SUMMARY
The molecular processes that contribute to degenerative diseases are not well understood. Recent observations suggest that some degenerative diseases are promoted by the accumulation of nuclear or cytoplasmic RNA-protein (RNP) aggregates, which can be related to endogenous RNP granules. RNP aggregates arise commonly in degenerative diseases because RNA binding proteins commonly self-assemble, in part through prion-domains, which can form self-propagating β-amyloids. RNP aggregates may be toxic due to multiple perturbations of post-transcriptional control, thereby disrupting the normal ribostasis of the cell. This suggests that understanding and modulating RNP assembly or clearance may be effective approaches to develop therapies for these diseases.
Introduction
Degenerative diseases are a poorly understood and largely untreatable set of pathologies that take a heavy toll in disability and death and have far-reaching socioeconomic impacts. Thus, illuminating the molecular causes of degenerative diseases to inform therapeutic strategies is an important area of research.
Over the past decade, several observations have highlighted important connections between protein aggregation, RNA biology and a subset of degenerative diseases (Table 1). A conspicuous feature of some degenerative diseases, particularly neurodegenerative diseases, is cytoplasmic or nuclear aggregates of RNA binding proteins and/or RNA. For example, redistribution of RNA-binding proteins, most notably TDP-43, from the nucleus to cytoplasmic inclusions is a hallmark pathological feature of most sporadic and familial forms of ALS, FTLD-U, and is also present in a subset of other neurodegenerative diseases (Chen-Plotkin et al., 2010). Toxic RNAs can also cause pathologies as a mutant RNA arising from a hexanucleotide expansion in the C9ORF72 gene leads to that RNA’s accumulation in nuclear foci and underlies some cases of ALS (DeJesus-Hernandez et al., 2011; Renton et al., 2011). Similarly, a relatively short CGG expansion in the fragile-X gene (FMR1) leads to a toxic RNA that also accumulates in nuclear foci and causes a late onset neurodegenerative disorder termed fragile-X tremor ataxia syndrome (FXTAS; (Hagerman and Hagerman, 2004).
Table 1.
Examples of Protein-Folding and RNP Diseases
| Diseases | Mutated Protein/Gene | Details and References |
|---|---|---|
|
| ||
|
Protein Folding Diseases: caused by point mutations that destabilize protein domains
| ||
| Alzheimer’s Parkinsons | APP Alpha Synuclein (A53T, A30P and E46K mutations | (Schellenberg and Montine, 2012) (Li et al., 2001; Zarranz et al., 2004). |
| ALS (a) | SOD1 | (Rosen et al., 1993) |
| Myofibrillar myopathy | Myotilin, Desmin | (Baloh, 2011; Olive et al., 2009) |
|
| ||
|
RNP Diseases: caused by mutations in RNAs or RNA-binding proteins
| ||
| Spinocerebellar ataxias (SCA1–3) | Polyglutamine expansions in respective RNA-binding proteins. | PolyQ expansions enhance homotypic and heterotypic aggregation of complexes. (De Los Rios et al., 2012; Ross and Poirier, 2004) |
| SCA-8 | CUG expansion in a non-coding RNA | (Daughters et al., 2009; Mutsuddi et al., 2004) Fly models for this are suppressed by mutations in staufen and two other RNA-binding proteins) |
| SCA-10 | Possibly due to an aberrant, stable intronic RNA (AUUCU expansion) | (Matsuura et al., 2000; White et al., 2011) |
| ALS (b) | Atx2, TDP-43, FUS, (prion-like domain mutations), hnRNPA1, hnRNPA2B1, VCP (mutations cause increased granule assembly). | Mutations may result in increased aggregation. (Couthouis et al., 2012; Elden et al., 2010; Johnson et al., 2009; Johnson et al., 2010; Kim et al., 2013; Liu-Yesucevitz et al., 2011; Neumann et al., 2006; Sreedharan et al., 2008; Vance et al., 2009) |
| ALS (c) | Microsatellite (GGGGCC)n expansion in C9ORF72 RNA. [Sequestration of RNA-binding proteins in RNA foci is a likely mechanism contributing to disease] | (DeJesus-Hernandez et al., 2011; Renton et al., 2011) |
| DM1 and 2 | Aberrant RNA | (Davis et al., 1997; Mankodi et al., 2001) |
| FXTAS | Aberrant RNA | (Hashem et al., 2009; Iwahashi et al., 2006; Tassone et al., 2004). |
| Laminopathies | Lam-A or Lam-C mutations with defects in RNA granule assembly in the nucleus. | (Speese et al., 2012) |
| Oculopharyngeal muscular dystrophy | Polyalanine expansion in PABPN1 | (Brais et al., 1998) |
| Welander distal myopathy | TIA1 (prion-like domain mutation that increases stress granule assembly) | (Hackman et al., 2012) |
| Distal myopathy (vocal cord and pharyngeal weakness variant) | Matrin-3 | (Senderek et al., 2009) |
|
| ||
|
Possibly RNP-Folding Diseases: associated with RNP aggregates.
| ||
| Huntington’s | Htt. | P-body and dendritic RNA granule protein associated with Ago-2 (Savas et al., 2010; Savas et al., 2008). |
|
| ||
| Prion disease | Prp mutations | Increased aggregation, potentially with RNAs. (Solomon et al., 2012) |
|
| ||
| Others (Eg. Alzheimer’s) | Sporadic forms | RNA and TDP-43 frequently reported in associated inclusions. (Davidson et al., 2011; Tremblay et al., 2011; Wilson et al., 2011) |
RNP aggregates are not limited to neurodegeneration. For example, myotonic dystrophy types 1 and 2 are caused by microsatellite expansions in noncoding regions of the RNAs encoding DMPK and ZNF9, respectively. The pathology in both of these disorders is characterized by nuclear RNP aggregates that sequester splicing factors (Fardaei et al., 2002; Miller et al., 2000), and Welander distal myopathy is caused by a mutation in the RNA binding protein, TIA1, with accompanying cytosolic inclusion pathology (Hackman et al., 2012; Klar et al., 2013).
