Summary
Most human transcription factors bind a small subset of potential genomic sites and often use different subsets in different cell types. To identify mechanisms that govern cell type-specific transcription factor binding, we used an integrative approach to study estrogen receptor α (ER). We found that ER exhibits two distinct modes of binding. Shared sites, bound in multiple cell types, are characterized by high affinity estrogen response elements (EREs), inaccessible chromatin and a lack of DNA methylation, while cell-specific sites are characterized by a lack of EREs, co-occurrence with other transcription factors and cell type-specific chromatin accessibility and DNA methylation. These observations enabled accurate quantitative models of ER binding that suggest tethering of ER to one-third of cell-specific sites. The distinct properties of cell-specific binding were also observed with glucocorticoid receptor and for ER in primary mouse tissues, representing an elegant genomic encoding scheme for generating cell type-specific gene regulation.
Introduction
Transcription factors (TFs) drive changes in gene expression and the re-organization of chromatin by binding to specific genomic loci. While many possible binding sites exist in a mammalian genome, less than 0.01% of the genome is typically bound by a TF in a particular cell type (2011). This observation raises the question of how TFs choose their binding sites and what genomic features discriminate bound and unbound sites. The process of binding site selection is difficult to study for most TFs, because binding can result in changes to a variety of regulatory features making it difficult to separate cause and effect.
Type I nuclear receptors provide the opportunity to measure the genomic landscape that a TF encounters prior to selecting binding sites, because their ability to bind DNA is dependent on the presence of their hormone ligands. Here we focus on estrogen signaling, and estrogen receptor α (ER), which impacts gene regulation in numerous cell types. The primary role of estrogens is to direct the development and maintenance of the female reproductive system by promoting cellular proliferation (Butt et al., 2008) and differentiation (Yashwanth et al., 2006); they also influence the musculoskeletal, cardiovascular, immune, and central nervous systems (Heldring et al., 2007).
Consistent with the observation of differential outcomes in response to estrogen signaling, ER binding shows remarkable differences between cell types and individuals. ER-positive breast cancer cell lines exhibit differential ER binding (Hurtado et al., 2011; Joseph et al., 2010) and ER binding differs significantly between primary tumors from different individuals and is associated with prognosis (Ross-Innes et al., 2012). Disparity in ER binding has also been observed in cells ectopically expressing ER (Kong et al., 2011; Krum et al., 2008). In previous work, we found little overlap between ER binding sites in breast and endometrial cancer lines (Gertz et al., 2012). Thus, the effects of estrogens are tissue- and cell type-specific.
The presence of DNaseI hypersensitive sites (open chromatin) prior to induction has been associated with most ER, androgen receptor and glucocorticoid receptor binding sites, varying from 51% to 95% of binding sites in open chromatin prior to treatment with ligand (He et al., 2012; John et al., 2011; Joseph et al., 2010). The predictive power of chromatin accessibility in determining steroid hormone receptor binding sites may lie in the overall identification of loci bound by interacting TFs. For example, the forkhead factor FOXA1 acts as a pioneer factor by binding to many ER and AR binding sites prior to hormone treatment and preparing chromatin for nuclear receptor binding (Carroll et al., 2005; Lupien et al., 2008). FOXA1 and other TFs are essential for many ER binding sites and knock-down or over-expression of these TFs can result in a change to ER binding targets (Hurtado et al., 2011; Kong et al., 2011; Simpson et al., 2012). The interaction of ER with some interacting factors is thought to occur in a tethered configuration (Carroll et al., 2006; Heldring et al., 2011; Kushner et al., 2000; O’Lone et al., 2004), without ER directly contacting DNA, thus widening the possible targets for ER across the genome.
Cell type-specific ER binding has been studied mostly in breast cancer cells (Joseph et al., 2010; Ross-Innes et al., 2012) or in systems that ectopically express ER (Krum et al., 2008). Between breast cancer cell lines, FOXA1 binding and chromatin accessibility are strong predictors of ER binding site selection (Joseph et al., 2010), supporting the conclusion that FOXA1 plays an important role in ER’s interactions with the genome. However, FOXA1 and ER do not act together in all cell types. For example, endometrial cancer cells that express ER and proliferate in response to estradiol undergo growth inhibition when FOXA1 is introduced (Abe et al., 2012). Understanding how ER selects binding sites in the absence of FOXA1 is an important step in determining the full spectrum of utilized ER binding sites.
To study cell type-specific binding site selection, we investigated two cell types that endogenously express ER -- breast and endometrial cancer lines that show very distinct binding patterns (Gertz et al., 2012). Through integration of TF binding, RNA levels, chromatin accessibility, and DNA methylation data, we found distinct properties of cell-specific and shared sites (bound by ER in both cell lines). Shared sites rely on high affinity matches to estrogen response elements (EREs), do not show a dependency on open chromatin and have low overlap with interacting factor binding sites. Cell-specific ER binding sites exhibit cell type-specific patterns of chromatin accessibility and DNA methylation. Cell-specific sites lack high affinity EREs and often overlap with interacting factor binding sites. This combination of ER binding sites creates a complex interplay between genome sequence and the cell type specificity of ER binding.
Results
Shared ER binding sites harbor high affinity EREs
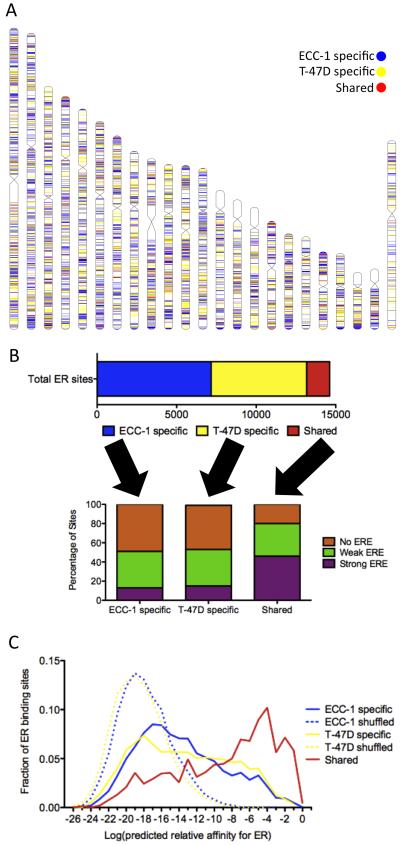
ER DNA binding has been shown to be highly cell type-specific. In previous work, we mapped the binding sites of ER by ChIP-seq in two estrogen responsive human cell lines: ECC-1, an endometrial cancer cell line, and T-47D, a breast cancer cell line. We identified ~8,000 reproducible binding sites induced by one-hour treatment with 10 nM 17β-estradiol (E2) in each cell line (Figure 1A). Out of the 14,620 total ER binding sites, only 1,466 (9.9%) are shared (bound in both cell lines) (Figure 1B).
