Abstract
Patients with Behçet's disease (BD) suffer from episodic inflammation often affecting the orogenital mucosa, skin, and eyes. To discover new BD-susceptibility loci, we performed a genome-wide association study (GWAS) of 779,465 SNPs with imputed genotypes in 1,209 Turkish BD patients and 1,278 controls. We identified novel associations at CCR1, STAT4, and KLRC4. Additionally, two SNPs in ERAP1, encoding ERAP1 p.Asp575Asn and p.Arg725Gln, recessively conferred disease risk. These findings replicated in 1,468 independent Turkish and/or 1,352 Japanese samples (combined meta-analysis p < 2 × 10−9). We also found evidence for interaction between HLA-B*51 and ERAP1 (p = 9 × 10−4). The CCR1 and STAT4 variants were associated with gene expression differences. Three risk loci shared with ankylosing spondylitis and psoriasis (MHC-I, ERAP1, and IL23R, and the MHC-I-ERAP1 interaction), as well as two loci shared with inflammatory bowel disease (IL23R and IL10) implicate shared pathogenic pathways in the spondyloarthritides and BD.
Behçet's disease (BD) is a form of vasculitis that manifests with orogenital ulcers, uveitis, skin inflammation, arthritis, enterocolitis, and inflammation in other organs1,2. BD is relatively common in Turkey, Japan, and modern-day countries that fall on Marco Polo's ancient Silk Routes, and is an important cause of vision loss in these countries2. Genetic risk factors contribute to disease-susceptibility. HLA-B*51 is the most strongly associated risk factor for BD, confirmed in multiple populations3-5. Although its association was established more than three decades ago, the role of HLA-B*51 in disease pathogenesis remains elusive5. In addition to HLA-B*51, two recent independent GWASs identified variants in regions encompassing MHC-I, IL10, and IL23R associated with BD in both the Turkish and Japanese populations6,7. However, the combined effects of these genetic factors do not fully explain the observed disease heritability.
The pathobiology of BD is also largely unknown. In 1974, based on clinical features, Moll et al. proposed the concept of “seronegative spondylarthritides”, and included BD along with ankylosing spondylitis (AS), psoriatic arthritis, reactive arthritis, and inflammatory bowel disease (IBD)8. Since then, the inclusion of BD within the spondyloarthropathy (SpA) category has been debated as BD patients rarely exhibit sacroiliitis, and BD is associated with HLA-B*51 rather than HLA-B*279-11. On the other hand, overlapping extra-articular clinical manifestations (inflammation in the eyes, skin, and intestine), genetic associations at MHC-I and IL23R6,7,12-14, and the effectiveness of tumor necrosis factor (TNF)-α blockade15,16, suggest shared pathogenesis between BD and SpA. Furthermore, IL10 and IL23R variants were found associated with both BD and IBD (Crohn's disease and ulcerative colitis), implicating common inflammatory pathways between BD and these other members of the SpA group.17,18
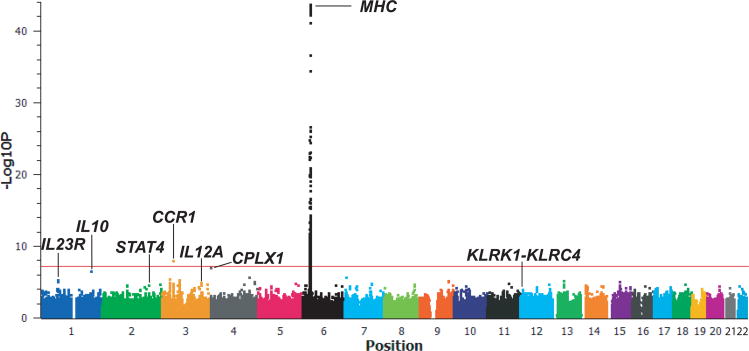
To identify novel genetic variants associated with BD, we imputed genotypes of autosomal SNPs in our GWAS collection of 1,209 cases and 1,278 controls using the previously genotyped SNPs6 and a reference panel of 96 Turkish controls genotyped for 814,474 SNPs (Online Methods). After quality control filtering, 779,465 autosomal imputed SNPs were subjected to statistical analysis by basic allele test using the best-guess imputed genotypes. In addition to the previously reported MHC-I, IL10, and IL23R discovery signals, we observed a strong signal in the CCR1 (C-C chemokine receptor type 1)-CCR3 locus, with a p-value that exceeded genome-wide significance, p<5 × 10-8 (Figure 1, rs7616215, p=1.29 × 10−8). Markers with p<3 × 10−5 are listed in Supplementary Table 1.
Figure 1. Manhattan plot of imputed SNPs.
The −log10P values for association of 779,465 autosomal imputed SNPs by basic allelic test in 1,209 Turkish BD cases and 1,278 controls. The results were segregated by chromosome and are shown by genomic position. The red horizontal line indicates genome-wide significance, p=5 × 10−8.