Here, we present a mechanistic framework for RNP-inclusion disease, which is based on the premise that RNP inclusions seen in (neuro)degenerative conditions are indicative of and arise from pathological changes in endogenous cellular pathways for RNP assembly and clearance. We suggest that the prevalence of such RNP inclusion diseases arises from the fact that RNPs normally form large RNP assemblies and therefore are poised for pathogenicity. These principles are highlighted in a model for how specific forms of amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration (FTLD) and multisystem proteinopathy (MSP) may originate from altered assembly or clearance of stress granules, a well-studied class of cytoplasmic RNP granule. Shared features of RNP assembly and misfolded protein handling pathways, such as the involvement of HSP70 and autophagy, may explain some overlap between mRNP-inclusion and misfolded protein disease pathologies. We present an altered ribostasis model to account for cell loss in RNP-inclusion diseases and discuss how understanding of endogenous RNP regulation may inform and instruct therapeutic strategies.
RNP aggregates seen in pathologies are related to endogenous RNP assemblies
An emerging theme is that the RNP inclusions seen in pathologies are related to endogenous RNP granules that are part of the normal metabolism of eukaryotic mRNAs and could reflect a disturbance in the assembly, disassembly or clearance of these structures (Figure 1). During transcription, nascent mRNAs in the nucleus assemble with a set of RNA binding proteins (Moore, 2005). These RNA binding proteins, including TDP-43, hnRNPA1, hnRNPA2B1 and others involved in neurodegenerative disease, affect pre-mRNA processing and are transported with the mRNA to the cytosol where they are removed by translation dependent and independent mechanisms for recycling into the nucleus (Moore, 2005). In the cytosol, when mRNAs are not engaged in translation they assemble into either P-bodies, which contain mRNAs, translation repressors, components of the miRNA silencing pathway, and the mRNA decay machinery (Parker and Sheth, 2007), or stress granules, which contain mRNAs associated with mRNA binding proteins and a subset of translation initiation factors. Stress granules accumulate to higher levels in cells or conditions, such as stress, where translation initiation is limited (Anderson and Kedersha, 2009; Buchan and Parker, 2009). Cytoplasmic mRNAs exchange between polysomes, P-bodies, and stress granules creating a dynamic cycle of mRNP exchanges that influences the translation and degradation of mRNAs (Figure 1). Stress granules can contain nuclear mRNA binding proteins suggesting that when entry into translation is compromised, nascent mRNPs accumulate in these structures before exchange of nuclear RNA binding proteins (Buchan and Parker, 2009).
Figure 1. mRNP remodeling and aggregation in the life cycle of an mRNA.
Transcribed RNAs form nuclear mRNPs. RNA-binding proteins associated with mRNA are transported to the cytosol, where they determine cytoplasmic localization, and translational competence of the mRNA. mRNAs begin to assemble translation complexes and then this process is stalled, the mRNPs accumulate as stress granules. mRNPs within stress granules can return to translation initiation and enter polysomes, or be targeted for autophagy. Following translation, mRNAs can exit translation and assemble a translationally repressed mRNP that can either be degraded or assemble into P-bodies. mRNAs within P-bodies can be subject to decapping and 5′ to 3′ degradation, or can exchange P-body components for stress granule components to re-enter translation. Stress granules are proposed to undergo pathological transitions wherein loose assemblies of prion-like domains can form irreversible β-amyloid structures.
Three observations suggest that the TDP-43 positive RNP aggregates seen in some ALS, FTLD-U patients are related to stress granules. First, inclusions seen in pathologies can contain other markers of stress granules (Liu-Yesucevitz et al., 2010; Wolozin, 2012). Second, the RNA-binding proteins that can become pathogenic when mutated (e.g. hnRNPA1, TDP-43, FUS, ATX2) all normally assemble into stress granules in response to stress stimuli (Dormann et al., 2010; Guil et al., 2006). Finally, the pathogenic alleles of several of these proteins lead to constitutive formation of stress granules in tissue culture cells, even in the absence of a stress response (Kim et al., 2013; Liu-Yesucevitz et al., 2010; Wolozin, 2012).
An analogous situation may exist in diseases characterized by aberrant accumulation of nuclear RNP aggregates, such as myotonic muscular dystrophy types 1 and 2, FXTAS, or with the C9ORF72-related ALS/FTD. These nuclear RNP inclusions may be related to a number of RNA-protein assemblies that normally form in the nucleus (Spector, 2006). One interesting possibility is that these nuclear RNA-foci are related to a specialized mega-RNP assembly containing lamin A, which plays a role in mRNA export by vesicle budding through the nuclear envelope (Speese et al., 2012). This is a particularly interesting possibility for the RNA foci seen in FXTAS since they are known to contain lamins A and C (Iwahashi et al., 2006).
The Assembly Properties of cytoplasmic RNP granules poise them for pathogenicity
An important point is that mRNP granules assemble through protein-protein interactions including interactions between “prion-like” domains. This was first observed with evidence that stress granules in mammalian cells assemble in part through multimerization of a prion-like domain in the TIA-1 protein (Gilks et al., 2004). A prion domain in the Lsm4 protein was also shown to be part of the assembly mechanism for yeast P-bodies (Decker et al., 2007). Moreover, many RNA binding proteins contain such prion-like domains, or other low complexity domains, that can mediate assembly into higher order structures in vivo and in vitro (Decker et al., 2007; Gilks et al., 2004; Kato et al., 2012; Kim et al., 2013; Reijns et al., 2008); Li et al., 2013). Thus, an emerging principle is that cytoplasmic RNP granules such as stress granules and P-bodies assemble due to the presence of prion-like polymerization domains on many RNA binding proteins, and these domains reversibly assemble and disassemble in the normal metabolism of cytoplasmic mRNPs (Figure 2).
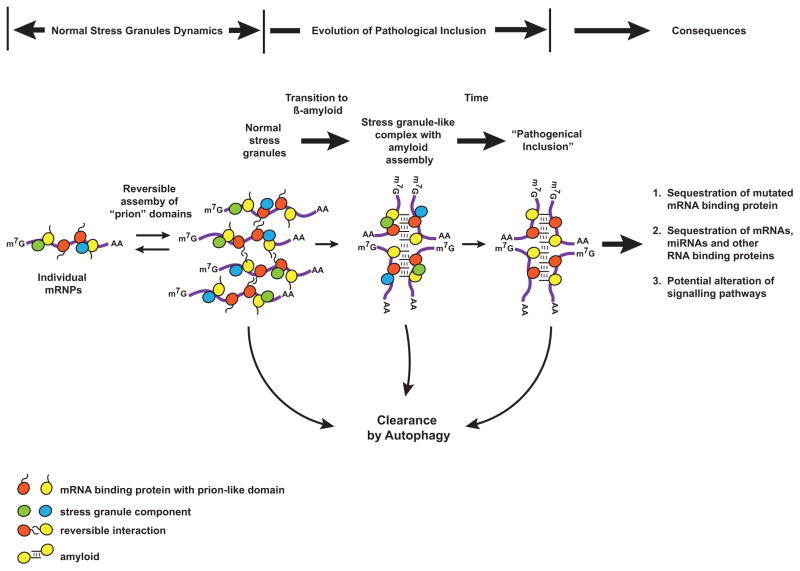
Figure 2. Normal Stress granule dynamics and possible evolution of pathogenic inclusions.
