Abstract
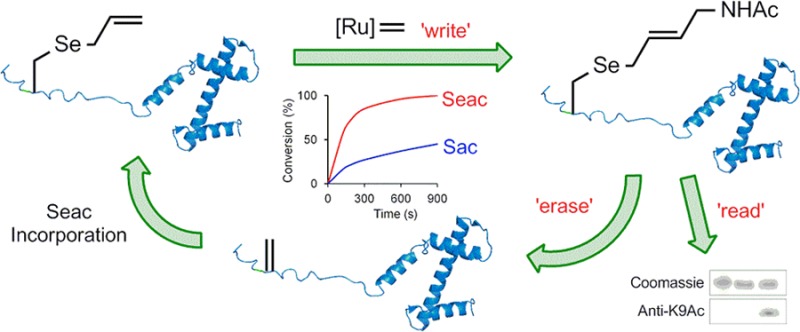
Cross-metathesis (CM) has recently emerged as a viable strategy for protein modification. Here, efficient protein CM has been demonstrated through biomimetic chemical access to Se-allyl-selenocysteine (Seac), a metathesis-reactive amino acid substrate, via dehydroalanine. On-protein reaction kinetics reveal a rapid reaction with rate constants of Seac-mediated-CM comparable or superior to off-protein rates of many current bioconjugations. This use of Se-relayed Seac CM on proteins has now enabled reactions with substrates (allyl GlcNAc, N-allyl acetamide) that were previously not possible for the corresponding sulfur analogue. This CM strategy was applied to histone proteins to install a mimic of acetylated lysine (KAc, an epigenetic marker). The resulting synthetic H3 was successfully recognized by antibody that binds natural H3-K9Ac. Moreover, Cope-type selenoxide elimination allowed this putative marker (and function) to be chemically expunged, regenerating an H3 that can be rewritten to complete a chemically enabled “write (CM)–erase (ox)–rewrite (CM)” cycle.
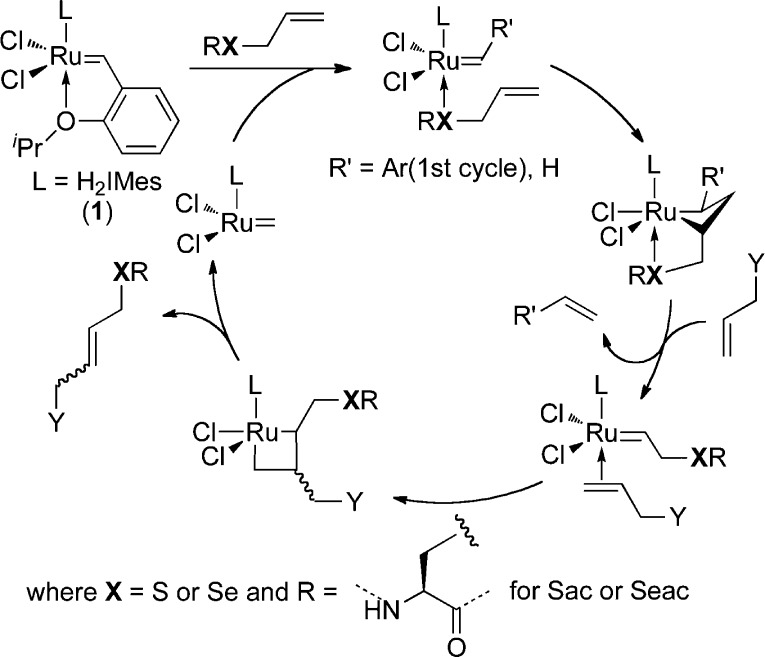
Developing strategies for site-specific protein modifications is an ongoing challenge in chemical biology.1−3 For over a decade, allylic alcohols have been identified as reactive in olefin metathesis.4,5 However, only in recent years has a generalized allylic heteroatom effect begun to emerge in cross-metathesis (CM).5 A mechanism involving pre-coordination to the metal center of catalyst 1 has been proposed to explain the observed reactivity of privileged substrates, such as amino acid S-allyl-cysteine (Sac, Scheme 1).6 This “heteroatom-relay” has enabled CM to be applied as a bioconjugation technique in selective protein modifications,7−11 and so greatly increased the benchmark for complexity in the substrate scope. Such putative beneficial effects of allylic chalcogens suggest a likely increase in the reactivity of higher chalcogens as Lewis bases that may be more preferred, softer ligands for ruthenium complexes. Indeed, simple allyl selenides are reactive substrates in aqueous CM.11 We show here that installation into peptides and proteins of the simplest amino acid residue containing an allyl selenide moiety, the unnatural amino acid Se-allyl-selenocysteine (Seac), enables rapid and efficient CM.