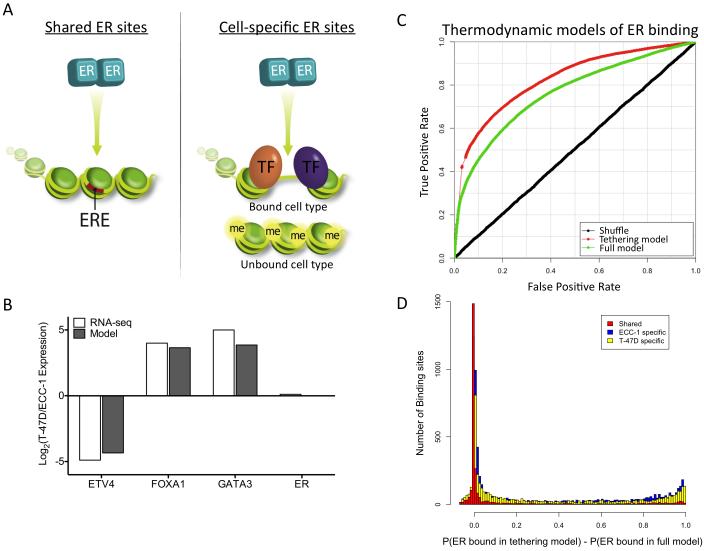
Figure 1. Shared and cell-specific sites have unique sequence properties.
A) The locations of ECC-1 specific (blue), T-47D specific (yellow) and shared (red) ER binding sites are distributed across human chromosomes with an overall lack of shared sites. B) Less than 10% of the total set of identified ER binding sites is consistently bound in ECC-1 and T-47D (top panel). Less than 20% of cell-specific ER binding sites contain EREs, while 46% of shared sites harbor EREs (bottom panel). C) The distribution of the predicted relative affinity for the best match to an ERE is shown for each ECC-1 specific (solid blue), T-47D specific (solid yellow) and shared (solid red) ER binding sites as well as ECC-1 and T-47D ER binding sites in which the order of the nucleotides has been shuffled (dashed lines). All three sets of observed binding sites (solid lines) are enriched over background (dashed lines); however, shared sites represent the highest affinity matches to an ERE. See also Figure S1.
De novo motif finding identified very similar motifs that match the canonical estrogen response element (ERE; GGTCANNNTGACC) in both ECC-1 and T-47D. To explore the distribution of EREs, we scanned each bound site and identified the highest affinity match to an ERE. We compared the distribution of EREs in ECC-1 and T-47D ER binding sites to a background distribution calculated by shuffling their nucleotide sequences (Figure 1C). There is an enrichment for EREs over background in both cell lines (P<2.2×10−16; Wilcoxon); however, not all ER binding sites have high affinity matches to an ERE. Of the 14,620 total ER bound sites, 2,475 (17%) harbor significant matches to an ERE as defined by Patser (Hertz and Stormo, 1999), while 38% have weak EREs, which are not significant matches to an ERE but can contain half sites, and 45% of sites do not have any sequence that resembles an ERE. The lack of EREs is not an artifact of weak signal, as there is no correlation between predicted ERE strength and ChIP-seq signal (R2=0.009).
The lack of EREs is most pronounced in cell-specific ER binding sites (Figure 1B). Less than 15% of cell-specific sites exhibit a strong ERE, while 46% of sites bound by ER in both cell lines have high affinity EREs. This pattern can also be observed in the distribution of ERE strength (Figure 1C), with shared site motifs shifted significantly towards stronger EREs (P<2.2×10−16; Wilcoxon). These results indicate that cell-specific and shared ER binding sites have distinct underlying DNA sequence patterns.
Shared sites are not only enriched for strong EREs, but those EREs are higher affinity than the significant EREs in cell-specific sites (P<2.2×10−16; Wilcoxon). Strong EREs found in shared sites are predicted to be 3-fold higher affinity on average than strong EREs found in cell-specific sites (Figure S1A). This pattern is more pronounced when ECC-1 and T-47D ER binding sites are compared with previously published MCF-7 ER binding sites (Welboren et al., 2009) (Figure S1B), where sites bound in all three cell lines exhibit the highest affinity EREs (P<2.2×10−16; Wilcoxon).
The prevalence of the highest affinity EREs in shared sites led us to look genome-wide at bound and unbound EREs. Using our definition of a strong ERE, there are 26,452 matches across the human genome. Only 2,475, 9.4% of all EREs are bound in either cell line. To assess the predictive ability of ERE strength, we ranked each ERE by the predicted relative affinity (based only on DNA sequence) and analyzed the specificity and sensitivity using a receiver operator characteristic (ROC) curve (Figure S1C). Consistent with the observation that the highest affinity EREs are bound in multiple cell types, we found that the strength of the DNA sequence match to an ERE could accurately predict binding, producing a ROC area under the curve (AUC) of 0.73. This simple approach was able to predict 50% of the bound EREs with a 15% false positive rate and because the very high end of the distribution of ERE strength was the most accurate, 20% of bound EREs can be predicted with a false positive rate of only 1%.
Cell-specific ER binding is associated with cell-specific chromatin accessibility
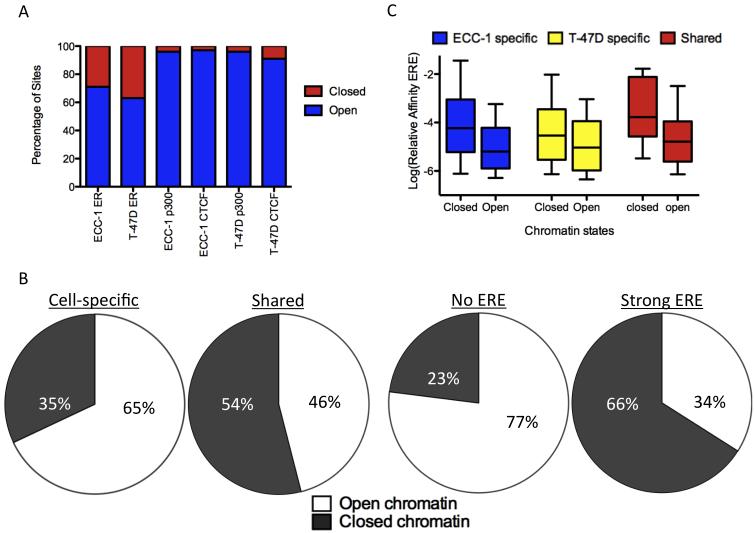
To explore the relationship between chromatin accessibility and cell-specific ER binding we performed DNase-seq, which maps DNaseI hypersensitive sites using massively parallel sequencing (Boyle et al., 2008). We analyzed ECC-1 and T-47D prior to treatment with E2 to observe the chromatin landscape that ER encounters upon induction with hormone. A comprehensive set of more than 100,000 DNaseI hypersensitive sites (hereafter called “open chromatin”) was identified in each cell line, covering 95% of CTCF and p300 binding sites (Figure 2A).
Figure 2. Relationship between chromatin accessibility and cell-specific binding.