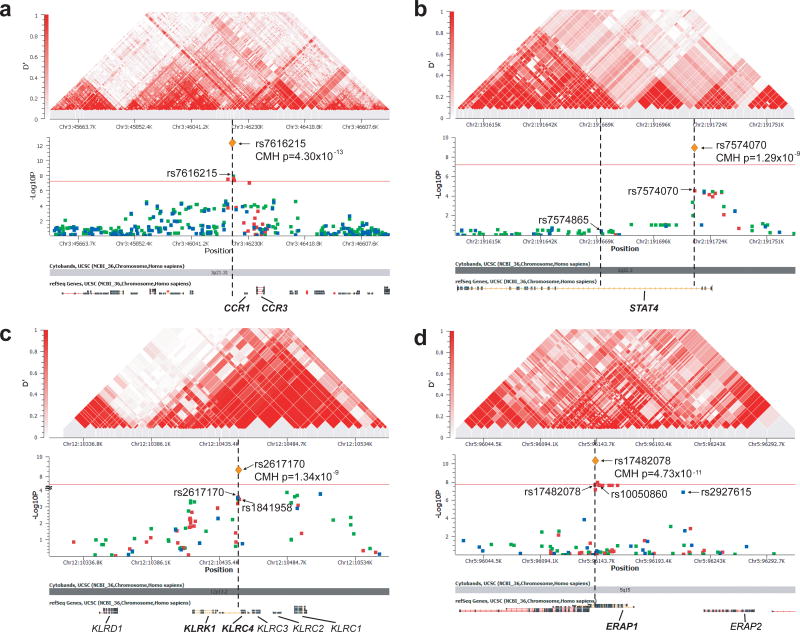
We selected 21 SNPs from the novel loci identified by imputation and one SNP from the previously reported Japanese GWAS7 for genotyping in a Turkish replication collection, comprising newly collected 838 Turkish cases and 630 controls (Supplementary Table 2). Four promising loci, STAT4 (signal transducer and activator of transcription 4), KLRC4 (killer cell lectin-like receptor subfamily C, member 4), the CCR1-CCR3 locus, and IL12A (interleukin-12 alpha chain) were then selected for validation of the imputed data (by direct genotyping) and fine-mapping studies in the original Turkish GWAS samples. As shown in Figure 2a-c, we found the strongest signals at rs7616215 3′ of CCR1, at rs7574070 in intron 3 of STAT4, and at nonsynonymous SNPs rs2617170 and rs1841958, encoding KLRC4 p.Asn104Ser and p.Ile129Ser. In a meta-analysis, we combined the Turkish GWAS and replication data, and if polymorphic, the Japanese replication data (from the reported GWAS collection with 612 cases and 740 controls7) using Cochran-Mantel-Haenszel tests (Table 1 and Figure 2). Three loci (CCR1-CCR3, STAT4, and KLRK1-KLRC1) were associated with BD at genome-wide significance (p=1.34 × 10−9 to 4.30 × 10−13). Additionally, the IL12A locus exhibited suggestive association (p=6 × 10−7). Of the 612 Japanese cases ascertained using the Japanese diagnostic criteria7, 496 also fulfilled the International Study Group Criteria19. An analysis including only cases that met the International Study Group criteria revealed genome-wide significance for the same three loci despite the reduced numbers (Supplementary Table 3).
Figure 2. Regional association plots.
Regional association plots (-log10 P values) and LD structures of the disease-associated regions surrounding (a) CCR1-CCR3, (b) STAT4, (c) KLRK1-KLRC1, and (d) ERAP1-ERAP2. The associations in (a-c) are for basic allelic tests, whereas the associations in (d) are for a recessive model test. Data are from the discovery collection with the genome-wide analysis genotypes (blue), imputed genotypes (green) and the fine-mapping genotypes (red). Orange diamonds show the meta-analyses results described in Table 1. Red horizontal lines in (a-c) indicate genome-wide significance (p=5 × 10−8) and in (d) indicates genome-wide significance accounting for three genetic models (p=1.67 × 10−8). SNP rs7574865 in (b) is a STAT4 intronic SNP associated with multiple autoimmune diseases. The LD structures of the same regions are shown in the upper panels, with red filled squares linking pairs of markers that indicate the strength of LD by intensity of fill: D′ = 1 (intense red) to D′ = 0 (no fill).
Table 1.
Cochran-Mantel-Haenszel meta-analysis of BD associated loci.
| Series* | Method | Diagnosis | Subject n | C | T | MAF(%) | OR (95% CI) | P value | |
|---|---|---|---|---|---|---|---|---|---|
| CCR1-CCR3 rs7616215 | Turkish Discovery | TOF-MS | BD | 1180 | 630 | 1730 | 27 | 0.71 (0.63-0.80) | 2.17 × 10−8 |
| Control | 1258 | 855 | 1661 | 34 | |||||
| Japanese replication | Affy 500K | BD | 610 | 160 | 1060 | 13.1 | 0.82 (0.66-1.02) | 0.079 | |
| Control | 692 | 215 | 1169 | 15.5 | |||||
| Turkish replication | TOF-MS | BD | 816 | 441 | 1191 | 27 | 0.70 (0.60-0.82) | 1.09 × 10−5 | |
| Control | 630 | 436 | 824 | 34.6 | |||||
| Combined | BD | 2606 | 0.72 (0.66-0.79) | 4.30 × 10−13 | |||||
| Control | 2580 | ||||||||
|
| |||||||||
| Series | Method | Diagnosis | Subject n | A | C | MAF(%) | OR (95% CI) | P value | |
| STAT4 rs7574070 | Turkish Discovery | TOF-MS | BD | 1166 | 1128 | 1204 | 48.4 | 1.28 (1.14-1.43) | 1.92 × 10−5 |
| Control | 1250 | 1059 | 1441 | 42.4 | |||||
| Japanese replication | Affy 500K | BD | 602 | 618 | 586 | 51.3 | 1.18 (1.01-1.37) | 0.034 | |
| Control | 716 | 676 | 756 | 47.2 | |||||
| Turkish replication | TOF-MS | BD | 815 | 803 | 827 | 49.3 | 1.34 (1.15-1.55) | 1.12 × 10−4 | |
| Control | 630 | 530 | 730 | 42.1 | |||||
| Combined | BD | 2583 | 1.27 (1.17-1.37) | 1.29 × 10−9 | |||||
