Abstract
The development of small molecules that bind DNA sequence specifically has the potential to modulate gene expression in a general way. One mode of DNA binding is intercalation, or the insertion of molecules between DNA base pairs. We have developed a modular polyintercalation system in which intercalating naphthalene diimide (NDI) units are connected by flexible linkers that alternate between the minor and major grooves of DNA when bound. We recently reported a threading tetraintercalator with a dissociation half-life of 16 days, the longest reported to date, from its preferred 14 bp binding site. Herein, three new tetraintercalator derivatives were synthesized with one, two, and three additional methylene units in the central major groove-binding linker. These molecules displayed dissociation half-lives of 57, 27, and 18 days, respectively, from the 14 bp site. The optimal major groove-binding linker was used in the design of an NDI hexaintercalator that was analyzed by gel-shift assays, DNase I footprinting, and UV-visible spectroscopy. The hexaintercalator bound its entire 22 bp binding site, the longest reported specific binding site for a synthetic, non-nucleic acid based DNA binding molecule, but with a significantly faster dissociation rate compared to the tetraintercalators.
INTRODUCTION
Intercalation, or the insertion of (typically) planar aromatic rings between DNA base pairs, is one type of well-studied DNA binding motif. In particular, threading intercalation involves an intercalating molecule that inserts between base pairs while having substituents lying simultaneously in the major and minor DNA grooves.1,2 Other DNA binding molecules include triplex-forming oligonucleotides3 (TFO), peptide nucleic acids4,5 (PNA), artificial zinc-finger proteins,6 and minor groove-binding polyamides.7,8 An advantage of threading intercalators is their slow dissociation from DNA due to the extensive molecular rearrangements that necessarily accompany unbinding, a desirable characteristic for molecules intended to alter DNA transcription. In fact, we recently reported a threading tetraintercalator composed of naphthalene diimide (NDI) intercalating units connected by peptide and aliphatic linkers that bound a specific 14 bp site with a dissociation half-life of 16 days,9 the longest reported for a non-base pairing DNA binding molecule. PNAs and triplex-forming bicyclic oligonucleotides have been described with long lifetimes and high affinities.10,11 While threading mono- and bisintercalators continue to be of interest,12–15 reports of longer polyintercalators, threading or not, are rare.16–21
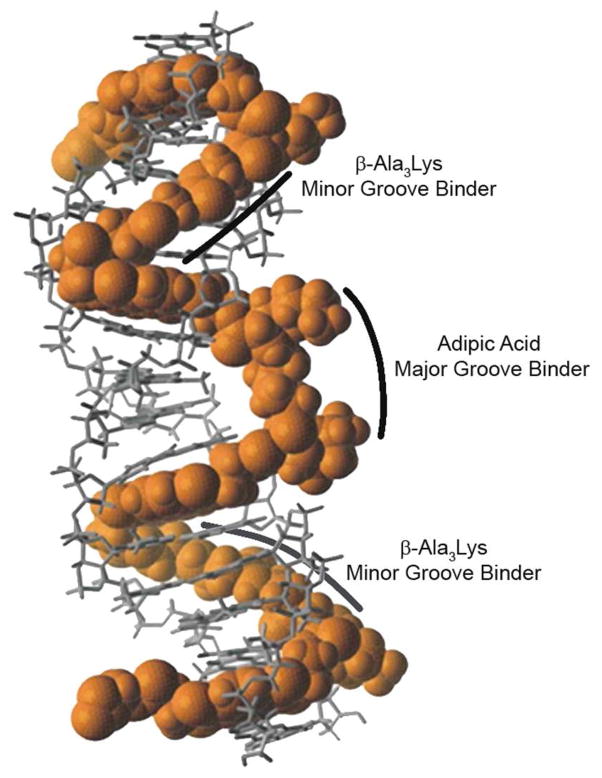
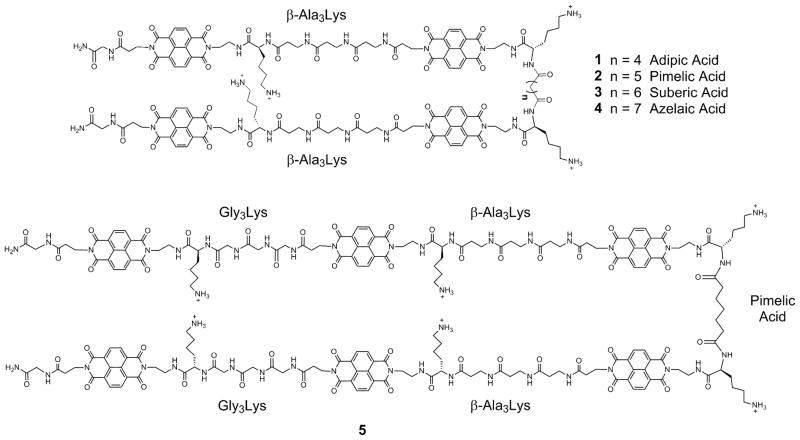
We have been exploring a modular approach to the construction of polyintercalating molecules in which bisintercalators of known specificity are combined to create tetraintercalators capable of recognizing composite sequences of DNA. In particular, an NDI-based bisintercalator with a Gly3Lys peptide linker bound the sequence 5′-GGTACC-3′, with the linker residing in the major groove,22 while a bisintercalator with a β-Ala3Lys linker bound the sequence 5′-GATAAG-3′ with the linker residing in the minor groove.23,24 Tetraintercalator 1 (Figure 1) is designed as a hybrid molecule, with linkers intended to reside in an alternating fashion in the order minor, major, and minor grooves. While the minor groove-binding portions of 1 are the β-Ala3Lys peptide linkers, the major groove-binding linker was derived from adipic acid rather than the Gly3Lys peptide linker in order to make the tetraintercalator C2 symmetric, yet retain the hydrophobic character and length of the Gly3Lys peptide. The 14 bp palindromic binding site identified as sequence A (5′-GATAAGTACTTATC-3′) is also a hybrid of the bisintercalator binding sites, allowing for the NMR structural determination of the entire C2 symmetric complex.25 The NMR-determined structure of the 1-DNA complex revealed that the aliphatic adipic acid major groove-binder, while the same length as the Gly3Lys peptide linker, made no hydrogen bond contacts within the major groove and likely caused the DNA helix to bend. In order to develop longer threading polyintercalators in the same manner with potentially slower dissociation rates, this major groove-binding linker was systematically lengthened.
