPs. In our search using the ARE database (http://brp.kfshrc.edu.sa/ARED/), we found at least eight AUUUA sequence motifs in the 3′-UTR of claudin-1 mRNA (Figure 1B). Therefore, we further determined whether putative sequence motifs located within the 3′-UTRs of claudin-1 mRNA help mediate HDAC-dependent posttranscriptional regulation of claudin-1 expression. To test this, we generated deletion mutants of claudin-1 3′-UTR (UTR1–UTR7; see Figure 1C). Resultant constructs were transiently transfected into SW480 and/or SW620 cells. Four hours posttransfection, the cells were exposed to 200 ng/ml TSA for 36 h to inhibit HDAC activity and resultant luciferase activity was examined. The pGL3–claudin-1–UTR1 (857–3344 bp full-length 3′-UTR; wild-type) demonstrated a 30–40% decrease in the relative luciferase activity, supporting its potential involvement in claudin-1 mRNA stabilization. The TSA-dependent decrease in luciferase activity was abrogated in the deletion mutants 6 or 7 [pGL3–claudin-1–UTR6 (857–1241 bp) (14); pGL3–claudin-1–UTR7 (857–1128 bp); Figure 1D].
HuR binds to the AREs in claudin-1 3′-UTR to regulate its mRNA stability
We further determined the potential basis for these observed differential luciferase reporter activities in response to TSA treatment. Of note, the decrease in luciferase activity in response to TSA treatment was observed in pGL–claudin-1–UTR5 (as shown in Figure 1D) but not in pGL3–claudin-1–UTR6 and thus suggested the presence of TSA-responsive elements in the region of UTR5, which is deleted in UTR6 (1241–1382 bp). Further analysis confirmed the presence of two AREs in this region (Figure 2A). To determine the importance of these AREs, we mutated these sites as described in Figure 2A. Use of either mutated site 1 or mutated site 2 resulted in the abrogation of the response to TSA treatment (Figure 2B), suggesting a role of these ARE sites in TSA-dependent regulation of claudin-1 expression.
Fig. 2.
Effect of the mutation of AREs within claudin-1 3′-UTR region UTR5 on promoter activity. (A) Schematic representation of AU-rich sequences in which two sites (ATTTA into AGGTA) were mutated; (B) SW480 and SW620 cells were transiently transfected with pGL3–claudin-1–UTR5 and mutated 3′-UTR5 fragments (mutated site 1 and mutated site 2) or pGL3 basic, along with the reference reporter (phRL-TK). Four hours later, cells were treated with methanol (control) or TSA (400ng/ml) for 36 h. Cells were then lysed and luciferase activity was measured. Each bar represents the mean ± SD of at least three independent experiments. ***P < 0.001.
Because AREs affect mRNA stability through their interaction with RBPs, we further examined whether HuR, a widely studied RBP, binds to the AREs in claudin-1 3′-UTR. HuR stabilizes ARE-containing mRNAs and its expression is highly upregulated in colon cancer (15). In this regard, we performed immunoprecipitation, using anti-HuR antibody, to detect a physical association of HuR with claudin-1 3′-UTR, following the protocol described previously (15). Reverse transcription–PCR readily amplified the product using primers for the flanking region encompassing UTR1, UTR2, UTR3, UTR4 or UTR5 fragments of the claudin-1 3′-UTR (Figure 3A). In contrast, primers flanking the sequences of claudin-1 3′-UTR6 and 3′-UTR7 did not amplify any product from the same immunoprecipitate (Figure 3A) and thus suggested that AREs present in UTR5 (and deleted in UTR6) are important for the binding of HuR with claudin-1 3′-UTR.
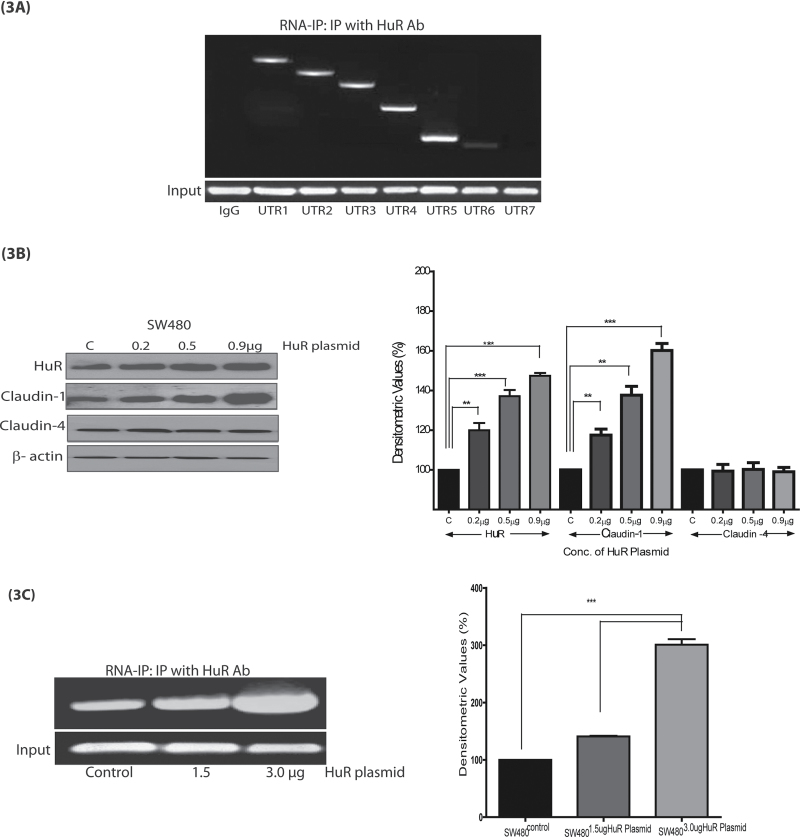
Fig. 3.