Scheme 1. Allyl Chalcogen-Assisted Cross-Metathesis.
Possible approaches to incorporating Seac into proteins were considered that relied on installation of the Se heteroatom either co- (either using Seac directly or from a precursor such as selenocysteine (Sec)) or post-translationally. Direct co-translational incorporation of Seac has not yet been demonstrated, likely due to rapid oxidative cellular metabolism, also observed for selenomethionine and S-methyl-selenocysteine.12−14 Despite the precedence for incorporation of Sec into recombinant proteins in Escherichia coli, this approach requires more complex genetic manipulation of both DNA and RNA15,16 that limits widespread utility. Moreover, our attempts to convert Sec to Seac via a desulfurative 2,3-sigmatropic rearrangement failed in model systems (see Supporting Information (SI) p S9). Notably, a recent study has revealed a biosynthetic mechanism for selenocysteine generation via conjugate C–Se bond formation from a dehydroalanyl-tRNASec intermediate.17 Inspired by nature, we therefore considered an alternative biomimetic route to incorporate Seac into proteins via dehydroalanine (Dha), which has recently emerged as a versatile chemical handle for protein modifications.18−22
First, the generation of an appropriate conjugate, allyl selenolate nucleophile, was explored on model Dha 3 (Scheme 2a). In situ generation of allyl selenolate via cleavage of methyl benzoselenoate 2a failed (Table S1). Next, we considered reductive access. Diallyl diselenide (2b) is unstable and exists in mixture with diallyl selenide.23 Moreover, allyl selenolate has previously only been reductively generated under aprotic conditions.23 Nevertheless, treatment of 2b with a stoichiometric amount of NaBH4 in degassed MeOH generated allyl selenolate; subsequent addition of this solution to Dha 3 in water yielded 53% of the desired Seac product 4 (Table S1). Allyl selenocyanate (2c), a potentially more stable,24 alternative allyl selenolate source, was investigated next. Pleasingly, due to cleaner pre-reduction, the desired addition product 4 was obtained with an improved yield of 86% after 1 h at room temperature (Scheme 2a). Notably, although allyl selenolate is sensitive to acidic conditions (allylselenol quickly decomposes to 2b in the absence of radical inhibitor23), under buffered basic conditions the lifetime of allyl selenolate is apparently long enough to successfully react with Dha in aqueous media.
Scheme 2. Biomimetic Synthesis of Se-Allyl-selenocysteine via Conjugate Addition of Allyl Selenolate to Dehydroalanine (Table S1).