| Control | 2596 | ||||||||
|
| |||||||||
| Series | Method | Diagnosis | Subject n | T | C | MAF(%) | OR (95% CI) | P value | |
| KLRC4 rs2617170 (KLRC4p.Asn104Ser) | Turkish Discovery | TOF-MS | BD | 1144 | 734 | 1554 | 32.1 | 0.80 (0.71-0.90) | 2.54 × 10−4 |
| Control | 1257 | 933 | 1581 | 37.1 | |||||
| Japanese replication | Affy 500K | BD | 580 | 361 | 799 | 31.1 | 0.75 (0.64-0.89) | 8.36 × 10−4 | |
| Control | 657 | 493 | 821 | 37.5 | |||||
| Turkish replication | TaqMan | BD | 821 | 510 | 1132 | 31.1 | 0.77 (0.66-0.89) | 7.61 × 10−4 | |
| Control | 628 | 465 | 791 | 37 | |||||
| Combined | BD | 2545 | 0.78 (0.72-0.85) | 1.34 × 10−9 | |||||
| Control | 2542 | ||||||||
|
| |||||||||
| Series | Method | Diagnosis | Subject n | A | G | MAF(%) | OR (95% CI) | P value | |
| IL12A rs17810546 | Turkish Discovery | Illumina chip | BD | 1209 | 208 | 2210 | 8.60 | 1.64 (1.31-2.04) | 1.20 × 10−5 |
| Control | 1278 | 139 | 2417 | 5.44 | |||||
| Turkish replication | TOF-MS | BD | 821 | 136 | 1504 | 8.29 | 1.41 (1.06-1.89) | 0.020 | |
| Control | 626 | 75 | 1173 | 6.00 | |||||
| Combined | BD | 2029 | 1.55 (1.30-1.85) | 6.01 × 10−7 | |||||
| Control | 1902 | ||||||||
|
| |||||||||
| Series | Method | Diagnosis | Subject n | TT | CC+CT | TT(%) | OR (95% CI) | P value | |
| ERAP1 rs17482078 (ERAP1 p.Arg725Gln) | Turkish Discovery | TOF-MS | Uveitis BD | 420 | 32 | 388 | 7.62 | 4.21 (2.45-7.23) | 2.03 × 10−8 |
| Control | 1249 | 24 | 1225 | 1.92 | |||||
| Turkish replication | TOF-MS | Uveitis BD | 370 | 21 | 349 | 5.67 | 5.36 (2.46-11.7) | 2.44 × 10−5 | |
| Control | 630 | 7 | 623 | 1.11 | |||||
| Combined | Uveitis BD | 790 | 4.56 (2.88-7.22) | 4.73 × 10−11 | |||||
| Control | 1879 | ||||||||
TOF-MS, time-of-flight mass-spectrometry; MAF, minor allele frequency, OR, odds ratio; 95%CI, 95% confidence interval. Meta-analysis results are denoted in bold. Japanese samples were not included in meta-analysis of the IL12A rs17810546 and ERAP1 rs17482078 variants because they are not polymorphic or they demonstrate too low frequency in the Japanese population.
In an attempt to reduce genetic heterogeneity, we performed genome-wide association tests in the subset of GWAS discovery patients with uveitis (435 cases, 1,278 controls)6. Neither the basic allelic test nor the dominant model test showed associations outside of the MHC. However, when we applied a recessive model, one SNP in the ERAP1-ERAP2 locus exhibited association with a p-value close to genome-wide significance level (rs2927615, p=1.02 × 10−7). We performed fine-mapping of this region in the uveitis subset of the GWAS discovery collection and identified rs10050860 and rs17482078, encoding ERAP1 p.Asp575Asn and p.Arg725Gln, which conferred risk for BD with uveitis in a recessive model (Figure 2d). A meta-analysis of p.Arg725Gln combining the Turkish discovery collection and the Turkish replication collection (with 370 BD cases with uveitis and 630 controls) exceeded the three model threshold for genome-wide significance and revealed a large effect size of the homozygous p.Arg725Gln genotype on BD with uveitis (odds ratio=4.56, p=4.73 × 10−11, Table 1 and Figure 2d).
Because a recessive model was required to detect the ERAP1 association in BD patients with uveitis, we tested whether the recessive model would reveal the ERAP1 p.Arg725Gln association with BD susceptibility in the combined uveitis and non-uveitis samples. A meta-analysis of the GWAS and replication collections found significant association of the homozygous p.Arg725Gln genotype with BD susceptibility (p=4.35 × 10−8, Supplementary Table 4). The minor allele frequency of rs2927615 (a variant in strong linkage disequilibrium [LD] with p.Arg725Gln) was too low in the Japanese population (1.8% in BD cases, 2.0% in controls) to evaluate recessive effects. Furthermore, none of the Japanese GWAS SNPs from the regions encompassing ERAP1 or IL12A (rs17810546 was not polymorphic in the Japanese population) were associated with BD (Supplementary Table 5).