Cytoplasmic RNP granules such as stress granules and P-bodies assemble due to the presence of prion-like polymerization domains on many RNA binding proteins. In the normal metabolism of cytoplasmic mRNPs these granules reversibly assemble and disassemble, or persistent stress granules may be cleared by autophagy, controlling the content of the translatable pool of mRNAs. We suggest that stable amyloid assemblies may be formed stochastically from normal stress granules and, further, that mutations affecting prion-like domains, or an increased number or persistence of stress granules, increases the probability of this transition. Over time these stable, persistent mRNP assemblies could evolve into the pathological inclusions characteristic of disease and impair ribostasis by sequestration of RNAs or RNA-binding proteins, or by altering signaling pathways.
The high prevalence of prion-like domains in RNA binding proteins creates a large genetic target for pathogenic mutations that create hyperaggregation variants with downstream toxic effects. For example, pathogenic mutations in the prion-like domains of hnRNPA1 and hnRNPA2B1 are predicted to increase its aggregation potential, show increased aggregation in vitro, and lead to hyperassembly of stress granules in cultured cells (Kim et al., 2013). Similarly, expansion of a polyglutamine domain in the Atx2 protein, which is a subclass of prion-like domains, leads to hyperaggregation of the Atx2 proteins, which can be a risk factor in ALS (Elden et al., 2010), and when extreme enough leads to spinocerebellar ataxia 2 (Orr and Zoghbi, 2007). The role of RNA binding proteins with prion-like domains in degenerative pathologies may be quite common as 10 of the top 20 human RNA binding proteins predicted to have such domains have been linked to some form of degenerative disease (Li et al., 2013). This prevalence suggests that hyperassembly of RNP aggregates related to stress granules may be a causative event in disease progression.
Additional evidence pointing to inappropriate stress granule formation as being toxic comes from the analysis of the VCP, or p97, protein. VCP is a highly conserved AAA-ATPase that plays a role, through multiple different adaptors, in segregating or rearranging a variety of complexes containing ubiquinated proteins (Meyer et al., 2012; Yamanaka et al., 2012). Mutations in VCP can be causative in multisystem proteinopathy (MSP) and ALS and give rise to the same spectrum of pathologies as dominant hyperaggregation mutations in TDP-43, hnRNPA1, or hnRNPA2B1 (Dewey et al., 2013). Three observations suggest this overlap in pathologies is due to a molecular overlap affecting stress granule dynamics. First, pathogenic mutations in VCP lead to the constitutive accumulation of TDP-43, hnRNPA1, hnRNPA2B1 and other stress granule markers in cytoplasmic foci (Dewey et al., 2013). Second, RNAi-mediated knockdown of VCP in mammalian cells or loss of function mutations in the yeast ortholog, CDC48 lead to the persistence or constitutive formation of stress granules due to defects in stress granule clearance (Buchan et al., 2013). Finally, over-expression of VCP harboring disease mutations inhibits the clearance of stress granules, at least in part by limiting their removal by autophagy (Buchan et al., 2013).
Since the same spectrum of diseases is caused by a variety of mutations in RNA binding proteins that promote RNP aggregation and/or stress granule formation, as well as mutations in VCP/Cdc48 that reduce stress granule clearance, a reasonable hypothesis is that hyper formation or persistence of stress granules is an underlying causative event in these diseases.
A working model for ALS, FTLD and MSP
The current observations suggest a working model whereby the pathology of these diseases is driven by the assembly of specific RNA binding proteins into self-propagating hyperstable amyloids (Figure 2), which form RNP aggregates and disrupt ribostasis eventually leading to cell death. In principle, amyloids could form during the normal assembly of stress granules and be continually disassembled by cellular mechanisms (e.g. chaperonins). However, since stress granules are highly dynamic (t1/2~30 sec by FRAP; Buchan and Parker, 2009), and prion-like domains can also form looser more reversible interactions (Eisenberg and Jucker, 2012), we favor a model wherein hyperstable amyloid assemblies are not part of the normal assembly mechanisms of stress granules, and instead can form stochastically from the looser interactions between prion-like domains that would predominate within normal stress granules. In this view, the frequency of amyloid formation would be affected by mutations in RNA binding proteins that increase the tendency to form a stable amyloid structure (Kim et al., 2013). In addition, since amyloid initiation is a stochastic event dependent on concentration and time (Eisenberg and Jucker, 2012), any alteration that increases the size or persistence of the high local concentration of RNA binding proteins in stress granules, such as defects in clearance mechanisms due to VCP mutations (Buchan et al., 2013) is expected to yield an increased frequency of amyloid initiation. Moreover, environmental insults that promote ALS can be understood in this context as triggering stress granule formation. For example, examination of the brains of boxers and trauma patients reveals the accumulation of TDP-43 in cytoplasmic aggregates (King et al., 2010; McKee et al., 2010; Stern et al., 2011), which may be remnants of trauma induced stress granule formation.
RNP aggregates that contain amyloid structures may no longer be capable of being disassembled by normal mechanisms and could only be cleared by autophagy. The self-perpetuation of such toxic assemblies might be tolerated in younger individuals but as autophagy declines with age, the clearance of such assemblies would slow down until a toxic level was reached, where disease would be triggered.
An interesting, and important area of future research, is the potential role that cell to cell transmission of RNA-protein aggregates plays in disease progression. This is an issue since it now appears that many “protein-folding” diseases such as Alzheimer’s disease, Parkinson’s disease, Tauopathies, and Huntington’s disease can be transmitted between cells in a “prionoid” type of mechanism that is poorly understood (Aguzzi and Rajendran, 2009). RNA-protein aggregates forming in these diseases could play a similar role, either by being taken up by neighboring cells after cell death and lysis, or by specific mechanisms that transfer material between cells. For example, RNA and protein complexes can be secreted from cells in exosomes (Raposo and Stoorvogel, 2013), which if taken up by neighboring cells might also transfer a self-perpetuating RNA-protein complex.
Does RNA matter?