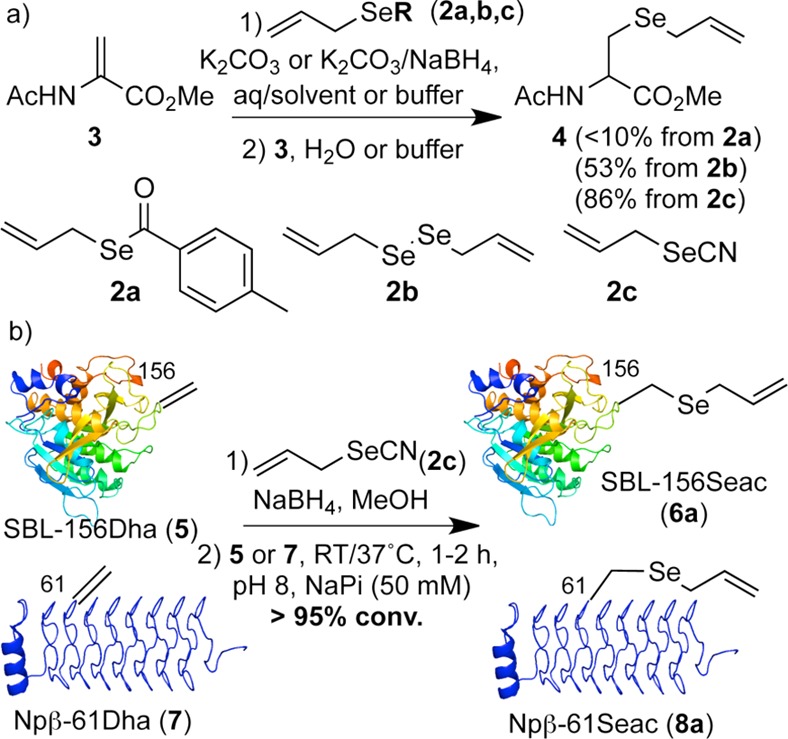
With promising results observed on amino acid models, we next explored installation of Seac via this biomimetic strategy in two differing protein structural motifs/folds: three-layer α/β-Rossman-fold protein subtilisin from Bacillus lentus (SBL) and all-β-helix protein 275–276 from Nostoc punctiforme (termed Npβ),25 respectively. Accordingly, Dha substrate proteins SBL-156Dha (5) and Npβ-61Dha (7) were synthesized from corresponding cysteine mutants following reported bis-alkylative elimination procedures.21 Use of selenolate derived from 2a or 2b failed on proteins and led to only a mixture of products. However, allyl selenolate solution generated from 2c allowed successful biomimetic conjugate addition (Scheme 2b); LC-MS analysis revealed >95% conversion of SBL-156Dha (5) to the desired product SBL-156Seac (6a) after 1 h at room temperature. Even addition to the more sterically demanding26 61 site in Npβ-61Dha (7) single Dha mutant under slightly elevated temperature (37 °C) afforded expected Seac-containing protein with >95% conversion after 2 h (8a, see SI). Importantly, resulting SBL-derivative proteins retained fold and functional (peptidase) activity (see SI p S23). Generation of Dha and addition likely leads to the formation of a 1:1 dr of adducts.26
With model Seac-tagged protein 6a in hand, CM with allyl alcohol was tested using catalyst 1 under previously optimized reaction conditions11 (which can be used at pH 4–8). This reaction was strikingly rapid and was complete after only 15 min at room temperature (Scheme 3). In comparison, the sulfur-equivalent protein substrate SBL-156Sac (9a) required 4 h to reach completion under the same reaction conditions. Rapid protein modification reactions are rare but powerful.27,28 We conducted a quantitative comparison of the enhanced rate observed here for CM of SBL-156Seac with popular bioconjugation techniques such as Staudinger ligation,29,30 azide–alkyne cycloaddition,31,32 and inverse electron-demand Diels–Alder cycloaddition,33−36 which have typically been studied in small-molecule models (“off-protein”). Under pseudo-first-order conditions with respect to the protein, starting material and product were monitored by LC-MS at several time points during reactions (Figure 1a).37 Data analyzed using nonlinear, single-exponential regression gave pseudo-first-order rate constants (Figure 1b).
Scheme 3. Enhanced CM of Seac-SBL 6a, cf. Sac-SBL 9a.
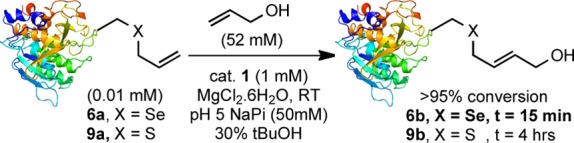
Figure 1.