ERAP1 is an endoplasmic reticulum expressed amino peptidase that functions to trim peptides for loading onto MHC Class I20. Previous GWASs have established associations of ERAP1 variants in psoriasis21, 22 and AS12. ERAP1 p.Asp575Asn and p.Arg725Gln, which are in strong LD, confer protection against these diseases through reduced peptide trimming and antigen presentation by MHC-Class I12,22-24. Of note, recent reports have shown that these ERAP1 variants confer protection preferentially in HLA-B*27 positive individuals in AS23 and HLA-C*06 positive individuals in psoriasis22, suggesting that peptide processing and binding/presentation mechanisms contribute to the pathogenesis of these diseases.
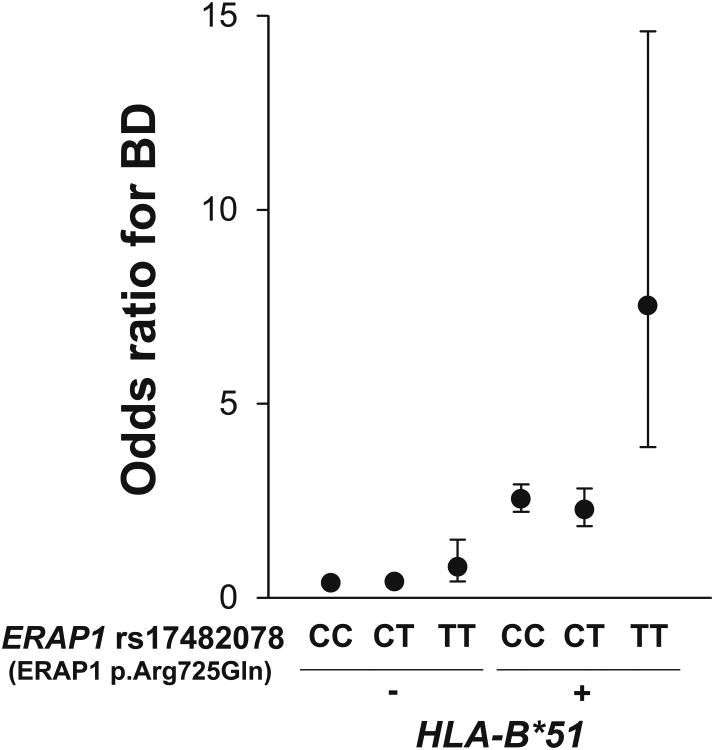
We therefore tested for an interaction between HLA-B*51 and ERAP1 in BD. The ERAP1 variants preferentially conferred risk for BD in HLA-B*51 positive individuals (p-value for interaction=0.0009 from a logistic likelihood ratio test comparing the full model including a multiplicative interaction term with the reduced model without interaction term) in the combined Turkish GWAS and replication (including uveitis and non-uveitis) samples (Figure 3). Furthermore, ERAP1 p.Arg725Gln homozygosity was associated with an odds ratio for BD of 3.78 [95% CI 1.94-7.35] in the HLA-B*51 positive individuals versus an odds ratio of 1.48 [95% CI 0.78-2.80] in the HLA-B*51 negative individuals. This finding indicates that the disease-associated peptidase variant contributes to disease susceptibility through an interaction with the HLA-B*51 protein.
Figure 3. Epistasis between HLA-B*51 and ERAP1 rs17482078 (ERAP1 p.Arg725Gln) coding variant in BD.
Epistasis was analyzed in the combined Turkish GWAS and replication samples. HLA-B*51+ indicates individuals either heterozygous or homozygous for HLA-B*51. In the replication samples, the surrogate marker rs2848713 (r2=0.68 with HLA-B*51 in Turkish GWAS) was used to predict HLA-B*51 positivity. Odds ratios for BD were determined comparing the frequency of the two marker genotypes in cases versus controls. The odds ratios shown are relative to the lowest disease-risk genotype group (HLA-B*51 negative and ERAP1 rs17482078 CC). Error bars represent the 95% confidence intervals. In an analysis restricted to the HLA-B*51 positive individuals, the ERAP1 TT genotype had an odds ratio of 3.78 (95% CI: 1.94-7.35), whereas in the HLA-B*51 negative individuals, the TT genotype odds ratio was 1.48 (95% CI: 0.78-2.80).
Homozygosity for the ERAP1 variants is associated with increased risk for BD, but decreased risk for AS23 and psoriasis22. The difference between risk and protection among these three diseases may depend on the variability and binding affinities of peptides loaded onto the respective MHC Class I molecules, which can affect their stability and function. Indeed, repertoires of MHC-bound peptides are altered in ERAP1 deficient mice25. ERAP1 p.Arg725Gln-related alterations might affect the repertoire of peptides that bind to HLA-B*51, which is known for its promiscuous peptide binding features5,26. The recessive nature of the ERAP1 effect in BD (one wild type copy in heterozygotes is sufficient to obscure the risk effect of the mutant allele) suggests that homozygotes fail to produce one or more disease-protective peptides.
The KLRC4 BD-associated SNP (Table 1) is within a haplotype block containing five natural killer (NK) cell receptor genes (KLRK1, KLRC1-4) (Figure 2c). Two non-synonymous variants in KLRC4, rs1841958 and rs2617170, encoding KLRC4 p.Ile29Ser and Asn104Ser, are found on the BD-protective haplotype of this LD block. This haplotype has been associated with reduced peripheral blood leukocyte cytotoxicity and increased incidence of cancer27. Conditional logistic regression analysis, conditioning on the KLRC4 Asn104Ser variant, showed no additional independent association signals within this LD block (Supplementary Figure 1a). KLRC4, also called NKG2F, encodes a c-type lectin receptor whose function is largely unknown. A possible clue to its function may be found in a related family member, NKG2D, encoded by KLRK1 and also located within the disease-associated haplotype block. NKG2D is expressed on NK cells and γδ T cells, and can act as a co-stimulatory molecule for CD4+ and CD8+ T cells28,29. Interestingly, a ligand of the NKG2D receptor is MICA (the MHC Class I chain-related protein A)28. The MICA gene is located within the MHC region and SNPs within the MICA locus are in linkage disequilibrium with HLA-B*516. The importance of NK receptors in BD pathogenesis is also supported by the observation that the strongest linkage peak in Turkish familial BD (LOD score of 3.94) is found at chr12p12-13, which includes the KLRK region locus30.
