1 Introduction
Over the past two decades, there has been significant growth in our understanding of the genetics underlying aging, catalyzed by the discovery that single-gene mutations can dramatically extend lifespan in the model organism C. elegans (Friedman and Johnson, 1988)(Kenyon et al., 1993). Importantly, one such mutation discovered to double the worm’s lifespan is the C. elegans insulin receptor (daf-2) (Kenyon et al., 1993); this effect is completely dependent on the gene daf-16 (Kenyon et al., 1993). The insulin/IGF-1 signaling (IIS) pathway’s components are highly conserved in worms, flies, and mammals, and the FOXO transcription factor, DAF-16, is the major downstream effector of DAF-2, C. elegans’ sole insulin/IGF-1-like receptor (Tatar et al., 2003).
In addition to its well-known regulation of longevity, DAF-16/FOXO is crucial for many important processes, including development, stress resistance, thermotolerance, pathogen resistance, and metabolism (Vowels and Thomas, 1992; Baugh and Sternberg, 2006; Murakami et al., 1996; Henderson and Johnson, 2001; Garsin et al., 2003; Ogg et al., 1997; Kimura et al., 1997). The transcriptional targets of DAF-16, which presumably execute its many roles, include stress-response, antimicrobial, autophagic, and metabolic genes, as well as many genes of unknown function (Murphy et al., 2003; McElwee et al., 2003; Lee et al., 2003; Oh et al., 2006), and many of the genes most highly regulated by DAF-16 influence aging (Murphy et al., 2003). Thus, the convergence of IIS signaling on these diverse outputs raises the question of how DAF-16 specificity is achieved. Many of the recent breakthroughs in the field involve the identification of new DAF-16/FOXO co-factors and their effects on DAF-16/FOXO functional output. Here we discuss developments in the area of DAF-16/FOXO co-factors that may help DAF-16 regulate these diverse functions.
2 Insulin signaling regulation of DAF-16
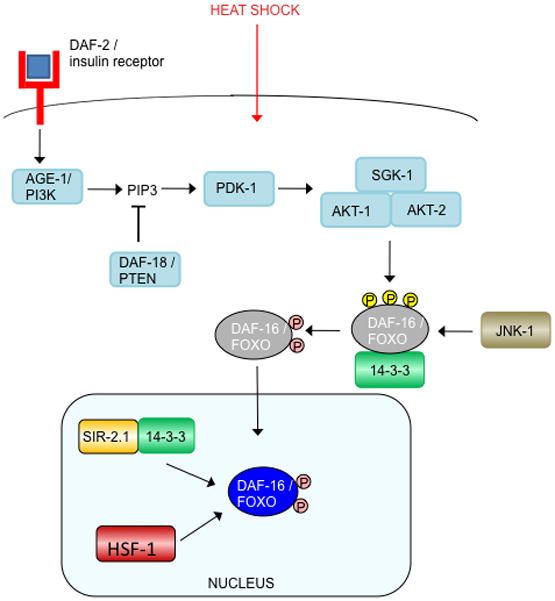
daf-2 was originally identified for its mutant’s dauer constitutive (Daf-C) phenotype, forming the alternative larval state inappropriately under growth-promoting conditions, while daf-16 was identified in a Daf-D (dauer defective) mutant that suppressed the daf-2 mutant’s dauer formation (Vowels and Thomas, 1992; Gottlieb and Ruvkun, 1994). In conditions suitable for growth and reproduction, activation of DAF-2 by binding of an agonist insulin-like ligand initiates a phosphorylation cascade that ultimately inhibits DAF-16 activity. Remarkably, there are 40 insulin-like molecules encoded in the C. elegans genome, some of which may function as DAF-2 agonists, and others as antagonists (Pierce et al., 2001; Li et al., 2003; Kawano et al., 2000; Murphy et al., 2007). Activated DAF-2 phosphorylates the phosphoinositide 3-kinase, AGE-1, generating PIP3, which, in turn, recruits the kinases AKT-1, AKT-2, SGK-1, and PDK-1 to the plasma membrane, where PDK-1 phosphorylates AKT and SGK-1 (Hertweck et al., 2004; Paradis et al., 1999). The AKT-1/AKT-2/SGK-1 complex phosphorylates the forkhead transcription factor DAF-16 (Paradis and Ruvkun, 1998; Hertweck et al., 2004), sequestering it in the cytoplasm (Ogg et al., 1997; Lin et al., 1997), thus preventing DAF-16 from activating or repressing transcription of its target genes in the nucleus (Figure 1). Inhibitors of this cascade include DAF-18, the phosphoinositide 3-phosphatase PTEN, which antagonizes AGE-1 by dephosphorylating PIP3 (Ogg and Ruvkun, 1998), and PPTR-1, a regulatory subunit of PP2A that dephosphorylates AKT-1 (Padmanabhan et al., 2009).
Fig. 1. Insulin signaling regulation of DAF-16/FOXO.

Activated DAF-2 initiates a phosphorylation cascade that ultimately phosphorylates DAF-16, thereby excluding it from the nucleus and inhibiting its activity. As noted in the text, it remains unclear whether DAF-16 interacts with both 14–3-3 proteins FTT-2 and PAR-5, so here we have labeled its interacting partner “14–3-3.”