A) DNaseI hypersensitivity data is comprehensive, capturing >90% of p300 and CTCF binding sites in each cell type, while indicating that ~65% of ER binding sites are found in open chromatin prior to treatment with E2. B) Most ER binding sites that are cell-specific and lack strong matches to an ERE are found in open chromatin prior to E2 treatment, while ER binding sites that are shared between the two cell lines and have strong EREs are found in closed chromatin prior to treatment. C) Bound EREs found in closed chromatin prior to E2 treatment are stronger matches to an ERE than sites found in open chromatin for ECC-1 specific (blue), T-47D specific (yellow) and shared (red) ER binding sites. See also Figure S2.
In ECC-1 and T-47D, 72% and 59% of ER binding sites reside in open chromatin prior to treatment, respectively. When ER binding sites are divided into ECC-1 specific, T-47D specific and shared sites, most ER binding sites have a matching pattern of open chromatin. For example, 44% of T-47D specific sites are found in open chromatin that is specific to T-47D, compared to 11% residing in open chromatin shared between the cell lines and <1% found in ECC-1 specific open chromatin (Figure S2A). This association suggests that chromatin accessibility prior to treatment plays a large role in cell type-specific ER binding.
Comparison of cell-specific and shared ER bound sites revealed large differences in patterns of chromatin accessibility. In total, 65% of cell-specific ER sites are found in chromatin that is open prior to induction versus 46% of shared ER sites that are found in shared open chromatin (Figure 2B; P<7.8×10−12, Fisher’s exact test). Underlying the association between chromatin accessibility and cell type-specific binding is the presence of EREs; 77% of ER binding sites without an ERE are found in open chromatin, while only 34% of sites with a strong ERE are located in open chromatin (Figure 2B; P<2.2×10− 16, Fisher’s exact test). ER binding sites with a weak ERE overlap with open chromatin at a rate of 64%, behaving similar to sites without an ERE. The reliance of ER binding sites without an ERE on open chromatin explains the patterns observed between cell-specific and shared sites.
Due to the strong relationship between chromatin accessibility and the presence of EREs, we further examined the EREs found in open and closed chromatin. We found a significant increase in predicted ERE strength for sites in closed chromatin as compared to open chromatin (Figure 2C; P<2.2×10−16 Wilcoxon). These results suggest that bound EREs found in closed chromatin prior to treatment are required to be high affinity, while EREs in open chromatin can be lower affinity ER binding sites. Based on this observation, we created a simple model to predict bound versus unbound EREs that incorporates both the strength of an ERE and chromatin accessibility. We took the predicted relative affinity and scaled it by a constant amount if the site was found in open chromatin. When the scaling factor is fit, the model is significantly better than a model that uses ERE strength alone. The ROC AUC increases from 0.73 to 0.79 (Figure S1C; P<2.2×10−16, DeLong’s test). The model also predicts that accessible chromatin increases the affinity of ER for an ERE 29-fold.
While DNaseI hypersensitivity patterns in cells that lack E2 give insight into the chromatin landscape that ER encounters upon stimulation, we also performed DNase-seq after one-hour treatment with E2 in both ECC-1 and T-47D to determine how ER activation affects chromatin accessibility. We found that ER binding leads to chromatin accessibility only at some loci, consistent with previous work in MCF-7 (He et al., 2012). After E2 treatment, 80% of T-47D sites and 77% of ECC-1 ER binding sites are found in open chromatin, increased from 59% and 72% before treatment, respectively (Figure S2B). We found that sites containing an ERE were more likely to open after binding, increasing from 34% in open chromatin prior to treatment to 60% in open chromatin after treatment compared to an increase from 72% open chromatin prior to treatment to 83% in open chromatin after treatment for non-ERE sites. This indicates that ER is only able to set up accessible chromatin at a subset of sites and those sites are more likely to contain an ERE.
Differential DNA methylation associates with cell-specific ER binding sites
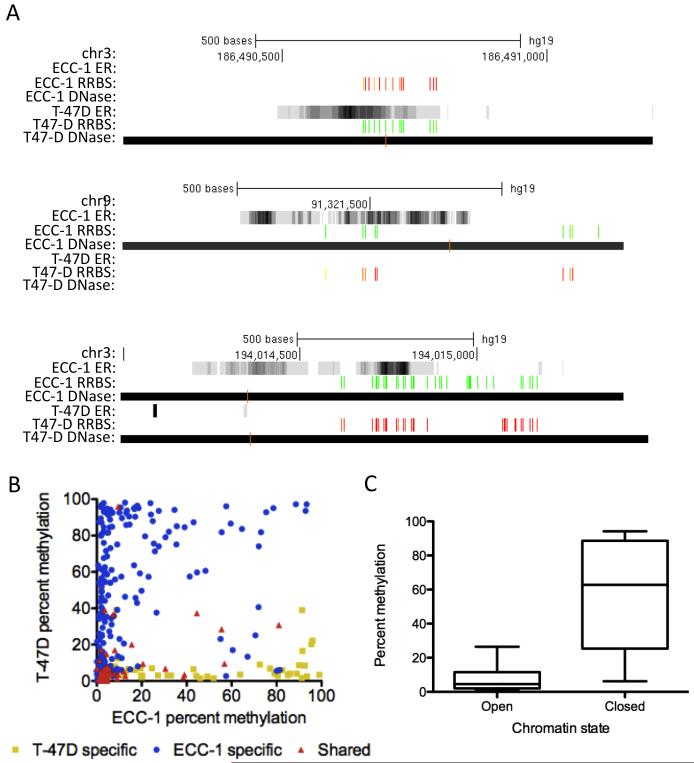
The association between chromatin accessibility and cell type-specific ER binding led us to explore the connection between DNA methylation, an epigenetic mark, and ER binding site selection. To examine DNA methylation, we performed reduced representation bisulfite sequencing (RRBS) (Gertz et al., 2011; Meissner et al., 2008; Varley et al., 2013) in both ECC-1 and T-47D prior to treatment with E2. This method allowed us to analyze 2,011 CpGs present in 480 ER binding sites (within 50 bp of binding site summits). We found that DNA methylation was often cell type-specific at ER binding sites and there was a lack of DNA methylation in the bound cell type (Figure 3A), consistent with previous observations concerning glucocorticoid receptor binding (Wiench et al., 2011). Figure 3B shows the average percent methylation in ECC-1 and T-47D of CpGs found in each ER binding site. The cell type specificity in binding is mirrored in the DNA methylation measurements. For example, CpGs within T-47D specific binding sites tend to show methylation in ECC-1 and lack methylation in T-47D. We also found that CpGs found within shared sites lack methylation in both ECC-1 and T-47D. Our data suggest that the presence of DNA methylation prior to treatment with E2 precludes ER binding.
Figure 3. Cell type-specific DNA methylation is found at cell-specific ER binding sites.
A) Typical examples of ER binding signal (grayscale read density, black is highest read coverage), DNase I hypersensitive sites (black bars) and DNA methylation (green: unmethylated, red: methylated). B) ECC-1 specific (blue), T-47D specific (yellow) and shared (red) ER binding sites have cell type-specific DNA methylation with the unbound cell type exhibiting methylation. C) DNA methylation is rarely present in open chromatin, while CpGs in closed chromatin can be either methylated or unmethylated. See also Figure S3.
