Abstract
Cholangiocarcinoma (CCA) cells paradoxically express the death ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) and thus rely on potent survival signals to circumvent cell death by TRAIL. Hedgehog (Hh) signaling is an important survival pathway in CCA. Herein, we further examine the mechanisms whereby Hh signaling mediates apoptosis resistance in CCA, revealing a pivotal role for the cell division regulating serine/threonine kinase polo-like kinase 2 (PLK2). We employed 50 human CCA samples (25 intrahepatic and 25 extrahepatic CCA) as well as human KMCH-1, Mz-CHA-1, and HUCCT-1 CCA cells for these studies. In vivo experiments were conducted using a syngeneic rat orthotopic CCA model. In human samples, polo-like kinase (PLK)1/2/3-immunoreactive cancer cells were present in the preponderance of intra- and extrahepatic CCA specimens. Inhibition of Hh signaling by cyclopamine reduced PLK2, but not PLK1 or PLK3, messenger RNA and protein expression in vehicle-treated and sonic Hh–treated CCA cells, confirming our previous microarray study. PLK2 regulation by Hh signaling appears to be direct, because the Hh transcription factors, glioma-associated oncogene 1 and 2, bind to the PLK2 promotor. Moreover, inhibition of PLK2 by the PLK inhibitor, BI 6727 (volasertib), or PLK2 knockdown was proapoptotic in CCA cells. BI 6727 administration or PLK2 knockdown decreased cellular protein levels of antiapoptotic myeloid cell leukemia 1 (Mcl-1), an effect reversed by the proteasome inhibitor, MG-132. Finally, BI 6727 administration reduced Mcl-1 protein expression in CCA cells, resulting in CCA cell apoptosis and tumor suppression in vivo.
Conclusion
PLK2 appears to be an important mediator of Hh survival signaling. These results suggest PLK inhibitors to be of therapeutic value for treatment of human CCA.
Cholangiocarcinoma (CCA) is the most common biliary cancer and its incidence is increasing in Western countries.1 CCA is a highly lethal malignancy with limited therapeutic options.2–4 Human CCA in vivo paradoxically express the death ligand tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) as well as its cognate receptors and are resistant to cell death by TRAIL.5–7 Thus, CCA cells most likely are dependent on potent survival signals. However, the mechanisms of CCA apoptosis resistance are complex, and further insight is needed to help develop more effective therapies.
CCAs are highly desmoplastic neoplasms with a tumor microenvironment plentiful in myofibroblasts (MFBs). We have recently reported that cross-talk between MFBs and CCA cells coactivates Hedgehog (Hh) signaling—an important survival pathway in CCA.8–10 Hh signaling is initiated by any of the following three ligands: indian; desert; or sonic Hh (SHH). These ligands bind to the Hh receptor, patched, resulting in activation of the Hh mediator, smoothened, and, subsequently, the transcription factors, glioma-associated oncogene (GLI)1, 2, and 3.11 SHH is expressed in CCA cells,8,12 and in a recent messenger RNA (mRNA) expression analysis employing CCA cells, Hh signaling was also suggested to positively regulate the cell-division–modulating enzyme, kinase polo-like kinase (PLK)2.8
PLK2 (or SNK) is one of five mammalian PLK family members that orchestrate a wide range of critical cell-cycle events.13–15 Besides PLK2, PLK1 (or STPK13), PLK3 (or CNK, FNK, and PRK), PLK4 (or SAK and STK18), and PLK5 have been identified. 14,15 All PLK proteins share a similar structure, with a canonical serine/threonine kinase domain at the N-terminus and a regulatory polo-box domain at the C-terminus13; however, PLK4 has a notably divergent structure, as compared to other PLK proteins and PLK5, because it lacks kinase activity.14,15 Approximately 80% of human cancers express high levels of PLK transcripts in tumor cells (these PLK transcripts are mostly absent in surrounding healthy tissues), and PLK overexpression is often associated with poor prognosis and lower overall survival.16 Though PLK1 has been extensively studied and has become an attractive candidate for anticancer drug development, the roles of the other PLK proteins, including PLK2, are less well understood.15
PLK inhibition in esophageal squamous cell carcinoma and osteosarcoma was reported to decrease protein levels of myeloid cell leukemia-1 (Mcl-1).17,18 This is of particular interest because Mcl-1, a potent antiapoptotic member of the B-cell lymphoma (Bcl)−2 protein family, has been identified as a survival factor in CCA.19–21 Given the pivotal role of Mcl-1 in mediating CCA resistance to TRAIL-induced apoptosis,19–21 PLK inhibition is a potential strategy for targeted treatment of this devastating disease.
The aim of this study was to examine the role for an Hh and PLK signaling coactivation network in mediating CCA cell resistance to TRAIL cytotoxicity. The results suggest that PLK2 mediates Hh survival signaling by inhibition of Mcl-1 proteasomal degradation, representing an important link between the Hh pathway and robust Mcl-1 expression in CCA cells. These observations have implications for treatment of human CCA.
Materials and Methods
Materials
Recombinant human (rh)SHH, rhTRAIL, rhPDGF-BB (platelet-derived growth factor BB; all from R&D Systems, Minneapolis, MN), MG-132 (Merck, Rockland, MA), cyclopamine (LC Laboratories, Woburn, MA), and GDC-0449 (Selleck, Houston, TX) were prepared according to the suppliers’ protocols. BI 6727/volasertib, a potent selective PLK inhibitor,22 was purchased from Active Biochem (Maplewood, NJ), dissolved in dimethyl sulfoxide (1 mmol/L stock solution; Sigma-Aldrich, St. Louis, MO), and subsequently diluted in cell-culture medium for use in in vitro experiments. The SHH-neutralizing antibody (Ab), 5E1, was obtained from the Developmental Studies Hybridoma Bank (Department of Biology, University of Iowa, Iowa City, IA). The construct encoding for S peptide-tagged human Mcl-1 mutant resistant to proteasomal degradation as a result of sequential mutagenesis of the established Mcl-1 ubiquitination sites (amino acids 5, 40, 136, 194, and 197) from lysine to arginine was generated as previously described.23
Cell Lines/Culture and Human Samples
The human CCA cell lines, KMCH-1, Mz-CHA-1, and HUCCT-1, as well as the erythroblastic leukemia viral oncogene homolog (ErbB-2)/neu transformed malignant rat cholangiocyte cell line, BDEneu (in vivo experiment), were cultured as previously described.8,24–26 Intra-(n=25) and extrahepatic (n=25) human CCA samples were collected with institutional review board approval according to the principles embodied in the Declaration of Helsinki.
Generation of a Stable Transfectant Expressing PLK1, PLK2, or PLK3 Short Hairpin RNA
Short hairpin RNA (shRNA) lentiviral plasmids for PLK1 and PLK3 were obtained from Thermo Fisher Scientific/ Open Biosystems (Oligo ID: V2LHS_241437, Gen-Bank accession no.: NM_005030 and Oligo ID: V2LHS_172853, GenBank accession no.: NM_004073, respectively; Huntsville, AL). PLK2 shRNA lentiviral plasmids were obtained from Sigma-Aldrich (GenBank accession no.: NM_006622.2). KMCH-1 cells were transfected using OptiMEM I (Gibco-Invitrogen, Carlsbad, CA) containing 6 μL/mL of Lipofectamine (Invitrogen), 1 μg/mL of plasmid DNA, and 6 μL/mL of Plus reagent (Invitrogen). Forty-eight hours after transfection, fresh Dulbecco’s modified Eagle’s medium, containing 0.5 μg/mL of puromycin, was added. Surviving clones were separated using cloning rings and individually cultured. A clone with a scrambled shRNA was employed as a control (stable scrambled KMCH-1 cells). Expression/knockdown of PLK1, PLK2, or PLK3 in clones was assessed by immunoblotting analysis.