3.1 DAF-16 may act in specific tissues to execute different outputs
As C. elegans is a multi-tissued organism, it is important to establish where DAF-16 may act to execute its diverse functions. daf-2 acts cell-non-autonomously to specify life span and formation of the stress-resistant dauer developmental stage (Apfeld and Kenyon, 1998), although the sites of action of DAF-2 and DAF-16 remain controversial. Restoration of DAF-2 and AGE-1 expression in neurons, but not the intestine, is sufficient to shorten the life span of long-lived daf-2 and age-1 mutant animals, respectively (Wolkow et al., 2000). However, subsequent results showed that AGE-1 in the intestine can shorten the life span of an age-1 mutant (Iser et al., 2007). The reason for this discrepancy is unclear, but may result from diffierences in transgenes or expression levels.
At the level of DAF-16 activity, Libina et al. showed that DAF-16 activity in the intestine partially restores the life span extension (60%) of daf-16;daf-2 mutants, suggesting that the intestine is an important site of DAF-16’s role in longevity, whereas neuronal DAF-16 effects a much lesser life span extension (Libina et al., 2003). By contrast, DAF-16 expressed in the neurons was sufficient to promote dauer formation, whereas DAF-16 in the intestine had very little effect on dauer formation (Libina et al., 2003), which indicates that some DAF-16 functionality may arise from its activity in specific tissues. Iser, et al. showed that tissue-restricted daf-16 expression in the intestine or neurons of a daf-16;age-1 mutant does not restore life span as strongly as does daf-16 expressed in multiple tissues (Iser et al., 2007), perhaps suggesting that DAF-16 may act in multiple tissues in a graded fashion to specify longevity. Finally, tissue-restricted daf-16 expression in the hypodermis resulted in embryonic or larval lethality, suggesting that expression of daf-16 in the hypodermis may be lethal unless accompanied by expression elsewhere. This result underscores the importance of appropriate DAF-16 regulation for the animal.
Such tissue-specific insulin receptor and FOXO activity is unlikely to be restricted to C. elegans. For example, in mice, loss of the insulin receptor in adipose (fat) tissue (the “FIRKO mouse”) extends life span (Blüher et al., 2003). However, loss of insulin signaling is not generally beneficial, as its loss in the liver causes diabetes (Michael et al., 2000). In flies, overexpression of dFOXO in the adipose tissue extends life span (Giannakou et al., 2004). The fact that both FOXO and insulin/IGF-1 receptors have been identified as genes linked to extreme longevity in recent human centenarian studies (Willcox et al., 2008; Flachsbart et al., 2009; Suh et al., 2008) suggests that discovering the downstream, tissue-specific FOXO-regulated targets, as in model organisms, will be important for understanding FOXO’s role in human longevity and aging.
3.2 DAF-16 may receive signals from the gonad
The C. elegans reproductive system regulates longevity signaling from the somatic gonad and the germ line, two tissues with opposing effects on longevity. Longevity is extended by germ line removal, an effect that is dependent on daf-16, but this extension is inhibited when both the somatic gonad and germ line are removed (Hsin and Kenyon, 1999). Removing the germ cells results in nuclear accumulation of DAF-16 in intestinal cells (Lin et al., 2001), and DAF-16 activity only in the intestine is completely sufficient for this germ line-mediated longevity (Libina et al., 2003). This effect requires the activity of KRI-1, an intestinally expressed ankyrin-repeat protein orthologous to the human disease gene KRIT1/CCM1 (Berman and Kenyon, 2006). The nuclear hormone receptor DAF-12 (Antebi et al., 2000) and the cytochrome P450 DAF-9 (which is postulated to help synthesize a lipophilic ligand for DAF-12 (Motola et al., 2006)) are also required for nuclear localization of DAF-16 (Berman and Kenyon, 2006). These results suggest that the germline communicates with DAF-16 in the intestine via endocrine signaling, and that the response to this signaling requires KRI-1. Similar regulation of germline-mediated longevity has been recently uncovered in flies (Flatt et al., 2008).
Thus, DAF-16 carries out specific roles both directly in and in response to signaling from different tissues, resulting in specific biological outputs, and such activity is likely at least partially conserved in higher organisms.
4 DAF-16 co-regulators and co-factors
While specific DAF-16 functions arise from activity in different tissues, DAF-16 is not differentially excluded from specific tissues during different life stages (Henderson and Johnson, 2001; Ogg et al., 1997). Thus, the question arises, how does DAF-16 choose its tissue-specific targets to effect these different outputs? Several lines of evidence suggest that DAF-16 does not act alone, but rather requires other molecules for its activity. First, simply increasing the amount of DAF-16 is not sufficient to confer its effect on longevity, as overexpression of DAF-16 in wild-type worms only increases life span slightly (Henderson and Johnson, 2001). Second, although nuclear localization of DAF-16 is necessary for target activation, nuclear localization is not sufficient to increase lifespan (Lin et al., 2001). Third, the canonical DAF-16 Binding Element (DBE) (Furuyama et al., 2000) is present in the 5-kb upstream of 78% of C. elegans genes (Kenyon and Murphy, 2006), yet only a small number of these genes are DAF-16 targets in young adulthood (Murphy et al., 2003). Thus, the presence of the DBE is not sufficient to predict DAF-16-induced expression, which suggests that there may be other factors to help DAF-16 to discriminate among potential targets. In this section, we describe putative DAF-16 cofactors and coregulators (Fig. 2), and discuss how they may contribute to DAF-16’s multiple roles.
Fig. 2. DAF-16 interacts with specific proteins under different stimuli.