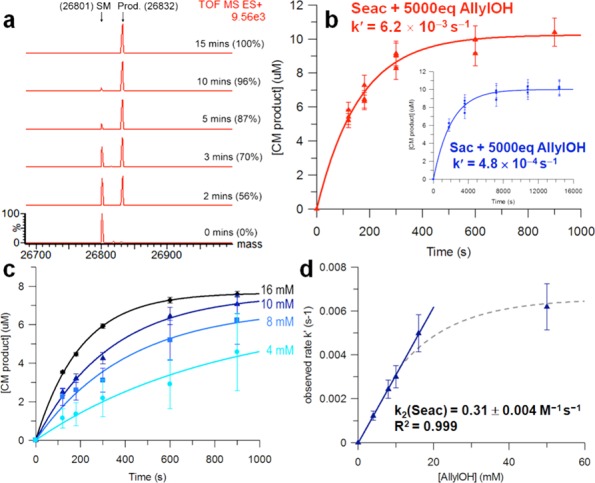
Kinetic studies of CM between SBL-156Seac (6a) and allyl alcohol. (a) LC-MS of reaction showing conversions over time. (b) Changes in [product] as a function of time. Inset shows kinetics of CM with SBL-156Sac (9a); note difference in time scale. Conditions: [protein] = 0.01 mM, [cat.] = 1 mM, [allyl alcohol] = 52 mM, [MgCl2·6H2O] = 26 mM in 3:7 tBuOH/NaPi (50 mM, pH 5) at room temperature. (c) CM of 6a with various concentrations of allyl alcohol. (d) Determination of the second-order CM rate constant for 6a.
Determination of pseudo-first-order rate constants at various allyl alcohol concentrations and analysis of the results using linear regression revealed second-order rate constants for Seac-mediated CM. The on-protein second-order rate constant for the CM of allyl alcohol with Seac-protein was determined to be 0.31 ± 0.004 M–1 s–1 (Figure 1c,d), whereas the value for on-protein Sac-mediated CM in an identical protein context (site 156) was some 10-fold lower (0.031 ± 0.0015 M–1 s–1) under the same conditions (Figures S21 and S22). Interestingly, Figure 1d indicated an initially linear increase in observed rate with respect to allyl alcohol concentration and gradually plateaued at concentrations >20 mM; this suggests that the catalyst plays a critical role under these conditions, presumably becoming “saturated” at high concentrations of allyl alcohol, leading to a shift in rate-limiting step (see also SI section 2.10 for off-protein catalyst dependency). Preliminary computations (using methyl allyl chalcogenides, see SI section 3) suggest a reaction profile in which the Ru-carbene intermediate after “Se-relay” is unusually stable and enhanced in stability over “S-relay”; this intermediate is also accessed through a lowered transition state.
When comparing the second-order rate constants with those from widely used bioconjugation techniques in chemical biology, the rate of on-protein Seac-assisted CM was found to be ∼150-fold faster than that of off-protein Staudinger ligation (k = 0.002 M–1 s–1)38 and comparable to faster variants of off-protein strain-promoted azide–alkyne cycloaddition (k = 0.002–0.9 M–1 s–1)39−41 and certain tetrazine-based cycloadditions (k > 0.15 M–1 s–1).27 Notably, however, recent select examples of catalyzed oxime ligations42 and tetrazine-based reactions are still more rapid (Figure 2).27,28,43,44 It seems likely that local environment strongly affects on-protein reactions compared with off-protein; indeed, here, crowded (e.g., Npβ-61Seac) sites showed negligible reactivity (see SI). However, detailed on-protein measurements are rare,42,44 so other relevant direct comparisons are not possible. Although here directly comparable reaction conditions could not be achieved, we extrapolate (see SI section 2.10) off-protein rates for Seac-mediated CM that are at least comparable and may be enhanced, consistent with CM’s steric sensitivity.5
Figure 2.