HuR affects claudin-1 expression and mRNA stability. (A) Association of HuR with claudin-1 3′-UTR regions (UTR1, UTR2, UTR3, UTR4 and UTR5). Whole-cell lysates from SW480 cells were used for immunoprecipitation (IP) using anti-HuR antibody or non-specific IgG. Total RNA was extracted from immunoprecipitated complexes, reverse transcribed and subjected to PCR analysis using primers to amplify UTR1–UTR7 regions of claudin-1 3′-UTR. Resulting PCR products were visualized in agarose gels. Negative control was IgG (mouse). Glyceraldehyde 3-phosphate dehydrogenase expression was determined (input) to ensure that equal amounts of samples were taken during the RNA immunoprecipitation experiment; (B) SW480 cells were transiently transfected for 48 h with HuR expression plasmid (control, 0.2, 0.5 and 0.9 μg) and were then immunoblotted for HuR and claudin-1 protein expression; (C) SW480 cells were transiently transfected for 48 h with HuR plasmid (control, 1.5 or 3 μg) and RNA immunoprecipitation was performed using anti-HuR antibody. Total RNA was extracted from the immunoprecipitated complexes, reverse transcribed and subjected to PCR analysis using claudin-1 3′- UTR5 primers and densitometric analysis; (D) HuR extends the half-life (t 1/2) of claudin-1 mRNA in TSA-treated SW480 cells. Control or HuR-overexpressing cells, after serum starvation for 24h, were pretreated with TSA 200ng/ml for 4 h and then with actinomycin D (10 µg/ml) for 0, 4, 8, 16, 24, 36 and 48 h to inhibit nascent RNA synthesis. Total RNA was isolated at the indicated times after actinomycin D treatment and levels of claudin-1 mRNA were measured by quantitative reverse transcription–PCR analysis; and (E) Effect of silencing of HuR expression on claudin-1 expression. Cells (SW480 and SW620) were infected with lentiviral supernatants encoding scrambled shRNA at a multiplicity of infection of 60 plaque-forming units (pfus)/cell. Whole-cell lysates were prepared after 24 and 48 h and expression of the indicated proteins was analyzed by immunobloting. β-Actin was used as loading control. Corresponding densitometry analysis is provided, where each bar represents the mean ± SD of three experiments. **P < 0.01, ***P < 0.001.
Based on this finding, we examined the effect of HuR overexpression on claudin-1 expression. Immunoblot analysis using the cell lysate from SW480 cells transfected with increasing concentrations of HuR indeed showed a dose-dependent increase in both HuR (~3-fold) and claudin-1 (~5-fold) proteins (Figure 3B). In the same lysates, claudin-4 protein levels remained unchanged and thus supported the specificity of the effect of HuR on claudin-1 expression. Further, RNA immunoprecipitation using samples from HuR-transfected (increasing concentrations: 1.5 and 3 μg) in SW480 cells also demonstrated a dose-dependent increase in HuR binding to claudin-1 3′-UTR (Figure 3C).
Next, we examined whether the HuR-dependent increase in claudin-1 expression is due to the increase in claudin-1 mRNA stability and whether it plays a role in the HDAC-dependent modulation of claudin-1 expression. In this regard, we determined claudin-1 mRNA turnover in cells overexpressing HuR with or without TSA treatment. SW480 cells were transfected with either pCDNA3 (SW480P) or pcDNA3–HuR expression plasmid (SW480HuR). Twenty-four hours posttransfection, cells were exposed to TSA (200ng/ml). To inhibit transcription, we added actinomycin D (10 µg/ml) 4 h after the addition of TSA. Samples were collected at 0, 4, 8, 16, 24, 36 and 48 h after actinomycin D treatment and were subjected to quantitative reverse transcription–PCR analysis. As shown in Figure 3D, the estimated half-life (t 1/2) of claudin-1 mRNA in the control cells (SW480control, transfected with empty expression plasmid) exposed to actinomycin D alone was 20 h and it decreased to 11 h following the combined treatment with TSA and actinomycin D. These findings were in accordance with those from our previous studies (3). In contrast, t 1/2 for claudin-1 mRNA increased to ~45 h in cells overexpressing HuR (SW480HuR cells). Also, the t 1/2 of claudin-1 mRNA increased from 11 to 30 h in SW480HuR cells that were exposed to actinomycin D and TSA (Figure 3D). Taken together, the outcome from the above studies supported the role of HuR in the regulation of claudin-1 mRNA stability.