Several observations argue that at least part of the pathology of these mutations involves their ability to bind RNA. First, this is implied by the plethora of mutations in RNA binding proteins that are causative in similar pathologies (Li et al., 2013). Second, in yeast and Drosophila model systems, mutations that disrupt the RNA binding ability of FUS, or TDP-43 reduces or prevents their toxicity (Daigle et al., 2013; Johnson et al., 2009; Sun et al., 2011; Voigt et al., 2010). Third, in yeast or Drosophila, other mutations that alter RNA physiology can enhance or suppress the toxicity of pathogenic mutations. For example, overexpression of Pbp1, the yeast ortholog of Atx2, enhances TDP-43 toxicity in yeast and increases aggregates of RNA binding proteins (Swisher and Parker, 2012), while loss of Pbp1 suppresses TDP-43 toxicity and reduces stress granule formation (Buchan et al., 2008); analogous interactions between the two proteins are also seen in the Drosophila eye (Elden et al., 2010). The Ataxin-2 and TDP-43 interaction is probably significant to neurodegeneration because genetic variants in both genes have been independently associated with human ALS, and because the two proteins co-localize in stress granules as well as in pathological inclusion bodies observed in patients (Bonini and Gitler, 2011). Similarly, mutations in the orthologs of hnRNPA1 or hnRNPA2B1 in Drosophila suppress the toxicity of pathogenic VCP mutations (Ritson et al., 2010). Taken together, these observations argue that toxicity in these conditions is caused by the formation of RNA-protein aggregates. Thus, a critical question is why an increased load of RNA-proteins aggregates leads to toxicity?
Toxicity and Altered Ribostasis
We suggest that aberrant RNP aggregates are pathological, at least in part, because they disrupt RNA homeostasis or “ribostasis”. We define ribostasis as the appropriate production and regulation of the cellular transcriptome, which has downstream effects on protein homeostasis. This is analogous to the concept of protein folding diseases where age-dependent changes in proteostasis contribute to some pathologies by saturating proteostatic mechanisms, resulting in altered proteomes and a variety of physiological changes that eventually result in cell death (Balch et al., 2008; Douglas and Dillin, 2010; Garber, 2009; Kopito and Ron, 2000; Muchowski and Wacker, 2005; Vij, 2011). Despite the parallels to proteostatic mechanisms, ribostasis should be considered to be a distinct aspect of cell homeostasis for two reasons. First, the mechanisms that underlie biogenesis, maturation, localization, translational control and degradation of RNA use distinct enzymatic and molecular machinery. Second, cellular phenotypes and pathologies caused by alterations in ribostasis may have features distinct from those caused by primary defects in mechanisms of protein handling (see below).
Formation or persistence of mRNP aggregates is anticipated to alter ribostasis by three different overlapping mechanisms, which may synergize to trigger cell toxicity (Figure 2). First, mRNP hyperaggregation could reduce the functional pool of the mutated aggregation-prone mRNA binding protein. Second, by sequestering associated mRNAs, RNP aggregates could alter the population of mRNAs available for translation. Third, RNP aggregates could sequester other mRNA binding regulatory factors including other mRNA binding proteins, miRNAs, and even ncRNAs such that they are not available for their normal functions. Sequestration of mRNA regulatory factors would be expected to alter the translation, localization or decay of many mRNAs, as well as affecting mRNA biogenesis. For example, sequestration of the splicing factor MBNL1 into nuclear RNP foci in myotonic dystrophies 1 and 2 results in alterations of splicing (Fardaei et al., 2002; Mankodi et al., 2001; Miller et al., 2000), and TDP-43 mislocalization can alter its own mRNAs regulation (Igaz et al., 2011). Thus, altered ribostasis can alter both the biogenesis and function of mRNAs, potentially creating a positive feedback loop where abnormal RNAs and altered protein production leads to more misregulation of cellular processes, eventually leading to systemic inflammatory signaling and cell death. In addition, aberrant mRNP aggregates are expected to influence other aspects of RNA metabolism since some of the aggregated proteins (e.g. FUS or TDP-43) can be required for transcription and/or snRNP assembly (Ishihara et al., 2013; Schwartz et al., 2012).
Aberrant mRNP aggregates are also likely to alter cell survival by affecting signaling pathways. This is a possibility since RNP aggregates can impact cell-signaling pathways that alter mediate the cellular response to environmental inputs and/or lead to cell death (Buchan and Parker, 2009; Thomas et al., 2010). In addition, it is possible that the aberrant RNP aggregate itself is sufficient to trigger a cellular response, which might lead to cell death. For example, progression of prion disease-related PrP pathology in mice requires phosphorylation of eIF2 (Moreno et al., 2012), a translation initiation factor whose phosphorylation triggers the accumulation of stress granules (reviewed in Buchan and Parker, 2009). Any such alterations in cell signaling might synergize with perturbation of ribostasis to trigger cell death.
Cross Talk between Protein Folding and RNP-Assembly Aggregates
An important point is that there is cross talk between mechanisms that maintain proteostasis and ribostasis, which may explain the clinical overlap in diseases associated with perturbations of these two homeostatic mechanisms. For example, muscles of patients with classic protein-aggregate myofibrillar myopathies caused by mutations in Desmin or the chaperone DNAJB6 contain cytoplasmic aggregates of the RNA binding protein TDP-43, a component of stress granules (Baloh, 2011; Olive et al., 2009). Thus, secondary ribostasis defects may be seen in cells primarily compromised by an increased misfolded protein load. Such cross talk could occur either due to the depletion of cytoplasmic chaperones such as HSP70, which regulate both protein and RNP aggregation in vivo (Gilks et al., 2004), or through recruitment of misfolded domains to common aggregates. An important corollary, that primary defects in ribostasis should not only increase RNP aggregation but also influence the protein-folding pathways remains to be rigorously tested, e.g. by examining the possible activation of unfolded protein response following perturbations to RNA regulatory pathways.
Cross talk between proteostasis and ribostasis is also evident in the emerging role of autophagy in clearing RNP aggregates from cells (Zhang et al., 2009). Thus, it is expected that impairment in autophagy either through age-related decline or genetic mutation (e.g. VCP mutations) not only perturbs proteostasis, but disrupts the normal clearance of stress granules resulting in perturbed ribostasis. The existence of these pathways emphasizes that some common pathologies may arise through distinct trigger mechanisms, and indicates the need for caution in classifying neurodegenerative diseases as arising from protein-folding or mRNP assembly defects based solely on limited characterization of inclusion body pathology.