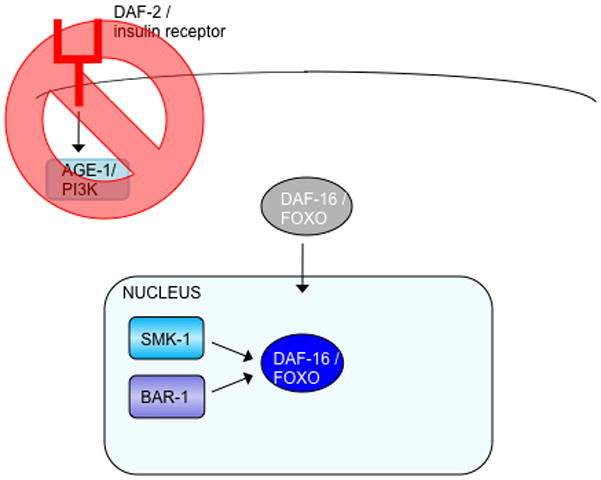

(a) Reduced insulin signaling relieves the phosphorylation of DAF-16, allowing it to enter the nucleus and interact with SMK-1 and BAR-1.
(b) In response to oxidative stress, JNK-1 activates DAF-16 by phosphorylation. (Note that the phosphatase that may remove the inhibitory AKT/SGK phosphates to allow nuclear translocation is unknown, but the serine/threonine phosphatase PP2A dephosphorylates mammalian FOXO1 (Yan et al., 2008).) Activated DAF-16 accumulates in the nucleus, where it interacts with SIR-2.1/14–3-3, SMK-1, BAR-1, SKN-1, and HCF-1.
(c). Heat shock causes JNK-1 to phosphorylate and activate DAF-16, which in turn, enters the nucleus and interacts with SIR-2.1/14–3-3 and HSF-1.
4.1 JNK-1
c-Jun N-terminal kinase (JNK), a stress-activated MAPK family member, is a positive regulator of FOXO activity in worms and mammals. Overexpression of the mammalian JNK ortholog jnk-1 extends C. elegans lifespan and increases resistance to heat and oxidative stress in a daf-16-dependent manner (Oh et al., 2005). JNK directly phosphorylates DAF-16 in vitro, and genetic and biochemical data suggest that jnk-1 acts in parallel to the insulin signaling pathway to regulate DAF-16, but in an opposite manner; that is, phosphorylation of DAF-16 by JNK promotes nuclear localization of DAF-16 and its subsequent activation (Fig. 2). Similarly, JNK phosphorylates the mammalian homolog FOXO4 in vivo, resulting in FOXO4 translocation from the cytoplasm to the nucleus and transcriptional activation (Essers et al., 2004).
4.2 CST-1
Like JNK, the conserved mammalian Ste20-like kinase, MST1, and its C. elegans homolog, CST-1, activate FOXO in response to oxidative stress. In mammalian neuronal cells, MST1 phosphorylates FOXO3 at a conserved residue in the DNA-binding domain, triggering dissociation of FOXO3 and 14–3–3 proteins, accumulation of FOXO3 in the nucleus, and neuronal cell death (Lehtinen et al., 2006). Similarly, MST1 phosphorylates C. elegans DAF-16 at the corresponding conserved site. Interestingly, while MST1-FOXO activation mammalian promotes neuronal cell death, the worm homolog CST-1 promotes longevity. Like JNK-1, overexpression of CST-1 extends lifespan in a daf-16-dependent manner, and further extends the longevity of daf-2 knockdown animals, suggesting that CST-1 may also act in parallel to the insulin signaling pathway to regulate DAF-16 activity (Lehtinen et al., 2006).
4.3 SIR-2
First discovered in yeast for their roles in epigenetic silencing (Rine and Herskowitz, 1987), sirtuins are NAD+-dependent deacetylases whose overexpression increases longevity in yeast, worms, and flies (Kaeberlein et al., 1999; Tissenbaum and Guarente, 2001; Rogina and Helfand, 2004). Similar to overexpression, chemical activators of sirtuins, including the well-known compound resveratrol, extend life span in worms and flies (Wood et al., 2004). Expression of extra copies of, sir-2.1, one of the four C. elegans Sir2 homologs, increases life span and resistance to thermal and oxidative stress, in a daf-16/FOXO-dependent manner (Tissenbaum and Guarente, 2001).
The mammalian ortholog of sir-2, SIRT1, deacetylates FOXO3a and FOXO4 (Brunet et al., 2004; van der Horst et al., 2004). As C. elegans DAF-16 shares several lysines that have been identified as acetylated residues in mammalian FOXO3a (Brunet et al., 2004), it is likely that a similar interaction exists between C. elegans SIR-2.1 and DAF-16. Interestingly, like the insulin-mediated phosphorylation and nuclear localization of DAF-16, the presence of FOXO3 in the nucleus alone, by mutation of the three Akt phosphorylation sites, is not sufficient to promote interaction between SIRT1 and FOXO3a in the absence of stress signals (Brunet et al., 2004). This suggests that stress-inducible FOXO3a acetylation and phosphorylation events may promote this interaction, and further demonstrates the complexity of FOXO control.
Notably, SIRT1 has a dual effect on FOXO3a function: SIRT1 increases FOXO3a’s induction of cell cycle arrest and resistance to oxidative stress, but inhibits FOXO3a’s induction of apoptosis (Brunet et al., 2004). It will be interesting to explore whether C. elegans SIR-2.1 can also differentially modulate aspects of DAF-16 function in a context-dependent manner.