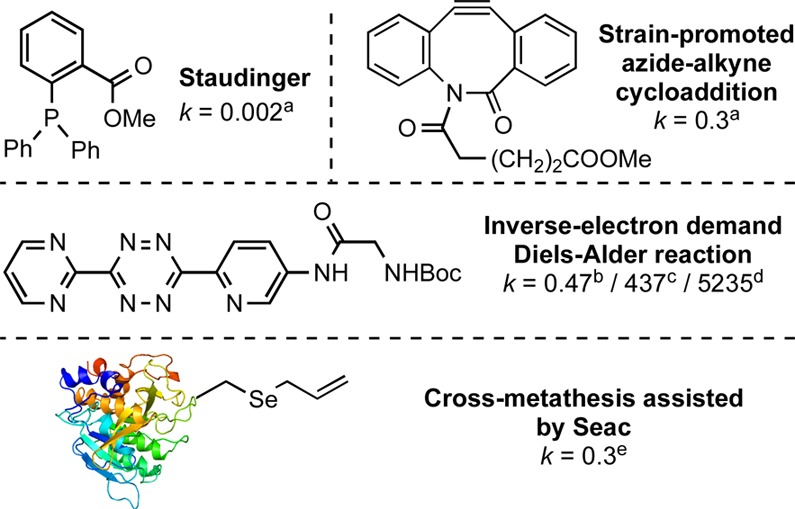
Selected rate constants (M–1 s–1) of conjugations. All rate constants are second-order in units of M–1 s–1 with abenzyl azide, bnorbornenol, cbicyclo[6.1.0]non-4-yn-9-ylmethanol, dtrans-cyclooctenol, and eallyl alcohol.
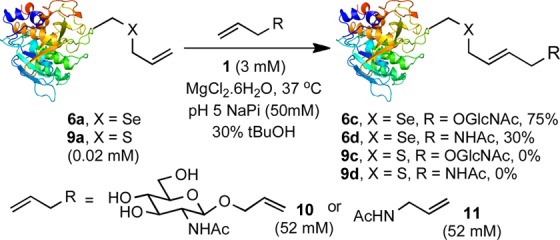
Next, having established usefully enhanced on-protein rates, we examined substrate breadth. Model Seac-tagged protein SBL-156Seac 6a was tested with challenging metathesis substrate GlcNAc 10; this olefin is essentially unreactive in sulfur-relayed CM with SBL-156Sac (9a). Pleasingly, reaction reached 75% conversion after 1.5 h at 37 °C (Scheme 4). Another biologically relevant substrate, N-allyl acetamide (11), was used in CM with protein 6a. Acetamide 11 has been suggested to poison catalyst 1, and also fails with SBL-156Sac (9a).11,45,46 Despite poisoning,47 after 20 min at 37 °C, LC-MS revealed conversion to desired 6d (Scheme 4), albeit in 30% yield.
Scheme 4. Cross-Metathesis of SBL 6a and 9a with Challenging Substrates 10 and 11.
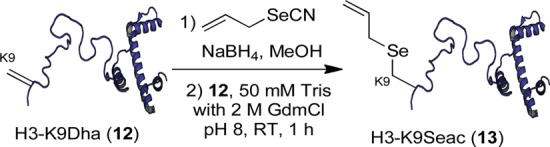
After demonstrating CM on SBL-Seac, we tested Se-relayed CM on more functionally relevant protein systems. Histone proteins are key chromatin components; post-translational modifications (e.g. K-Ac) are critical regulators of structure and function.48,49 First, Seac was installed into key epigenetic site 9 of histone H3. After generation of H3-9Dha,22 subsequent allyl selenolate addition to Dha 12 proceeded successfully to yield Seac protein 13 after 1 h at room temperature (Scheme 5).
Scheme 5. Synthesis of H3-9Seac (13).