To confirm the role of HuR in the regulation of claudin-1 expression, we performed loss-of-function studies. Silencing of HuR expression was achieved using target-specific shRNA transfection in SW480 and SW620 cells. Efficiency and specificity of HuR shRNA was determined by immunoblot analysis using antigen-specific antibody. As shown in Figure 3E, expressions of both HuR and claudin-1 were inhibited in cells expressing HuR shRNA, whereas claudin-4 expression remained unchanged. Taken together, our data strongly support the important role of HuR in the regulation of claudin-1 mRNA stability in colon cancer cells.
TSA treatment affects HuR and TTP expression in an inverse manner to decrease claudin-1 expression
Outcomes from a number of studies suggest that HuR and TTP act reciprocally to regulate the stability/expression of ARE-containing mRNAs. Because our studies demonstrated the role of HuR as an inducer of claudin-1 mRNA stability, we determined the effect of HDAC inhibition (TSA treatment) on the expressions of HuR and TTP in SW480 and SW620 cells (Figure 4A). TSA treatment, as expected, resulted in sharp decreases in the expressions of HuR (~4-fold versus control) and claudin-1 (~5-fold versus control) protein; however, it increased TTP expression in a dose-dependent manner (2-fold versus control). Similar to our earlier observations, claudin-4 expression remained unaltered. TSA treatment had similar effects on claudin-1, HuR and TTP expression in other colon cancer cell lines, HT29 and DLD-1 (Supplementary Figure 2, available at Carcinogenesis Online). Further analysis using real-time reverse transcription–PCR demonstrated that TSA treatment modulated mRNA expression of HuR and TTP (Supplementary Figure 3, available at Carcinogenesis Online).
Fig. 4.
Effect of the histone deacetylase inhibitor TSA on claudin-1 and RBP expression. (A) SW480 and SW620 cells were treated with increasing concentrations of TSA for 36 h and resultant lysates were immunoblotted with anti-HuR, anti-TTP, anti-claudin-1 or anti-claudin-4 antibodies. β-Actin was used as loading control. Graph represents the corresponding densitometry analysis, where each bar represents the mean ± SD of the values from three experiments; (B) Effect of TSA on the expressions of claudin-1, claudin-4, HuR and TTP in colon cancer cell (SW480), breast cancer cell (MDA-MB-468) and renal cells (MDCK-II). **P < 0.01, ***P < 0.001.
We further asked whether these findings were specific to colonic claudin-1 expression. In this regard, we determined the effect of TSA treatment on the expressions of claudin-1, HuR and TTP in MDA-MB-468, a human breast cancer cell line, and MDCK-II, a renal tubular epithelial cell line. SW480 cells served as positive control. As shown in Figure 4B, TSA treatment did not affect claudin-1 expression in either of the cell lines of non-colonic origin (Figure 4B and Supplementary Figure 4, available at Carcinogenesis Online) and thus suggested a colon cancer cell-specific role of HuR/TTP in the regulation of claudin-1 expression.