Figure 1.
NMR-determined structure of threading tetraintercalator 1 with 14 bp sequence A.25
We report here the design, synthesis, and binding studies of tetraintercalators 2 – 4 (Figure 2) with major groove-binding aliphatic linkers lengthened by one, two, and three methylene units, respectively. Kinetic analyses of 2 – 4 revealed a major groove binder lengthened by one methylene unit, corresponding to a pimelic acid cross-linker and tetraintercalator 2, to be optimal. The dissociation half-life of 2 also breaks our previous record and has the slowest reported dissociation rate from DNA for a non-nucleic acid molecule. Using this optimal pimelic acid cross-linker, symmetric NDI hexaintercalator 5 (Figure 2) was synthesized and analyzed by gel-shift assays, DNase I footprinting, and UV-visible spectroscopy. While 5 occupies its entire predicted 22 bp binding site, a new standard for synthetic, non-nucleic acid DNA binding molecules, its dissociation rate was significantly faster than those of similar tetraintercalators, implying an overall less stable complex with DNA.
Figure 2.
Structures of NDI tetraintercalators 1 – 4 and hexaintercalator 5. The identities of the groove-binding linkers are labeled.
RESULTS
Design and Synthesis of 2 – 4
The previous structural analysis of the 1-DNA complex indicated some degree of distortion of the DNA duplex structure around the central region. We hypothesized that the central adipic acid unit of 1 might be shorter than is optimal, and the resulting distortion of the DNA structure with an inadequate length linker might come at the expense of some binding energy. Pimelic acid, suberic acid and azelaic acid were used as central linkers in 2 – 4 to investigate systematically the influence of central linker length. Note that these linkers maintain the hydrophobic character of adipic acid, but the increased flexibility introduced by the additional methylene units could raise the entropic cost of binding.
Tetraintercalators 2 – 4 were synthesized as previously described25 using Fmoc-based solid phase peptide synthesis (SPPS) incorporating orthogonal t-Boc protecting groups for the lysine side chains. The Fmoc-(β-Ala)3-OH linker was synthesized prior to SPPS in order to maximize yields.21
Kinetic Analysis of Lengthened Tetraintercalators
Gel-shift association and dissociation assays were performed for 2 – 4 with 24-mer oligo A containing 14 bp sequence A (Figure 3), expected to be the preferred binding sequence based on previous results.9,21 These experiments exploit the retarded migration seen by polyacrylamide gel electrophoresis (PAGE) when polyintercalator is tightly bound to an oligonucleotide. For these experiments, a stoichiometric amount of intercalator and [32P]-labeled DNA duplex were allowed to equilibrate by heating the DNA to 60 °C, adding intercalator, allowing the solution to cool slowly, and then holding at room temperature overnight. Dissociation results from this type of incubation match those of extended 37 °C incubations.21 A recent study with a ruthenium monointercalator also suggested that the activation energy for intercalation is lowered with heat denatured DNA, resulting in faster intercalation processes.26 Complete association between polyintercalator and duplex was confirmed by native PAGE, as all of the radioactively labeled duplex migrated at the expected slower rate. A 100-fold excess of unlabeled DNA was added to the preassembled complex in order to “trap” any intercalator once it dissociated from labeled DNA. Autoradiography was used to measure the ratio of bound (slower moving band) to unbound (faster moving band) radioactively labeled DNA as a function of time, and the data was fitted to a mono-exponential decay equation. The dissociation rate constants and half-lives of 2 – 4 are reported in Table 1.
Figure 3.