We also observed that DNA methylation is generally absent in open chromatin (Figure 3C). Closed chromatin is significantly more methylated than open chromatin on the whole (P<2.2×10−16; Wilcoxon); however, closed chromatin can also be maintained in an unmethylated state. The presence of unmethylated closed chromatin seems to be important for ER binding, as many shared sites are found in unmethylated closed chromatin. Overall, DNA methylation and chromatin accessibility, prior to treatment with E2, are good indicators of the cell type specificity of ER binding.
To determine if DNA methylation was associated with ER’s selection of a distinct set of EREs across the genome, we analyzed the presence of DNA methylation around bound and unbound EREs. There was a marked decrease in DNA methylation nearby bound EREs compared to unbound EREs (Figure S3A; P<5.4×10−14, Wilcoxon), in agreement with our cell type-specific observations described above. Because the strength of an ERE predicts the likelihood of being bound, we hypothesized that the presence of DNA methylation is more prevalent at higher affinity unbound EREs compared to lower affinity unbound EREs. We found that higher affinity unbound EREs were more likely to be methylated (Figure S3B, P<1.1×10−4, Wilcoxon), with the median percent methylation differing by 40% between higher affinity unbound EREs and lower affinity unbound EREs. These results suggest that DNA methylation is precluding ER binding at some high affinity EREs; however, the effect is not complete and some unmethylated high affinity EREs remain unbound.
Cell-specific ER binding sites exhibit interacting factor co-occupancy
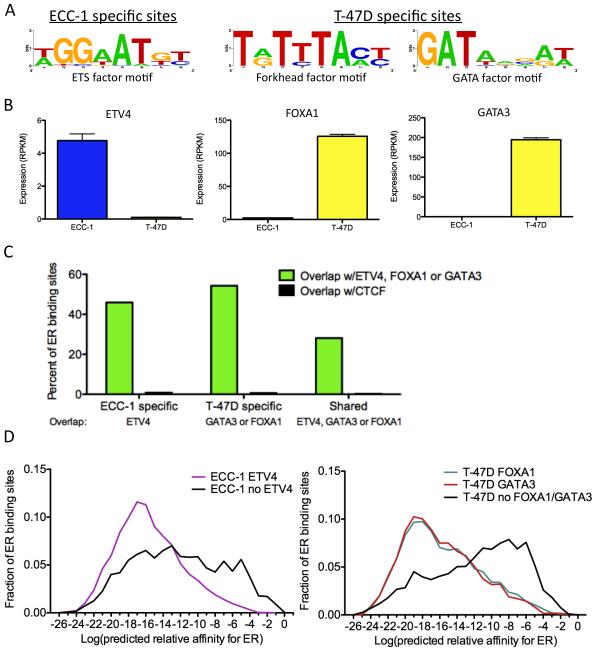
Chromatin accessibility patterns and DNA methylation levels are dictated in large part by sequence-specific TFs (Kang et al., 2002; Stadler et al., 2011). The strong association between cell type-specific ER binding and chromatin accessibility/DNA methylation led us to explore factors that may be setting up cell-specific ER binding sites. To identify these factors, we performed de novo motif discovery on ER binding sites, after canonical EREs were masked out, and identified three prevalent motifs within the ER binding sites: an ETS binding motif in ECC-1, and a forkhead factor motif and a GATA factor motif in T-47D (Figure 4A).
Figure 4. Transcription factors co-occur with ER at cell-specific binding sites.
A) Motifs were identified in ECC-1 (left panel) and T-47D (right panel) ER binding sites after EREs were masked out. B) The expression levels from RNA-seq data of transcription factors that recognize each family of motifs were analyzed and identified ETV4, FOXA1 and GATA3 as potential ER interacting factors. Error bars represent s.e.m. C) Cell-specific ER binding sites overlap ETV4, FOXA1 and GATA3 binding sites measured by ChIP-seq at a higher rate than shared ER binding sites. In each case, the overlap is significantly higher than CTCF, which serves as a negative control. D) ER binding sites that co-occur with these transcription factors in ECC-1 cells (left panel) and T-47D cells (right panel) lack strong matches to an ERE. See also Table S1.
We used expression measurements from RNA-seq data that we previously collected in these cell lines (Gertz et al., 2012) to determine differentially expressed TFs associated with each motif. In ECC-1, the ETS factor ETV4 (also known as PEA3) shows differential expression between the cell types (Figure 4B). ETV4 has been shown to cooperate with estrogen receptor β to activate expression of IL-8 (Chen et al., 2011); however, we could not identify previously reported evidence of an interaction between ETV4 and ER. In T-47D, forkhead factor FOXA1 and GATA factor GATA3 are differentially expressed between cell types (Figure 4B). Both FOXA1 and GATA3 are known to play a role in ER binding and are thought to be major drivers of ER binding site selection in breast cancer cells (Carroll et al., 2005; Joseph et al., 2010). To examine co-occupancy of these TFs and ER in each cell type, we performed ChIP-seq for each factor in their respective cell line, as well as FOXA1 in ECC-1, where FOXA1 is expressed at a low but detectable level (RPKM = 3.2 vs. 165).
There was high overlap between post-treatment ER bound sites and pre-treatment binding sites of these additional TFs (Table S1). Using CTCF binding as an empirical null model to evaluate rates of binding site overlap in each line, each factor showed substantially more co-occupancy than expected by chance. ETV4 co-occurred at 45% of ECC-1-specific ER binding sites, while GATA3 co-occurred with 46% and FOXA1 co-occurred with 40% of T-47D-specific ER binding sites. In ECC-1, FOXA1 is expressed ~50-fold lower compared to T-47D and resulted in an ~12-fold fewer binding sites. The reduced number of binding sites led to less overlap (5%) between ER and FOXA1 in ECC-1. Shared sites were less likely to overlap with these three TFs, as only 28% co-occurred with FOXA1, GATA3 or ETV4 (Figure 4C; P<2.2×10−16, Fisher’s exact test), suggesting a reduced dependence on TF co-occurrence among shared sites. These results indicate that cell-specific ER binding is associated with cell type-specific interacting TF binding prior to treatment.
We also found that ER binding sites that co-occur with other TFs were strongly depleted in EREs (Figure 4D; P<2.2×10−16; Wilcoxon). In ECC-1, only 8% of ER binding sites with a strong ERE overlapped ETV4 binding, while 62% of ER binding sites without an ERE co-occurred with ETV4. In T-47D, 9% of ER sites with an ERE overlapped either FOXA1 or GATA3, while 69% of ER binding sites without an ERE overlapped FOXA1 or GATA3. The presence of FOXA1, GATA3 and ETV4 at sites that lack EREs suggests two mechanisms of ER binding site selection: one set of sites that relies on strong matches to a canonical ERE and another set relies on the presence of co-localized factors prior to treatment with E2.