4.4 14–3–3
The 14–3–3 proteins are members of a highly conserved family present in all eukaryotes examined (van Hemert et al., 2001). They can act as localization anchors or as adaptor proteins with scaffold activity that facilitates protein-protein interactions (van Hemert et al., 2001). The C. elegans genome encodes two 14–3-3 genes, par-5 (also known as ftt-1) and ftt-2 (Wang and Shakes, 1997). Demonstrating a role for 14–3-3 proteins in regulation of DAF-16, reduced expression of ftt-2 enhances dauer formation, and also promotes DAF-16 nuclear localization and activation of several target genes (Li et al., 2007). FTT-2 directly binds to DAF-16, and this association is reduced under daf-2(−) conditions, consistent with the model that in reduced insulin signaling conditions, DAF-16 is dephosphorylated and dissociates from 14–3-3 in the cytoplasm. This regulation is conserved, as mammalian 14–3-3ς also binds and sequesters FOXO3a in the cytoplasm (Brunet et al., 1999).
Whether PAR-5 regulates DAF-16 remains unclear, due to the high sequence identity between ftt-2 and par-5 and resultant cross-recognition by par-5 RNAi clones and antibodies. Berdichevsky et al. found that, like ftt-2, reducing par-5 results in nuclear accumulation of DAF-16 (Berdichevsky et al., 2006). Additionally, Li et al., noted that a low level of PAR-5 also co-immunoprecipitated with DAF-16; this could be due to the PAR-5 antibody cross-reacting with FTT-2, or alternatively, PAR-5 may in fact interact with DAF-16. Interestingly, ftt-2 RNAi (Li et al., 2007; Berdichevsky et al., 2006) and par-5 RNAi (Berdichevsky et al., 2008) shorten the life span of wild-type worms, but neither affect the life span of daf-2(−) animals. The SIR-2 overexpression/DAF-16-dependent increases in life span and heat and oxidative stress resistance are dependent on ftt-2, and par-5 may function in life span extension (Berdichevsky et al., 2006). Using immunoprecipitation/mass spectrometry and yeast two-hybrid assays, respectively, Wang et al. showed that SIR-2 interacts with PAR-5 and FTT-2 (Wang et al., 2006), and in response to heat stress, but not low insulin-like signaling, SIR-2.1 and DAF-16 physically interact in the nucleus, dependent on ftt-2 (Berdichevsky et al., 2006). 14–3-3 proteins bridge the interaction between SIR-2.1 and DAF-16 in response to stress; thus it is possible that, although there is more DAF-16 in the nucleus of ftt-2, and perhaps par-5, knockdown worms, this is countered by dissociation of the 14–3-3/SIR-2.1/DAF-16 complex and perhaps other protein complexes.
4.5 HSF-1
Heat-shock factor (HSF) induces transcription of chaperones and proteases in response to heat and other stressors. In C. elegans, reducing the activity of HSF-1 causes a rapid-tissue-aging phenotype and shortens life span (Garigan et al., 2002). hsf-1 is required for daf-2 dauer formation, longevity, and pathogen resistance (Morley and Morimoto, 2004; Hsu et al., 2003; Singh and Aballay, 2006). Conversely, HSF-1 overexpression increases heat resistance and extends life span in a DAF-16-dependent manner (Hsu et al., 2003), suggesting that DAF-16 and HSF-1 act together to promote longevity. Indeed, HSF-1 and DAF-16 act together to induce small heat-shock protein genes, all of which have consensus DAF-16 and HSF-1 binding sites in upstream regions. Yet, HSF-1 inactivation does not prevent DAF-16 accumulation in the nuclei of daf-2 mutants, or the expression of the stress-responsive DAF-16 target genes sod-3 and mtl-1 (Hsu et al., 2003). Likewise, the heat-inducibility of several HSF-1 targets is not diminished by loss of daf-16. Thus, HSF-1 and DAF-16 appear to function together to turn on a specific subset of genes.
Interestingly, while reduction of hsf-1 does not affect nuclear localization of DAF-16 under reduced IIS conditions, RNAi knockdown of hsf-1 and its target Hsp70/hsp-1 delay nuclear export of DAF-16 during recovery from heat shock (Singh and Aballay, 2009), showing that HSF-1 may have a role in controlling DAF-16 activity. The mechanism of this regulatory interaction remains to be elucidated.
4.6 β-catenin
Best known for mediating Wnt signaling by binding to members of the T-cell factor (TCF) family of transcription factors, the C. elegans β-catenin homolog, bar-1, also interacts with DAF-16. Loss of bar-1 reduces the activity of DAF-16 in promoting dauer formation. Furthermore, like daf-16 mutants, bar-1 mutants are short lived and are more sensitive to oxidative stress (Essers et al., 2005). bar-1 is required for oxidative stress-induced expression of the direct DAF-16 target sod-3 and physically binds to DAF-16. Thus, these results demonstrate a direct role for β-catenin in regulating DAF-16/FOXO function.
This β-catenin interaction with FOXO is evolutionarily conserved in mammalian cells. As in C. elegans, mammalian β-catenin physically interacts with the homologs FOXO1 and FOXO3a, and binding between β-catenin and FOXO is enhanced in response to oxidative stress. β-catenin in mammalian cells increases the expression of FOXO activity reporters as well as an endogenous cell cycle target gene (Essers et al., 2005). Moreover, at the functional level, β-catenin enhances the ability of FOXO to inhibit cell cycle progression. By contrast, β-catenin-TCF signaling has a role in cancer development. Thus, an exciting possibility is that FOXO could compete with TCF for β-catenin interaction, and thereby inhibit cell cycle and tumor progression. In support of this model, binding between β-catenin and TCF is reduced by FOXO overexpression and oxidative stress (Hoogeboom et al., 2008).