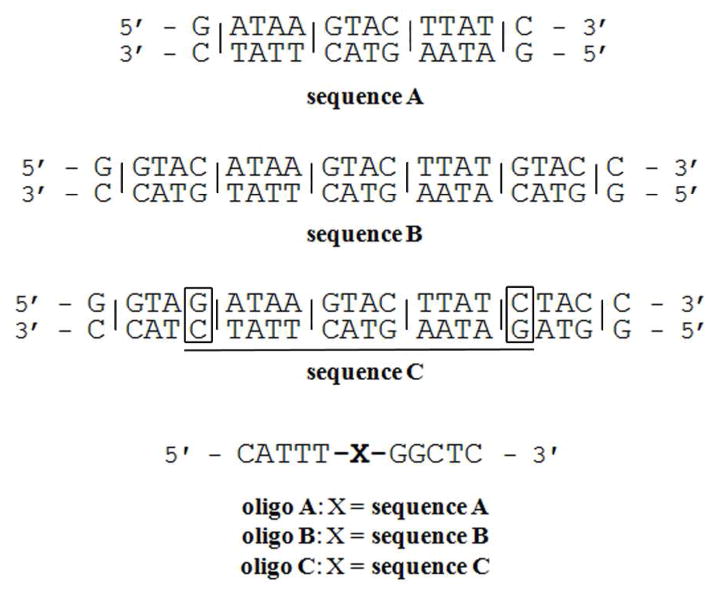
Sequence A is the 14 bp binding site for tetraintercalators 1 – 4. Sequences B and C are the 22 bp designed binding sites for hexaintercalator 5. Lines between base pairs indicate expected intercalation sites. Sequence C differs from sequence B by 2 bp, highlighted in the boxes, and also contains sequence A as underlined. Oligos A – C are the oligonucleotides used for gel-shift assays and UVvisible spectroscopy experiments.
Table 1.
Association and Dissociation Rate Constants of Tetraintercalators 1 – 4 with oligo A
| ka × 103(M−1s−1) | kd × 10−7(s−1) | t1/2(d) | |
|---|---|---|---|
|
|
|||
| 1a | 10 ± 5 | 5.0 ± 0.5 | 16 |
| 2 | 8 ± 3 | 1.4 ± 0.2 | 57 |
| 3 | 8 ± 3 | 3.0 ± 0.5 | 27 |
| 4 | 1 ± 0.2 | 4.4 ± 0.9 | 18 |
See ref 9.
The intercalators ranked from fastest to slowest dissociation rate are 1 ≈ 4 > 3 > 2, with 2 demonstrating a dissociation half-life of 57 days. Increasing the length of the tetraintercalator major groove-binder by one methylene unit in 2 resulted in an almost 4-fold decrease in dissociation rate compared to 1. Tetraintercalator 3, lengthened by two methylene units, also has a 2-fold slower dissociation rate compared to 1. Interestingly, the dissociation rate constant of 4, lengthened by three methylene units, is indistinguishable by this method to that of 1. In each case, dissociations were monitored for 65 days, until the radiolabel signal was so weak as to be non-quantifiable. Because of the extremely slow dissociation rate of 2, this only corresponds to a little over one half-life (Supporting Information), so the reported value must be considered only our preliminary best estimate. Nevertheless, it is clear that 2 is the slowest dissociating molecule of the group. Binding to a control sequence (5′-CATTTAACAACATGTTGTTGGCTC-3′) lacking any predicted polyintercalator binding sites was also investigated, but 2 – 4 showed no binding by gelshift, confirming the sequence-specific nature of binding.
Gel-shift assays were also used to obtain estimated association rate constants for 2 – 4, as described previously9 (Table 1). Stoichiometric amounts of radiolabeled 24-mer oligo A and tetraintercalator were combined at concentrations of 0.5 μM, 1.0 μM, and 1.5 μM, and representative time points were taken for analysis by native PAGE. Association rates were calculated by using the integrated rate equation for stoichiometric binding.27 The association rates of 2 and 3 were observed to be similar to that of 1, with that of 4 being slower. In our experience, this method is prone to significant errors, yet it can be used to provide an estimate of relative association rates.
Design and Synthesis of 5
Hexaintercalator 5 was designed to bind to a 22 bp binding site by threading alternatively through the major, minor, major, minor, major grooves of DNA (Figure 2). In 5, the β-Ala3Lys peptide linkers were used as the minor groove-binding linkers, while the first and last major groove-binding linkers were composed of Gly3Lys. The middle major groove-binding linker was the aliphatic cross-linker pimelic acid previously identified as the optimum central major groove-binding linker in the context of tetraintercalators 1 – 4. Hexaintercalator 5 was synthesized in the same fashion as the tetraintercalators, and the Fmoc-(β-Ala)3-OH and Fmoc-(Gly)3-OH linkers were synthesized prior to SPPS in order to maximize yields.21
Two possible 22 bp palindromic binding sites were designed for 5 (Figure 3). Sequence B is the unaltered combination of the major and minor groove-binding bisintercalator binding sites. One potential issue with sequence B is the presence of two CA intercalation steps (for the second and fifth NDI units), as our intercalators have shown higher affinities when all intercalation steps are purine-purine.21,22,24 For this reason, sequence C, differing by 2 bp, was designed to maintain all purinepurine intercalation sites and also preserves the original tetraintercalator binding site, sequence A.