The disease-associated variants in the 3′ flanking region of CCR1-CCR3 (rs7616215) and within the third intron of STAT4 (rs7574070) are noncoding and are not in strong LD with any coding variants (Figure 2a-b). The CCR1-CCR3 locus contains a cluster of chemokine receptor genes within the LD block. Logistic regression analysis conditioning on covariate rs7616215 revealed only a single association signal in the region (Supplementary Figure 1b). ENCODE data indicated that rs7616215 and rs7574070 are located within DNase I hypersensitivity and histone 3 lysine 4 methylation sites, suggesting effects on transcription.
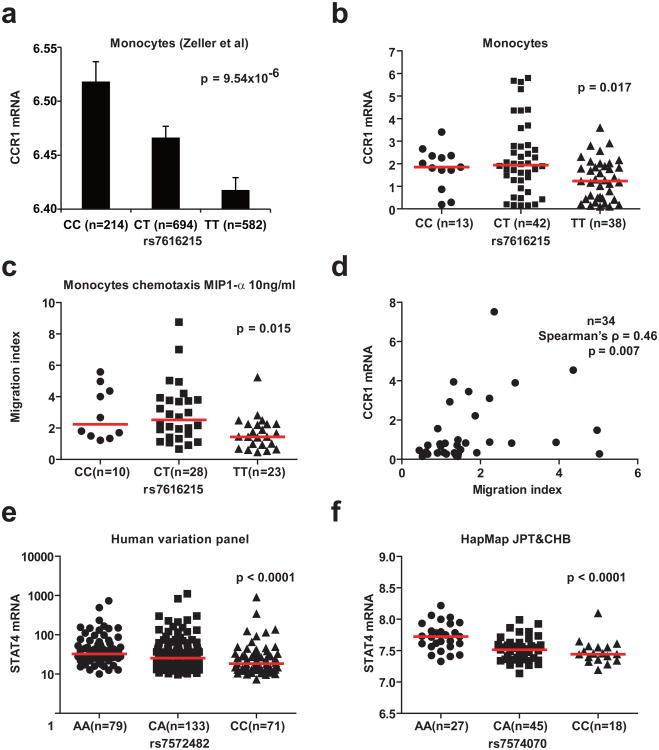
Indeed, CCR1 mRNA expression was higher in primary human monocytes from healthy donors with the disease protective C allele (Figure 4a, Zeller et al.31, p = 9.5 × 10−6, and 4b, p = 0.017). The BD-associated variant was not, however, found associated with expression of the nearby gene CCR3.31 Concordant with expression data, migration of monocytes in response to a gradient of the CCR1 ligand MIP1-α was higher in C allele individuals (Figure 4c, p = 0.015). Comparison between CCR1 mRNA expression level and migration index within matched samples (n=34) showed significant correlation (Spearman's ρ = 0.46, p= 0.007, Figure 4d). Thus, CCR1 expression and monocyte chemotaxis were reduced in individuals with the disease risk allele suggesting that impaired clearance of pathogens may contribute to BD pathogenesis. Future experiments will be required to further elucidate the importance of the observed differences.
Figure 4. Association of CCR1 and STAT4 variants with gene expression levels and function in human cells.
(a) Mean CCR1 mRNA expression in human primary monocytes according to rs7616215 genotype (data from Zeller et al31). Error bars represent the standard error of the mean. (b) CCR1 mRNA expression in human primary monocytes from our study according to rs7616215 genotype. (c) Chemotaxis of human primary monocytes against a gradient of MIP1-α according to rs7616215 genotype. “Migration index” indicates number of migrated cells in the presence of MIP1-α divided by the number of migrated cells in the absence of MIP1-α from the same sample. (d) Correlation between monocyte CCR1 expression and chemotactic activity. Comparison between CCR1 mRNA level and chemotaxis migration index were determined in monocytes isolated from the same blood sample. Spearman correlation test was performed. (e) STAT4 mRNA expression in Human Variation Panel samples (GEO GSE24277). rs7572482 is a surrogate marker for rs7574070 (r2=1, D′=1 in HapMap CEU samples). (f) STAT4 mRNA expression in HapMap JPT and CHB samples according to rs7574070 genotype (GEO GSE6536). In panels b, c, e, and f, the horizontal bars indicate median values and the p-values shown are from the Kruskal-Wallis rank sum test.
STAT4 mRNA expression was higher in individuals with the risk allele A (Figure 4e and 4f). The BD-associated variant rs7574070 (and its surrogate rs7572482) is in poor LD with the previously reported autoimmune disease-associated STAT4 variant, rs757486532; in fact it is located two LD blocks away, suggesting the associations are independent, although both variants are located within the large third intron of STAT4 (Figure 2b). Both variants are associated with increased expression of STAT4 (ref 33 and data presented here), but the genetically distinct disease-associations suggest different STAT4 regulatory mechanisms in BD compared with rheumatoid arthritis, systemic lupus erythematosus, and other autoimmune diseases. Smaller effect sizes observed for the associations of the CCR1 and STAT4 variants in the Japanese replication (Table 1) could be explained by “the winner's curse”34.
