Abstract
The tensor-based morphometry (TBM) has been widely used in characterizing tissue volume difference between populations at voxel level. We present a novel computational framework for investigating the white matter connectivity using TBM. Unlike other diffusion tensor imaging (DTI) based white matter connectivity studies, we do not use DTI but only T1-weighted magnetic resonance imaging (MRI). To construct brain network graphs, we have developed a new data-driven approach called the ε-neighbor method that does not need any predetermined parcellation. The proposed pipeline is applied in detecting the topological alteration of the white matter connectivity in maltreated children.
Index Terms: tensor-based morphometry, structural connectivity, brain network, maltreatment, Jacobian determinant
1. INTRODUCTION
The human brain exhibits one of the most complex networks. This anatomical substrate supports the emergence of the coherent physiological activities in the distant brain regions that make up a functional network [1]. Unlike extensively studied functional brain networks, structural connectivity is not often explored till the introduction of diffusion tensor imaging (DTI) which is often used to investigate the structure of axonal fiber bundles in human brains in vivo.
Recently, there has been an attempt of using cross-correlation of cortical thickness as a way to investigate cortico-cortical connectivity [2, 3]. Besides the cortical thickness, it is possible to correlate other voxel-wise morphometric measures such as the Jacobian determinant (JD) obtained from the tensor-based morphometry (TBM) framework. The JD measures the change in the volume of a voxel in deforming the template brain to match an individual brain [4]. By correlating the JD at different voxels, we can quantify how the local volume in one voxel is correlated to the local volume in other voxels. Thus it can be directly used in constructing the whole brain map of corresponding structures.
In this paper, we propose to construct a structural connectivity map based on the cross-correlation of the JD for the first time. The main advantage of the proposed technique is that it does not require DTI but still able to construct the population specific connectivity maps only using T1-weighted MRI. Since MRI has been extensively collected than DTI so far in clinical applications, it would be highly beneficial if we can exploit this massive database of MRI.
The second advantage is that it can build connectivity maps over the whole brain. While the cortical thickness-based connectivity is restricted to the gray matter only, neural development such as myelination or glial proliferation contributes the white matter volume [5]. Our approach can build a connectivity map of the whole brain enabling such an investigation that involves in the white matter.
The proposed framework is applied to the brain networks of the children who have been maltreated in the early stages of life and have been institutionalized in orphanages in East Europe and China but are now living with adopted families in the USA (Post-Institutionalized; PI). It is known that individuals who experience such an early adversity are at heightened risk for a various mental and physical problems. In the physically abused children, smaller local volume was found in the orbitofrontal cortex, which is known as central to social and emotional regulation [6]. Rodent models show that the chronic stress occurs the cytoarchitectural changes in the frontal cortex [7]. Thus we expect decreased white matter connectivity in the PIs in the regions including the frontal cortex.
2. METHODS
2.1. Subjects and MRI image
T1-weighted MRIs were collected using a 3T GE SIGNA scanner for 32 PI and 33 normal control (NC) subjects. Two groups are matched in terms of age (PI: 11.19 ± 1.73 years, NC: 11.48 ± 1.62 years; T (63)=0.71, p > 0.47). But the gender ratios (girls over all; PI: 0.59, NC: 0.39) and the whole brain volume (PI: 1,808.2 ± 117.6 cm3, NC: 1,690.0 ± 156.4 cm3; T (63)= 3.44, p < 0.001) are different. A study specific template was constructed using symmetric normalization (SyN) through Advanced Normalization Tools (ANTS) [8].
Computational load quadratically increases as the number of nodes increases. Even with 336363 white matter voxels in the template as in this case, there will be total 113140000000 cross-correlations to compute. Thus, to reduce the computational load, the 1mm-resolution JD maps are subsampled at every 5 mm after spatial smoothening with a Gaussian kernel with 3 mm FWHM. Subsequently 2692 nodes are obtained (reduction ratio= 0.0080). This is more than sufficient number of nodes for modeling white matter connectivity and substantially larger than most of connectivity studies that use between 50–100 nodes.