Kinetic Analysis of 5
Gel-shift assays were used to determine dissociation and association rate constants of 5 with 32-mer oligos B and C, containing sequences B and C respectively (Table 2). Hexaintercalator 5 dissociated slower from oligo C than oligo B, with dissociation halflives corresponding to 5.3 and 2.1 days, respectively. The dissociation rates of 5 from its designed binding sites were significantly faster than those measured for 1 – 4 with oligo A. A control sequence (5′-CATTTTACTAACAACATGTTGTTAGTAGGCTC-3′) was also tested, and although partial association was seen by gel-shift, 5 was completely dissociated within 3 hrs. The association rates for 5 with oligo B and C were the same, but slower than the rates for 1 – 3 binding to oligo A.
Table 2.
Association and Dissociation Rate Constants of Hexaintercalator 5 with oligos B and C
| ka × 103(M−1s−1) | kd × 10−6(s−1) | t1/2(d) | |
|---|---|---|---|
|
|
|||
| Oligo B | 1 ± 0.4 | 3.8 ± 0.8 | 2.1 |
| Oligo C | 1 ± 0.5 | 1.5 ± 0.3 | 5.3 |
DNase I Footprinting
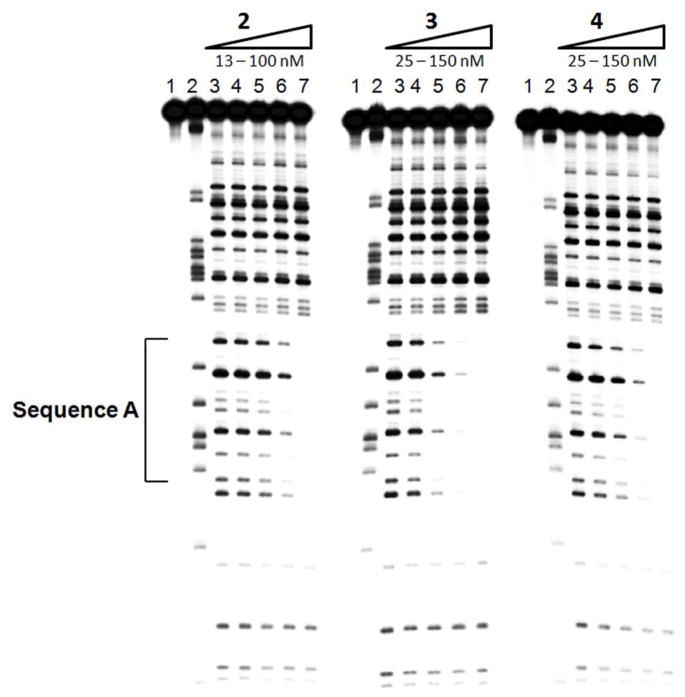
Binding of tetraintercalators 2 – 4 to a synthetic oligonucleotide containing sequence A was evaluated by concentration-dependent DNase I footprinting (Figure 4). Incubations for footprinting were performed by first heating radiolabeled DNA, adding the intercalator, and allowing the incubation to slowly cool to room temperature overnight. For all three tetraintercalators, a footprint begins to appear at 50 nM. The binding site is completely occupied at 100 nM by 2 and 3, and at 150 nM for 4. While the footprints are very similar, the strength of the footprints qualitatively follows the pattern of dissociation rate constants, with 2 showing the strongest footprint, followed by 3, and then 4. This result matches DNase I footprinting of 1 with sequence A, which showed a footprint starting at 50 nM and full occupation at 100 nM.21 At these concentrations, no non-specific binding was observed for 2 – 4.
Figure 4.
DNase I footprints of tetraintercalators 2 – 4 with sequence A. Lane 1 contains undigested DNA. Lane 2 contains an adenine-specific cleavage reaction.34 Lane 3 contains digested DNA with no intercalator. Lanes 4 – 7 contain increasing concentration of intercalator. For 2, lanes 4 – 7 contain 13, 25, 50, and 100 nM intercalator. For 3 and 4, lanes 4 – 7 contain 25, 50, 100, and 150 nM intercalator.
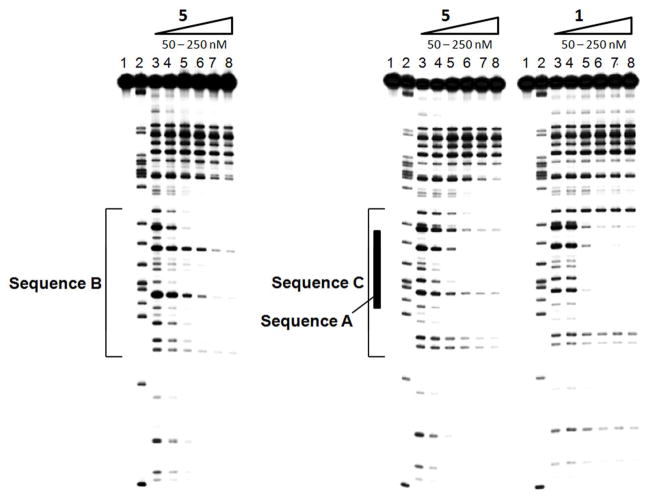
Hexaintercalator 5 was evaluated using concentration-dependent DNase I footprinting assays with synthetic oligonucleotides containing sequences B and C (Figure 5). A footprint begins to show for both sequences at 100 nM with full occupation at 200 nM. While both footprints are similar, the footprint of sequence C is qualitatively stronger than that of sequence B, consistent with the relative observed dissociation rate constants. It is noteworthy that 5 seems to occupy all of sequence B and sequence C. Some non-specific binding is also present at higher concentrations in GC-rich regions.