Thermodynamic model of ER binding sites indicates ER tethering model
The distinct types of ER binding sites represent a simple and elegant way to encode cell type specificity (Figure 5A). Shared sites are encoded by high affinity canonical EREs that are active in both open and closed chromatin, but require a lack of DNA methylation. Cell-specific sites lack high affinity canonical EREs and instead contain motifs of interacting factors while residing in cell type-specific open chromatin that lacks DNA methylation.
Figure 5. Model of cell type specificity in ER binding site selection.
A) The model depicts the differences between shared and cell-specific binding sites with respect to the presence of EREs (red bars), chromatin accessibility, DNA methylation (“me” yellow circles) and co-occurring transcription factors (“TF” orange and purple ovals). B) Model predictions (gray) of the relative expression of each factor between cell lines are highly correlated with observed transcript levels from RNA-seq experiments (white). C) ROC curves show that the thermodynamic model performs significantly better when a tethering model of ER binding is implemented. D) The distributions of differences between the probability of ER binding in the tethering model and the full model are significantly different between the shared binding sites (red) and either the ECC-1 specific (blue) or T-47D specific (yellow) binding sites. See also Figure S4.
To explore the predictive ability of this simple model of ER binding site selection, we constructed a thermodynamic model capable of predicting cell type-specific ER binding. Thermodynamic models of gene regulation assume that transcription factor binding and polymerase recruitment are driven solely by the free energy of protein:DNA and protein:protein interactions (Shea and Ackers, 1985). We used a previously implemented thermodynamic model that is capable of incorporating changes in TF concentration and differences in affinity between genomic loci (Gertz and Cohen, 2009; Gertz et al., 2009). The model uses as input the DNA sequence of each binding site, the recognition motifs for each TF (ER, ETV4, FOXA and GATA3) and DNaseI hypersensitivity data in ECC-1 and T-47D without E2 treatment to fit parameters that describe the free energies of possible protein:DNA and protein:protein interactions. These parameters include the difference in TF concentration between ECC-1 and T-47D, the impact of chromatin accessibility on binding affinities and the interaction energies of each TF pair (described in detail in Supplemental Experimental Procedures). With these parameters the model can then predict the steady state probability that ER is bound to each site in each cell type.
The thermodynamic model was able to accurately predict the cell type specificity of ER binding based only on TF motifs, chromatin accessibility and genome sequence (Figure S4). The ROC AUC for this model is 0.75. Because the binding sites of FOXA1, GATA3 and ETV4 were measured, we were also able to create a “fixed state” model in which the observed interacting TF binding, as opposed to interacting factor motif strength, was incorporated. The fixed state model had a slight increase in predictive power (AUC=0.76), suggesting that interacting factor binding is mostly captured by motif strength and chromatin accessibility.
The parameters of the model also represent predictions about the system. For example, the model predicts the difference in TF concentration between ECC-1 and T-47D for each factor. To assess this prediction, we compared the difference in TF concentration predicted by the model and the difference in transcript levels measured by RNA-seq (Gertz et al., 2012). We observed a very high correlation between RNA-seq levels and the model predictions (R2 = 0.996; Figure 5B). Because RNA-seq information was not used to create the thermodynamic model, these results serve as an external validation of the model parameters.
Since we observed a strong depletion of canonical EREs within sites that overlap with FOXA1, GATA3 or ETV4, we postulated that ER might not be binding DNA directly at these sites. Therefore, we modified the thermodynamic model to not require ER to bind DNA when co-occurring TFs are present. This has the effect of forcing ER to bind through protein:protein interactions only at those sites that are co-occupied. This model was significantly better, with an AUC of 0.83 (Figure 5C; P<2.2×10−16, DeLong’s test). The improvement in predictive ability supports a tethering mechanism that brings ER to some binding sites via protein:protein, rather than protein:DNA, interactions (Heldring et al., 2011).
In order to explore the predicted prevalence of tethering at ER binding sites, we examined how the full model and the tethering model differed at individual binding sites. In total, the models suggest that 4,380 out of 14,682 (29.8%) ER bound sites show evidence of tethering (the predicted probability of ER binding is greater than 0.5 higher in the tethered model than in the full thermodynamic model). These 4,380 ER bound sites represent loci that have no discernible ERE and does not include sites with strong or weak EREs. The modeling results also suggest that cell-specific sites are significantly more likely to utilize tethering compared to shared ER binding sites, 32% vs. 12%, respectively (Figure 5D; P<2.2×10−16, Wilcoxon). These findings suggest that about one-third of ER binding sites involve tethering with a strong bias towards cell-specific binding sites.
It is important to point out that potential ER:DNA interactions are treated as a continuous quantitative variable in this model, which allows for weak matches to a motif to contribute to binding (Gertz et al., 2009). For example, the difference in affinity between a site with a strong ERE and a weak ERE is similar to the difference between a site with no ERE and a site with a weak ERE. This distinction between a weak ERE and no ERE is substantial, as sites with a weak ERE do not require tethering to create a high probability of ER binding. Our results highlight the important role that weak or half sites play in orchestrating ER binding.
Similar properties of cell-specific ER binding in primary mouse tissues
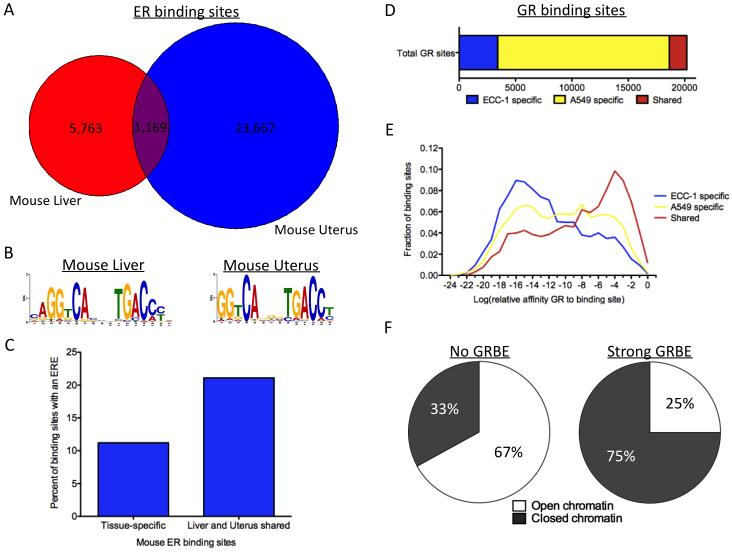
The model describing cell type specificity in ER binding was developed using human cancer cell lines and we were interested to see if the findings would hold true in normal primary tissue. We recently developed a method for performing ChIP-seq in frozen tissue samples (Savic et al., manuscript in preparation) and applied the technique to mapping ER binding in liver and uterus tissue from C57BL/6J mice. We identified 6,932 ER bound sites in liver and 24,836 ER bound sites in uterus. Sites bound by ER in liver were found nearby genes involved in metabolism (Figure S5A), while ER bound sites in uterus were found near genes involved in vasculature and muscular development (Figure S5B; enrichments found using GREAT (McLean et al., 2010)). The ER binding sites we identified in uterus have considerable overlap (57%) with ER binding sites identified in mouse uteri in a previous study (Hewitt et al., 2012).