4.7 SMK-1
The highly conserved SMEK (suppressor of MEK) nuclear factor was identified in Dictyostelium discoideum as a suppressor of mek1 cell polarization and motility defects (Mendoza et al., 2005), and a role in longevity was discovered in C. elegans soon thereafter. Similar to daf-16 mutation, reduced smk-1 completely suppresses the extended longevity of a daf-2 mutant and slightly shortens the life span of wild-type worms (Wolff et al., 2006). Also like daf-16, smk-1 is required for the long life span of germ-line ablated animals. Moreover, SMK-1 is required for DAF-16-mediated innate immune, UV and oxidative stress response, but not for thermotolerance. Corresponding to these physiological data, molecular data indicate that smk-1 RNAi reduces expression of oxidative stress response (sod-3, ctl-1) and pathogen response (lys-8) genes, but does not reduce the expression of a heat-stress-inducible gene (hsp-12.6). Notably, SMK-1 is also required for the transcriptional repression of daf-15/raptor by DAF-16, suggesting that SMK-1 mediates transcriptional activator and repressor activities of DAF-16. Although it remains to be determined whether SMK-1 forms a complex with DAF-16, SMK-1 temporally and spatially co-localizes with DAF-16 in the nuclei of intestinal cells, head and hail neurons, and hypodermal cells (Wolff et al., 2006). Taken together, these data suggest the possibility that SMK-1 is nuclear coregulator that acts specifically to modulate a subset of DAF-16’s function.
4.8 HCF-1
Whereas the previously discussed putative cofactors represent positive regulators of DAF-16, the C. elegans homolog of host cell factor 1 (HCF-1) functions as a negative regulator of DAF-16 (Li et al., 2008). HCF-1 inactivation extends life span and may function in a pathway in parallel to IIS, as loss of hcf-1 and loss of daf-2/age-1 act synergistically to extend lifespan. While loss of HCF-1 increases resistance to oxidative and heavy metal stress, both effects that require DAF-16, it does not affect heat shock survival. Consistent with these uncoupled physiological outputs, HCF-1 regulates the DAF-16-mediated transcription of a subset of previously identified DAF-16 target genes. Loss of hcf-1 leads to enrichment of DAF-16 on the promoters of several DAF-16-upregulated target genes, suggesting that in the absence of HCF-1, more DAF-16 is able to bind to the promoters of its targets. HCF-1 is ubiquitously expressed, is constitutively localized to nuclei, and physically associates with DAF-16, providing evidence that HCF-1 functions as a DAF-16 co-factor or co-regulator (Li et al., 2008).
4.9 SKN-1
The C. elegans transcription factor SKN-1/Nrf defends against oxidative stress by activating the conserved phase 2 detoxification system (An and Blackwell, 2003). Additionally, SKN-1 upregulates numerous other genes involved in detoxification, cellular repair, pathogen resistance, and others, and downregulates a set of genes that reduce stress resistance and life span (Oliveira et al., 2009). SKN-1 is expressed in the intestine, where it mediates the phase 2 stress response, and in the ASI neurons (An and Blackwell, 2003). Similar to the regulation of DAF-16 by insulin signaling, the IIS kinases phosphorylate SKN-1, and reduced IIS leads to constitutive SKN-1 accumulation in intestinal nuclei and target gene activation (Tullet et al., 2008). Furthermore, SKN-1 contributes to the longevity and stress resistance phenotypes of reduced insulin signaling. These results raise the possibility that SKN-1 acts together with DAF-16 in regulating some processes and target genes. In support of this, DAF-16 contributes to the induction of a subset of SKN-1 targets under reduced insulin signaling conditions (Tullet et al., 2008). Further work will elucidate the sites and nature of SKN-1/DAF-16 interaction.
4.10 The DAF-16-Associated Element Binding Factor
An unbiased search for sequence motifs overrepresented in upstream regions of DAF-16 targets identified the in vitro-defined canonical DAF-16-binding element (DBE; TGTTTAC) (Furuyama et al., 2000), and a new GATA sequence, the DAF-16 Associated Element (DAE; CTTATCA) (Murphy et al., 2003). Both of these motifs are present in the promoters of up- and downregulated targets. Furthermore, the upstream regions of dauer-enriched, age-1-regulated genes, and age-modulated genes are also enriched for a GATA motif that is similar to that of the DAE ([T/C/G]GATAA[C/G][A/G]) (Budovskaya et al., 2008). Because the DAE differs significantly from the DBE, it is difficult to imagine how DAF-16 would also bind to the DAE; rather, a known transcription factor or currently unknown transcription factor may bind this motif and regulate these genes.
There are fourteen GATA transcription factors in C. elegans (Budovskaya et al., 2008) that are candidate DAE binding factors. Among these, RNAi knockdown of the elt-3 GATA transcription factor suppresses the longevity of daf-2 mutants (Budovskaya et al., 2008). Thus, elt-3 functions to promote life span, making it an attractive candidate for the unknown DAE binding factor. elt-3 is expressed in hypodermal cells, the intestine, the pharyngeal-intestinal valve cells, and the intestinal-rectal valve cells, and shows a complex pattern of age-related change in these tissues. Reduction of elt-3 attenuates the expression of three age-regulated genes in the tissues where elt-3 and the target is co-expressed, and mutation of the GATA DNA sites of these reporters reduces their expression. One of these genes, sod-3, is a known direct DAF-16 target (Honda and Honda, 1999) and expression of the other two increases in a reduced IIS condition. Thus, elt-3 is a reasonable candidate DAE binding factor, but it further work must be done to determine whether elt-3 directly binds these GATA sites, for example, by using ChIP-seq to compare ELT-3 targets to direct DAF-16 targets (Oh et al., 2006). Additionally, given the variations in the GATA motif reported by Budovskaya et al., it is possible that more than one GATA factor is modulating expression of these genes; indeed, the GATA factors egr-1 and egl-27 were also found to suppress the longevity of daf-2 mutants (Budovskaya et al., 2008).