Real-Time Polymerase Chain Reaction
Total RNA was extracted from cells or tissue using the RNeasy Plus Mini Kit (Qiagen, Hilden, Germany) and was reverse-transcribed with Moloney leukemia virus reverse transcriptase (RT) and random primers (Invitrogen, Camarillo, CA). Quantitation of the complementary DNA template was performed with real-time polymerase chain reaction (PCR; LightCycler; Roche, Indianapolis, IN) using SYBR green (Roche) as a fluorophore. 19 Oligonucleotide sequences and expected product sizes for primer pairs used for quantitative RT-PCR analysis are depicted in Supporting Table 1. As an internal control, primers for 18S ribosomal RNA (rRNA; Ambion, Austin, TX) were employed. Using gel-purified amplicons, a standard curve was generated to calculate the copy number/μL. Target mRNA expression of each sample was calculated as the copy ratio of target mRNA to 18S rRNA and then normalized to the target mRNA expression of vehicle controls.
Immunoblotting Analysis
Whole cell lysates were obtained and processed as previously described.18 Primary antisera used were actin (1:2,000; C-11; Santa Cruz Biotechnology, Santa Cruz, CA), PLK1 (1 μg/mL; catalog no.: 05–844; Merck Millipore, Darmstadt, Germany), PLK2 (1 μg/mL; ab34811; Abcam, Cambridge, MA), PLK3 (1:1,000; catalog no.: D14F12; Cell Signaling Technology, Danvers, MA), Mcl-1 (1:1,000; sc-819; Santa Cruz Biotechnology), and Bcl-2 (1:1,000; sc-492; Santa Cruz Biotechnology). The mouse anti-S peptide Ab was a generous gift from S.H. Kaufmann (Oncology Research, Mayo Clinic, Rochester, MN). Horseradish-peroxidase–conjugated secondary Abs for rabbit (sc-2004; Santa Cruz Biotechnology), goat (sc-2020; Santa Cruz Biotechnology), and mouse (sc-2031; Santa Cruz Biotechnology) were incubated at a dilution of 1:2,000 for 1 hour at room temperature. Proteins were visualized using enhanced chemiluminescence reagents (ECL/ECL-Prime; Amersham Biosciences, Buckinghamshire, UK) and Kodak X-OMAT films (Eastman Kodak Company, Rochester, NY).
Quantitation of Apoptosis
Apoptosis in CCA cells was quantified by assessing characteristic nuclear changes of apoptosis after staining with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma-Aldrich) using fluorescence microscopy.27 Terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assays (cell coculture and rat liver samples) were carried out using the In Situ Cell Death Detection Kit (Roche), according to the supplier’s protocol and as previously described.8 Caspase-3/7 activity was quantitated using the ApoONE Homogenous Caspase-3/-7 Assay (Promega, Madison, WI), according to manufacturer’s recommendations.27
Animal Experiments
All animal studies were performed in accord with and approved by the local institutional animal care and use committee. In vivo intrahepatic cell implantation was carried out in male adult Fischer 344 rats (Harlan, Indianapolis, IN) with initial body weights of 190–220 (cyclopamine study) and 227–251 g (BI 6727 study), as previously described.8,24–26 Animals were treated with vehicle (cyclopamine), as previously described,8 or BI 6727 (3 injections of 10 mg/kg body weight [b.w.; 0.5 mL] intraperitoneally [IP] every other day; the first injection was given on postoperative day 7, and the third injection was given on postoperative day 11) formulated in hydrochloric acid (0.1 N) diluted with 0.9% NaCl.22 Twenty-four (cyclopamine study) and forty-eight hours (BI 6727 study) after receiving the last injection, rats were euthanized and livers were removed for further analysis. To assess the number of metastases-free and metastases-bearing rats, abdominal cavities, retroperitoneal spaces, and thoracic cavities were thoroughly examined as previously described.8,24,25
Statistical Analysis
Data are expressed as the mean±standard error of the mean (SEM) and represent at least three independent experiments. Differences in experiments with two groups were compared using the two-tailed Student t test or the chi-square test (analysis of metastasis) as well as Mann Whitney’s test (analysis of PLK1/2/3 expression in human CCA samples). Differences in experiments with more than two groups were compared using analysis of variance with Bonferroni’s post-hoc correction. Differences were considered as significant at levels of P<0.05.
Supplemental Methods
Chromatin immunoprecipitation (ChIP), immunohistochemistry (IHC) for PLK1, PLK2, and PLK3, as well as immunofluorescence (IF) microscopy for PLK2, Mcl-1, and cytokeratin 7 (CK-7) are described in the Supporting Materials.
Results
PLK1, PLK2, and PLK3 Are Prominently Expressed in Human CCA
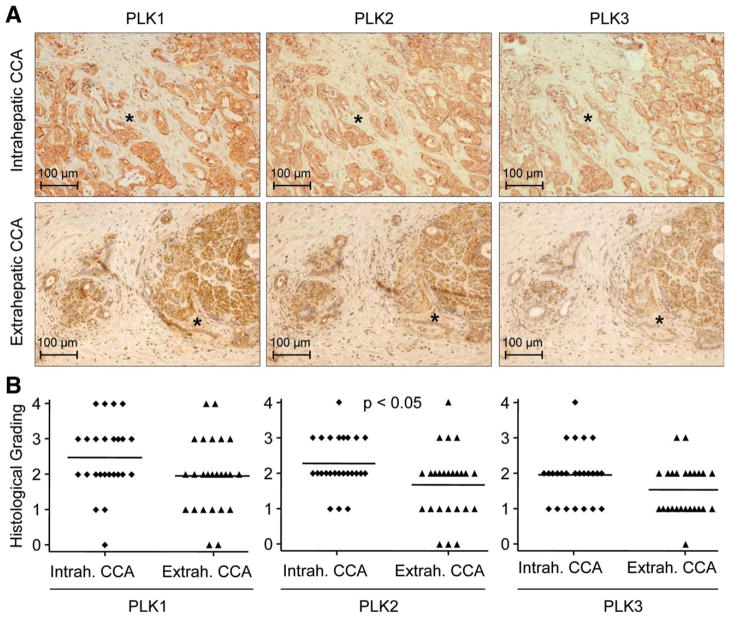
Initially, we examined the expression of PLK1, PLK2, and PLK3 (because of their structural and functional divergences from other PLK family members, PLK4 and PLK5 were excluded from this study14,15) in 50 CCA samples (25 intrahepatic and 25 extrahepatic CCA) by IHC (Fig. 1). PLK1, PLK2, and PLK3 immunoreactive cells were present in the preponderance of the intrahepatic and extrahepatic CCA specimens (Fig. 1A). Histological grading revealed that in extrahepatic and intrahepatic CCA samples, PLK2 was slightly less expressed than PLK1, whereas more cells stained positive for PLK2 than for PLK3 (Fig. 1B and Supporting Fig. 1). PLK1, PLK2, and PLK3 expression was more pronounced in intrahepatic, as compared to extrahepatic, CCA, and for PLK2 expression, this difference was statistically significant (Fig. 1B). In intrahepatic and extrahepatic CCA, predominantly cancer cells/glands stained positive for PLK proteins; however, some stromal cells within the tumor microenvironment also displayed PLK1, PLK2, and PLK3 immunoreactivity. Moreover, PLK2 mRNA was overexpressed in CCA, as compared to paired normal liver tissue from the same patients (Supporting Fig. 2). Thus, PLK1, PLK2, and PLK3 are abundantly expressed in the majority of human CCA specimens.
Fig. 1.