Figure 5.
DNase I footprints of hexaintercalator 5 with sequences B and C and tetraintercalator 1 with sequence C. The region where sequence A is located within sequence C is marked. Lane 1 contains undigested DNA. Lane 2 contains an adeninespecific cleavage reaction.34 Lane 3 contains digested DNA with no intercalator. Lanes 4 – 8 contain 50, 100, 150, 100, and 250 nM intercalator.
Because sequence C also contains sequence A, a concentration-dependent DNase I footprinting analysis of tetraintercalator 1 was carried out with sequence C (Figure 5). In these studies, sequence A is mostly occupied by 1 at 100 nM. Importantly, the footprints of 1 and 5 with sequence C look quite different. Digestion bands flanking the central sequence A are quite apparent in the footprint of 1, revealing that 1 does not occupy all 22 bp of sequence C. In contrast, these same bands diminish in a concentration dependent manner in the footprint of 5, indicating that 5 is occupying those regions, and hence the entirety of the 22 bp site of sequence C.
Hypochromism
The extinction coefficients of 1 and 5 were measured in water with and without oligo A and oligo C, respectively (Table 3). In both cases, absorbance maxima were observed at 385 nm in water and 383 nm in the presence of binding site DNA. Intercalators 1 and 5 displayed equal degrees of hypochromism, with a 28% decrease in absorbance when combined with their preferred binding site DNA, respectively. For comparison, the extinction coefficient of 5 with control DNA, the same used in the dissociation experiments, displayed only 19% hypochromism.
Table 3.
Extinction Coefficients of NDI Polyintercalators
λmax = 385 nm.
λmax = 383 nm, in buffer (10 mM PIPES, pH = 7.0, 100 mM NaCl, 1 mM EDTA). For 1 and 5, a 1:1 molar ratio of oligo A and oligo C was used, respectively.
See ref 29.
DISCUSSION
The lengthening of the major groove-binding aliphatic linker of the previously reported 1 by one methylene unit resulted in 2, which displayed a 4-fold slower dissociation rate compared to 1, corresponding to a half-life of 57 days. This eclipses the previous record set by 1, and establishes 2 as the slowest non-base pairing dissociating molecule from DNA yet reported. Viewed another way, pimelic acid represents the optimal length for an aliphatic major groove-binding linker in our NDI-based polyintercalation system. Although there is no structural characterization of 2 bound to sequence A available yet, it is assumed that 2 – 4 bind in a threading polyintercalating fashion analogous to the structurally characterized 1-DNA complex.25 Consistent with this assumption, the very slow dissociation rates displayed by 2 – 4 essentially preclude any other mode of binding. The extremely slow dissociation rates seen with threading polyintercalators are not surprising given the molecular rearrangements that must occur during dissociation from DNA. Note that dissociation and association of 1 – 4 with sequence A are assumed to occur in multiple steps, but the assays used in this work report the limiting ones in each direction.
As measured by gel-shift assays, the association rate constants of 1 – 3 are essentially the same, yet that for 4 is noticeably slower (Table 1). As a result, the differences in dissociation rate constants for 1 – 3 implies that 2 forms a more stable complex with sequence A compared to the complexes of 1 and 3. Recalling that some distortion of the DNA was seen in the NMR structural analysis of 1 bound to sequence A,25 it is reasonable to assume that the pimelic acid linker of 2 introduces less distortion upon binding compared to 1, resulting in a roughly 4-fold more stable complex. The suberic acid and azelaic acid linkers in 3 and 4 appear too long compared to pimelic acid. Nevertheless, the suberic acid linker in 3 still leads to a 2-fold slower dissociation rate compared to 1, which contains the adipic acid linker. The reason for the slower association rate observed with 4 is not clear, although the increased flexibility of the azelaic acid linker likely plays a role.
The DNase I footprints of 2 – 4 with sequence A show that the 14 bp binding site is fully occupied, and the qualitative strengths of the footprints track with relative dissociation rate constants (Figure 4). Importantly, at the concentrations in which the tetraintercalators are fully bound to sequence A, no non-specific binding is observed. Although the mechanism by which threading polyintercalators sample binding sites on long stretches of DNA is unknown, the kinetic analysis presented here verifies that relative sequence specificity derives from differences between the dissociation rate constants from the preferred 14 bp sequence and other random sequences. An examination of the association and dissociation rate constants of 2 – 4 would result in overall dissociation constants (Kd) in the picomolar range. However, footprinting methods could not be used for direct determination of Kd values for these high affinity molecules due to the low concentration of DNA required and the time to reach equilibrium.28 Nevertheless, we still assume that binding in the context of larger sequences compared to short oligonucleotides is complicated by additional mechanistic steps required to finally reside at the preferred binding site.