5 Perspectives: Challenges and Solutions
C. elegans DAF-16/FOXO is a key node in a diverse array of physiological processes, including development, aging, immunity, and stress response. Because DAF-16/FOXO plays many roles, its fine-tuned regulation is critical for eliciting appropriate responses in response to different stimuli. As we have described here, recent work has uncovered several conserved interactions that modulate DAF-16 activity in a specific subset of its many functions. While the interactions that regulate DAF-16/FOXO are beginning to be uncovered, more work needs to be done to understand how multiple signals are integrated to elicit an appropriate context-dependent response, and the tissues that are involved in each of these responses must be identified. DAF-16’s interactions with co-factors may also specify its choice of transcriptional targets; thus the identification of those targets, the associated DNA motifs, and the associated co-factors will help uncover the mechanisms underlying the regulation of DAF-16 activity. In addition to standard genetic, biochemical, and expression microarray techniques, the recent development of such approaches as massively parallel sequencing combined with chromatin immunoprecipitation and high-throughput protein mass spectrometry should help fill these gaps in our knowledge of DAF-16/FOXO interactions.
Acknowledgments
JL is supported by a National Science Foundation Pre-doctoral Fellowship. CTM is a Richard B. Fisher Preceptor in Integrative Genomics, Lewis-Sigler Institute, Princeton.
References
- An JH, Blackwell TK. SKN-1 links C. elegans mesendodermal specification to a conserved oxidative stress response. Genes Dev. 2003;17:1882–1893. doi: 10.1101/gad.1107803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antebi A, Yeh WH, Tait D, Hedgecock EM, Riddle DL. daf-12 encodes a nuclear receptor that regulates the dauer diapauses and developmental age in C. elegans. Genes Dev. 2000;14:1512–1527. [PMC free article] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C. Cell nonautonomy of C. elegans daf-2 function in the regulation of diapause and life span. Cell. 1998;95:199–210. doi: 10.1016/s0092-8674(00)81751-1. [DOI] [PubMed] [Google Scholar]
- Baugh LR, Sternberg PW. DAF-16/FOXO regulates transcription of cki-1/Cip/Kip and repression of lin-4 during C. elegans L1 arrest. Curr Biol. 2006;16:780–785. doi: 10.1016/j.cub.2006.03.021. [DOI] [PubMed] [Google Scholar]
- Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14–3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165–1177. doi: 10.1016/j.cell.2006.04.036. [DOI] [PubMed] [Google Scholar]
- Berman JR, Kenyon C. Germ-cell loss extends C. elegans life span through regulation of DAF-16 by kri-1 and lipophilic-hormone signaling. Cell. 2006;124:1055–1068. doi: 10.1016/j.cell.2006.01.039. [DOI] [PubMed] [Google Scholar]
- Blüher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng H, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Budovskaya YV, Wu K, Southworth LK, Jiang M, Tedesco P, Johnson TE, Kim SK. An elt-3/elt-5/elt-6 GATA transcription circuit guides aging in C. elegans. Cell. 2008;134:291–303. doi: 10.1016/j.cell.2008.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers MA, Weijzen S, de Vries-Smits AM, Saarloos I, de Ruiter ND, Bos JL, Burgering BMT. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004;23:4802–4812. doi: 10.1038/sj.emboj.7600476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essers MAG, de Vries-Smits LMM, Barker N, Polderman PE, Burgering BMT, Korswagen HC. Functional interaction between β-catenin and FOXO in oxidative stress signaling. Science. 2005;308:1181–1184. doi: 10.1126/science.1109083. [DOI] [PubMed] [Google Scholar]
- Flachsbart F, Caliebe A, Kleindorp R, Blanché H, von Eller-Eberstein H, Nikolaus S, Schreiber S, Nebel A. Association of FOXO3A variation with human longevity confirmed in German centenarians. Proc Natl Acad Sci U S A. 2009;106:2700–2705. doi: 10.1073/pnas.0809594106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatt T, Min KJ, D’Alterio C, Villa-Cuesta E, Cumbers J, Lehmann R, Jones DL, Tatar M. Drosophila germ-line modulation of insulin signaling and lifespan. Proc Natl Acad Sci U S A. 2008;105:6368–6373. doi: 10.1073/pnas.0709128105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Johnson TE. A mutation in the age-1 gene in Caenorhabditis elegans lengthens life and reduces hermaphrodite fertility. Genetics. 1988;118:75–86. doi: 10.1093/genetics/118.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuyama T, Nakazawa T, Nakano I, Mori N. Identification of the diffierential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem J. 2000;349:629–634. doi: 10.1042/0264-6021:3490629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu A, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garsin DA, Villanueva JM, Begun J, Kim DH, Sifri CD, Calderwood SB, Ruvkun G, Ausubel FM. Long-lived C. elegans daf-2 mutants are resistant to bacterial pathogens. Science. 2003;300:1921. doi: 10.1126/science.1080147. [DOI] [PubMed] [Google Scholar]