PLK proteins are abundantly expressed in human CCA. (A) PLK1 (left), PLK2 (middle), and PLK3 (right) expression in 25 intrahepatic (upper) and 25 extrahepatic (lower) human CCA samples was examined by IHC. Photomicrographs were taken in 200× magnification. Asterisks indicate similar positions within tumors because PLK IHC was performed on neighboring slides. Note that, predominantly, tumorous glands, but also some stromal cells, exhibit PLK immunoreactivity (brown). Counterstaining was performed with hematoxylin (blue). (B) PLK1/2/3 protein expression quantitation of intra-c and extrahepatic CCA samples by histological grading (grade 0=no protein expression; grade 4=high protein expression; examples of representative grade 0–4 CCA samples are given in Supporting Fig. 1).
PLK2 Is Regulated by Hedgehog Signaling in Human CCA Cells
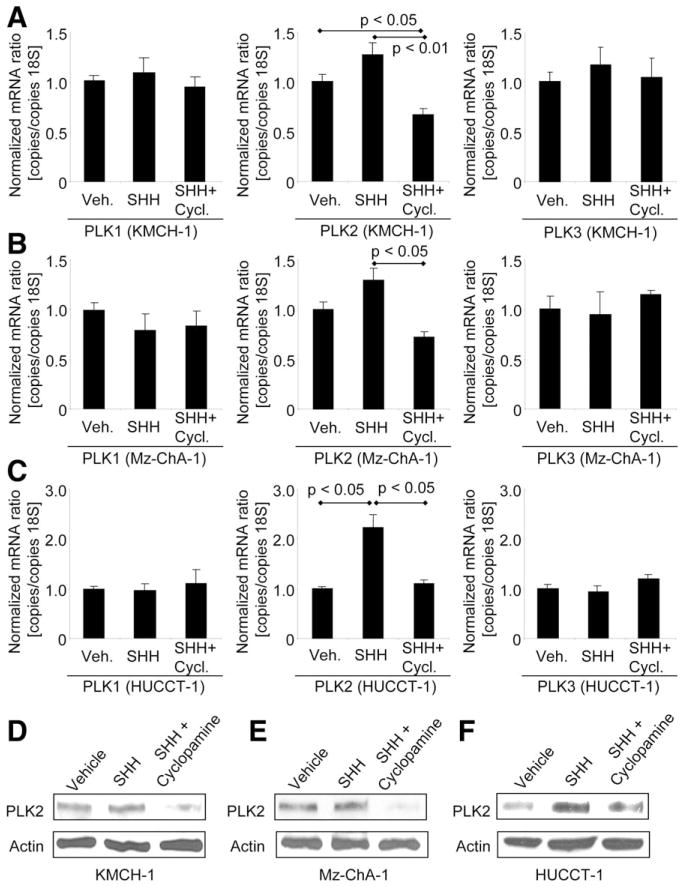
Having confirmed an abundant expression of PLK1, 2, and 3 proteins in human CCA, we next examined whether PLK signaling can be regulated by the Hh survival pathway, as implicated by a microarray expression analysis in CCA cell lines.8 Consistent with this previous study,8 inhibition of Hh signaling with cyclopamine (an inhibitor of the Hh mediator, smoothened) reduced PLK2, but not PLK1 or PLK3, mRNA expression, as compared to SHH-only–treated CCA cells (Fig. 2A [KMCH-1 cells], 2B [Mz-ChA-1 cells], and 2C [HUCCT-1 cells]). This observation was confirmed, on the protein level, by immunoblotting analysis for PLK1, 2, and 3 protein expression in similarly treated CCA cells (Fig. 2D [KMCH-1 cells], 2E [Mz-ChA-1 cells], and 2F [HUCCT-1 cells]) for PLK2 protein expression; PLK1 and PLK3 protein expression of all three CCA cell lines are depicted in Supporting Fig. 3A. Consistent with these findings, more specific Hh signaling inhibition with SHH-neutralizing Ab 5E1 or the small molecule, smoothened, inhibitor, GDC-0449, also reduced PLK2 protein expression, as compared to SHH-only–treated KMCH-1, MzChA-1, and HUCCT-1 cells (Supporting Fig. 3B). Given the positive regulation of Hh signaling by PDGF-BB in CCA,8 we also assessed the effect of PDGF-BB on PLK2 protein expression. Indeed, PLK2 protein levels increased in the presence of PDGF-BB (Supporting Fig. 3C). Thus, PLK2, but not PLK1 or PLK3, appears to be positively regulated by Hh signaling.
Fig. 2.
Hh-signaling inhibition reduces PLK2 expression in CCA cells. Cells were treated as indicated in serum-free medium with vehicle or rhSHH (500 ng/mL, 4 hours) in the presence or absence of Hh-signaling inhibitor cyclopamine (10 μM, 4 hours). (A–C) Quantitative RT-PCR analysis for PLK1/2/3 mRNA expression was performed in the human CCA cell lines, KMCH-1 (A), Mz-ChA-1 (B), and HUCCT-1 (C). Mean±SEM; n=3. (D and E) Treatment of KMCH-1 (D), Mz-ChA-1 (E), and HUCCT-1 (F) cells was followed by immunoblotting analysis for PLK2 protein expression (actin=loading control).
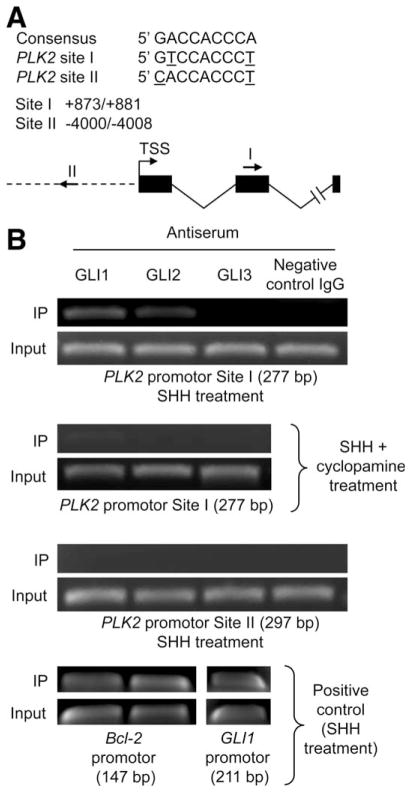
To further investigate how Hh signaling promotes PLK2 expression, we examined whether the hedgehog transcription factors, GLI1, GLI2, and/or GLI3, bind to the PLK2 promoter. After identification of two putative GLI-binding sites (sites I and II) within the PLK2 promoter region containing two mismatches, compared to the consensus sequence, GACCACCCA28 (Fig. 3A), we performed a ChIP assay in SHH-stimulated KMCH-1 cells (with or without cyclopamine) employing Abs to GLI1, GLI2, and GLI3. Indeed, binding of GLI1 and GLI2 was exclusively observed in SHH-only–treated cells using primers that amplify site I (277 base pairs [bp]; Fig. 3B, first and second panels), but not site II (297 bp; Fig. 3B, third panel). A control immunoglobulin G (IgG) did not yield a product, additionally demonstrating specificity of the antisera used. As a positive control, we employed the Bcl-2 promoter, which contains well-recognized GLI1/2-binding sites,29 and the GLI1 promotor, which contains well-recognized GLI3 binding sites.30 These studies identified the expected occupation by GLI1 and GLI2 (Bcl-2 promoter: 147 bp) as well as GLI3 (GLI1 promoter: 211 bp; Fig. 3B, forth panel). Taken together, these data suggest that Hh signaling may directly regulate PLK2 expression by binding of the GLI1 and GLI2 transcription factors to the PLK2 promoter.
Fig. 3.