2.2. Partial correlation maps
Fig. 1 illustrates the proposed pipeline. Between 2692 nodes, we link two nodes if the partial correlation of the JDs is statistically signifi-cant at a certain threshold. The JD is defined as the determinant of the displacement gradient matrix ∂U/∂x [4] as
Fig. 1.
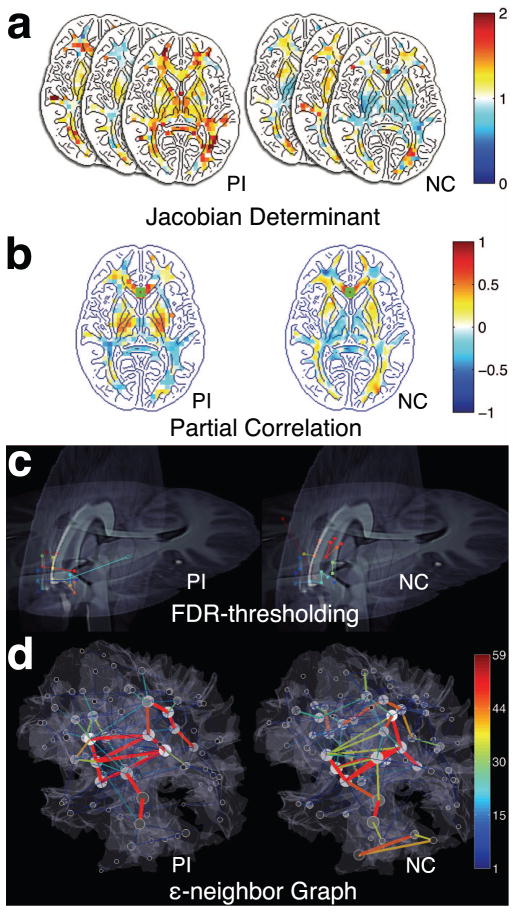
Framework of the proposed analysis applied to post-institutionalized (PI) children and normal control (NC). (a) Jacobian determinant maps of individuals projected on the template. (b) partial correlation maps seeded at the genu (marked with green squares) (c) FDR-thresholding on partial correlation is used to establish edges of the connectivity network. Only edges connecting near the genu are visualized. The different pairings are marked with different colors. (d) The proposed ε-neighbor graphs of connectivity. Only positive correlations are shown here. The gray shading of nodes indicates the node degree. The size of nodes represents the number of nodes that are merged in the ε-neighbor construction.
| (1) |
where U is the displacement matrix and x is the coordinate vector. To remove the possible confounding effect of age, gender and brain volume, we used the partial correlation obtained from fitting general linear models (GLM). Let z = (1, age, gender, volume) be the nuisance covariate vector. Then we modeled the JD on the i-th node as
| (2) |
where λi = (λi1, …, λi4)′ is the unknown parameter vector and ε i is the correlated zero mean Gaussian noise. The residual of the fit is given by ri = Ji − zλ̂i, where λ̂i are the least-squares estimation. It can be shown that the partial correlation ρij between Ji and Jj while factoring out the effect of the nuisance covariates z is simply given by the correlation between the residuals ri and rj [9]. The partial correlation ρij is then estimated using the Pearson correlation as
| (3) |
where n is the number of subjects in each group.
2.3. FDR thresholding of partial correlations
In order to obtain the deterministic network graph, we have thresholded the partial correlations using the false discovery rate (FDR) thresholding. The distribution of correlations can be approximated using the Fisher transform: [10]:
| (4) |
The null hypothesis H0 is that there is no link between the nodes i and j, i.e. ρij = 0.
The family-wise error rate (FWER) would be highly inflated with 2692 × 2691/2 possible tests between all nodes. Thus we applied the FDR with q = 0.01 under a weak assumption of dependency. If the resulting FDR-threshold is given by s, the adjacency matrix A = (aij ) is given by aij = 1 if zij ≥ s and a ij = 0 otherwise, with the diagonal terms aii = 0.