In order to extend our threading polyintercalator design beyond a tetraintercalator, NDI hexaintercalator 5 was synthesized with the pimelic acid cross-linker. Taking advantage of the modular nature of our threading polyintercalator approach, hexaintercalator 5 was designed to bind a 22 bp DNA binding site in a threading manner by alternating through the major-minor-majorminor-major grooves. Both gel-shift dissociation rate analysis and DNase I footprinting reveal that sequence C is preferred by 5 compared to sequence B. This result matches with prediction, since sequence C maintains all intercalation sites as purine-purine steps, known to be preferred by NDI-based intercalators. In addition, sequence C also contains 14 bp sequence A, which is known to be a high affinity site for the interior minormajor-minor groove threading portion of 5. The satisfying sequence specificity seen with threading polyintercalators is highlighted by the fact that even though sequence C differs from sequence B by only 2 of 22 bp, 5 is able to distinguish between the two sequences by more than 2-fold.
Surprisingly, 5 was found to dissociate from the 22 bp sequence C an order of magnitude faster than tetraintercalator 2 dissociating from its 14 bp preferred site, sequence A. An obvious explanation for the increase in dissociation rate constants would be that 5 is not fully hexaintercalated into the putative 22 bp binding sites in sequences B and C. However, DNase I footprinting results were not consistent with a lack of full intercalation. For sequences B and C, 5 shows a footprint over the entirety of both binding sites. This result contrasts with 1 binding sequence C, because while 1 fully occupies the central portion of sequence C, which is sequence A, it does not cause attenuation of the periphery digestion bands in sequence C. The footprint of 5 with sequence C does prevent cleavage by DNase I at those sites, indicating full hexaintercalation.
Examination of the extinction coefficients of 1 and 5 with and without their respective binding sites also supports the claim that 5 is bound and fully intercalated into sequence C. Upon intercalating DNA, NDI exhibits significant hypochromism.29 The degree of hypochromism observed when 1 is bound to sequence A (which is known to be fully intercalated) and 5 with sequence C are both 28%. When 5 was incubated with a control DNA sequence, which showed some binding during gel-shift assays but is very unlikely fully bound, only 19% hypochromism was seen. Like the DNase I footprints, taken together, these spectroscopic results are consistent with full hexaintercalation by 5.
The reasons for the slower association rates measured for 5 binding to both sequences B and C compared to 1 binding sequence A are not completely understood, but there are a number of possible explanations. For one, 5 is a longer and more flexible molecule, which would predict a larger entropic penalty for binding to DNA. At the other end of the spectrum, the six NDI units in 5, relative to the four NDI units in 1, are likely self-stacked and these interactions must be disrupted prior to intercalation into DNA. Another possible problem with the design of 5 is that the linkers may not yet be precisely the correct lengths, a problem that will amplify over longer structures.
Assuming hexaintercalator 5 is fully intercalated into the 22 bp sequences B and C, these complexes represent the longest reported binding sites for synthetic, nonnucleic acid based DNA binding molecules. Triple helixforming oligonucleotides and peptide nucleic acids, which both rely on nucleic acid bases for specificity, have targeted longer sequences,30,31 and artificial zinc-finger proteins, constructed from naturally occurring proteins, have been produced to bind up to 50 bp.32,33
CONCLUSION
NDI tetraintercalator 2 with a central major groove pimelic acid linker displays an extremely slow dissociation rate from its preferred 14 bp binding site, corresponding to a half-life of 57 days, a record for synthetic DNA binding molecules. This result supports the hypothesis that the adipic acid linker in 1 was indeed too short to be optimum. Taken further, tetraintercalators 3 and 4, with two and three additional methylene units, respectively, also display dissociation rates that are as slow or slower relative to 1, indicating that the threading polyintercalation system better accommodates linkers that are too long as opposed to too short.
The modular nature of NDI-based threading polyintercalators was exploited in the design of 5, which appears to fully occupy a 22 bp binding site, a record for non-nucleic acid based DNA recognition by synthetic molecules. Although in an absolute sense the approximately 5 day half-life for 5 dissociating from its preferred sequence C can still be considered remarkably slow, it is considerably faster than predicted by previous studies with structurally analogous tetraintercalators. Future work, including structural studies, will investigate the reasons for the faster than predicted dissociation in an effort to create molecules that bind long sequences with even slower dissociation.
EXPERIMENTAL
Synthesis of 2 – 5
Solvents and starting materials were used without further purification. Organic chemicals and solvents were obtained from Sigma-Aldrich or Fisher, and resins and amino acids were obtained from Novabiochem. The Fmoc-Lys(Boc)-NDI-OH monomer, Fmoc-(β-Ala)3-OH, and Fmoc-(Gly)3-OH were all synthesized as previously described.21 Fmoc-based SPPS was performed as reported.25 Cross-linking on the resin was performed with pimelic acid for 2 and 5, suberic acid for 3, and azelaic acid for 4. Intercalators 2 and 4 were purified by reverse-phase preparative HPLC on a Waters system using a Vydac C18 column in a H2O/acetonitrile solvent system with 0.1% trifluoroacetic acid. Intercalators 3 and 5 were initially purified by ion exchange on an AKTA FPLC (GE) with a GE HiTrap SP FF cation exchange column in H2O with 0.2% formic acid eluting with NaCl. Salts were removed with a Waters Sep-Pak C18 cartridge. The desired fractions were lyophilized and further purified by HPLC as before. Intercalators were characterized by high-resolution electrospray ionization mass spectrometry. 2: 10% yield; HRMS-ESI C129H153N30O34 +3 [M+H]+3 calcd 888.70497, found 888.70554. 3: 2% yield; HRMS-ESI C130H154N30O34 +2 [M+H]+2 calcd 1339.56200, found 1339.56200. 4: 3% yield; HRMS-ESI C131H156N30O34 +2 [M+H]+2 calcd 1346.56946, found 1346.56819. 5: 2% yield, HRMS-ESI C191H221N46O52 +3 [M+H]+3 calcd 1330.20155, found 1330.20145.