GLI1 and GLI2 bind to a predicted GLI-binding site in the PLK2 promoter region. (A) Two putative GLI-binding sites that contain two mismatches, compared to the consensus sequence, were identified in the PLK2 promoter region. Nucleotide positions were counted from the transcription start site (TSS). Positions and directions of both potential binding sites are illustrated by arrows (I and II). (B) KMCH-1 cells treated with rhSHH (500 ng/mL, 5 hours)±cyclopamine (10μM, 5 hours) were employed for this study. ChIP using antiserum to GLI1, GLI2, and GLI3 or a sheep negative control IgG was performed, followed by PCR using primer flanking site I (277 bp) or II (297 bp) within the PLK2 promoter region. As positive controls, ChIP was performed using primers flanking the Bcl-2 promoter GLI-binding sites (GLI1 and GLI2; 147 bp) or primers flanking the GLI3-binding site within the GLI1 promoter (211 bp).
PLK2 Inhibition Is Proapoptotic in CCA Cells
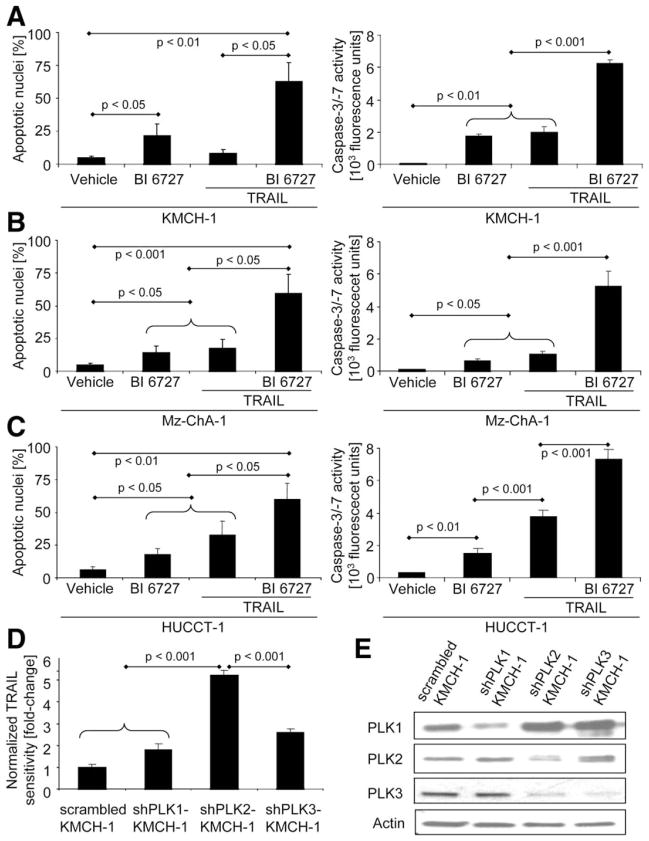
Next, we examined whether inhibition of Hh target gene PLK2 with the potent PLK inhibitor, BI 6727 (volasertib), promotes (TRAIL-induced) apoptosis in CCA cells. As measured by either cell morphology (Fig. 4AC, left) or biochemically (Fig. 4A–C, right), BI 6727 and, especially, BI 6727 plus TRAIL significantly induced apoptosis in KMCH-1 (Fig. 4A), Mz-ChA-1 (Fig. 4B), as well as HUCCT-1 (Fig. 4C) cells. Because BI 6727 not only inhibits PLK2, but also PLK1 and PLK3,22 we selectively silenced PLK1, PLK2, or PLK3 in KMCH-1 cells by an shRNA technique to more specifically investigate the effect of PLK inhibition on CCA cell apoptosis (Fig. 4D; knockdown of PLKs was confirmed by immunoblotting analysis [Fig. 4E]). Stable knockdown of PLK2 sensitized KMCH-1 to TRAIL-induced apoptosis, as compared to control KMCH-1 cells (Fig. 4D). Knocking down PLK1 and PLK3 as well was also proapoptotic; however, this effect was significantly less pronounced (Fig. 4D). Thus, PLK2 can mediate Hh cytoprotection against TRAIL-induced apoptosis in CCA cells.
Fig. 4.
PLK2 inhibition is proapoptotic in CCA cells. (A–C) KMCH-1 (A), Mz-ChA-1 (B), and HUCCT-1 (C) cells were treated as indicated with vehicle, PLK inhibitor BI 6727 (200 nM, 24 hours), rhTRAIL (2.5 ng/mL, 8 hours), or BI 6727 (200 nM, 24 hours) plus rhTRAIL (2.5 ng/mL, 8 hours). Apoptosis was measured by DAPI staining with quantitation of apoptotic nuclei by fluorescence microscopy (left; mean±SEM; n=3) or fluorescent analysis of caspase-3/-7 activity (right; mean±SEM; n=5). (D) Stable scrambled, shPLK1-, shPLK2-, and shPLK3-KMCH-1 cells were treated with vehicle or rhTRAIL (2.5 ng/mL, 8 hours), and apoptosis was measured by DAPI staining with quantitation of apoptotic nuclei by fluorescence microscopy. Sensitivity to TRAIL (apoptosis ratio TRAIL-treated versus vehicle-treated cells) was normalized to stable scrambled KMCH cells (A; mean±SEM; n=3). (E) PLK1/2/3 expression/knockdown in stable scrambled, shPLK1-, shPLK2-, and shPLK3-KMCH-1 cells was assessed by immunoblotting analysis (actin=loading control). Note that stable PLK3 knockdown also affects PLK2 expression.
Proapoptotic Effects of PLK2 Inhibition Are Mediated by Mcl-1 Degradation
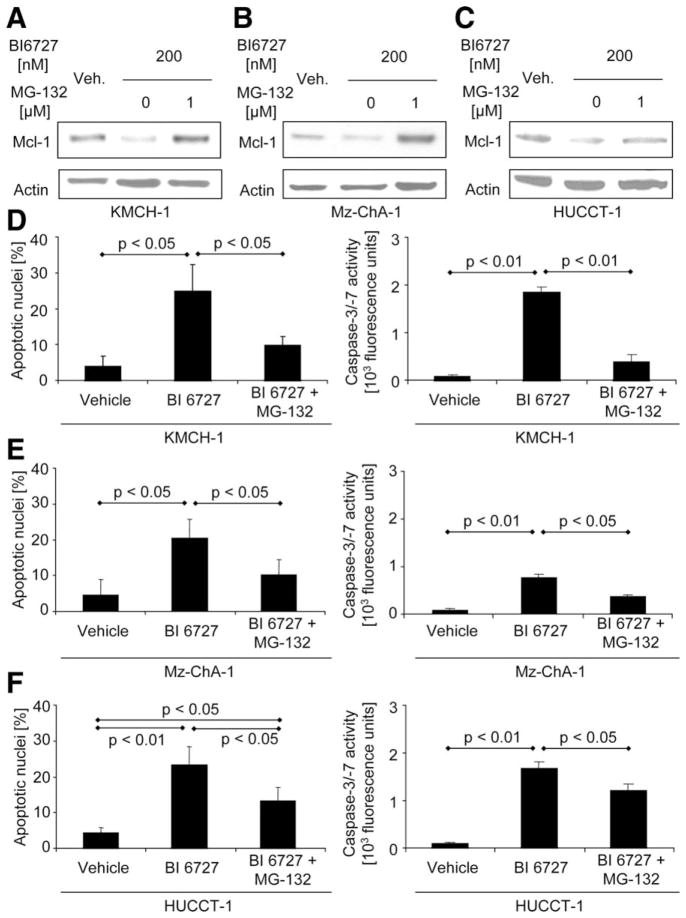
We next sought to investigate the mechanism whereby PLK2 inhibition exerts its proapoptotic effects. Because cell-cycle enzymes can regulate Mcl-1 expression31 and Mcl-1 as well as Bcl-2 are down-regulated by PLK inhibition in osteosarcoma and esophageal carcinoma cells,17,18 we explored the effect of PLK inhibition on cellular Mcl-1 and Bcl-2 protein levels. Indeed, BI 6727 administration rapidly decreased Mcl-1 (but not Bcl-2) protein levels in KMCH-1 cells (Supporting Fig. 4A). To assess whether decreased Mcl-1 protein levels are the result of proteasomal degradation, we employed the potent proteasome inhibitor, MG-132, or transfected KMCH-1 cells with a proteasome-resistant Mcl-1 mutant. Mcl-1 down-regulation by BI 6727 is likely mediated by proteasomal degradation, because MG-132 (Fig. 5A [KMCH-1 cells], 5B [Mz-ChA-1 cells], and 5C [HUCCT-1 cells]) or transfection with the proteasome-resistant Mcl-1 mutant (Supporting Fig. 4B) completely abrogated this effect. Moreover, inhibition of proteasomal degradation in the presence of BI 6727 increased Mcl-1 protein levels, as compared to vehicle controls (Fig. 5A,B and Supporting Fig. 4B), suggesting that Mcl-1 up-regulation is an initial protective cell response to BI 6727 (which, in BI 6727-only–treated cells, is overwhelmed by proteasomal Mcl-1 degradation). Accordingly, BI 6727-treated KMCH-1 cells display a compensatory up-regulation of Mcl-1 mRNA levels (Supporting Fig. 5A). Consistent with these observations, MG-132 also blocked BI 6727-induced apoptosis in KMCH-1 (Fig. 5D), Mz-ChA-1 (Fig. 5E), and HUCCT-1 (Fig. 5F) cells, as assessed morphologically (Fig. 5D–F, left) or biochemically (Fig. 5D–F, right). Moreover, stable knockdown of PLK2 by the shRNA technique also reduced Mcl-1 protein levels, as displayed by immunoblotting analysis (Supporting Fig. 4C). These data suggest that PLK2 inhibition reduces Mcl-1 protein levels in a posttranslational manner by proteasomal degradation, thereby promoting apoptosis.