2.4. ε-neighbor graph simplification
Though the obtained adjacency matrices via the FDR thresholding are sparse, almost ten thousands edges still encumber biological interpretation. Furthermore, isolated single connections consisting of two nodes are more likely false positives. Therefore, we need to perform a network simplification without distorting underlying network topology. For this purpose, we have adapted the ε-neighbor scheme [11], which was originally applied in constructing network graphs out of numerous white matter tracts obtained from DTI. The algorithm condenses a given complex graph to a much simpler graph iteratively.
From the FDR-thresholding, we obtain collection of significant edges ei1ei2 linking two nodes ei1 and ei2. Suppose we have constructed the graph
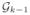 using edges e11e12, · · ·, ek−1,1ek−1,2 only. Then at the k-th iteration, we construct the graph
using edges e11e12, · · ·, ek−1,1ek−1,2 only. Then at the k-th iteration, we construct the graph
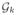 using ek1ek2 somehow. In order to do this, we need to define the ε-neighbor of a graph. Let us define the distance d(p,
using ek1ek2 somehow. In order to do this, we need to define the ε-neighbor of a graph. Let us define the distance d(p,
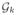 ) of a node p to the graph
) of a node p to the graph
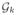 as
as
| (5) |
If d(p,
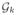 ) ≤ ε for some radius ε, the node p is called the ε-neighbor of
) ≤ ε for some radius ε, the node p is called the ε-neighbor of
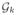 .
.
Initially the graph
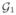 = {
= {
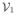 ,
,
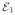 } starts with two nodes
} starts with two nodes
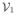 = {e11, e12} and a single edge
= {e11, e12} and a single edge
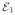 = {e11e12}. At the 2nd iteration, we check if the new nodes e21 and e22 are the ε-neighbor of
= {e11e12}. At the 2nd iteration, we check if the new nodes e21 and e22 are the ε-neighbor of
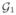 . We will merge a new node to the existing node in
. We will merge a new node to the existing node in
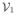 if the new node is the ε neighbor of a node in
if the new node is the ε neighbor of a node in
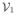 . The idea is best illustrated with a toy example given in Fig. 2.
. The idea is best illustrated with a toy example given in Fig. 2.
Fig. 2.
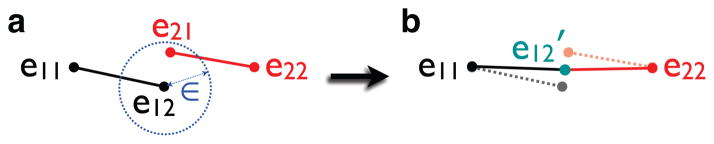
Schematic illustration of the ε-neighbor updating scheme. (a) Initially the graph
 consists of one edge e11e12 (black). At the next stage, we determine how to connect the new edge e21e22 (red) to the existing graph
consists of one edge e11e12 (black). At the next stage, we determine how to connect the new edge e21e22 (red) to the existing graph
 . The node e21 is within the ε radius (blue) of the node e12. So e21 is the ε-neighbor of
. The node e21 is within the ε radius (blue) of the node e12. So e21 is the ε-neighbor of
 and has to be merged with e12. (b) The coordinates of the merged node e12 is updated to e12′ (green) and the new edge e12′e22 is included in
and has to be merged with e12. (b) The coordinates of the merged node e12 is updated to e12′ (green) and the new edge e12′e22 is included in
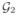 .
.