Similar to ECC-1 and T-47D, there was a high degree of cell type specificity in ER binding between liver and uterus. In total, 30,599 ER bound sites were identified in liver and uterus; however, only 1,169 (3.8%) sites were bound in both tissues (Figure 6A). The canonical ERE was identified in both liver and uterus ER binding sites (Figure 6B). Consistent with results from cancer cell lines, ER binding sites shared between liver and uterus were significantly more likely to have strong matches to an ERE compared to tissue-specific ER binding sites (Figure 6C), 21.2% vs. 11.4%, respectively (P<2.2×10−16, Fisher’s exact test). These results indicate that the cell type-specific patterns observed in human cancer cell lines can also be found in primary mouse tissues, which suggests a generality to the model of ER binding site selection.
Figure 6. Cell-specific sites observed in primary mouse tissues as well as for the glucocorticoid receptor.
A) ER binding shows a high degree of cell type specificity between mouse liver (red) and uterus (blue). B) The canonical ERE motif was identified using ER binding sites found in either liver or uterus. C) Similar to results in human cancer cell lines, tissue-specific ER binding sites are significantly less likely to harbor EREs than shared sites bound by ER in both liver and uterus. D) Very few GR binding sites are bound in both ECC-1 and A549. E) The distribution of the best sequence match to an GRBE is shown for each ECC-1 specific (blue), A549 specific (yellow) and shared (red) GR binding site. F) Sites without a strong match to a GRBE are more likely to be found in open chromatin prior to treatment with dexamethasone than sites without a strong match to a GRBE. See also Figure S5.
Glucocorticoid receptor exhibits similar characteristics compared to ER
To determine if the properties of cell type specificity in ER binding hold for other nuclear receptors, we analyzed genome-wide binding of glucocorticoid receptor (GR) and chromatin accessibility patterns in ECC-1 and A549, a lung carcinoma cell line. We found a large cell type-specific component to GR binding between ECC-1 and A549 with only 1,553 (7.7%) shared binding sites out of 20,190 total GR binding sites (Figure 6D). The shared GR binding sites are significantly enriched for GR binding elements (GRBEs) compared to cell-specific GR binding sites (Figure 6E; P<2.2×10−16; Fisher’s exact test). We also discovered that GR binding sites with a GRBE are much more likely to be found in closed chromatin prior to treatment with dexamethasone than sites without a GRBE. Only 25% (1,152 out of 4,609) of bound sites with a significant GRBE were found in open chromatin prior to treatment with dexamethasone, while 67% (10,414 out of 15,581) of sites without a significant GRBE were found in open chromatin prior to treatment (Figure 6F; P<2.2×10−16; Fisher’s exact test). These results are consistent with our findings for ER and suggest that our model of ER binding site selection may be applicable to other nuclear receptors.
Cell type specificity of ER binding matches cell type specificity of gene expression
While there are two modes of ER binding, it is unclear whether each type of binding site is equally likely to impact gene regulation. Because cell-specific sites are pre-occupied before estrogen treatment, it is possible that ER binding may not impact gene regulation to the same extent as at shared sites. We approached this question from two angles: 1) comparing ER binding to genome-wide expression changes and 2) testing the ability of cell-specific and shared ER binding sites to enhance transcription in reporter assays.
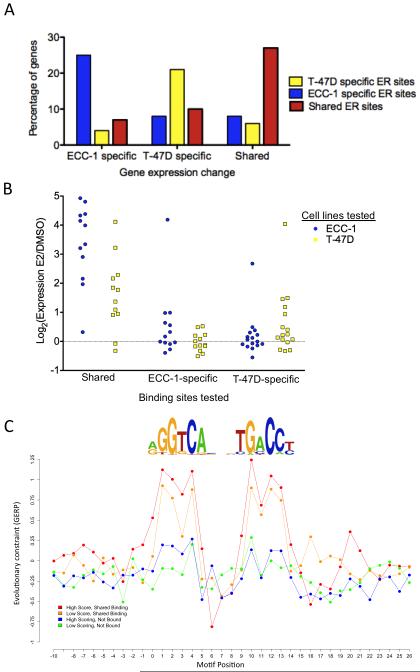
We previously identified 987 genes in ECC-1 and 611 genes in T-47D whose expression was significantly affected by estradiol treatment (Gertz et al., 2012). Overall, cell type-specific gene expression changes were often found near cell type-specific ER binding (Figure 7A), with an average of 25% of affected genes having an ER binding site with the same cell type specificity within 50 kbp of the transcription start site. Overlap between gene expression changes and ER binding revealed that sites with and without an ERE are equally likely to be nearby genes that respond to E2 (Figure S6A). ER binding sites near affected genes represent a random sampling of sites with respect to the presence of an ERE (P=0.3162, Fisher’s Exact Test). We also analyzed the magnitude of the expression changes for genes near ER sites with and without an ERE and found no difference in either cell type (Figure S6B; P=0.497, Wilcoxon). These findings are consistent with each type of ER binding site having an equal chance of impacting gene regulation.
Figure 7. Evidence supporting the model of ER binding site selection.
A) ER binding sites found within 50 kbp of the transcription start site of genes that show cell type-specific gene expression changes most often share the same cell type specificity in binding. B) Luciferase enhancer assays indicate that shared sites with an ERE show strong context-independent activity and cell-specific ER binding sites without EREs drive less activity; however, that activity is cell type-specific. C) High affinity bound EREs (red) are under more evolutionary constraint than low affinity bound EREs (orange) and bound EREs are under more evolutionary constraint than unbound EREs (blue and green). The levels of constraint are highly correlated with the information content of the ERE motif. See also Figure S6 and Table S2.
To explore the regulatory activity of ER binding sites, we cloned 12 shared sites with an ERE, 13 ECC-1-specific and 16 T-47D-specific ER binding sites without an ERE upstream of a minimal promoter driving luciferase expression. We transfected each construct into ECC-1 and T-47D cells treated with E2 or DMSO, and measured luciferase activity (all results are shown in Figure S6C-H). Of the 12 shared sites, 11 (92%) significantly change expression in response to E2 in ECC-1 and 10 (83%) changed significantly in T-47D. Only 4 of 13 (31%) cell-specific ECC-1 sites and 5 out of the 16 (31%) cell-specific T-47D sites were significantly activated by E2 in their respective cell lines. Shared and cell-specific ER sites that significantly responded to E2 had very different magnitudes of expression changes (Figure 7B), with shared sites resulting in larger increases in expression (P<10−4, t-test). Cell-specific elements maintained cell type-specific activity in our assays; only one cell-specific ER binding site was active in both cell types, a T-47D-specific ER binding site exhibited a significant E2 response in ECC-1 with 3-fold lower magnitude compared to T-47D. In agreement with our model of ER binding site selection, cell-specific sites seem to rely more on context for activity, while shared ER binding sites show activity in a context-independent manner.