In conclusion, this study adds substantially to the understanding of genetic factors that contribute to BD susceptibility (the new loci are CCR1-CCR3, STAT4, KLRK1-KLRC4, and ERAP1). Furthermore, the results support an emerging concept delineating common pathogenic mechanisms for BD and the SpA. BD, AS, and psoriasis are inflammatory disorders affecting the skin, eyes, and joints, with significant MHC Class I associations (B*51 for BD, B*27 for AS, and C*06 for psoriasis). Recent genetic studies implicate variants of IL23R, encoding an upstream molecule in Th17 activation, in susceptibility to all three disorders. The present work adds ERAP1 to the list of shared genetic factors and furthermore, interactions between MHC Class I and ERAP1 are also found in all three of these diseases. ERAP1 trims peptides for proper loading onto Class I antigens, thus suggesting that peptide-MHC Class I interactions contribute to all three of these diseases. These data suggest the existence of shared inflammatory pathways among these diseases leading to the possibility of common therapeutic strategies, while raising questions about the specific disease characteristics, which may be related to their different MHC Class I associations.
Online methods
Patients
1,215 Turkish Behçet's disease (BD) cases and 1,278 genetically matched controls used in previous GWAS were studied6. Individuals who also met the Tel-Hashomer clinical criteria for the diagnosis of familial Mediterranean fever (FMF)35 were excluded (n=6). For replication, an additional similarly collected 838 Turkish cases (none fulfilled FMF criteria) and 630 controls, and 612 Japanese BD cases and 740 control samples enrolled in the previous GWAS7 were included. Turkish BD patients fulfilled the International Study Group diagnostic criteria for Behçet's disease19. All Japanese BD patients fulfilled the Japanese BD diagnostic criteria7 and 496 of them also fulfilled the International Study Group criteria. All study participants provided written informed consent, and the study was approved by the Ethics Committees of each investigative institution.
Genotype imputation
We imputed genotypes of 1,209 BD cases and 1,278 controls using MACH v1.0. For a reference panel, we used 96 Turkish healthy controls who participated in the previous BD GWAS using the HumanHap370CNV chip (Illumina) and additionally genotyped on the Human OMNI 1M chip (Illumina, San Diego, CA). For quality control, we excluded SNPs from the reference panel if they had a minor allele frequency less than 5%, deviated from Hardy–Weinberg equilibrium (P<0.0001), or had a call rate below 95%, yielding 814,474 SNPs for the imputation. Quality scores for the imputation are shown in Supplementary Table 6. SNPs with Rsq<0.3 were excluded from the association analysis. A total of 779,465 imputed SNPs were included in the genome-wide association analysis.
Validation and fine-mapping
The Turkish GWAS imputation provided the discovery data. Twenty-two SNPs from the novel genetic loci were selected for evaluation in the Turkish replication samples. Three of these SNPs with P < 0.001 and one SNP from the phenotypic subset analysis identified four regions for validation and fine-mapping by i-PLEX assays (TOF-MS, Sequenom, San Diego, CA) using the original Turkish GWAS samples6. The IL12A locus with two SNPs with P < 0.05 in the replication samples was also investigated, but failed to reach genome-wide significance. For variants that failed TOF-MS design or reaction, TaqMan genotyping was performed (Applied Biosystems, Foster City, CA). Genotyping was performed in an unbiased fashion by masking the phenotype of the samples. For the fine-mapping, we used the Tagger SNP selection tool from HapMap to select SNPs to augment the coverage of the GWAS SNPs with the intent to obtain 100% coverage of the HapMap Phase III SNPs with greater than 5% minor allele frequency in the CEU HapMap population with pairwise r2 > 0.8. Although already tagged, additional SNPs with r2 > 0.8 with the most significantly associated SNP of the region were also included. Genotyping of the same samples used for the Turkish GWAS discovery collection was performed6. After quality control, the resulting coverage for the CCR1 locus (chr3:45441901-46908964, hg18) was 92%, the STAT4 locus (chr2:191602386-191769025) was 84%, the KLRK1-KLRC1 locus (chr12:10329925-10557292) was 92%, and the ERAP1-2 locus (chr5:96026703-96305246) was 94%. The most significantly associated marker from each region was used for the replications. The Japanese replication collection genotypes were from the Japanese GWAS7. The Turkish replication collection samples were genotyped by TOF-MS or TaqMan assay.
mRNA expression data and migration assay
CCR1 mRNA expression data were extracted from the report by Zeller and colleagues, which includes genome-wide SNP data along with mRNA expression array data from monocytes of n = 1490 European ancestry individuals31. SNP rs7616215 showed association with CCR1 mRNA expression (p=9.54×10−6), whereas CCR3 data were not reported, indicating that the association of rs7616215 with CCR3 is less significant (p>5×10−5). STAT4 mRNA expression data in lymphoblastoid samples were obtained from Gene Expression Omnibus (GEO) datasets36, 37. Primary human monocytes from unrelated healthy volunteers were isolated from peripheral blood mononuclear cells by MACS Human Monocyte Isolation Kit (Miltenyi Biotec, Gladbach, Germany). RNA was isolated from PBMCs by RNeasy kit (Qiagen, Valencia, CA) and preserved at −80 C° until used. cDNAs were prepared from DNase I (Invitrogen, Carlsbad, CA)-pretreated-RNA using SuperScript II according to manufacturer's protocol (Invitrogen). Q-PCR gene expression assay for human CCR1 (Hs00174298_m1) was purchased from Applied Biosystems (Foster City, CA). Human GAPDH (4310884E) served as an internal control. Multiplex PCR (GAPDH with CCR1) was performed in triplicate or quadruplicate as in the manufacturer's protocol (Applied Biosystems). The ΔΔCT method was used for the analysis (n=93).