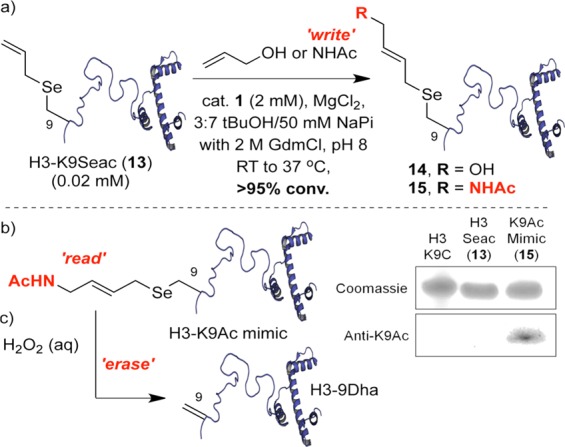
With H3-9Seac (13) in hand, we carried out model CM with allyl alcohol. Coloration implying metal coordination to 13 was observed; such coordination6,50 can necessitate metal scavenging prior to MS analysis. Use of 3-mercaptopropionic acid51,50 allowed ready MS monitoring of H3 CM reactions. Pleasingly, this revealed CM of H3-K9Seac and allyl alcohol with nearly full conversion to CM-modified H3 14 (Scheme 6a). This ability to modify site 9 suggested a strategy for chemical recapitulation (under benign conditions) of a “write–read–erase” cycle that is observed (enzyme-mediated) in histone epigenetics.48,49 As a “write” step we used the more challenging N-allyl acetamide olefin in CM; complete conversion was observed to 15 (Scheme 6a). As a “read” step, CM-modified H3 (15) was analyzed by an antibody raised to natural epigenetic marker N-acetyl lysine at position 9 of H3. We were pleased to find that this antibody successfully cross-reacted and recognized the CM-installed modification at position 9 as a K9Ac PTM mimic (Scheme 6b). Next, as an “erase” step the CM-installed K9Ac mimic “mark” was removed with full conversion using Cope-type elimination through the generation of labile selenoxide using mild peroxide oxidation to regenerate Dha. Met-containing H3 may be prone to side reaction;52 this was tested and confirmed through the use of a Met-free H3 variant in which the mark was cleanly expunged without side reaction (see SI sections 2.11–2.14). Finally, a full chemical “write–read–erase–rewrite–read” cycle was performed (SI section 2.12) to demonstrate iterative histone “switching”.
Scheme 6. CM Reactions on H3-9Seac and the Chemical “Write–Read–Erase” Cycle via a K9Ac Mimic “Mark” and Anti-K9Ac Western Blots of H3 Proteins (Also See SI).
In conclusion, we have demonstrated efficient chemical incorporation of Seac into proteins. This has facilitated rapid cross-metathesis for protein modifications; determination of on-protein rates shows that these outstrip or are comparable to many of the so-called “click” reactions in chemical biology. Using such allyl selenide (Seac) proteins, we were able to access a broader substrate scope in Se-relayed CM than was possible with allyl sulfides. Direct access to Seac in proteins creates opportunities for potential PTM mimicry via CM as demonstrated here: a K9Ac mimic was successfully installed, recognized, and removed (write–read–erase) in histone H3. Although, to the best of our knowledge, such a chemical “write–read–erase” approach has not been explored before, one could envisage a system based on creation, e.g., of disulfide-linked modification mimics53 and subsequent reductive cleavage. This synthetic manipulation of biology also highlights opportunities for new metathesis catalysts; uses of CM on protein/cell systems may be enabled by tuned solubilities, compatibilities, and permeabilities. We note too that this work provides access to a protected (allylated) Sec in proteins (that could be revealed using, e.g., Pd(0))54 and also further motivates developments in genetic incorporation of allyl chalcogenide amino acids into proteins to access more generally the wider biological applications of cross-metathesis.
Acknowledgments
We thank EPSRC (Y.A.L., L.L.), CRUK (L.L.), Rhodes Trust (B.B.), and EU FP7 Marie Curie programme (O.B.) for funding. B.G.D. is a Royal Society Wolfson Merit Research Award Recipient.
Supporting Information Available
Full procedures and protein ESI-MS. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Present Address
† O.B.: Departament de Química Analítica i Orgànica, Universitat Rovira i Virgili, C/Marcel·lí Domingo s/n, Tarragona, Spain.
The authors declare no competing financial interest.