HDAC inhibitors modulate the binding of HuR and TTP to claudin-1 3′-UTR
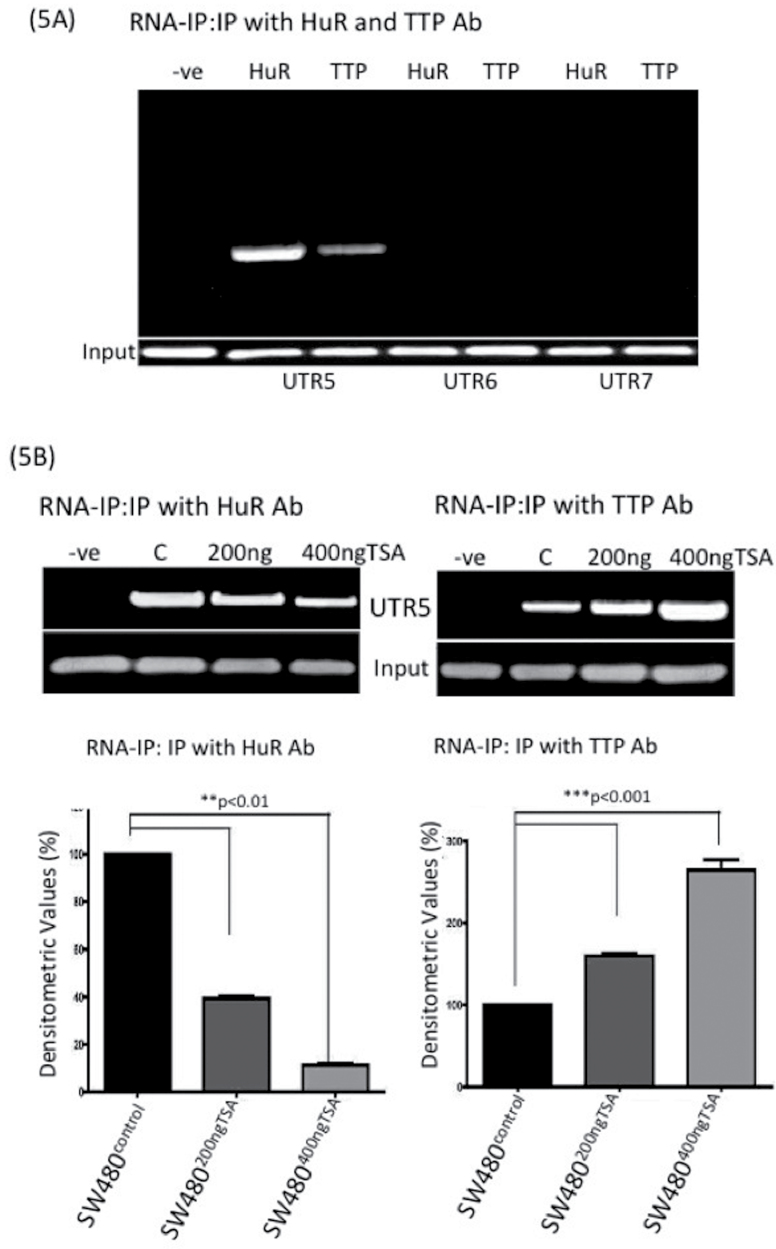
As described herein, HuR binds to claudin-1 3′-UTR. Because our findings suggested a potential inverse correlation between HuR and TTP expressions in response to the TSA treatment in colon cancer cells, we determined whether TTP also binds to claudin-1 3′-UTR. We determined the binding of HuR and TTP to the claudin-1 3′-UTR regions UTR5, UTR6 or UTR7 by RNA immunoprecipitation using anti-HuR or anti-TTP antibody for the pulldown. We were able to enrich claudin-1 3′-UTR5 region in the immunoprecipitate using the anti-TTP antibody and the anti-HuR antibody, which supported an association between TTP or HuR and claudin-1 3′-UTR (Figure 5A). Furthermore, we failed to amplify the UTR6 or UTR7 regions of claudin-1 3′-UTR by PCR in the immunoprecipitate using either anti-TTP or anti-HuR antibody, which suggested that neither TTP nor HuR binds to the UTR6 and/or UTR7 region (Figure 5A). Overall, our data suggested that HuR and TTP potentially bind to the ARE elements in the UTR5 region of claudin-1 3′-UTR, which is lost in UTR6 (Figure 1C). To further determine how TSA treatment affects the binding of HuR and/or TTP in the UTR5 region of claudin-1 3′-UTR, we examined their binding with claudin-1 3′-UTR in colon cancer cells subjected to TSA treatment. As expected, TSA treatment resulted in a decrease in the binding of HuR to the UTR5 in a dose-dependent manner. In contrast, binding of the TTP increased in the same samples in a dose-dependent manner (Figure 5B). Taken together, the above findings suggested that HDAC inhibition potentially destabilizes claudin-1 mRNA stability through the inhibition of the binding of HuR to claudin-1 3′-UTR and by increasing the binding of TTP, an mRNA-destabilizing protein.
Fig. 5.
HuR and TTP proteins associate with claudin-1 3′-UTR. (A) SW480 cells were treated with TSA (200 ng/ml) for 36 h and subjected to immunoprecipitation using anti-HuR or anti-TTP antibodies. Total RNA was then extracted from the immunoprecipitated complexes, reverse transcribed and subjected to PCR analysis using claudin-1 3′-UTR primers encompassing UTR5, UTR6 or UTR7 region and, subsequently, corresponding densitometry analysis; (B) Association of HuR and TTP with claudin-1 3′-UTR (UTR5 region) following TSA treatment (200 and 400 ng/ml). SW480 whole-cell lysates were immunoprecipitated with anti-HuR or anti-TTP antibodies or non-specific IgG (negative control). Glyceraldehyde 3-phosphate dehydrogenase expression was determined (input) before IP to ensure that equal amounts of samples were taken for immunoprecipitation. Corresponding densitometry analysis is provided, where each bar represents the mean ± SD of three experiments. **P < 0.01, ***P < 0.001.