Fig. 5.
Proapoptotic effects of PLK2 inhibition are mediated by Mcl-1 degradation. (A–C) KMCH-1 (A), Mz-ChA-1 (B), and HUCCT-1 (C) cells were treated as indicated with vehicle or BI 6727 (200 nM, 24 hours) in the absence or presence of the potent proteasome inhibitor, MG-132 (1 μM, 24 hours), followed by immunoblotting analysis for Mcl-1 protein expression (actin=loading control). (D–F) KMCH-1 (D), Mz-ChA-1 (E), and HUCCT-1 (F) cells were treated as indicated with vehicle or BI 6727 (200 nM, 24 hours) in the absence or presence of MG-132 (1 μM, 24 hours). Apoptosis was measured by DAPI staining with quantitation of apoptotic nuclei by fluorescence microscopy (left; mean±SEM; n=3) or fluorescent analysis of caspase-3/-7 activity (right; mean±SEM; n=5).
PLK Inhibition Promotes CCA Cell Apoptosis and Is Tumor Suppressive In Vivo
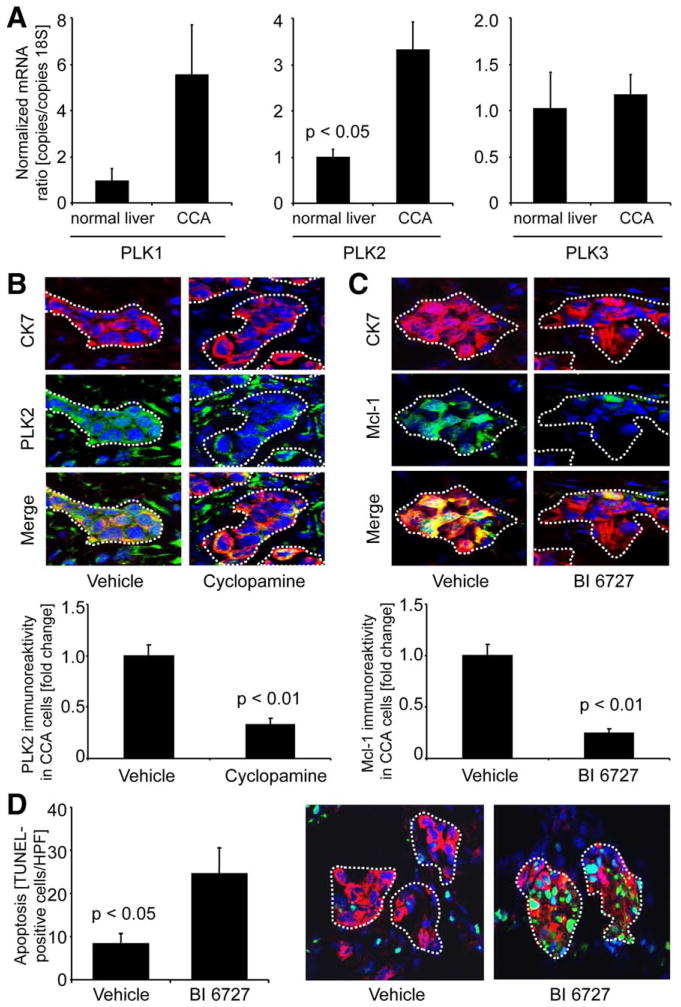
To determine whether the proapoptotic in vitro effect of PLK-signaling inhibition by BI 6727 is translatable to an in vivo model, we employed a syngeneic rat orthotopic CCA model (BDEneu malignant cells injected into liver of male Fischer 344 rats).3,8,24,26 This preclinical rodent model of CCA recapitulates many characteristic features of human CCA, including the paradoxical expression of TRAIL.24 First, we sought to further validate this model by assessing mRNA expression of PLK1–3 in CCA specimens, as compared to normal rat liver tissue, by quantitative RT-PCR. PLK2 mRNA levels in CCA were significantly higher, as compared to normal rat livers (Fig. 6A). On the protein level, PLK2, similar to the human disease (Fig. 1), was abundantly expressed in tumorous glands (identified by CK-7 costaining; CK-7 is a biliary epithelial cell marker expressed by CCA cells) and also by occasional stromal cells within the tumor microenvironment (Fig. 6B). Administration of Hh inhibitor cyclopamine significantly decreased PLK2 expression in tumorous glands (dotted lines), but not stromal cells, in this in vivo CCA model (Fig. 6B, middle photomicrographs and bar graph). Also consistent with our in vitro observations, treatment with PLK inhibitor BI 6727 in vivo reduced Mcl-1 protein expression in CCA cells (Fig. 6C, middle photomicrographs and bar graph). PLK2 mRNA levels were also increased in CCA specimens of BI 6727-treated rats, as compared to controls (Supporting Fig. 5B). Finally, CCA cell apoptosis was increased in animals treated with BI 6727, as compared to vehicle-treated rats (Fig. 6D; CCA cell apoptosis was confirmed by demonstrating colocalization of TUNEL-positive cell nests with tumorous glands displaying CK-7). Thus, this preclinical rodent model of CCA recapitulates characteristic features observed in human CCA tissue and in vitro studies, including the proapoptotic effect of PLK inhibition on CCA cells.
Fig. 6.