Supplementary Material
References
- Sletten E. M.; Bertozzi C. R. Angew. Chem., Int. Ed. 2009, 48, 6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker J. M.; Bernardes G. J. L.; Davis B. G. Acc. Chem. Res. 2011, 44, 730. [DOI] [PubMed] [Google Scholar]
- Takaoka Y.; Ojida A.; Hamachi I. Angew. Chem., Int. Ed. 2013, 52, 4088. [DOI] [PubMed] [Google Scholar]
- Hoye T. R.; Zhao H. Org. Lett. 1999, 1, 1123. [DOI] [PubMed] [Google Scholar]
- Lin Y. A.; Davis B. G. Beilstein J. Org. Chem. 2010, 6, 1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. A.; Chalker J. M.; Floyd N.; Bernardes G. J. L.; Davis B. G. J. Am. Chem. Soc. 2008, 130, 9642. [DOI] [PubMed] [Google Scholar]
- Brik A. Adv. Synth. Catal. 2008, 350, 1661. [Google Scholar]
- Kirshenbaum K.; Arora P. S. Nat. Chem. Biol. 2008, 4, 527. [DOI] [PubMed] [Google Scholar]
- Lin Y. A.; Chalker J. M.; Davis B. G. ChemBioChem 2009, 10, 959. [DOI] [PubMed] [Google Scholar]
- Ai H.-W.; Shen W.; Brustad E.; Schultz P. G. Angew. Chem., Int. Ed. 2010, 49, 935. [DOI] [PubMed] [Google Scholar]
- Lin Y. A.; Chalker J. M.; Davis B. G. J. Am. Chem. Soc. 2010, 132, 16805. [DOI] [PubMed] [Google Scholar]
- Foster S. J.; Kraus R. J.; Ganther H. E. Arch. Biochem. Biophys. 1986, 251, 77. [DOI] [PubMed] [Google Scholar]
- Ganther H. E. Carcinogenesis 1999, 20, 1657. [DOI] [PubMed] [Google Scholar]
- Stolz J. F.; Basu P.; Santini J. M.; Oremland R. S. Annu. Rev. Microbiol. 2006, 60, 107. [DOI] [PubMed] [Google Scholar]
- Arnér E. S. J.; Sarioglu H.; Lottspeich F.; Holmgren A.; Böck A. J. Mol. Biol. 1999, 292, 1003. [DOI] [PubMed] [Google Scholar]
- Aldag C.; Bröcker M. J.; Hohn M. J.; Prat L. H., G.; Plummer A.; Söll D. Angew. Chem., Int. Ed. 2013, 52, 1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palioura S.; Sherrer R. L.; Steitz T. A.; Söll D.; Simonovic M. Science 2009, 325, 321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levengood M.; van der Donk W. A. Nat. Protoc. 2007, 1, 3001. [DOI] [PubMed] [Google Scholar]
- Wang J.; Schiller S. M.; Schultz P. G. Angew. Chem., Int. Ed. 2007, 46, 6849. [DOI] [PubMed] [Google Scholar]
- Bernardes G. J. L.; Chalker J. M.; Errey J. C.; Davis B. G. J. Am. Chem. Soc. 2008, 130, 5052. [DOI] [PubMed] [Google Scholar]
- Chalker J. M.; Gunnoo S. B.; Boutureira O.; Gerstberger S. C.; Fernández-González M.; Bernardes G. J. L.; Griffin L.; Hailu H.; Schofield C. J.; Davis B. G. Chem. Sci. 2011, 2, 1666. [Google Scholar]
- Chalker J. M.; Lercher L.; Rose N. R.; Schofield C. J.; Davis B. G. Angew. Chem., Int. Ed. 2012, 51, 1835. [DOI] [PubMed] [Google Scholar]
- Riague E. H.; Guillemin J.-C. Organometallics 2001, 21, 68. [Google Scholar]
- Crich D.; Krishnamurthy V.; Brebion F.; Karatholuvhu M.; Subramanian V.; Hutton T. K. J. Am. Chem. Soc. 2007, 129, 10282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetting M. W.; Hegde S. S.; Hazleton K. Z.; Blanchard J. S. Protein Sci. 2007, 16, 755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-González M.; Boutureira O.; Bernardes G. J. L.; Chalker J.; Young M.; Errey J.; Davis B. G. Chem. Sci. 2010, 1, 709. [Google Scholar]
- Lang K.; Davis L.; Torres-Kolbus J.; Chou C.; Deiters A.; Chin J. W. Nat. Chem. 2012, 4, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang K.; Davis L.; Wallace S.; Mahesh M.; Cox D. J.; Blackman M. L.; Fox J. M.; Chin J. W. J. Am. Chem. Soc. 2012, 134, 10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhn M.; Breinbauer R. Angew. Chem., Int. Ed. 2004, 43, 3106. [DOI] [PubMed] [Google Scholar]
- Saxon E.; Bertozzi C. R. Science 2000, 287, 2007. [DOI] [PubMed] [Google Scholar]
- Hong V.; Steinmetz N. F.; Manchester M.; Finn M. G. Bioconjugate Chem. 2010, 21, 1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hest J. C. M.; van Delft F. L. ChemBioChem 2011, 12, 1309. [DOI] [PubMed] [Google Scholar]
- Blackman M. L.; Royzen M.; Fox J. M. J. Am. Chem. Soc. 2008, 130, 13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj N. K.; Weissleder R.; Hilderbrand S. A. Bioconjugate Chem. 2008, 19, 2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch J.; Wießler M.; Jäschke A. J. Am. Chem. Soc. 2010, 132, 8846. [DOI] [PubMed] [Google Scholar]
- Devaraj N. K.; Weissleder R. Acc. Chem. Res. 2011, 44, 816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliquots taken, quenched (Sephadex G-25) prior to LC-MS. No precipitation observed; % conversion used for concentrations. For validation of ESI-MS in protein reaction monitoring see ref (11).