For the cell-specific sites that showed significant expression responses to E2, we sought to determine if the co-occurring TFs were necessary for enhancer activity. We assayed the response to E2 for 3 ECC-1 specific, 3 T-47D specific and 3 shared ER binding sites after treatment with siRNAs targeting ETV4 in ECC-1 and FOXA1 and GATA3 in T-47D. The enhancer activity of all six cell-specific sites was abolished upon knockdown of the co-occurring TFs, while the shared sites were unaffected (Figure S6I-J). These results show that ER relies on the presence of ETV4, FOXA1 and GATA3 for enhancer activity at some cell-specific sites.
Bound EREs are evolutionarily constrained
Another prediction based on our model of ER binding site selection is that the highest affinity EREs would be under strong evolutionary constraint in shared sites since they are specifically required for appropriate function. Using a nucleotide-level measure of mammalian sequence evolution (Cooper et al., 2005), we found that higher affinity EREs exhibit significantly higher levels of evolutionary conservation at critical positions in the ERE relative to flanking sites (Figure 7C). We also observed that both higher and lower affinity bound EREs are more constrained than unbound EREs, suggesting evolutionary maintenance of EREs in shared ER binding sites. These findings demonstrate that shared ER binding sites are under strong selective pressure to retain high affinity EREs.
Discussion
Through the integration of experimental measurements of TF binding, chromatin accessibility, DNA methylation and mRNA levels with genome sequence analysis, we have constructed a model that explains how cell type specificity in ER binding is mechanistically regulated. We found distinct features that separate cell-specific ER binding sites and ER binding sites occupied in multiple cell types. ER sites bound in both cell types contain evolutionarily conserved high affinity EREs, lack DNA methylation in each cell type and seem to be indifferent to chromatin accessibility. Cell-specific ER binding sites are accompanied by cell type-specific chromatin accessibility and DNA methylation, lack high affinity EREs and are most likely driven by the presence of multiple interacting TFs. These two types of ER binding sites imply an elegant and flexible system for encoding ER binding sites in the genome.
We were able to quantitatively capture the properties of this qualitative model using a thermodynamic model of gene regulation. With only genome sequence, chromatin accessibility data and TF motifs, the model can accurately predict cell type-specific ER binding. While the model explains how ER binding sites are selected between cell types, it is still unclear why a certain small subset of EREs is bound. Interestingly, relative to bound high affinity EREs, unbound high affinity ERE sites show minimal evidence for evolutionary constraint, suggesting that many appeared randomly and are not maintained as functional binding sites. These results are congruent with previous observations suggesting that selection on enhancer activity can preserve the presence of binding motifs (Pennacchio et al., 2006).
We found that most ER binding, especially at cell-specific sites, takes place in chromatin that is accessible prior to treatment with E2. This finding is consistent with previous work on inducible TFs showing that chromatin patterns direct TF binding (Guertin and Lis, 2010; John et al., 2011; Wu et al., 2011). However, there are ER binding sites that are not accessible prior to E2 treatment, which harbor high affinity EREs. ER may be acting as a pioneer at these sites as they become accessible after ER binding. A limited pioneering role for GR at sites with GRBEs has been observed, with >80% of GR binding sites that become accessible after treatment harboring a GRBE (John et al., 2011). Our results with both ER and GR are consistent with this finding and suggest that nuclear receptors are able to bind high affinity recognition motifs in inaccessible chromatin.
In addition to chromatin accessibility directing ER to suboptimal EREs in a cell type-specific manner, tethering of ER to binding sites through protein:protein interactions also plays a role in cell type specificity. Quantitative models that allowed for tethering of ER were significantly more accurate than models that forced ER to bind DNA. This increase in accuracy came mostly from cell-specific sites. The prevalence of tethering predicted by the thermodynamic models is similar to a recent study that used an ER DNA binding domain mutant to determine that 25% of gene expression changes rely on tethering (Stender et al., 2010). Together the combination of chromatin accessibility patterns and the presence of TFs that can tether ER to binding sites provide a template for the creation of cell-specific ER binding sites.
Cell-specific ER binding sites without EREs and shared ER binding sites with EREs seem equally likely to associate with gene expression changes of similar magnitudes. When ER binding sites are taken out of their genomic context and placed upstream of a minimal promoter in an enhancer assay, large differences between the types of binding sites are observed. Shared sites with high affinity EREs exhibit large responses to E2, while cell-specific sites without EREs drive modest responses and are less likely to activate expression. Cell-specific sites that do respond are dependent on the presence of interacting factors FOXA1, GATA3 and ETV4. Our findings suggest that cell-specific sites depend on context for their full activity and shared sites act in a context-independent manner.
Experimental Procedures
Cell growth and inductions
ECC-1 (ATCC CRL-2923) and T-47D (ATCC HTB-133) cells were grown and induced with E2 as previously described (Gertz et al., 2012). A549 was grown as previously described (Reddy et al., 2009). Cell growth and induction details are in the Supplemental Experimental Procedures.
DNase I hypersensitivity measurements
DNase-seq was performed and analyzed as previously described (Boyle et al., 2008; Song and Crawford, 2010) (see details in the Supplemental Experimental Procedures).
ChIP-seq and analysis
Cell line ChIP-seq was performed as previously described (Reddy et al., 2009). For ER ChIP-seq in mouse tissues, flash-frozen liver and uterus dissected from 8-week old female C57BL/6J mice were obtained from The Jackson Laboratories. ChIP-seq and analysis details are in the Supplemental Experimental Procedures.
DNA methylation analysis with RRBS
Reduced representation bisulfite sequencing (RRBS) was performed and analyzed as previously described (Gertz et al., 2011; Varley et al., 2013) (see details in the Supplemental Experimental Procedures).
Thermodynamic modeling
The thermodynamic modeling was implemented as previously described (Gertz and Cohen, 2009; Gertz et al., 2009). Thermodynamic modeling details are in the Supplemental Experimental Procedures.
Enhancer assays
To create enhancer vectors, a 500 bp fragment centered on the summit of each ER binding site was selected. ON-TARGETplus SMART pools of siRNA (Thermo Scientific) were used to perform knockdown. Enhancer assay details are in the Supplemental Experimental Procedures.
Evolutionary constraint analysis
To analyze sequence conservation of ERE motifs, we obtained evolutionary rate information from Genomic Evolutionary Rate Profiling (GERP) (Cooper et al., 2005; Davydov et al., 2010). Evolutionary constraint analysis details are in the Supplemental Experimental Procedures.
Supplementary Material
Highlights.