- Giannakou ME, Goss M, Jnger MA, Hafen E, Leevers SJ, Partridge L. Long-lived Drosophila with overexpressed dFOXO in adult fat body. Science. 2004;305:361. doi: 10.1126/science.1098219. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Ruvkun G. daf-2, daf-16, and daf-23: genetically interacting genes controlling Dauer formation in Caenorhabditis elegans. Genetics. 1994;137:107–120. doi: 10.1093/genetics/137.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson ST, Johnson TE. daf-16 integrates developmental and environmental inputs to mediate aging in the nematode Caenorhabditis elegans. Curr Biol. 2001;11:1975–1980. doi: 10.1016/s0960-9822(01)00594-2. [DOI] [PubMed] [Google Scholar]
- Hertweck M, Gbel C, Baumeister R. C. elegans SGK-1 is the critical component in the Akt/PKB kinase complex to control stress response and life span. Dev Cell. 2004;6:577–588. doi: 10.1016/s1534-5807(04)00095-4. [DOI] [PubMed] [Google Scholar]
- Honda Y, Honda S. The daf-2 gene network for longevity regulates oxidative stress resistance and Mnsuperoxide dismutase gene expression in Caenorhabditis elegans. FASEB J. 1999;13:1385–1393. [PubMed] [Google Scholar]
- Hoogeboom D, Essers MAG, Polderman PE, Voets E, Smits LMM, Burgering BMT. Interaction of FOXO with β-catenin inhibits β-catenin/T cell factor activity. J Biol Chem. 2008;283:9224–9230. doi: 10.1074/jbc.M706638200. [DOI] [PubMed] [Google Scholar]
- Hsin H, Kenyon C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature. 1999;399:362–366. doi: 10.1038/20694. [DOI] [PubMed] [Google Scholar]
- Hsu A, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- Iser WB, Gami MS, Wolkow CA. Insulin signaling in Caenorhabditis elegans regulates both endocrine-like and cell-autonomous outputs. Dev Biol. 2007;303:434–447. doi: 10.1016/j.ydbio.2006.04.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano T, Ito Y, Ishiguro M, Takuwa K, Nakajima T, Kimura Y. Molecular cloning and characterization of a new insulin/IGF-like peptide of the nematode Caenorhabditis elegans. Biochem Biophys Res Commun. 2000;273:431–436. doi: 10.1006/bbrc.2000.2971. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Chang J, Gensch E, Rudner A, Tabtiang R. A C. elegans mutant that lives twice as long as wild type. Nature. 1993;366:461–464. doi: 10.1038/366461a0. [DOI] [PubMed] [Google Scholar]
- Kenyon C, Murphy CT. Enrichment of regulatory motifs upstream of predicted DAF-16 targets. Nat Genet. 2006;38:397–398. doi: 10.1038/ng0406-397. [DOI] [PubMed] [Google Scholar]
- Kimura KH, Tissenbaum A, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- Lee SS, Kennedy S, Tolonen AC, Ruvkun G. DAF-16 target genes that control C. elegans life-span and metabolism. Science. 2003;300:644–647. doi: 10.1126/science.1083614. [DOI] [PubMed] [Google Scholar]
- Lehtinen MK, Yuan Z, Boag PR, Yang Y, Villn J, Becker EB, DiBacco S, de la Iglesia N, Gygi S, Blackwell TK, Bonni A. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell. 2006;125:987–1001. doi: 10.1016/j.cell.2006.03.046. [DOI] [PubMed] [Google Scholar]
- Li J, Ebata A, Dong Y, Rizki G, Iwata T, Lee SS. Caenorhabditis elegans HCF-1 functions in longevity maintenance as a DAF-16 regulator. PLoS Biol. 2008;6:e233. doi: 10.1371/journal.pbio.0060233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Tewari M, Vidal M, Lee SS. The 14–3-3 protein FTT-2 regulates DAF-16 in Caenorhabditis elegans. Dev Biol. 2007;301:82–91. doi: 10.1016/j.ydbio.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kennedy SG, Ruvkun G. daf-28 encodes a C. elegans insulin superfamily member that is regulated by environmental cues and acts in the DAF-2 signaling pathway. Genes Dev. 2003;17:844–858. doi: 10.1101/gad.1066503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libina N, Berman JR, Kenyon C. Tissue-specific activities of C. elegans DAF-16 in the regulation of lifespan. Cell. 2003;115:489–502. doi: 10.1016/s0092-8674(03)00889-4. [DOI] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- McElwee J, Bubb K, Thomas JH. Transcriptional outputs of the Caenorhabditis elegans forkhead protein DAF-16. Aging Cell. 2003;2:111–121. doi: 10.1046/j.1474-9728.2003.00043.x. [DOI] [PubMed] [Google Scholar]
- Mendoza MC, Du F, Iranfar N, Tang N, Ma H, Loomis WF, Firtel RA. Loss of SMEK, a novel, conserved protein, suppresses mek1 null cell polarity, chemotaxis, and gene expression defects. Mol Cell Biol. 2005;25:7839–7853. doi: 10.1128/MCB.25.17.7839-7853.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael MD, Kulkarni RN, Postic C, Previs SF, Shulman GI, Magnuson MA, Kahn CR. Loss of insulin signaling in hepatocytes leads to severe insulin resistance and progressive hepatic dysfunction. Mol Cell. 2000;6:87–97. [PubMed] [Google Scholar]
- Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, Xu EH, Auchus RJ, Antebi A, Mangelsdorf DJ. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Murakami S, Johnson TE. A genetic pathway conferring life extension and resistance to UV stress in Caenorhabditis elegans. Genetics. 1996;143:1207–1218. doi: 10.1093/genetics/143.3.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, Lee S, Kenyon C. Tissue entrainment by feedback regulation of insulin gene expression in the endoderm of Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2007;104:19046–19050. doi: 10.1073/pnas.0709613104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy CT, McCarroll SA, Bargmann CI, Fraser A, Kamath RS, Ahringer J, Li H, Kenyon C. Genes that act downstream of DAF-16 to influence the lifespan of Caenorhabditis elegans. Nature. 2003;424:277–283. doi: 10.1038/nature01789. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Ogg S, Ruvkun G. The C. elegans PTEN homolog, DAF-18, acts in the insulin receptor-like metabolic signaling pathway. Mol Cell. 1998;2:887–893. doi: 10.1016/s1097-2765(00)80303-2. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Dixit BL, Raha T, Green MR, Tissenbaum HA. Identification of direct DAF-16 targets controlling longevity, metabolism and diapause by chromatin immunoprecipitation. Nature Genetics. 2006;38:251–257. doi: 10.1038/ng1723. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira RP, Abate JP, Dilks K, Landis J, Ashraf J, Murphy CT, Blackwell TK. Condition-adapted stress and longevity gene regulation by Caenorhabditis elegans SKN-1/Nrf. Aging Cell. 2009;8:524–541. doi: 10.1111/j.1474-9726.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabhan S, Mukhopadhyay A, Narasimhan SD, Tesz G, Czech MP, Tissenbaum HA. A PP2A regulatory subunit regulates C. elegans insulin/IGF-1 signaling by modulating AKT-1 phosphorylation. Cell. 2009;136:939–951. doi: 10.1016/j.cell.2009.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ailion M, Toker A, Thomas JH, Ruvkun G. A PDK1 homolog is necessary and sufficient to transduce AGE-1 PI3 kinase signals that regulate diapause in Caenorhabditis elegans. Genes Dev. 1999;13:1438–1452. doi: 10.1101/gad.13.11.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Ruvkun G. Caenorhabditis elegans Akt/PKB transduces insulin receptor-like signals from AGE-1 PI3 kinase to the DAF-16 transcription factor. Genes Dev. 1998;12:2488–2498. doi: 10.1101/gad.12.16.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce SB, Costa M, Wisotzkey R, Devadhar S, Homburger SA, Buchman AR, Ferguson KC, Heller J, Platt DM, Pasquinelli AA, Liu LX, Doberstein SK, Ruvkun G. Regulation of DAF-2 receptor signaling by human insulin and ins-1, a member of the unusually large and diverse C. elegans insulin gene family. Genes Dev. 2001;15:672–686. doi: 10.1101/gad.867301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Aballay A. Heat-shock transcription factor (HSF)-1 pathway required for Caenorhabditis elegans immunity. Proc Natl Acad Sci U S A. 2006;103:13092–13097. doi: 10.1073/pnas.0604050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Aballay A. Regulation of DAF-16-mediated innate immunity in C. elegans. J Biol Chem. 2009;103:13092–13097. doi: 10.1074/jbc.M109.060905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh Y, Atzmon G, Cho MO, Hwang D, Liu B, Leahy DJ, Barzilai N, Cohen P. Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc Natl Acad Sci U S A. 2008;105:3438–3442. doi: 10.1073/pnas.0705467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tullet JMA, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst A, Tertoolen LGJ, de Vries-Smits LMM, Frye RA, Medema RH, Burgering BMT. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1) J Biol Chem. 2004;279:28873–28879. doi: 10.1074/jbc.M401138200. [DOI] [PubMed] [Google Scholar]
- van Hemert MJ, Steensma HY, van Heusden GP. 14–3-3 proteins: key regulators of cell division, signalling and apoptosis. BioEssays. 2001;23:936–946. doi: 10.1002/bies.1134. [DOI] [PubMed] [Google Scholar]
- Vowels JJ, Thomas JH. Genetic analysis of chemosensory control of dauer formation in Caenorhabditis elegans. Genetics. 1992;130:105–123. doi: 10.1093/genetics/130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Shakes DC. Expression patterns and transcript processing of ftt-1 and ftt-2, two C. elegans 14–3-3 homologues. J Mol Biol. 1997;268:619–630. doi: 10.1006/jmbi.1997.1002. [DOI] [PubMed] [Google Scholar]
- Wang Y, Oh SW, Deplancke B, Luo J, Walhout AJ, Tissenbaum HA. C. elegans 14–3-3 proteins regulate life span and interact with SIR-2.1 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:741–747. doi: 10.1016/j.mad.2006.05.005. [DOI] [PubMed] [Google Scholar]
- Wilcox BJ, Donlon TA, He Q, Chen R, Grove JS, Yano K, Masaki KH, Willcox DC, Rodriguez B, Curb JD. FOXO3A genotype is strongly associated with human longevity. Proc Natl Acad Sci U S A. 2008;105:13987–13992. doi: 10.1073/pnas.0801030105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff S, Ma H, Burch D, Maciel GA, Hunter T, Dillin A. SMK-1, an essential regulator of DAF-16-mediated longevity. Cell. 2006;124:1039–1053. doi: 10.1016/j.cell.2005.12.042. [DOI] [PubMed] [Google Scholar]
- Wolkow CA, Kimura KD, Lee MS, Ruvkun G. Regulation of C. elegans life-span by insulinlike signaling in the nervous system. Science. 2000;290:147–150. doi: 10.1126/science.290.5489.147. [DOI] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- Yan L, Lavin VA, Moser LR, Cul Q, Kanles C, Yang E. PP2A regulates the pro-apoptotic activity of FOXO1. J Biol Chem. 2008;283:7411–7420. doi: 10.1074/jbc.M708083200. [DOI] [PMC free article] [PubMed] [Google Scholar]