Why cell-type specific degenerative disease?
A prominent but enigmatic feature of age-related degenerative diseases is cell type-selective degeneration, with brain and muscle most frequently affected. Moreover, individual diseases have characteristic patterns of cell type involvement. For example, ALS typically shows the greatest impact on motor neurons of the frontal cortex, brainstem and spinal cord; whereas degeneration in FTD is typically restricted to the cerebrum, but spread over a broader area of cortex involving the frontal and temporal lobes. Remarkably, these cell type-selective degeneration patterns persist even in dominantly inherited disease where the mutant protein (e.g. TDP-43) is ubiquitously expressed. Thus, an important question is why these defects in RNP or protein folding mechanisms have a disproportionate impact on certain cells? And, how and why do these show their remarkable dependence on age? The answer to these questions must lie in some of the unique properties of the vulnerable cell types, which might relate to: a) their longevity; b) their unusual cell biology and cytoarchitecture; and c) their connectivity (Figure 3). One possibility is that since neurons and muscle cells are unusually long-lived cells, they may have increased vulnerability to time-dependent accumulation of sublethal damage. Whereas shorter-lived cells turn over before such damage reaches a pathological level, perhaps longer-lived cells accumulate age-related damage to pathological levels resulting in cellular dysfunction. Alternatively, non-neuronal cells may also die in response to RNP or protein-folding defects but, because they are efficiently replaced by stem cells, may not cause visible organismal dysfunction.
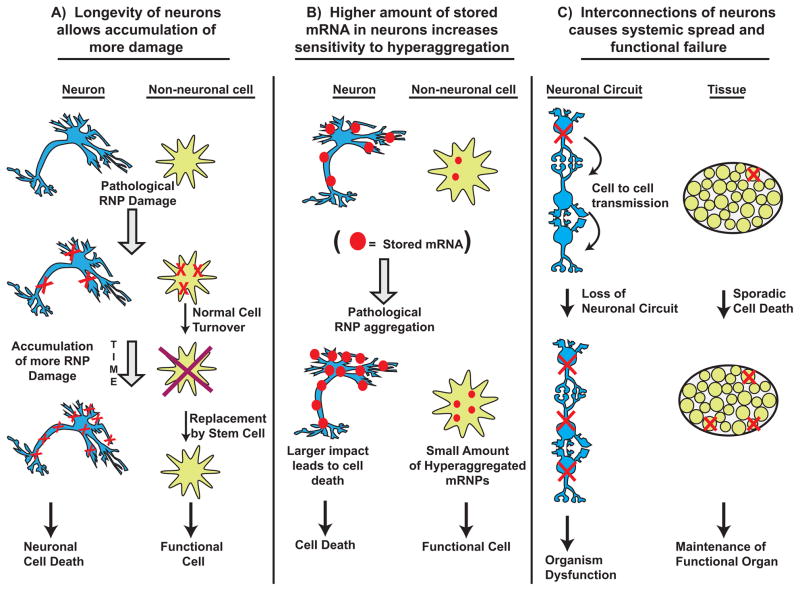
Figure 3. Three models to account for the special sensitivity of neurons to altered ribostasis caused by enhanced levels of mRNA aggregation.
A. Neurons are long-lived cell and therefore accumulate more damage over their lifetimes. In addition, because they are not efficiently replaced by stem cells, neuronal loss is more consequential that the death of a non-neuronal cell like a fibroblast. B. Neurons have higher levels of translationally repressed mRNAs than other cells, particularly in their dendrites and axons. They may therefore be more easily tripped into a pathological state: either by being more responsive to aggregation-inducing conditions, or by being more sensitive to altered RNA regulation. C. The connectivity of neurons results in: (a) enhanced spread of mRNP aggregation from a triggering neuron to a synaptically connected one; or (b) a neuron being particularly susceptible to cell death, when it has lost a synaptic partner on which in may depend for survival signals.
The unusual cellular architecture of neurons and muscle, with accompanying increased requirement for spatially restricted gene expression, may also contribute to their selective vulnerability. For example, neurons contain mRNP granules related to stress granules and P-bodies, both in the soma and at synapses (Barbee et al., 2006). Moreover, components of these stress granules are required for synaptic plasticity, at least in model systems (Kwak et al., 2008; McCann et al., 2011). Neurons also contain a related set of RNP granules referred to as neuronal RNA transport granules that deliver translationally repressed mRNAs to synaptic sites in dendrites, where synaptic activity promotes their localized translation (Batish et al., 2012; Holt and Bullock, 2009). The translation of these mRNAs at synapses, presumably necessary for normal neuronal function, may place unusual demands on local mRNA synthesis mechanisms to maintain the synaptic proteome or respond to synaptic activity (Cajigas et al., 2012). In addition, it is possible that retrograde communication to the nucleus that arises from local translation of synaptic mRNAs could mediate cell survival or cell maintenance signaling (Lin and Holt, 2008). Consistent with a scenario in which altered translational control promotes cell death by blocking synaptically derived survival signals: (i) the SCA2-associated, stress-granule protein Atx2 is required for normal translational control in Drosophila synapses (McCann et al., 2011); and (ii) synaptic degeneration has been found to precede neuronal loss in systems where this issue has been analyzed (Mallucci, 2009; Selkoe, 2002). Likewise, in skeletal muscle cells the development and maintenance of certain specialized structures such as the postsynaptic neuromuscular junction and possibly the contractile apparatus requires spatial control of local protein translation (Liu et al., 2012a; Sanes and Lichtman, 2001).
Finally, tissue degeneration that characterizes neurodegenerative disease is not solely cell autonomous, and involves mechanisms for tissue-wide spread of disease that may include the dissemination of toxic aggregates between synaptically connected neurons (Brundin et al., 2010; de Calignon et al., 2012; Lee et al., 2010; Liu et al., 2012b), which might also contribute to the sensitivity of the nervous system to degenerative diseases.