Suppose e21 is the ε-neighbor of
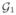 . We assume that e12 is the closet node to e21. Then we merge e21 to e12 and update the vertex and edge sets as
. We assume that e12 is the closet node to e21. Then we merge e21 to e12 and update the vertex and edge sets as
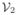 = {e11, e12, e22},
= {e11, e12, e22},
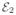 = {e11e12, e12e22}. Other possible scenarios are given in [11]. This merging and deletion process is iteratively performed. However, the original ε-neighbor construction method as presented in [11] does not produce a unique graph and it depends on the initial choice of edge set
= {e11e12, e12e22}. Other possible scenarios are given in [11]. This merging and deletion process is iteratively performed. However, the original ε-neighbor construction method as presented in [11] does not produce a unique graph and it depends on the initial choice of edge set
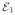 . To guarantee the stability, we decided to update the coordinates of the pre-existing node when a merging occurs. In the toy example, the coordinates of e12 are updated to e12′ as
. To guarantee the stability, we decided to update the coordinates of the pre-existing node when a merging occurs. In the toy example, the coordinates of e12 are updated to e12′ as
| (6) |
where is the total number of nodes that are merged to the existing node eij at the k-th iteration. If the merging happens, we have to update as well. So we have . For this study, ε was set to be 21 mm to investigate the connectivity at macro-scale level.
3. RESULTS
The FRD-thresholding produces graphs with 2692 nodes. The ε-neighbor method simplifies the graphs with only 88 nodes and 241 edges for the PIs, and 86 nodes and 276 edges for the NCs. In terms of the number of nodes, the ε-neighbor method achieves the compression rate of 3.27 % while still preserving the overall topological structures in the graphs with 2692 nodes.
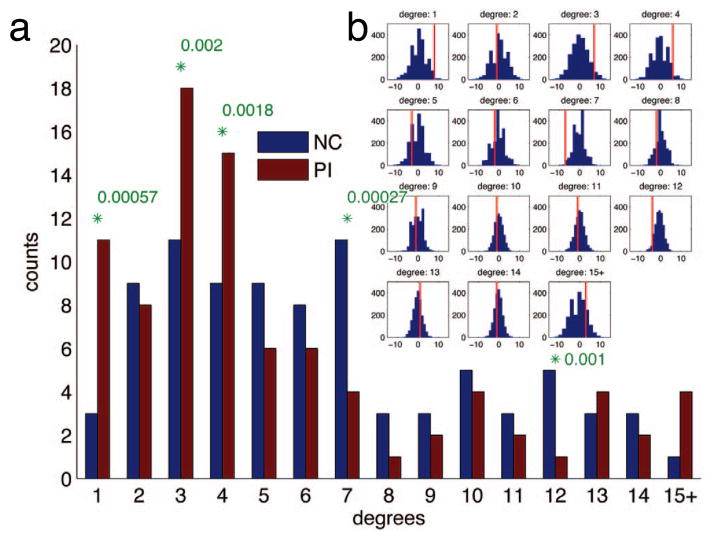
We used the degree of nodes as a discriminating feature between the two groups. The degree distributions of ε-neighbor graphs are shown in Fig. 3. The counts in the high degrees are prone to noise thus those exceeding degree 14 are summed into a single bin [1]. Since the underlying distribution is unknown, the significance was tested using a non-parametric permutation test. We randomly permuted the group identifiers for 2000 times and proceeded with the graph construction procedures. Since we need to perform multiple tests for 15 degree bins simultaneously, Bonferroni correction threshold for an individual test was set at 0.05/15 = 0.0033. There are significantly more nodes with the low degrees (1, 3 and 4) in the PIs than the NCs and more nodes with the high degrees (7 and 12) in the NCs than the PIs. Since the numbers of nodes are expected to be similar across groups, it suggests that the nodes with the high degrees are affected in the PIs resulting more low degree nodes. It also implicates weakened connectivity in the PIs in accordance with the previous literature [7].
Fig. 3.
Permutation tests on degree distributions. (a) Degree distributions. The significant differences between the PIs and the NCs marked with green asterisks with p-values (Bonferroni corrected at α=0.05). (b) The null distribution obtained by 2000 permutation tests. X-axis is for the difference of degrees between the PIs and the NCs. Y-axis is for the number of counts. Red vertical lines note the actual differences.
The anatomical patterns of the ε-neighbor graphs are shown in Fig. 4. While the inter-hemispherical edges connecting homologous sub-cortical regions are commonly found in the both groups, the differences in the edge concentration are observed in the regions that include the cerebellar, the brainstem and the regions around the anterior cingulate gyri. In addition, the extension of the edges that reach to the dorsal lateral prefrontal cortex and the medial temporal cortex seems to be limited in the PIs than the NCs suggesting consistency with the reduced local volume in those regions [6].