Gel-Shift Assays
DNA was radiolabeled and gel-shift assays were performed as previously reported.21 All DNA was purchased from Integrated DNA Technologies (IDT) and purified by gel filtration with a PD MidiTrap G-25 cartridge (GE). For 2 – 4, 24-mer oligo A containing the 14 bp binding site was used, and for 5, 32-mer oligos B and C containing the respective 22 bp sequences of interest were used. Dissociations were followed for 5 halflives or until the radiolabel was no longer viable (about 65 days). The data was analyzed by ImageQuant TL v2005 and mono-exponential decay equation fits were obtained using KaleidaGraph v4.1 (Supporting Information).
DNase I Footprinting
Radiolabeled DNA used for footprinting experiments was obtained as previously reported21 (Supporting Information). The length of DNA used for footprinting with sequence A was 95 bp, and with sequences B and C was 103 bp. Incubations (20 mM sodium phosphate buffer, 2 mM MgCl2, pH = 7.5) of intercalators with radiolabeled DNA were performed by first heating the DNA to 80°C for 5 min, adding the appropriate concentration of intercalator, and allowing the incubation to cool at 0.5°C/min until reaching 25°C. Incubations then sat overnight at 25°C. The DNA was digested with 3.0 U/mL DNase I for 4 min, and fragments were separated on an 8% denaturing polyacrylamide gel until the bromophenol blue marker reached the end of the gel. The adenine-specific cleavage reaction was performed as previously described.34
Extinction Coefficient Determination
The concentration of a solution of 5 in H2O was determined by using NMR analysis. A known amount of a 1000 μg/mL standard solution of acetaldehyde in H2O (SPEX CertiPrep) was mixed with a solution of 5 in 9:1 H2O:D2O. The NMR sample was prepared to have a ratio of acetaldehyde:5 of approximately 54:1. The acetaldehyde formed a significant amount of hydrate,35 so the actual concentration of acetaldehyde was extrapolated from integration comparisons of the aldehyde proton quartet and methyl doublet to the corresponding peaks of the hydrate. Integration comparisons of the aldehyde proton quartet and all amide and aromatic protons in 5 (54 H) led to the determination of the concentration of the stock solution of 5, which was then used for extinction coefficient determination at λmax = 385 nm in water. For extinction coefficients of bound complexes, a 1:1 molar ratio of intercalator to oligo A or oligo C was incubated at 37 °C in 10 mM PIPES (pH = 7.0), 100 mM NaCl, and 1 mM EDTA.
Supplementary Material
Acknowledgments
This work was supported by the Robert A. Welch Foundation (F1188) and the National Institutes of Health (GM-069647). We thank Steven Sorey for his help with the 1H NMR spectra.
Footnotes
DNA sequence details for DNase I footprinting, dissociation plots. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
References
- 1.Yen SF, Gabbay EJ, Wilson WD. Biochemistry. 1982;21:2070–2076. doi: 10.1021/bi00538a014. [DOI] [PubMed] [Google Scholar]
- 2.Tanious FA, Yen SF, Wilson WD. Biochemistry. 1991;30:1813–1819. doi: 10.1021/bi00221a013. [DOI] [PubMed] [Google Scholar]
- 3.Duca M, Vekhoff P, Oussedik K, Halby L, Arimondo PB. Nucleic Acids Res. 2008;36:5123–5138. doi: 10.1093/nar/gkn493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen PE. ChemBioChem. 2010;11:2073–2076. doi: 10.1002/cbic.201000346. [DOI] [PubMed] [Google Scholar]
- 5.Corey DR. Int J Pept Res Ther. 2003;10:347–352. [Google Scholar]
- 6.Negi S, Imanishi M, Matsumoto M, Sugiura Y. Chem Eur J. 2008;14:3236–3249. doi: 10.1002/chem.200701320. [DOI] [PubMed] [Google Scholar]