Effects of Hh and PLK inhibition are recapitulated in vivo. A syngeneic rat orthotopic CCA model (BDEneu cells; Fischer 344 rats) was employed for this study. (A) CCA and normal liver specimens of untreated rats were analyzed for mRNA expression of PLK1 (left), PLK2 (middle), and PLK3 (right) by quantitative RT-PCR. Mean±SEM; n=3. (B) CCA specimens of cyclopamine-treated (2.5 mg/kg b.w. IP daily for 1 week; first injection: postoperative day 7; seventh injection: postoperative day 13) or vehicle-treated rats were analyzed for PLK2 (green) expression of tumor cells (identified by costaining for CCA cell marker CK-7; red) by IF microscopy. Merged images depict colocalized CK-7/PLK2 protein expression in yellow (green-red overlay). PLK2 immunoreactivity in CCA cells was quantitated using the software, ImageJ 1.44o (B lower; mean±SEM, n=7). (C) CCA specimens of BI 6727-treated (3 injections of 10 mg/kg b.w. IP every other day; first injection: postoperative day 7; third injection: postoperative day 11) or vehicle-treated rats were analyzed for Mcl-1 (green) expression of CCA cells (similar CK-7 costaining and quantitation [mean±SEM; n=9] as in B). (D) Apoptotic nuclei were assessed in CCA samples of vehicle-treated (left photomicrograph) and BI 6727-treated (right photomicrograph) rats by TUNEL staining (green), and the identity of TUNEL-positive cells was confirmed by CK-7 costaining (red). Quantitation of TUNEL-positive cells (expressed as number per high power field [HPF], D left) demonstrates that in BI 6727-treated animals, CCA cell apoptosis was increased, as compared to controls (mean±SEM; n=9). In all photomicrographs, nuclei are counterstained with DAPI (blue) and CCA glands within the tumor stroma are illustrated by white dotted lines.
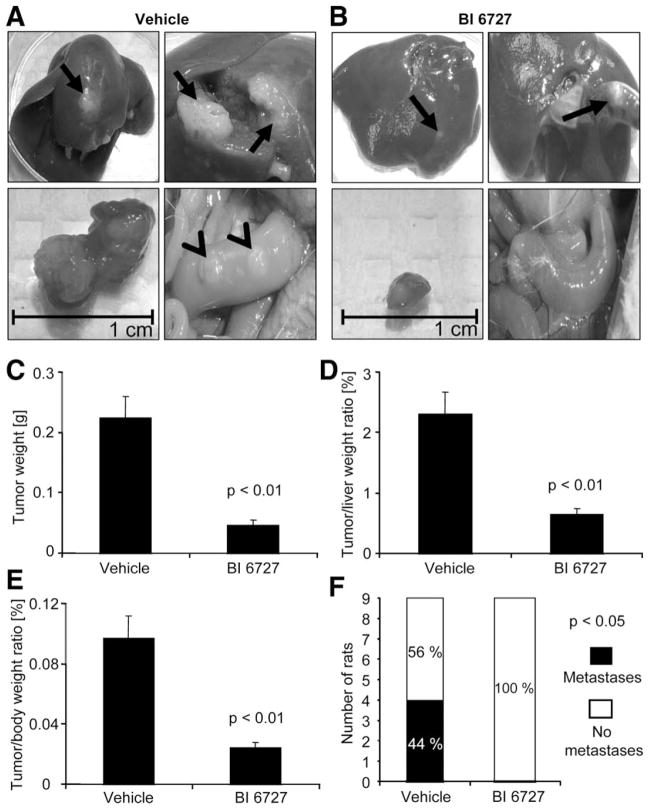
Consistent with the proapoptotic effects of PLK2 observed in the CCA in vivo model, PLK2 was also effective in reducing tumor size and metastasis (Fig. 7A–F). Indeed, tumor weight and tumor/liver weight ratios were significantly decreased in BI 6727-treated rats (Fig. 7A–E). Additionally, 100% of rats treated with BI 6727 displayed no extrahepatic metastases, whereas only 56% of vehicle-treated animals were free of metastases (4 of 9 animals in this group showed tumors predominantly occurring in the greater omentum and peritoneum; Fig. 7A [lower right photomicrograph] and F). Taken together, these data suggest that BI 6727 decreases tumor growth as well as metastasis in an in vivo rodent model of CCA.
Fig. 7.
PLK inhibition is tumor suppressive in vivo. A syngeneic rat orthotopic model of CCA (BDEneu cells; Fischer 344 rats) was employed for this study. In BI 6727-treated (3 injections of 10 mg/kg b.w. IPy every other day; first injection: postoperative day 7; third injection: postoperative day 11) or vehicle-treated rats tumor/liver/body weight and extrahepatic metastasis were assessed 13 days after tumor cell implantation into the left lateral liver lobe. (A and B) Representative photomicrographs display explanted livers (left upper), gross pathological tumor appearance within livers (right upper), extirpated tumors with scale (left lower), and abdominal cavity (right lower) of vehicle-treated (A) or BI 6727-treated (B) rats (arrows indicate liver tumors and arrowheads extrahepatic metastases). (C–E) Changes in tumor weight (C), tumor/liver weight ratios (D), and tumor/body weight ratios (E) are depicted as bar graphs. Mean±SEM; n=9. (F) Stacked column plot indicates numbers of animals with and without extrahepatic metastases for vehicle-treated and BI 6727-treated groups (P<0.05 by chi-square test).
Discussion
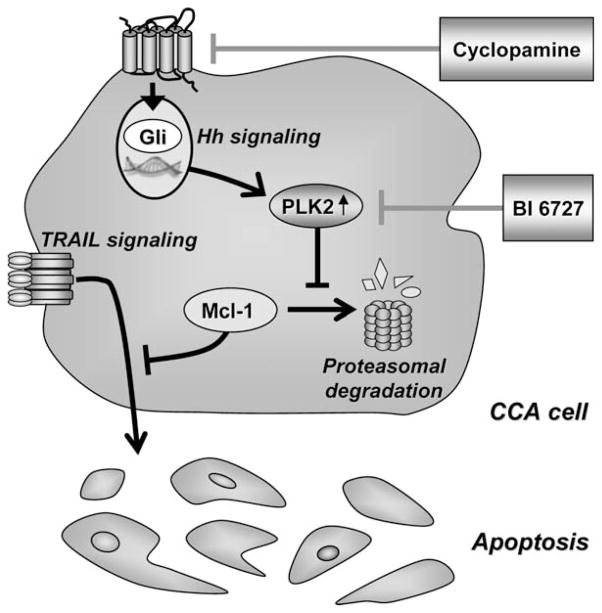
This study provides new mechanistic insights regarding an Hh-signaling and PLK-signaling coactivation network in CCA. These data indicate that (1) Hh signaling directly regulates PLK2 mRNA and protein expression, (2) PLK2 promotes Mcl-1 stabilization, providing resistance to cell death by TRAIL, and (3) PLK inhibition is tumor suppressive in an orthotopic syngeneic rodent in vivo CCA model. These findings are illustrated in Fig. 8 and discussed in greater detail below.
Fig. 8.
Schematic diagram illustrating the role of Hh and PLK2 signaling in promoting CCA cell resistance to endogenous TRAIL cytotoxicity. Hh or PLK inhibitors, such as cyclopamine or BI 6727, block Hh/ PLK2 survival signaling and thus promote apoptosis in CCA cells.
We recently reported that MFB-to-cancer cell paracrine signaling imparts survival signals for CCA by coactivation of the Hh-signaling pathway.8 Indeed, Hh signaling inhibition by cyclopamine increased susceptibility of CCA cells to apoptotic stimuli.8 In the same study, a microarray mRNA expression analysis also suggested that Hh signaling positively regulates cell-cycle enzyme PLK2 in CCA cells.8 Therefore, we further examined PLK2 regulation by Hh signaling and explored whether targeting of PLK2 signaling would be similarly effective in restoring CCA cell susceptibility to TRAIL-induced apoptosis. Because cancer cells frequently develop resistance to smoothened inhibitors, 32,33 blocking PLK2 may be a promising new therapeutic approach for targeting this cancer survival pathway downstream of smoothened. In vitro, PLK2 was found to be directly regulated by the Hh pathway. PLK2 cytoprotection against TRAIL-induced apoptosis was abrogated in the presence of PLK inhibitor BI 6727 or when PLK2 was selectively knocked down in KMCH-1 cells. Moreover, PLK2-mediated cytoprotection was, at least in part, a result of stabilization of Mcl-1, a crucial survival factor for CCA.19–21 These observations suggest that PLK2 represents an important link between Hh survival signaling and Mcl-1 protein expression in CCA cells. Thus, PLK inhibition might be a promising strategy for treatment of CCA.