A monocyte migration assay was performed with 24-well Transwell 5 μm polycarbonate membrane chambers (Costar, Corning, NY). 1% bovine serum albumin RPMI1640 medium was used for incubation of the cells. 5×104 cells were seeded in the upper chamber and the CCR1 chemokine MIP1-α (0 or 10ng/ml) (R&D, Minneapolis, MN) was placed in lower chamber. After incubation for 2 hours, cells that migrated into the membrane were fixed and stained by DiffQuik (Siemens, Newark, DE). Cells were counted in 5 high power fields on membranes from each of two duplicate wells. The relative change in cell migration in response to the chemokine was determined by dividing the migrated cell count obtained with MIP1-α by the cell count obtained from the same sample without MIP1-α (n=61).
Statistical analysis
Genome-wide SNP association tests were performed based on allelic tests by comparing the allele frequencies between BD cases and controls, using Golden Helix SVS 7.5.2 software (Golden Helix, Bozeman, MT). P<5×10−8 was considered genome-wide significance. For the uveitis subset analysis, as well as the basic allelic test, we applied dominant and recessive genetic model tests and therefore employed 1.67×10−8 for genome-wide significance. Conditional analyses were performed by fitting the logistic regression model with SNPs rs2617170 (for KLRK1-KLRC1) or rs7616215 (for CCR1-CCR3) as covariates. For meta-analysis, Cochran-Mantel-Haenszel tests were performed. To test for the interactive effects, we fit the log likelihood of the full model, including additive terms of the main effects and a multiplicative term of the interaction effect, versus a reduced model of the additive terms of the main effects only. P value was calculated by likelihood ratio tests between full and reduced models based on 1 degree of freedom Chi-square test statistics. For comparisons in expression and chemotaxis data, the Kruskal-Wallis rank sum test as a non-parametric version of one-way ANOVA and Spearman's rank correlation test were performed.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Programs of the National Human Genome Research Institute and the National Institute of Arthritis and Musculoskeletal and Skin Diseases, and the Center for Human Immunology, Autoimmunity and Inflammation, of the National Institutes of Health, USA, and by the Istanbul University Research Fund, and Research on Specific Disease of the Health Science Research Grants from the Japanese Ministry of Health, Labor, and Welfare, and the Japan Rheumatism Foundation. We thank Dr. Alexander Wilson (Genometrics Section, Inherited Disease Research Branch, National Human Genome Research Institute, National Institutes of Health, Baltimore, Maryland, USA) for helpful comments on this manuscript.
Footnotes
Author Contributions: Study design: Y.K., G.B., A.G., M.J.O., E.F.R., D.L.K. Analysis: Y.K., G.B., Y.Kim, A.M., A.G., E.F.R., D.L.K. Sample procurement and data generation: Y.K., G.B., Y.I., N.M., I.T., E.S., Y.O., F.S.S., B.E., H.I., Z. E., A.C., N.A., D.U., C.S., A.U., M.T., Y.Kim, G.M.W., M.J.O., A.M, A.G., E.F.R., D.L.K. Writing: Y.K., G.B., Y.Kim, M.J.O., A.G., E.F.R., D.L.K. All the authors read and approved the final version of the manuscript.
Competing Financial Interests: The authors declare no competing financial interests.
URLs: International HapMap project, http://HapMap.ncbi.nlm.nih.gov/ ENCODE, http://genome.ucsc.edu/ ENCODE/MACH1.0, http://www.sph.umich.edu/csg/abecasis/MACH/index.html, GEO, http://www.ncbi.nlm.nih.gov/geo (accession numbers: GSE6536 and GSE24277)
Contributor Information
Ahmet Gül, Email: dr.agul001@gmail.com.
Elaine F. Remmers, Email: remmerse@mail.nih.gov.
Daniel L. Kastner, Email: kastnerd@mail.nih.gov.