Fig. 4.
Local connectivity patterns of the ε-neighbor graphs. Only positive correlations are shown in a lateral view. Edges are color-coded by the number of merged connections implying the strength of connections. The gray shading of nodes indicates the degree and the size of nodes represents the number of nodes that are merged.
4. DISCUSSIONS
We have presented a novel structural connectivity mapping technique that uses only T1-weighted MRI. The constructed partial correlation maps (Fig. 1) look very similar to the probabilistic connectivity maps obtained from DTI. Further research is needed for validating the closeness of the partial correlation maps to the probabilistic connectivity maps.
The network graphs showed significantly different degree distributions in the PIs implying abnormal connectivity. The anatomical pattern of the white matter connectivity seems to be locally different across groups. However, it should be more thoroughly validated in a further study.
In this paper, we have mainly focused on developing the connectivity mapping technique via the TBM framework and the ε-neighbor graph simplification.
Acknowledgments
This work was supported by National Institutes of Health Research Grants MH61285 and MH68858 to S.D.P., funded by the National Institute of Mental Health and the Childrens Bureau of the Administration on Children, Youth and Families as part of the Child Neglect Research Consortium, as well as National Institute of Mental Health Grant MH84051 to R.J.D. WCU Grant from the government of Korea to M.K.C. is also acknowledged.
References
- 1.Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nature Reviews Neuroscience. 2009;10(3):186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- 2.Lerch JP, Worsley K, Shaw WP, Greenstein DK, Len-root RK, Giedd J, Evans AC. Mapping anatomical correlations across cerebral cortex (MACACC) using cortical thickness from MRI. Neuroimage. 2006;31(3):993–1003. doi: 10.1016/j.neuroimage.2006.01.042. [DOI] [PubMed] [Google Scholar]
- 3.He Y, Chen Z, Evans A. Structural insights into aberrant topological patterns of large-scale cortical networks in alzheimer’s disease. Journal of Neuroscience. 2008;28(18):4756. doi: 10.1523/JNEUROSCI.0141-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chung MK, Worsley KJ, Paus T, Cherif DL, Collins C, Giedd J, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. NeuroImage. 2001;14:595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- 5.Groeschel S, Vollmer B, King MD, Connelly A. Developmental changes in cerebral grey and white matter volume from infancy to adulthood. International Journal of Developmental Neuroscience. 2010 doi: 10.1016/j.ijdevneu.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 6.Hanson JL, Chung MK, Avants BB, Shirtcliff EA, Gee JC, Davidson RJ, Pollak SD. Early stress is associated with alterations in the orbitofrontal cortex: A tensor-based morphometry investigation of brain structure and behavioral risk. Journal of Neuroscience. 2010;30(22):7466. doi: 10.1523/JNEUROSCI.0859-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnsten AFT. Stress signalling pathways that impair pre-frontal cortex structure and function. Nature Reviews Neuro-science. 2009;10(6):410–422. doi: 10.1038/nrn2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avants BB, Epstein CL, Grossman M, Gee JC. Symmetric diffeomorphic image registration with cross-correlation: Evaluating automated labeling of elderly and neurodegenerative brain. Medical image analysis. 2008;12(1):26. doi: 10.1016/j.media.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kolaczyk Eric D. Statistical Analysis of Network Data: Methods and Models. Springer Publishing Company, Incorporated; 2009. [Google Scholar]
- 10.Fisher RA. Frequency distribution of the values of the correlation coefficient in samples from an indefinitely large population. Biometrika. 1915;10(4):507–521. [Google Scholar]
- 11.Chung MK, Adluru N, Dalton KM, Alexander AL, Davidson RJ. Scalable brain network construction on white matter fibers. SPIE Medical Imaging. 2010 doi: 10.1117/12.874245. [DOI] [PMC free article] [PubMed] [Google Scholar]