The effect of PLK inhibition on CCA cell viability has also been investigated in a single earlier study.34 In their in vitro study, Thrum et al. reported that PLK inhibition reduces CCA cell proliferation. Herein, we expand the effects of PLK inhibition by demonstrating that it also is proapoptotic in vitro and in vivo. Moreover, we demonstrate that selective PLK2 inhibition has a more pronounced proapoptotic effect than selective PLK1 or PLK3 inhibition in CCA cells. Other than PLK2 or PLK3, PLK1 has been intensively studied and is also a potential target for anticancer therapy in other cell types.15 The roles of PLK2 and PLK3 are less well understood, and studies in hematologic diseases assume that PLK2 and PLK3 act as tumor suppressors through their interactions with p53 signaling.15,35 Consistent with these observations, PLK2 and PLK3 are minimally expressed in various human tumors.36 However, in CCA cells, PLK2 (and, to a lesser extent, PLK3) appears to have a cytoprotective function, which likely explain their abundant expression in human and rodent CCA specimens (Figs. 1 and 6B). As with MET, nuclear factor kappa B, β-cantenin, c-Jun N-terminal kinase, src homology 2 domain-containing tyrosine phosphatase 2, and signal transducer and activator of transcription 3, PLK2 (and PLK3) thus might belong to the emerging group of molecules that seem to have conflicting tumor-suppressive and oncogenic roles in carcinogenesis, depending on tumor type and state of tumor development.37
The orthotopic, syngeneic rodent CCA model used in the present study recapitulates the molecular signature and TRAIL expression of human CCA.24,26 It also provides a syngeneic tumor microenvironment, avoiding problems of immunocompromise and tumor stromal incompatibilities problematic in human xenograft models and mimics the cellular expression patterns of PLK2 found in the human disease. Moreover, BDEneu CCA cells injected into rat livers express all relevant Hh-signaling pathway factors.8 In an earlier study employing this in vivo model, Hh-signaling inhibition with smoothened inhibitor cyclopamine was reported to induce CCA cell death and inhibit tumor growth. In the present study, we extend these earlier observations by demonstrating that Hh inhibition also reduces PLK2 protein expression in vivo. In addition, we show that PLK inhibition leads to a decrease in Mcl-1 protein expression in CCA cells, also resulting in CCA cell apoptosis and reduced tumor growth in vivo. Our in vitro observations are most consistent with the proapoptotic and tumor-suppressive in vivo effects of BI 6727 mainly being mediated by PLK2 inhibition; however, we cannot exclude a contribution of PLK1 and PLK3 inhibition to the effects noted in BI 6727-treated animals.
In conclusion, targeting the Hh-signaling and PLK-signaling coactivation network appears to sensitize CCA cells to TRAIL-induced apoptosis. In addition to Hh inhibitors, these observations suggest PLK inhibitors to be of therapeutic value for treatment of human CCA.
Supplementary Material
Acknowledgments
The superb secretarial and technical service of Courtney Hoover and Lena Wingerter is gratefully acknowledged.
This work was supported by grants DFG FI 1630/3–1 (CDF), National Institutes of Health (NIH) DK59427 (to G.J.G.), NIH R01 CA 39225 (to A.E.S.), and the optical microscopy and clinical core of NIH DK84567. Mouse anti-S peptide antibody was provided by S.H. Kaufmann (Oncology Research, Mayo Clinic, Rochester, MN).
Abbreviations
- Ab
antibody
- Bcl
B-cell lymphoma
- bp
base pairs
- b.w
body weight
- CCA
cholangiocarcinoma
- ChIP
chromatin immunoprecipitation
- CK7
cytokeratin 7
- DAPI
4′,6-diamidino-2-phenylindole dihydrochloride
- ErbB-2
erythroblastic leukemia viral oncogene homolog
- GLI
glioma-associated oncogene
- Hh
hedgehog
- IF
immunofluorescence
- IgG
immunoglobulin G
- IHC
immunohistochemistry
- IP
intraperitoneally
- MBF
myofibroblast
- Mcl-1
myeloid cell leukemia-1
- mRNA
messenger RNA
- PCR
polymerase chain reaction
- PDGF
platelet-derived growth factor
- PLK
polo-like kinases
- rh
recombinant human
- rRNA
ribosomal RNA
- RT
reverse transcriptase
- SEM
standard error of the mean
- SHH
sonic hedgehog
- shRNA
short hairpin RNA
- TRAIL
tumor necrosis factor-related apoptosis-inducing ligand
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
Footnotes
Potential conflict of interest: Nothing to report.
Additional Supporting Information may be found in the online version of this article.
References
- 1.Blechacz B, Gores GJ. Cholangiocarcinoma: advances in pathogenesis, diagnosis, and treatment. Hepatology. 2008;48:308–321. doi: 10.1002/hep.22310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Groen PC, Gores GJ, LaRusso NF, Gunderson LL, Nagorney DM. Biliary tract cancers. N Engl J Med. 1999;341:1368–1378. doi: 10.1056/NEJM199910283411807. [DOI] [PubMed] [Google Scholar]
- 3.Fingas CD, Katsounas A, Kahraman A, Siffert W, Jochum C, Gerken G, et al. Prognostic assessment of three single-nucleotide polymorphisms (GNB3 825C>T, BCL2–938C>A, MCL1–386C>G) in extrahepatic cholangiocarcinoma. Cancer Invest. 2010;28:472–478. doi: 10.3109/07357900903095714. [DOI] [PubMed] [Google Scholar]
- 4.Roberts SK, Ludwig J, Larusso NF. The pathobiology of biliary epithelia. Gastroenterology. 1997;112:269–279. doi: 10.1016/s0016-5085(97)70244-0. [DOI] [PubMed] [Google Scholar]
- 5.Ishimura N, Isomoto H, Bronk SF, Gores GJ. Trail induces cell migration and invasion in apoptosis-resistant cholangiocarcinoma cells. Am J Physiol Gastrointest Liver Physiol. 2006;290:G129–G136. doi: 10.1152/ajpgi.00242.2005. [DOI] [PubMed] [Google Scholar]
- 6.Johnstone RW, Frew AJ, Smyth MJ. The TRAIL apoptotic pathway in cancer onset, progression, and therapy. Nat Rev Cancer. 2008;8:782–798. doi: 10.1038/nrc2465. [DOI] [PubMed] [Google Scholar]
- 7.Walczak H, Miller RE, Ariail K, Gliniak B, Griffith TS, Kubin M, et al. Tumoricidal activity of tumor necrosis factor-related apoptosisinducing ligand in vivo. Nat Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 8.Fingas CD, Bronk SF, Werneburg NW, Mott JL, Guicciardi ME, Cazanave SC, et al. Myofibroblast-derived PDGF-BB promotes hedgehog survival signaling in cholangiocarcinoma cells. Hepatology. 2011;54:2076–2088. doi: 10.1002/hep.24588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kurita S, Mott JL, Almada LL, Bronk SF, Werneburg NW, Sun SY, et al. GLI3-dependent repression of DR4 mediates hedgehog antagonism of TRAIL-induced apoptosis. Oncogene. 2010;29:4848–4858. doi: 10.1038/onc.2010.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurita S, Mott JL, Cazanave SC, Fingas CD, Guicciardi ME, Bronk SF, et al. Hedgehog inhibition promotes a switch from type II to type I cell death receptor signaling in cancer cells. PLoS One. 2011;6:e18330. doi: 10.1371/journal.pone.0018330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walterhouse DO, Yoon JW, Iannaccone PM. Developmental pathways: Sonic hedgehog-Patched-GLI. Environ Health Perspect. 1999;107:167–171. doi: 10.1289/ehp.99107167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Khatib M, Kalnytska A, Palagani V, Kossatz U, Manns MP, Malek NP, et al. Inhibition of hedgehog signaling attenuates carcinogenesis in vitro and increases necrosis of cholangiocellular carcinoma. Hepatology. 2013;57:1035–1045. doi: 10.1002/hep.26147. [DOI] [PubMed] [Google Scholar]