Modulation of claudin-1 mRNA transcription also contributes to the HDAC-dependent regulation of claudin-1 expression
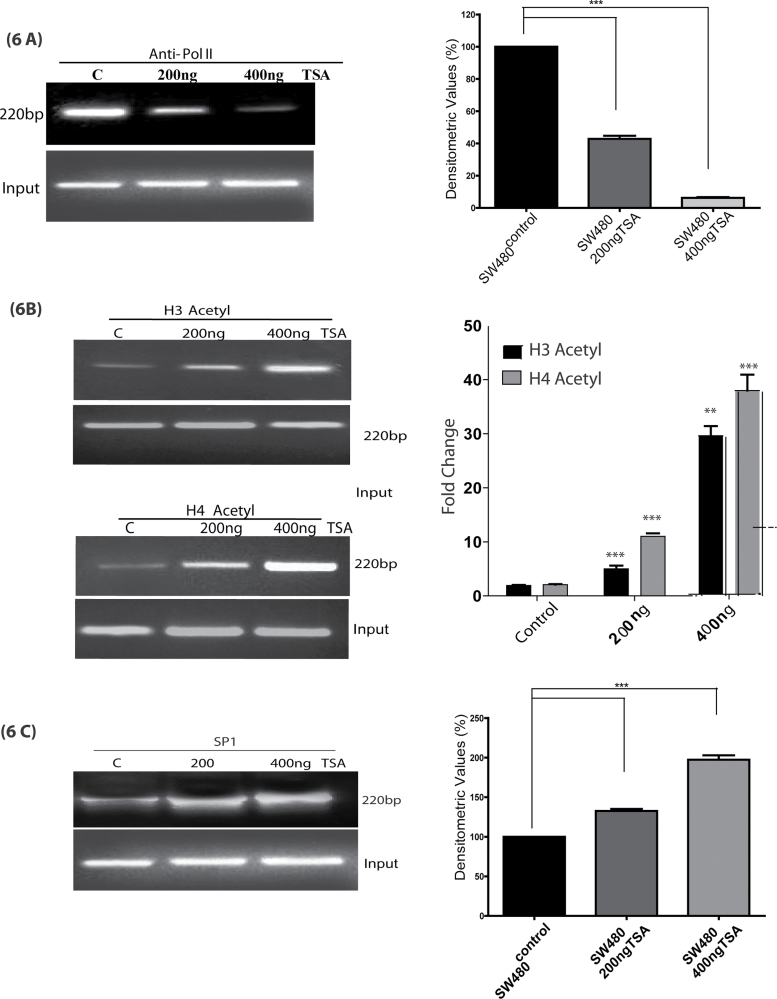
These results established a major role of the regulation of mRNA stability in HDAC-dependent inhibition of colonic claudin-1 expression. In our previous studies, we have shown that TSA treatment also decreases claudin-1 mRNA transcription to a modest 15–20% of the levels in control cells (3). However, this evaluation was based on simple transient transfection experiments using claudin-1 luciferase reporter construct. Notably, it has been pointed out that transient transfections may not be enough to rule out the potential HDAC-dependent transcriptional regulations as the system may not allow physiological and appropriate chromatin reconfiguration necessary for such evaluation (16). Histone acetylation is a critical component of chromatin remodeling and transcriptional regulation. Therefore, to further clarify the potential role of transcriptional regulation in HDAC-dependent regulation of claudin-1 expression, we determined whether TSA treatment affects the recruitment of RNA polymerase II. We used ChIP assay to analyze the effect of TSA on transcription rate, as described in Materials and methods. The results indicated an association of Pol II with the claudin-1 promoter. TSA treatment decreased the abundance of RNA polymerase II in the claudin-1 gene promoter and hampered the rate of transcription of claudin-1 in a dose-dependent manner (Figure 6A). Studies have shown that TSA and other HDAC inhibitors induce the accumulation of acetylated histones in human cells (16). Therefore, ChIP analysis was performed to examine the effect of HDAC inhibition on the acetylation of histone H3 and/or histone H4 in association with claudin-1 promoter. Chromatin fragments from human cancer cells cultured with or without TSA (200, 400 ng/ml) for 36 h were immunoprecipitated with antibodies specific to the acetylated histones H3 or H4. DNA was isolated from the resulting immunoprecipitates and PCR was performed using primers specific for the claudin-1 promoter (Figure 6B). Notably, acetylation of histones H3 and H4 associated with the claudin-1 promoter region was very low prior to the TSA treatment. However, acetylation of histones H3 and H4 increased significantly in the claudin-1 promoter following TSA treatment (Figure 6B) and thus supported the role of histone deacetylation in the transcriptional repression of claudin-1.
Fig. 6.
TSA regulates the hyperacetylation of histones and SP1 to claudin-1 promoter in SW480 cells. SW480 cells were either untreated (represented by C, control) or pretreated with TSA (200 and 400 ng/ml) for 36h and then cross-linked DNA–protein complexes were immunoprecipitated with (A) anti-RNA polymerase II (Pol II) (B) anti-acetyl H3, anti-acetyl H4 or (C) anti-SP1 antibody. Chromatin immunoprecipitation (ChIP) analysis was performed to determine binding to claudin-1 promoter using specific primers. Corresponding densitometry analysis is provided, where each bar represents the mean ± SD of three experiments. **P < 0.01, ***P < 0.001.