- 7.(a) Dervan PB. Bioorgan Med Chem. 2001;9:2215–2235. doi: 10.1016/s0968-0896(01)00262-0. [DOI] [PubMed] [Google Scholar]; (b) Meier JL, Yu AS, Korf I, Segal DJ, Dervan PB. J Am Chem Soc. 2012;134:17814–17822. doi: 10.1021/ja308888c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Satam V, Babu B, Chavda S, Savagian M, Sjoholm R, Tzou S, Liu Y, Kiakos K, Lin S, David Wilson W, Hartley JA, Lee M. Bioorgan Med Chem. 2012;20:693–701. doi: 10.1016/j.bmc.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 9.Holman GG, Zewail-Foote M, Smith AR, Johnson KA, Iverson BL. Nat Chem. 2011;3:875–881. doi: 10.1038/nchem.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosaganov YN, Stetsenko DA, Lubyako EN, Kvitko NP, Lazurkin YS, Nielsen PE. Biochemistry. 2000;39:11742–11747. doi: 10.1021/bi0006417. [DOI] [PubMed] [Google Scholar]
- 11.Chaudhuri NC, Kool ET. J Am Chem Soc. 1995;117:10434–10442. doi: 10.1021/ja00147a004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson JR, Wang Y, Eng MP, Kann N, Lincoln P, Andersson J. Chem Eur J. 2013;19:6246–6256. doi: 10.1002/chem.201300483. [DOI] [PubMed] [Google Scholar]
- 13.Howell LA, Gulam R, Mueller A, O’Connell MA, Searcey M. Bioorg Med Chem Lett. 2010;20:6956–6959. doi: 10.1016/j.bmcl.2010.09.128. [DOI] [PubMed] [Google Scholar]
- 14.He Z, Bu X, Eleftheriou A, Zihlif M, Qing Z, Stewart BW, Wakelin LPG. Bioorgan Med Chem. 2008;16:4390–4400. doi: 10.1016/j.bmc.2008.02.063. [DOI] [PubMed] [Google Scholar]
- 15.Sato S, Takenaka S. Anal Sci. 2012;28:643–649. doi: 10.2116/analsci.28.643. [DOI] [PubMed] [Google Scholar]
- 16.Laugaa P, Markovits J, Delbarre A, Le Pecq JB, Roques BP. Biochemistry. 1985;24:5567–5575. doi: 10.1021/bi00341a042. [DOI] [PubMed] [Google Scholar]
- 17.Wirth M, Buchardt O, Koch T, Nielsen PE, Norden B. J Am Chem Soc. 1988;110:932–939. [Google Scholar]
- 18.Ueyama H, Takagi M, Takenaka S. Analyst. 2002;127:886–888. doi: 10.1039/b204019k. [DOI] [PubMed] [Google Scholar]
- 19.Murr MM, Harting MT, Guelev V, Ren J, Chaires JB, Iverson BL. Bioorgan Med Chem. 2001;9:1141–1148. doi: 10.1016/s0968-0896(00)00335-7. [DOI] [PubMed] [Google Scholar]
- 20.Far S, Kossanyi A, Verchére-Béaur C, Gresh N, Taillandier E, Perrée-Fauvet M. Eur J Org Chem. 2004;2004:1781–1797. [Google Scholar]
- 21.Rhoden Smith A, Ikkanda BA, Holman GG, Iverson BL. Biochemistry. 2012;51:4445–4452. doi: 10.1021/bi300317n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guelev V, Lee J, Ward J, Sorey S, Hoffman DW, Iverson BL. Chem Biol. 2001;8:415–425. doi: 10.1016/s1074-5521(01)00013-8. [DOI] [PubMed] [Google Scholar]
- 23.Guelev VM, Harting MT, Lokey RS, Iverson BL. Chem Biol. 2000;7:1–8. doi: 10.1016/s1074-5521(00)00007-x. [DOI] [PubMed] [Google Scholar]
- 24.Guelev V, Sorey S, Hoffman DW, Iverson BL. J Am Chem Soc. 2002;124:2864–2865. doi: 10.1021/ja016834e. [DOI] [PubMed] [Google Scholar]
- 25.Lee J, Guelev V, Sorey S, Hoffman DW, Iverson BL. J Am Chem Soc. 2004;126:14036–14042. doi: 10.1021/ja046335o. [DOI] [PubMed] [Google Scholar]
- 26.Kogan M, Nordén B, Lincoln P, Nordell P. ChemBioChem. 2011;12:2001–2006. doi: 10.1002/cbic.201100182. [DOI] [PubMed] [Google Scholar]
- 27.Anslyn EV, Dougherty DA. Modern Physical Organic Chemistry. University Science Books; Sausalito, CA: 2006. [Google Scholar]
- 28.Hampshire AJ, Rusling DA, Broughton-Head VJ, Fox KR. Methods. 2007;42:128–140. doi: 10.1016/j.ymeth.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 29.Lokey RS, Kwok Y, Guelev V, Pursell CJ, Hurley LH, Iverson BL. J Am Chem Soc. 1997;119:7202–7210. [Google Scholar]
- 30.Vasquez KM, Wang G, Havre PA, Glazer PM. Nucleic Acids Res. 1999;27:1176–1181. doi: 10.1093/nar/27.4.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aoki H, Tao H. Analyst. 2005;130:1478–1482. doi: 10.1039/b507121f. [DOI] [PubMed] [Google Scholar]
- 32.Lloyd A, Plaisier CL, Carroll D, Drews GN. P Natl Acad Sci USA. 2005;102:2232–2237. doi: 10.1073/pnas.0409339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hirata T, Nomura W, Imanishi M, Sugiura Y. Bioorg Med Chem Lett. 2005;15:2197–2201. doi: 10.1016/j.bmcl.2005.03.040. [DOI] [PubMed] [Google Scholar]
- 34.Iverson BL, Dervan PB. Nucleic Acids Res. 1987;15:7823–7830. doi: 10.1093/nar/15.19.7823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Socrates G. J Org Chem. 1969;34:2958–2961. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.