- 13.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 14.de Carcer G, Manning G, Malumbres M. From Plk1 to Plk5: functional evolution of polo-like kinases. Cell Cycle. 2011;10:2255–2262. doi: 10.4161/cc.10.14.16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov. 2010;9:643–660. doi: 10.1038/nrd3184. [DOI] [PubMed] [Google Scholar]
- 16.Schoffski P. Polo-like kinase (PLK) inhibitors in preclinical and early clinical development in oncology. Oncologist. 2009;14:559–570. doi: 10.1634/theoncologist.2009-0010. [DOI] [PubMed] [Google Scholar]
- 17.Feng YB, Lin DC, Shi ZZ, Wang XC, Shen XM, Zhang Y, et al. Over-expression of PLK1 is associated with poor survival by inhibiting apoptosis via enhancement of survivin level in esophageal squamous cell carcinoma. Int J Cancer. 2009;124:578–588. doi: 10.1002/ijc.23990. [DOI] [PubMed] [Google Scholar]
- 18.Liu X, Choy E, Harmon D, Yang S, Yang C, Mankin H, et al. Inhibition of polo-like kinase 1 leads to the suppression of osteosarcoma cell growth in vitro and in vivo. Anticancer Drugs. 2011;22:444–453. doi: 10.1097/CAD.0b013e32834513f4. [DOI] [PubMed] [Google Scholar]
- 19.Isomoto H, Kobayashi S, Werneburg NW, Bronk SF, Guicciardi ME, Frank DA, et al. Interleukin 6 upregulates myeloid cell leukemia-1 expression through a STAT3 pathway in cholangiocarcinoma cells. Hepatology. 2005;42:1329–1338. doi: 10.1002/hep.20966. [DOI] [PubMed] [Google Scholar]
- 20.Okaro AC, Deery AR, Hutchins RR, Davidson BR. The expression of antiapoptotic proteins Bcl-2, Bcl-X(L), and Mcl-1 in benign, dysplastic, and malignant biliary epithelium. J Clin Pathol. 2001;54:927–932. doi: 10.1136/jcp.54.12.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taniai M, Grambihler A, Higuchi H, Werneburg N, Bronk SF, Farrugia DJ, et al. Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells. Cancer Res. 2004;64:3517–3524. doi: 10.1158/0008-5472.CAN-03-2770. [DOI] [PubMed] [Google Scholar]
- 22.Rudolph D, Steegmaier M, Hoffmann M, Grauert M, Baum A, Quant J, et al. BI 6727, a Polo-like kinase inhibitor with improved pharmacokinetic profile and broad antitumor activity. Clin Cancer Res. 2009;15:3094–3102. doi: 10.1158/1078-0432.CCR-08-2445. [DOI] [PubMed] [Google Scholar]
- 23.Masuoka HC, Mott J, Bronk SF, Werneburg NW, Akazawa Y, Kaufmann SH, et al. Mcl-1 degradation during hepatocyte lipoapoptosis. J Biol Chem. 2009;284:30039–30048. doi: 10.1074/jbc.M109.039545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fingas CD, Blechacz BR, Smoot RL, Guicciardi ME, Mott J, Bronk SF, et al. A smac mimetic reduces TNF related apoptosis inducing ligand (TRAIL)-induced invasion and metastasis of cholangiocarcinoma cells. Hepatology. 2010;52:550–561. doi: 10.1002/hep.23729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fingas CD, Mertens JC, Razumilava N, Bronk SF, Sirica AE, Gores GJ. Targeting PDGFR-beta in cholangiocarcinoma. Liver Int. 2012;32:400–409. doi: 10.1111/j.1478-3231.2011.02687.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sirica AE, Zhang Z, Lai GH, Asano T, Shen XN, Ward DJ, et al. A novel “patient-like” model of cholangiocarcinoma progression based on bile duct inoculation of tumorigenic rat cholangiocyte cell lines. Hepatology. 2008;47:1178–1190. doi: 10.1002/hep.22088. [DOI] [PubMed] [Google Scholar]
- 27.Malhi H, Barreyro FJ, Isomoto H, Bronk SF, Gores GJ. Free fatty acids sensitise hepatocytes to TRAIL mediated cytotoxicity. Gut. 2007;56:1124–1131. doi: 10.1136/gut.2006.118059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallikas O, Palin K, Sinjushina N, Rautiainen R, Partanen J, Ukkonen E, et al. Genome-wide prediction of mammalian enhancers based on analysis of transcription-factor binding affinity. Cell. 2006;124:47–59. doi: 10.1016/j.cell.2005.10.042. [DOI] [PubMed] [Google Scholar]
- 29.Regl G, Kasper M, Schnidar H, Eichberger T, Neill GW, Philpott MP, et al. Activation of the BCL2 promoter in response to Hedgehog/GLI signal transduction is predominantly mediated by GLI2. Cancer Res. 2004;64:7724–7731. doi: 10.1158/0008-5472.CAN-04-1085. [DOI] [PubMed] [Google Scholar]
- 30.Hu MC, Mo R, Bhella S, Wilson CW, Chuang PT, Hui CC, et al. GLI3-dependent transcriptional repression of Gli1, Gli2, and kidney patterning genes disrupts renal morphogenesis. Development. 2006;133:569–578. doi: 10.1242/dev.02220. [DOI] [PubMed] [Google Scholar]
- 31.Kobayashi S, Lee SH, Meng XW, Mott JL, Bronk SF, Werneburg NW, et al. Serine 64 phosphorylation enhances the antiapoptotic function of Mcl-1. J Biol Chem. 2007;282:18407–18417. doi: 10.1074/jbc.M610010200. [DOI] [PubMed] [Google Scholar]
- 32.Metcalfe C, de Sauvage FJ. Hedgehog fights back: mechanisms of acquired resistance against Smoothened antagonists. Cancer Res. 2011;71:5057–5061. doi: 10.1158/0008-5472.CAN-11-0923. [DOI] [PubMed] [Google Scholar]
- 33.Yauch RL, Dijkgraaf GJ, Alicke B, Januario T, Ahn CP, Holcomb T, et al. Smoothened mutation confers resistance to a Hedgehog pathway inhibitor in medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thrum S, Lorenz J, Mossner J, Wiedmann M. Polo-like kinase 1 inhibition as a new therapeutic modality in therapy of cholangiocarcinoma. Anticancer Res. 2011;31:3289–3299. [PubMed] [Google Scholar]
- 35.Coley HM, Hatzimichael E, Blagden S, McNeish I, Thompson A, Crook T, et al. Polo Like Kinase 2 Tumour Suppressor and cancer biomarker: new perspectives on drug sensitivity/resistance in ovarian cancer. Oncotarget. 2012;3:78–83. doi: 10.18632/oncotarget.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winkles JA, Alberts GF. Differential regulation of polo-like kinase 1, 2, 3, and 4 gene expression in mammalian cells and tissues. Oncogene. 2005;24:260–266. doi: 10.1038/sj.onc.1208219. [DOI] [PubMed] [Google Scholar]
- 37.Feng GS. Conflicting roles of molecules in hepatocarcinogenesis: paradigm or paradox. Cancer Cell. 2012;21:150–154. doi: 10.1016/j.ccr.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.