References
- 1.Sakane T, Takeno M, Suzuki N, Inaba G. Behçet's disease. N Engl J Med. 1999;341:1284–91. doi: 10.1056/NEJM199910213411707. [DOI] [PubMed] [Google Scholar]
- 2.Yazici H, Fresko I, Yurdakul S. Behçet's syndrome: disease manifestations, management, and advances in treatment. Nat Clin Pract Rheumatol. 2007;3:148–55. doi: 10.1038/ncprheum0436. [DOI] [PubMed] [Google Scholar]
- 3.Ono S, Aoki K, Sugiura S, Nakayama E, Itakura K. Letter: HL-A5 and Behçet's disease. Lancet. 1973;2:1383–4. doi: 10.1016/s0140-6736(73)93343-6. [DOI] [PubMed] [Google Scholar]
- 4.de Menthon M, Lavalley MP, Maldini C, Guillevin L, Mahr A. HLA-B51/B5 and the risk of Behçet's disease: a systematic review and meta-analysis of case-control genetic association studies. Arthritis Rheum. 2009;61:1287–96. doi: 10.1002/art.24642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gül A, Ohno S. HLA-B*51 and Behçet Disease. Ocul Immunol Inflamm. 2012;20:37–43. doi: 10.3109/09273948.2011.634978. [DOI] [PubMed] [Google Scholar]
- 6.Remmers EF, et al. Genome-wide association study identifies variants in the MHC class I, IL10, and IL23R-IL12RB2 regions associated with Behçet's disease. Nat Genet. 2010;42:698–702. doi: 10.1038/ng.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mizuki N, et al. Genome-wide association studies identify IL23R-IL12RB2 and IL10 as Behçet's disease susceptibility loci. Nat Genet. 2010;42:703–6. doi: 10.1038/ng.624. [DOI] [PubMed] [Google Scholar]
- 8.Moll JM, Haslock I, Macrae IF, Wright V. Associations between ankylosing spondylitis, psoriatic arthritis, Reiter's disease, the intestinal arthropathies, and Behçet's syndrome. Medicine (Baltimore) 1974;53:343–64. doi: 10.1097/00005792-197409000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Chang HK, et al. The comparison between Behçet's disease and spondyloarthritides: does Behçet's disease belong to the spondyloarthropathy complex? J Korean Med Sci. 2002;17:524–9. doi: 10.3346/jkms.2002.17.4.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yazici H, Tuzlaci M, Yurdakul S. A controlled survey of sacroiliitis in Behçet's disease. Ann Rheum Dis. 1981;40:558–9. doi: 10.1136/ard.40.6.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olivieri I, Salvarani C, Cantini F. Is Behçet's disease part of the spondyloarthritis complex? Journal Rheumatol. 1997;24:1870–2. [PubMed] [Google Scholar]
- 12.Consortium WTCC, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–37. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–3. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair RP, et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohno S, et al. Efficacy, safety, and pharmacokinetics of multiple administration of infliximab in Behçet's disease with refractory uveoretinitis. J Rheumatol. 2004;31:1362–8. [PubMed] [Google Scholar]
- 16.Dougados M, Baeten D. Spondyloarthritis. Lancet. 377:2127–37. doi: 10.1016/S0140-6736(11)60071-8. [DOI] [PubMed] [Google Scholar]
- 17.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genetics. 2010;42:1118–25. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genetics. 2011;43:246–52. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Criteria for diagnosis of Behçet's disease. International Study Group for Behcet's Disease. Lancet. 1990;335:1078–80. [PubMed] [Google Scholar]
- 20.Haroon N, Inman RD. Endoplasmic reticulum aminopeptidases: Biology and pathogenic potential. Nat Rev Rheumatol. 6:461–7. doi: 10.1038/nrrheum.2010.85. [DOI] [PubMed] [Google Scholar]
- 21.Sun LD, et al. Association analyses identify six new psoriasis susceptibility loci in the Chinese population. Nat Genet. 42:1005–9. doi: 10.1038/ng.690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strange A, et al. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet. 42:985–90. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Evans DM, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat Genet. 43:761–7. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kochan G, et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc Natl Acad Sci U S A. 2011;108:7745–50. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.York IA, Brehm MA, Zendzian S, Towne CF, Rock KL. Endoplasmic reticulum aminopeptidase 1 (ERAP1) trims MHC class I-presented peptides in vivo and plays an important role in immunodominance. Proc Natl Acad Sci U S A. 2006;103:9202–7. doi: 10.1073/pnas.0603095103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gebreselassie D, Spiegel H, Vukmanovic S. Sampling of major histocompatibility complex class I-associated peptidome suggests relatively looser global association of HLA-B*5101 with peptides. Hum Immunol. 2006;67:894–906. doi: 10.1016/j.humimm.2006.08.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayashi T, et al. Identification of the NKG2D haplotypes associated with natural cytotoxic activity of peripheral blood lymphocytes and cancer immunosurveillance. Cancer Res. 2006;66:563–70. doi: 10.1158/0008-5472.CAN-05-2776. [DOI] [PubMed] [Google Scholar]
- 28.Bauer S, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 29.Raulet DH. Roles of the NKG2D immunoreceptor and its ligands. Nat Rev Immunol. 2003;3:781–90. doi: 10.1038/nri1199. [DOI] [PubMed] [Google Scholar]
- 30.Karasneh J, Gül A, Ollier WE, Silman AJ, Worthington J. Whole-genome screening for susceptibility genes in multicase families with Behçet's disease. Arthritis Rheum. 2005;52:1836–42. doi: 10.1002/art.21060. [DOI] [PubMed] [Google Scholar]
- 31.Zeller T, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Remmers EF, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357:977–86. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abelson AK, et al. STAT4 associates with systemic lupus erythematosus through two independent effects that correlate with gene expression and act additively with IRF5 to increase risk. Ann Rheum Dis. 2009;68:1746–53. doi: 10.1136/ard.2008.097642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zollner S, Pritchard JK. Overcoming the winner's curse: estimating penetrance parameters from case-control data. Am J Hum Genet. 2007;80:605–15. doi: 10.1086/512821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Livneh A, et al. Criteria for the diagnosis of familial Mediterranean fever. Arthritis Rheum. 1997;40:1879–85. doi: 10.1002/art.1780401023. [DOI] [PubMed] [Google Scholar]
- 36.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–53. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niu N, et al. Radiation pharmacogenomics: a genome-wide association approach to identify radiation response biomarkers using human lymphoblastoid cell lines. Genome Res. 2010;20:1482–92. doi: 10.1101/gr.107672.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.