- Lin F. L.; Hoyt H. M.; van Halbeek H.; Bergman R. G.; Bertozzi C. R. J. Am. Chem. Soc. 2005, 127, 2686. [DOI] [PubMed] [Google Scholar]
- Debets M. F.; van Berkel S. S.; Dommerholt J.; Dirks A. J.; Rutjes F. P. J. T.; van Delft F. L. Acc. Chem. Res. 2011, 44, 805. [DOI] [PubMed] [Google Scholar]
- Debets M. F.; van Berkel S. S.; Schoffelen S.; Rutjes F. P. J. T.; van Hest J. C. M.; van Delft F. L. Chem. Commun. 2010, 46, 97. [DOI] [PubMed] [Google Scholar]
- Jewett J. C.; Sletten E. M.; Bertozzi C. R. J. Am. Chem. Soc. 2010, 132, 3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashidian M.; Mahmoodi M. M.; Shah R.; Dozier J. K.; Wagner C. R.; Distefano M. D. Bioconjugate Chem. 2013, 24, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plass T.; Milles S.; Koehler C.; Szymański J.; Mueller R.; Wießler M.; Schultz C.; Lemke E. A. Angew. Chem., Int. Ed. 2012, 51, 4166. [DOI] [PubMed] [Google Scholar]
- Seitchik J. L.; Peeler J. C.; Taylor M. T.; Blackman M. L.; Rhoads T. W.; Cooley R. B.; Refakis C.; Fox J. M.; Mehl R. A. J. Am. Chem. Soc. 2012, 134, 2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürstner A.; Langemann K. J. Am. Chem. Soc. 1997, 119, 9130. [Google Scholar]
- Fürstner A.; Seidel G.; Kindler N. Tetrahedron 1999, 55, 8215. [Google Scholar]
- The reaction mixture colored consistent with catalyst degradation and no further reaction was observed (see SI).
- Kouzarides T. Cell 2007, 128, 693. [DOI] [PubMed] [Google Scholar]
- Bannister A. J.; Kouzarides T. Cell Res. 2011, 21, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer C. D.; Davis B. G. Chem. Commun. 2011, 47, 1698. [DOI] [PubMed] [Google Scholar]
- Macdonald J. E.; Veinot J. G. C. Langmuir 2008, 24, 7169. [DOI] [PubMed] [Google Scholar]
- Guo J.; Wang J.; Lee J. S.; Schultz P. G. Angew. Chem., Int. Ed. 2008, 47, 6399. [DOI] [PubMed] [Google Scholar]
- Chatterjee C.; McGinty R. K.; Fierz B.; Muir T. M. Nat. Chem. Biol. 2010, 6, 267. [DOI] [PubMed] [Google Scholar]
- Shimizu I. In Handbook of Organopalladium Chemistry for Organic Synthesis; Negishi E.-I., Ed.; John Wiley & Sons, Inc.: New York, 2003; pp 1981–1994. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.