In further studies, using TF-SEARCH, a tool for searching putative transcription factor-binding sites, we detected potential binding sites for transcription factors previously associated with the response to HDAC inhibitors and that were known to be acetylated in other genes, especially Sp1 (17). Sp1 acts as both a negative and a positive regulator of gene transcription (18). We identified two consensus Sp1-binding domains at −178/−169bp (Sp1-1) and −147/−138bp (Sp1-2). Thereafter, ChIP assay was conducted using Sp1-specific antibody to illustrate the binding of Sp1 to claudin-1 promoter. Importantly, TSA treatment increased the binding of Sp1 to the claudin-1 promoter in a dose-dependent manner and thus suggested that the TSA-dependent induction of Sp1 helps inhibit claudin-1 transcription (Figure 6C).
Discussion
A robust increase in claudin-1 expression in colon cancer has been reported by multiple groups, including ours (19–21). We have further demonstrated the causal association of claudin-1 expression with colon cancer progression, invasion and metastasis (3,19). Thus, understanding the mechanisms that regulate claudin-1 expression in colon cancer could help provide therapeutic opportunities. Importantly, a common finding in our studies (published and/or currently unpublished) has been that manipulation of claudin-1 expression has an inverse effect on the differentiation status of colon cancer cells. In this regard, knockdown of claudin-1 expression in poorly differentiated and high-claudin-1-expressing SW620 cells induced epithelial characteristics, including E-cadherin expression, whereas its forced overexpression in low-claudin-1-expressing SW480 cells resulted in a more mesenchymal phenotype (19). Of interest, appropriate functioning of colonic epithelial cell differentiation is essential to maintain the normal mucosal homeostasis, and its dysregulation is associated with colonic pathogenesis, including neoplastic transformation and growth (19). Here, it is worthy to note that poorly differentiated colon cancer cells are often highly tumorigenic and produce metastatic lesions when subjected to mouse models of colon tumorigenesis. Thus, it is tempting to postulate that claudin-1 modulates the tumorigenic ability of colon cancer cells through the regulation of differentiation. Such a postulation gains support from our previous finding that pharmacological induction of epithelial differentiation in colon cancer cells, using HDAC inhibitors (sodium butyrate and/or TSA), is associated with a sharp decrease in claudin-1 expression (3). Notably, HDAC-dependent histone acetylation is an important mechanism in the regulation of cancer-related genes and inhibition of HDACs induces epithelial differentiation and decreases invasion.
Conceptually, hyperacetylation of histones following the inhibition of HDAC activity would be predicted to promote a general increase in gene expression. However, treatment of cells with HDAC inhibitors such as TSA changes the activity state of only a limited number of genes. Here, genes with upregulated activity include p21, p53, Bax and Bad (18,22). Conversely, a smaller number of genes are downregulated on HDAC inhibition and these include bcl-2, HIF-1α, vascular endothelial growth factor and p16 (23,24). Reasons for such selectivity are not clearly understood; however, genes downregulated on HDAC inhibition are primarily considered tumor promoters. Importantly, studies have reported epigenetic regulation of specific claudin family members in cancer (25–28).
Claudin-1 is a multifunctional tight junction protein and its regulation appears to be tissue specific and context dependent. Further, analysis of the published literature supports a complex regulation of claudin-1 expression, which requires input from multiple pathways affecting gene expression on both transcriptional and posttranscriptional levels (3,10,11). HDAC inhibitors inhibit claudin-1 expression primarily through the regulation of its mRNA stability. The best-characterized cis-acting sequences responsible for mRNA decay in mammalian cells are the AREs present within the 3′-UTRs of mRNAs (29). Various ARE-binding proteins that possess RNA-stabilizing (such as HuR) or RNA-destabilizing (such as TTP) functions are recruited by the sequence-specific motifs (7). Thus, presence of the highly conserved AREs within the 3′-UTR of the claudin-1 mRNA (Figure 1C) suggests claudin-1 to be among the 7% human gene pool and emphasizes that posttranscriptional regulation represents a key regulatory mechanism for the regulation of claudin-1 gene expression.
The HuR protein is a ubiquitously expressed protein, which can function in an mRNA-stabilizing capacity. When overexpressed in cells, HuR stabilizes ARE-containing transcripts and promotes their translation (30). In contrast, TTP promotes the rapid decay of ARE-containing mRNAs. Our results demonstrate that HuR binds to the AREs in the 3′-UTR of claudin-1 mRNA and regulates claudin-1 expression through the regulation of mRNA stability in colon cancer cells. Our data from ChIP analysis confirm the specific binding of HuR with claudin-1 mRNA and further demonstrate that this binding is restricted to the 81bp (1321–1402 bp) of claudin-1 3′-UTR. Additionally, both HuR-binding sites appeared important for the maintenance of claudin-1 mRNA stability. Our findings that ectopic overexpression of HuR increases the binding to claudin-1 promoter and stabilizes the half-life of claudin-1 mRNA further support the role of HuR in the regulation of claudin-1 mRNA stability.