Altered Ribostasis: Looking Forward
The involvement of RNP assembly perturbations in neurodegenerative diseases highlights future directions. It will be critical to understand how both normal and pathological RNP aggregates are assembled, disassembled and cleared in neurons and muscle cells. Equally important will be understanding the relationship between the normal RNP assemblies and the pathological aggregates that characterize end-stage disease. Do the pathological inclusions in ALS, FTD, IBM and related diseases evolve from normal RNP assemblies? In addition, for analysis of human exome and genome sequences from patients with familial and sporadic neurodegenerative disease, genes encoding components of RNP granules or factors that regulate their assembly, disassembly or clearance should be considered as candidate neurodegeneration susceptibility genes e.g. (Buchan et al., 2013; Ohn et al., 2008). Identifying disease-causative genes will offer new opportunities for diagnosis, which should improve the ability to predict patient-specific neurological symptoms and their rates of progression. Finally, it will be important to explore the concept of directly targeting RNP granule assembly, disassembly or clearance as a strategy of therapeutic intervention. Possible therapeutic approaches include drugs that: a) inhibit signaling/assembly mechanisms that promote toxic aggregate formation; b) activate mechanisms that antagonize aggregate formation; or c) promote clearance of toxic aggregates (Pandey et al., 2007), for instance by using drugs such as rapamycin to trigger a burst of autophagy which might have both prophylactic and therapeutic benefits.
An unresolved issue is how frequently perturbations in ribostasis will be involved in other pathologies. Reports have described the presence of RNA and RNA binding proteins in aggregates that form in prion disease, taupathies and Alzheimer’s raising the speculation that many of these aggregates will be RNA-protein assemblies (Ginsberg et al., 1998). Thus, an important future direction will be to discern which diseases involve RNA in the pathology and what those roles suggest for diagnostic and therapeutic interventions.
Acknowledgments
Our research is primarily funded by: Science Foundation Ireland (MR); the Target ALS Foundation and US NIH NS053825 (JPT); the HHMI and US NIH GM045443 (RP). We acknowledge useful discussions and feedback from members of our laboratories as well as many colleagues including Chris Weihl, Nancy Bonini, Mel Feaney and Leo Pallanck. MR acknowledges K. VijayRaghavan and the NCBS, Bangalore for their support.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguzzi A, Rajendran L. The transcellular spread of cytosolic amyloids, prions, and prionoids. Neuron. 2009;64:783–790. doi: 10.1016/j.neuron.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. Stress granules. Curr Biol. 2009;19:R397–398. doi: 10.1016/j.cub.2009.03.013. [DOI] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW. Adapting proteostasis for disease intervention. Science. 2008;319:916–919. doi: 10.1126/science.1141448. [DOI] [PubMed] [Google Scholar]
- Baloh RH. TDP-43: the relationship between protein aggregation and neurodegeneration in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. FEBS J. 2011;278:3539–3549. doi: 10.1111/j.1742-4658.2011.08256.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Estes PS, Cziko AM, Hillebrand J, Luedeman RA, Coller JM, Johnson N, Howlett IC, Geng C, Ueda R, et al. Staufen- and FMRP-containing neuronal RNPs are structurally and functionally related to somatic P bodies. Neuron. 2006;52:997–1009. doi: 10.1016/j.neuron.2006.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batish M, van den Bogaard P, Kramer FR, Tyagi S. Neuronal mRNAs travel singly into dendrites. Proc Natl Acad Sci U S A. 2012;109:4645–4650. doi: 10.1073/pnas.1111226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonini NM, Gitler AD. Model Organisms Reveal Insight into Human Neurodegenerative Disease: Ataxin-2 Intermediate-Length Polyglutamine Expansions Are a Risk Factor for ALS. J Mol Neurosci. 2011 doi: 10.1007/s12031-011-9548-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundin P, Melki R, Kopito R. Prion-like transmission of protein aggregates in neurodegenerative diseases. Nat Rev Mol Cell Biol. 2010;11:301–307. doi: 10.1038/nrm2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Kolatis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013 doi: 10.1016/j.cell.2013.05.037. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchan JR, Parker R. Eukaryotic stress granules: the ins and outs of translation. Mol Cell. 2009;36:932–941. doi: 10.1016/j.molcel.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cajigas IJ, Tushev G, Will TJ, tom Dieck S, Fuerst N, Schuman EM. The local transcriptome in the synaptic neuropil revealed by deep sequencing and high-resolution imaging. Neuron. 2012;74:453–466. doi: 10.1016/j.neuron.2012.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen-Plotkin AS, Lee VM, Trojanowski JQ. TAR DNA-binding protein 43 in neurodegenerative disease. Nat Rev Neurol. 2010;6:211–220. doi: 10.1038/nrneurol.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couthouis J, Hart MP, Erion R, King OD, Diaz Z, Nakaya T, Ibrahim F, Kim HJ, Mojsilovic-Petrovic J, Panossian S, et al. Evaluating the role of the FUS/TLS-related gene EWSR1 in amyotrophic lateral sclerosis. Hum Mol Genet. 2012 doi: 10.1093/hmg/dds116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle JG, Lanson NA, Jr, Smith RB, Casci I, Maltare A, Monaghan J, Nichols CD, Kryndushkin D, Shewmaker F, Pandey UB. RNA-binding ability of FUS regulates neurodegeneration, cytoplasmic mislocalization and incorporation into stress granules associated with FUS carrying ALS-linked mutations. Hum Mol Genet. 2013;22:1193–1205. doi: 10.1093/hmg/dds526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:e1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson YS, Raby S, Foulds PG, Robinson A, Thompson JC, Sikkink S, Yusuf I, Amin H, DuPlessis D, Troakes C, et al. TDP-43 pathological changes in early onset familial and sporadic Alzheimer’s disease, late onset Alzheimer’s disease and Down’s syndrome: association with age, hippocampal sclerosis and clinical phenotype. Acta Neuropathol. 2011;122:703–713. doi: 10.1007/s00401-011-0879-y. [DOI] [PubMed] [Google Scholar]
- Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci U S A. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Calignon A, Polydoro M, Suarez-Calvet M, William C, Adamowicz DH, Kopeikina KJ, Pitstick R, Sahara N, Ashe KH, Carlson GA, et al. Propagation of tau pathology in a model of early Alzheimer’s disease. Neuron. 2012;73:685–697. doi: 10.1016/j.neuron.2011.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Los Rios P, Hafner M, Pastore A. Explaining the length threshold of polyglutamine aggregation. J Phys Condens Matter. 2012;24:244105. doi: 10.1088/0953-8984/24/24/244105. [DOI] [PubMed] [Google Scholar]