Two types of estrogen receptor α binding sites: shared and cell-specific
Shared sites are encoded in the genome as high affinity estrogen response elements
Cell-specific sites rely on interacting factors and depend on genomic context
Cell-specific binding is predicted from DNA sequence and chromatin accessibility
Acknowledgements
We thank Greg Barsh, Barbara Wold, Michael Garabedian, Chris Gunter and members of the Myers lab for suggestions and helpful discussions. We also thank J.D. Frey for help with artwork. This work was funded by NHGRI ENCODE Grant U54 HG004576 (to R.M.M.) and NIH Pathways to Independence Award K99 HG006922 (to J.G.). J.G. was supported by Postdoctoral Fellowship PF-12-028-01-TBE from the American Cancer Society for a portion of this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abe Y, Ijichi N, Ikeda K, Kayano H, Horie-Inoue K, Takeda S, Inoue S. Forkhead box transcription factor, forkhead box A1, shows negative association with lymph node status in endometrial cancer, and represses cell proliferation and migration of endometrial cancer cells. Cancer Sci. 2012;103:806–812. doi: 10.1111/j.1349-7006.2012.02201.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–322. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt AJ, Caldon CE, McNeil CM, Swarbrick A, Musgrove EA, Sutherland RL. Cell cycle machinery: links with genesis and treatment of breast cancer. Advances in experimental medicine and biology. 2008;630:189–205. doi: 10.1007/978-0-387-78818-0_12. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Chen Y, Chen L, Li JY, Mukaida N, Wang Q, Yang C, Yin WJ, Zeng XH, Jin W, Shao ZM. ERbeta and PEA3 co-activate IL-8 expression and promote the invasion of breast cancer cells. Cancer Biol Ther. 2011;11:497–511. doi: 10.4161/cbt.11.5.14667. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome research. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davydov EV, Goode DL, Sirota M, Cooper GM, Sidow A, Batzoglou S. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Cohen BA. Environment-specific combinatorial cis-regulation in synthetic promoters. Molecular systems biology. 2009;5:244. doi: 10.1038/msb.2009.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Reddy TE, Varley KE, Garabedian MJ, Myers RM. Genistein and bisphenol A exposure cause estrogen receptor 1 to bind thousands of sites in a cell type-specific manner. Genome research. 2012 doi: 10.1101/gr.135681.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz J, Varley KE, Reddy TE, Bowling KM, Pauli F, Parker SL, Kucera KS, Willard HF, Myers RM. Analysis of DNA methylation in a three-generation family reveals widespread genetic influence on epigenetic regulation. PLoS genetics. 2011;7:e1002228. doi: 10.1371/journal.pgen.1002228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin MJ, Lis JT. Chromatin landscape dictates HSF binding to target DNA elements. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Chen MW, Jordan VC, Brown M, Liu XS. Differential DNase I hypersensitivity reveals factor-dependent chromatin dynamics. Genome research. 2012 doi: 10.1101/gr.133280.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N, Isaacs GD, Diehl AG, Sun M, Cheung E, Ranish JA, Kraus WL. Multiple Sequence-Specific DNA-Binding Proteins Mediate Estrogen Receptor Signaling through a Tethering Pathway. Molecular endocrinology (Baltimore, Md. 2011 doi: 10.1210/me.2010-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, et al. Estrogen receptors: how do they signal and what are their targets. Physiological reviews. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Hertz GZ, Stormo GD. Identifying DNA and protein patterns with statistically significant alignments of multiple sequences. Bioinformatics. 1999;15:563–577. doi: 10.1093/bioinformatics/15.7.563. [DOI] [PubMed] [Google Scholar]
- Hewitt SC, Li L, Grimm SA, Chen Y, Liu L, Li Y, Bushel PR, Fargo D, Korach KS. Research resource: whole-genome estrogen receptor alpha binding in mouse uterine tissue revealed by ChIP-seq. Molecular endocrinology (Baltimore, Md. 2012;26:887–898. doi: 10.1210/me.2011-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Ross-Innes CS, Schmidt D, Carroll JS. FOXA1 is a key determinant of estrogen receptor function and endocrine response. Nat Genet. 2011;43:27–33. doi: 10.1038/ng.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–268. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph R, Orlov YL, Huss M, Sun W, Kong SL, Ukil L, Pan YF, Li G, Lim M, Thomsen JS, et al. Integrative model of genomic factors for determining binding site selection by estrogen receptor-alpha. Molecular systems biology. 2010;6:456. doi: 10.1038/msb.2010.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang JG, Hamiche A, Wu C. GAL4 directs nucleosome sliding induced by NURF. The EMBO journal. 2002;21:1406–1413. doi: 10.1093/emboj/21.6.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong SL, Li G, Loh SL, Sung WK, Liu ET. Cellular reprogramming by the conjoint action of ERalpha, FOXA1, and GATA3 to a ligand-inducible growth state. Molecular systems biology. 2011;7:526. doi: 10.1038/msb.2011.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M. Unique ERalpha cistromes control cell type-specific gene regulation. Molecular endocrinology (Baltimore, Md. 2008;22:2393–2406. doi: 10.1210/me.2008-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner PJ, Agard DA, Greene GL, Scanlan TS, Shiau AK, Uht RM, Webb P. Estrogen receptor pathways to AP-1. The Journal of steroid biochemistry and molecular biology. 2000;74:311–317. doi: 10.1016/s0960-0760(00)00108-4. [DOI] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, Bejerano G. GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol. 2010;28:495–501. doi: 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Lone R, Frith MC, Karlsson EK, Hansen U. Genomic targets of nuclear estrogen receptors. Molecular endocrinology (Baltimore, Md. 2004;18:1859–1875. doi: 10.1210/me.2003-0044. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome research. 2009;19:2163–2171. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature. 2012;481:389–393. doi: 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea MA, Ackers GK. The OR control system of bacteriophage lambda. A physical-chemical model for gene regulation. J Mol Biol. 1985;181:211–230. doi: 10.1016/0022-2836(85)90086-5. [DOI] [PubMed] [Google Scholar]
- Simpson NE, Gertz J, Imberg K, Myers RM, Garabedian MJ. Research resource: enhanced genome-wide occupancy of estrogen receptor alpha by the cochaperone p23 in breast cancer cells. Molecular endocrinology (Baltimore, Md. 2012;26:194–202. doi: 10.1210/me.2011-1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Crawford GE. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5384. pdb prot5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011;480:490–495. doi: 10.1038/nature10716. [DOI] [PubMed] [Google Scholar]
- Stender JD, Kim K, Charn TH, Komm B, Chang KC, Kraus WL, Benner C, Glass CK, Katzenellenbogen BS. Genome-wide analysis of estrogen receptor alpha DNA binding and tethering mechanisms identifies Runx1 as a novel tethering factor in receptor-mediated transcriptional activation. Mol Cell Biol. 2010;30:3943–3955. doi: 10.1128/MCB.00118-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, et al. Dynamic DNA methylation across diverse human cell lines and tissues. Genome research. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. The EMBO journal. 2009;28:1418–1428. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, et al. DNA methylation status predicts cell type-specific enhancer activity. The EMBO journal. 2011;30:3028–3039. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Cheng Y, Keller CA, Ernst J, Kumar SA, Mishra T, Morrissey C, Dorman CM, Chen KB, Drautz D, et al. Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome research. 2011;21:1659–1671. doi: 10.1101/gr.125088.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashwanth R, Rama S, Anbalagan M, Rao AJ. Role of estrogen in regulation of cellular differentiation: a study using human placental and rat Leydig cells. Molecular and cellular endocrinology. 2006;246:114–120. doi: 10.1016/j.mce.2005.11.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.