As described, TTP promotes the rapid decay of ARE-containing mRNAs as binding of TTP to AREs targets the mRNA for rapid degradation through exosome recruitment and association with mRNA decay enzymes (31). It has been further reported that HuR and TTP often antagonize each other. Recently, it was also demonstrated that during the early stages of colorectal tumorigenesis, expression of the mRNA stability factor HuR increases, whereas expression of the decay factor TTP decreases (7). In the current study, association of HuR and TTP with the claudin-1 mRNA was tested by immunoprecipitation using anti-HuR and anti-TTP. Results showed that the association of the stability factor HuR with claudin-1 3′-UTR decreased, whereas association of TTP, the decay factor, increased with TSA treatment. This reciprocal regulation of HuR and TTP by TSA was evident at both protein and mRNA expression levels. Further studies will help decipher the molecular details of TSA-dependent regulation of HuR and TTP.
Taken together, our data strongly suggest that epigenetic regulation underlies the dysregulated expression of claudin-1 in colon cancer cells. Most importantly, we demonstrate for the first time the key roles of HuR and TTP expression and physical association with the specific regulatory sequences in claudin-1 3′-UTR help regulate claudin-1 expression in colon cancer cells. Our findings further elaborate the complex nature of the regulation of colonic claudin-1 expression as our data suggest that HDACs also regulate claudin-1 mRNA transcription. This conclusion gains strong support from our data using ChIP analysis, which showed enhancement of the acetylation status of histones H3 and H4 at the claudin-1 promoter (Figure 6B). Sp1 is a ubiquitously expressed zinc finger-containing DNA-binding protein that can activate or repress transcription in response to physiologic and pathological stimuli (32). Acetylation, sumoylation, ubiquitylation and glycosylation are among other posttranslational modifications that influence the transcriptional activity and stability of Sp1 (28). Recently, it was discovered that Sp1 acts as a transcriptional repressor to stanniocalcin-1 in TSA-treated colon cancer cells (33). The ChIP analysis in our study revealed that Sp1 binding increases on the claudin-1 proximal promoter in TSA-treated cells, whereas the transcriptional rate decreases in a dose-dependent manner. Our data gain support from other published studies, which have demonstrated the role of Sp1/3 in the regulation of the claudin-1 promoter activity (34,35). Together with these results, we come to the conclusion that TSA may exert a direct effect on the regulation of claudin-1 mRNA stability and transcriptional regulation.
In summary, we demonstrate that claudin-1 expression in colon cancer cells is regulated through the regulation of mRNA stability and transcription. We have further provided data that HuR and TTP physically associate with and possibly compete to regulate the HDAC-dependent decrease in claudin-1 expression. Considering the established role of claudin-1 in the regulation of colon cancer growth progression and the known tumor-suppressive effect of HDAC inhibition, our findings may provide a better understanding of the mechanism of how HDAC inhibition suppresses colon cancer and thus may open new therapeutic opportunities.
Supplementary material
Supplementary Figures 1–4 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (CA124977 to P.D., DK088902 to A.B.S.).
Conflict of Interest Statement: None declared.
Supplementary Material
Glossary
Abbreviations:
- ARE
AU-rich cis-acting element
- cDNA
complementary DNA
- ChIP
chromatin immunoprecipitation
- HDAC
histone deacetylase
- HuR
human antigen R
- mRNA
messenger RNA
- RBP
RNA-binding protein
- shRNA
short hairpin RNA
- TSA
trichostatin-A
- TTP
tristetraprolin
- UTR
untranslated region.
References
- 1. Sadeh R., et al. (2011). Genome-wide “re”-modeling of nucleosome positions. Cell, 147, 263–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carey N., et al. (2006). Histone deacetylase inhibitors: gathering pace. Curr. Opin. Pharmacol., 6, 369–375 [DOI] [PubMed] [Google Scholar]
- 3. Krishnan M., et al. (2010). HDAC inhibitors regulate claudin-1 expression in colon cancer cells through modulation of mRNA stability. Oncogene, 29, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Khera T.K., et al. (2010). Mechanisms of TNFα regulation in uveitis: focus on RNA-binding proteins. Prog. Retin. Eye Res., 29, 610–621 [DOI] [PubMed] [Google Scholar]
- 5. Siddiqui N., et al. (2012). mRNA export and cancer. Wiley Interdiscip. Rev. RNA, 3, 13–25 [DOI] [PubMed] [Google Scholar]
- 6. Palanisamy V., et al. (2012). Control of cytokine mRNA expression by RNA-binding proteins and microRNAs. J. Dent. Res., 91, 651–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anant S., et al. (2009). HuR and TTP: two RNA binding proteins that deliver message from the 3′ end. Gastroenterology, 136, 1495–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Blackshear P.J. (2002). Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. Trans., 30(Pt 6), 945–952 [DOI] [PubMed] [Google Scholar]