- Decker CJ, Teixeira D, Parker R. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J Cell Biol. 2007;179:437–449. doi: 10.1083/jcb.200704147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CM, Cenik B, Sephton CF, Johnson BA, Herz J, Yu G. TDP-43 aggregation in neurodegeneration: are stress granules the key? Brain Res. 2013;1462:16–25. doi: 10.1016/j.brainres.2012.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormann D, Rodde R, Edbauer D, Bentmann E, Fischer I, Hruscha A, Than ME, Mackenzie IR, Capell A, Schmid B, et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010;29:2841–2857. doi: 10.1038/emboj.2010.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas PM, Dillin A. Protein homeostasis and aging in neurodegeneration. J Cell Biol. 2010;190:719–729. doi: 10.1083/jcb.201005144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. doi: 10.1016/j.cell.2012.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Garber K. Proteostasis therapeutics. Nat Biotechnol. 2009;27:1070. doi: 10.1038/nbt1209-1070. [DOI] [PubMed] [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg SD, Galvin JE, Chiu TS, Lee VM, Masliah E, Trojanowski JQ. RNA sequestration to pathological lesions of neurodegenerative diseases. Acta Neuropathol. 1998;96:487–494. doi: 10.1007/s004010050923. [DOI] [PubMed] [Google Scholar]
- Guil S, Long JC, Caceres JF. hnRNP A1 relocalization to the stress granules reflects a role in the stress response. Mol Cell Biol. 2006;26:5744–5758. doi: 10.1128/MCB.00224-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackman P, Sarparanta J, Lehtinen S, Vihola A, Evila A, Jonson PH, Luque H, Kere J, Screen M, Chinnery PF, et al. Welander distal myopathy is caused by a mutation in the RNA-binding protein TIA1. Ann Neurol. 2012 doi: 10.1002/ana.23831. [DOI] [PubMed] [Google Scholar]
- Hagerman PJ, Hagerman RJ. The fragile-X premutation: a maturing perspective. Am J Hum Genet. 2004;74:805–816. doi: 10.1086/386296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem V, Galloway JN, Mori M, Willemsen R, Oostra BA, Paylor R, Nelson DL. Ectopic expression of CGG containing mRNA is neurotoxic in mammals. Hum Mol Genet. 2009;18:2443–2451. doi: 10.1093/hmg/ddp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL. Subcellular mRNA localization in animal cells and why it matters. Science. 2009;326:1212–1216. doi: 10.1126/science.1176488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz LM, Kwong LK, Lee EB, Chen-Plotkin A, Swanson E, Unger T, Malunda J, Xu Y, Winton MJ, Trojanowski JQ, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest. 2011;121:726–738. doi: 10.1172/JCI44867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara T, Ariizumi Y, Shiga A, Kato T, Tan CF, Sato T, Miki Y, Yokoo M, Fujino T, Koyama A, et al. Decreased number of Gemini of coiled bodies and U12 snRNA level in amyotrophic lateral sclerosis. Hum Mol Genet. 2013 doi: 10.1093/hmg/ddt262. [DOI] [PubMed] [Google Scholar]
- Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, Babineau B, Lebrilla CB, Hagerman RJ, Hagerman PJ. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. doi: 10.1016/j.cell.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A, Sweeney F, Bodi I, Troakes C, Maekawa S, Al-Sarraj S. Abnormal TDP-43 expression is identified in the neocortex in cases of dementia pugilistica, but is mainly confined to the limbic system when identified in high and moderate stages of Alzheimer’s disease. Neuropathology. 2010;30:408–419. doi: 10.1111/j.1440-1789.2009.01085.x. [DOI] [PubMed] [Google Scholar]
- Klar J, Sobol M, Melberg A, Mabert K, Ameur A, Johansson AC, Feuk L, Entesarian M, Orlen H, Casar-Borota O, et al. Welander Distal Myopathy Caused by an Ancient Founder Mutation in TIA1 Associated with Perturbed Splicing. Hum Mutat. 2013;34:572–577. doi: 10.1002/humu.22282. [DOI] [PubMed] [Google Scholar]
- Kopito RR, Ron D. Conformational disease. Nat Cell Biol. 2000;2:E207–209. doi: 10.1038/35041139. [DOI] [PubMed] [Google Scholar]
- Kwak JE, Drier E, Barbee SA, Ramaswami M, Yin JC, Wickens M. GLD2 poly(A) polymerase is required for long-term memory. Proc Natl Acad Sci U S A. 2008;105:14644–14649. doi: 10.1073/pnas.0803185105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJ, Desplats P, Sigurdson C, Tsigelny I, Masliah E. Cell-to-cell transmission of non-prion protein aggregates. Nat Rev Neurol. 2010;6:702–706. doi: 10.1038/nrneurol.2010.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Uversky VN, Fink AL. Effect of familial Parkinson’s disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human alpha-synuclein. Biochemistry. 2001;40:11604–11613. doi: 10.1021/bi010616g. [DOI] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. Stress granules as crucibles of ALS pathogenesis. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin AC, Holt CE. Function and regulation of local axonal translation. Curr Opin Neurobiol. 2008;18:60–68. doi: 10.1016/j.conb.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xu Y, Stoleru D, Salic A. Imaging protein synthesis in cells and tissues with an alkyne analog of puromycin. Proc Natl Acad Sci U S A. 2012a;109:413–418. doi: 10.1073/pnas.1111561108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Drouet V, Wu JW, Witter MP, Small SA, Clelland C, Duff K. Trans-synaptic spread of tau pathology in vivo. PLoS One. 2012b;7:e31302. doi: 10.1371/journal.pone.0031302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bassell GJ, Gitler AD, Hart AC, Klann E, Richter JD, Warren ST, Wolozin B. Local RNA translation at the synapse and in disease. J Neurosci. 2011;31:16086–16093. doi: 10.1523/JNEUROSCI.4105-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Yesucevitz L, Bilgutay A, Zhang YJ, Vanderweyde T, Citro A, Mehta T, Zaarur N, McKee A, Bowser R, Sherman M, et al. Tar DNA binding protein-43 (TDP-43) associates with stress granules: analysis of cultured cells and pathological brain tissue. PLoS One. 2010;5:e13250. doi: 10.1371/journal.pone.0013250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallucci GR. Prion neurodegeneration: starts and stops at the synapse. Prion. 