- 9. Singh A.B., et al. (2010). Claudin family of proteins and cancer: an overview. J. Oncol., 2010, 541957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakagawa S., et al. (2011). Expression of CLDN1 in colorectal cancer: a novel marker for prognosis. Int. J. Oncol., 39, 791–796 [DOI] [PubMed] [Google Scholar]
- 11. Singh A.B., et al. (2011). Claudin-1 up-regulates the repressor ZEB-1 to inhibit E-cadherin expression in colon cancer cells. Gastroenterology, 141, 2140–2153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gilbert C., et al. (2006). RNA immunoprecipitation for determining RNA-protein associations in vivo. In Ausubel F.M., Brent R., Kingston R.E., Moore D.D., Seidman J.G., Smith J.A., Struhl K. (eds) Current Protocols in Molecular Biology. John Wiley & Sons, New York, Chapter 27, pp. 27.4.1–27.4.11 [DOI] [PubMed] [Google Scholar]
- 13. Rattenbacher B., et al. (2012). Evaluating posttranscriptional regulation of cytokine genes. Methods Mol. Biol., 820, 71–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kleinberg L., et al. (2008). Claudin upregulation in ovarian carcinoma effusions is associated with poor survival. Hum. Pathol., 39, 747–757 [DOI] [PubMed] [Google Scholar]
- 15. Young L.E., et al. (2009). The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology, 136, 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu W.S., et al. (2007). Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene, 26, 5541–5552 [DOI] [PubMed] [Google Scholar]
- 17. Waby J.S., et al. (2010). Sp1 acetylation is associated with loss of DNA binding at promoters associated with cell cycle arrest and cell death in a colon cell line. Mol. Cancer, 9, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sowa Y., et al. (1999). Histone deacetylase inhibitor activates the p21/WAF1/Cip1 gene promoter through the Sp1 sites. Ann. N. Y. Acad. Sci., 886, 195–199 [DOI] [PubMed] [Google Scholar]
- 19. Dhawan P., et al. (2005). Claudin-1 regulates cellular transformation and metastatic behavior in colon cancer. J. Clin. Invest., 115, 1765–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ouban A., et al. (2012). Claudin-1 expression in squamous cell carcinomas of different organs: comparative study of cancerous tissues and normal controls. Int. J. Surg. Pathol., 20, 132–138 [DOI] [PubMed] [Google Scholar]
- 21. Kinugasa T., et al. (2012). Increased claudin-1 protein expression in hepatic metastatic lesions of colorectal cancer. Anticancer Res., 32, 2309–2314 [PubMed] [Google Scholar]
- 22. Nakamura H., et al. (2011). Cooperative role of the RNA-binding proteins Hzf and HuR in p53 activation. Mol. Cell. Biol., 31, 1997–2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jóna Á., et al. (2011). The histone deacetylase inhibitor entinostat (SNDX-275) induces apoptosis in Hodgkin lymphoma cells and synergizes with Bcl-2 family inhibitors. Exp Hematol., 39, 1007–1017.e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Naldini A., et al. (2012). Downregulation of hypoxia-related responses by novel antitumor histone deacetylase inhibitors in MDAMB231 breast cancer cells. Anticancer. Agents Med. Chem., 12, 407–413 [DOI] [PubMed] [Google Scholar]
- 25. Nishikiori N., et al. (2008). Prevention of murine experimental corneal trauma by epigenetic events regulating claudin 6 and claudin 9. Jpn. J. Ophthalmol., 52, 195–203 [DOI] [PubMed] [Google Scholar]
- 26. Kwon M.J., et al. (2011). Claudin-4 overexpression is associated with epigenetic derepression in gastric carcinoma. Lab. Invest., 91, 1652–1667 [DOI] [PubMed] [Google Scholar]
- 27. Osanai M., et al. (2007). Epigenetic silencing of claudin-6 promotes anchorage-independent growth of breast carcinoma cells. Cancer Sci., 98, 1557–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Honda H., et al. (2006). Crucial roles of Sp1 and epigenetic modifications in the regulation of the CLDN4 promoter in ovarian cancer cells. J. Biol. Chem., 281, 21433–21444 [DOI] [PubMed] [Google Scholar]
- 29. Guhaniyogi J., et al. (2001). Regulation of mRNA stability in mammalian cells. Gene, 265, 11–23 [DOI] [PubMed] [Google Scholar]
- 30. Shultz J.C., et al. (2012). The flip-flop HuR: part of the problem or the solution in fighting cancer? J. Clin. Invest., 122, 16–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sanduja S., et al. (2012). The role of tristetraprolin in cancer and inflammation. Front. Biosci., 17, 174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li L., et al. (2010). The role of Sp1 and Sp3 in normal and cancer cell biology. Ann. Anat., 192, 275–283 [DOI] [PubMed] [Google Scholar]
- 33. Law A.Y., et al. (2011). Sp1 is a transcription repressor to stanniocalcin-1 expression in TSA-treated human colon cancer cells, HT29. J. Cell. Biochem., 112, 2089–2096 [DOI] [PubMed] [Google Scholar]
- 34. Dufresne J., et al. (2007). Activation of an SP binding site is crucial for the expression of claudin 1 in rat epididymal principal cells. Biol. Reprod., 76, 825–832 [DOI] [PubMed] [Google Scholar]
- 35. Lopardo T., et al. (2008). Claudin-1 is a p63 target gene with a crucial role in epithelial development. PLoS One, 3, e2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.