2009;3:195–201. doi: 10.4161/pri.3.4.9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Yamagata T, Burgess DL, Rasmussen A, Grewal RP, Watase K, Khajavi M, McCall AE, Davis CF, Zu L, et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- McCann C, Holohan EE, Das S, Dervan A, Larkin A, Lee JA, Rodrigues V, Parker R, Ramaswami M. The Ataxin-2 protein is required for microRNA function and synapse-specific long-term olfactory habituation. Proc Natl Acad Sci U S A. 2011;108:E655–662. doi: 10.1073/pnas.1107198108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee AC, Gavett BE, Stern RA, Nowinski CJ, Cantu RC, Kowall NW, Perl DP, Hedley-Whyte ET, Price B, Sullivan C, et al. TDP-43 proteinopathy and motor neuron disease in chronic traumatic encephalopathy. J Neuropathol Exp Neurol. 2010;69:918–929. doi: 10.1097/NEN.0b013e3181ee7d85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- Moreno JA, Radford H, Peretti D, Steinert JR, Verity N, Martin MG, Halliday M, Morgan J, Dinsdale D, Ortori CA, et al. Sustained translational repression by eIF2alpha-P mediates prion neurodegeneration. Nature. 2012;485:507–511. doi: 10.1038/nature11058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchowski PJ, Wacker JL. Modulation of neurodegeneration by molecular chaperones. Nat Rev Neurosci. 2005;6:11–22. doi: 10.1038/nrn1587. [DOI] [PubMed] [Google Scholar]
- Mutsuddi M, Marshall CM, Benzow KA, Koob MD, Rebay I. The spinocerebellar ataxia 8 noncoding RNA causes neurodegeneration and associates with staufen in Drosophila. Curr Biol. 2004;14:302–308. doi: 10.1016/j.cub.2004.01.034. [DOI] [PubMed] [Google Scholar]
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 2008;10:1224–1231. doi: 10.1038/ncb1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive M, Janue A, Moreno D, Gamez J, Torrejon-Escribano B, Ferrer I. TAR DNA-Binding protein 43 accumulation in protein aggregate myopathies. J Neuropathol Exp Neurol. 2009;68:262–273. doi: 10.1097/NEN.0b013e3181996d8f. [DOI] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Parker R, Sheth U. P bodies and the control of mRNA translation and degradation. Mol Cell. 2007;25:635–646. doi: 10.1016/j.molcel.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijns MA, Alexander RD, Spiller MP, Beggs JD. A role for Q/N-rich aggregation-prone regions in P-body localization. J Cell Sci. 2008;121:2463–2472. doi: 10.1242/jcs.024976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritson GP, Custer SK, Freibaum BD, Guinto JB, Geffel D, Moore J, Tang W, Winton MJ, Neumann M, Trojanowski JQ, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O’Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat Med. 2004;10(Suppl):S10–17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci. 2001;2:791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
- Savas JN, Ma B, Deinhardt K, Culver BP, Restituito S, Wu L, Belasco JG, Chao MV, Tanese N. A role for huntington disease protein in dendritic RNA granules. J Biol Chem. 2010;285:13142–13153. doi: 10.1074/jbc.M110.114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas JN, Makusky A, Ottosen S, Baillat D, Then F, Krainc D, Shiekhattar R, Markey SP, Tanese N. Huntington’s disease protein contributes to RNA-mediated gene silencing through association with Argonaute and P bodies. Proc Natl Acad Sci U S A. 2008;105:10820–10825. doi: 10.1073/pnas.0800658105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg GD, Montine TJ. The genetics and neuropathology of Alzheimer’s disease. Acta Neuropathol. 2012;124:305–323. doi: 10.1007/s00401-012-0996-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Ebmeier CC, Podell ER, Heimiller J, Taatjes DJ, Cech TR. FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes & development. 2012;26:2690–2695. doi: 10.1101/gad.204602.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selkoe DJ. Alzheimer’s disease is a synaptic failure. Science. 2002;298:789–791. doi: 10.1126/science.1074069. [DOI] [PubMed] [Google Scholar]
- Solomon IH, Schepker JA, Harris DA. Prion neurotoxicity: insights from prion protein mutants. Curr Issues Mol Biol. 2012;12:51–61. [PMC free article] [PubMed] [Google Scholar]
- Spector DL. SnapShot: Cellular bodies. Cell. 2006;127:1071. doi: 10.1016/j.cell.2006.11.026. [DOI] [PubMed] [Google Scholar]
- Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell. 2012;149:832–846. doi: 10.1016/j.cell.2012.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern RA, Riley DO, Daneshvar DH, Nowinski CJ, Cantu RC, McKee AC. Long-term consequences of repetitive brain trauma: chronic traumatic encephalopathy. PM R. 2011;3:S460–467. doi: 10.1016/j.pmrj.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassone F, Iwahashi C, Hagerman PJ. FMR1 RNA within the intranuclear inclusions of fragile X-associated tremor/ataxia syndrome (FXTAS) RNA Biol. 2004;1:103–105. doi: 10.4161/rna.1.2.1035. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2010;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremblay C, St-Amour I, Schneider J, Bennett DA, Calon F. Accumulation of transactive response DNA binding protein 43 in mild cognitive impairment and Alzheimer disease. J Neuropathol Exp Neurol. 2011;70:788–798. doi: 10.1097/NEN.0b013e31822c62cf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance C, Rogelj B, Hortobagyi T, De Vos KJ, Nishimura AL, Sreedharan J, Hu X, Smith B, Ruddy D, Wright P, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–1211. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vij N. The case for therapeutic proteostasis modulators. Expert Opin Ther Targets. 2011;15:233–236. doi: 10.1517/14728222.2011.553610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt A, Herholz D, Fiesel FC, Kaur K, Muller D, Karsten P, Weber SS, Kahle PJ, Marquardt T, Schulz JB. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PLoS One. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M, Xia G, Gao R, Wakamiya M, Sarkar PS, McFarland K, Ashizawa T. Transgenic mice with SCA10 pentanucleotide repeats show motor phenotype and susceptibility to seizure: a toxic RNA gain-of-function model. J Neurosci Res. 2011;90:706–714. doi: 10.1002/jnr.22786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson AC, Dugger BN, Dickson DW, Wang DS. TDP-43 in aging and Alzheimer’s disease - a review. Int J Clin Exp Pathol. 2011;4:147–155. [PMC free article] [PubMed] [Google Scholar]
- Wolozin B. Regulated protein aggregation: stress granules and neurodegeneration. Mol Neurodegener. 2012;7:56. doi: 10.1186/1750-1326-7-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Sasagawa Y, Ogura T. Recent advances in p97/VCP/Cdc48 cellular functions. Biochim Biophys Acta. 2012;1823:130–137. doi: 10.1016/j.bbamcr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, Vidal L, Hoenicka J, Rodriguez O, Atares B, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55:164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell. 2009;136:308–321. doi: 10.1016/j.cell.2008.12.022. [DOI] [PubMed] [Google Scholar]