Abstract
In neuroscience, collaboration and data sharing are undermined by concerns over the management of protected health information (PHI) and personal identifying information (PII) in neuroimage datasets. The HIPAA Privacy Rule mandates measures for the preservation of subject privacy in neuroimaging studies. Unfortunately for the researcher, the management of information privacy is a burdensome task. Wide scale data sharing of neuroimages is challenging for three primary reasons: (i) A dearth of tools to systematically expunge PHI/PII from neuroimage data sets, (ii) a facility for tracking patient identities in redacted datasets has not been produced, and (iii) a sanitization workflow remains conspicuously absent. This article describes the XNAT Redaction Toolkit—an integrated redaction workflow which extends a popular neuroimage data management toolkit to remove PHI/PII from neuroimages. Quickshear defacing is also presented as a complementary technique for deidentifying the image data itself. Together, these tools improve subject privacy through systematic removal of PII/PHI.
Keywords: HIPAA Privacy Rule, Neuroimage Data, Neuroscience, XNAT Redaction Toolkit
INTRODUCTION
The digitization of medical records has transformed healthcare in both clinical and research settings. Research organizations have the potential for global, simultaneous and instant access to subject data along with the tools needed to perform analysis on a large scale. Nowhere is this potential more evident than in the field of neuroscience. Inter-organizational data sharing encourages collaboration, reduces the upfront costs of neuroimage data acquisition efforts, and fulfills requirements for data sharing established by funding institutions and journals.
Even so, special concerns over subject privacy have a chilling effect on the sharing of neuroimage data sets. Neuroimage data formats contain meta data that can reveal subject identity. The image data itself can be used to reconstruct a photorealistic rendering of a subject’s face. Thus, neuroimage data sharing in an evolving regulatory environment presents new questions and obstacles that neuroscientists are ill-prepared to address.
In response, we describe the XNAT Redaction Toolkit -- an integrated redaction workflow extending a popular neuroimage data management toolkit to remove PHI/PII from neuroimages. Quickshear defacing is also presented as a complementary technique for deidentifying the image data itself. Together, these tools improve subject privacy through systematic removal of PII/PHI.
BACKGROUND
Data sharing facilitates collaboration and enables reproducibility, peer review, and meta-analysis studies. This is especially significant in neuroimaging studies, where subject enrollment tends to be low (Costafreda, 2009; Van Horn et al., 2009). Collaboration allows institutions to pool data to increase the number of scans and diversity of subjects, improving statistical power and reliability.
The notable success of the Alzheimer’s Disease Neuroimaging Initiative (ADNI), a multisite neuroimaging project, demonstrates the potential for global inter-organizational collaboration (Mueller et al., 2005; Kolata, 2010; Toga et al., 2010). Data has been collected from 57 sites and images distributed to more than 1,300 investigators to date (Weiner et al., 2012). ADNI data has led to over 250 publications, and the effort has inspired similar data sharing initiatives for other conditions.
Collaborative efforts of this scale, however, introduce new challenges. Technical solutions to data storage, transmission, management, and dissemination problems continue to evolve through the development of medical imaging data management tools like the eXtensible Neuroimage Archiving Toolkit (XNAT), enabling a certain ease of data access (Marcus et al., 2007). However, none of these sharing solutions offer adequate and systematic tools for the preservation of subject privacy. Within these systems, subjects and their associated data must be managed by deidentifying the records before being added to the storage solution. While automated tools have been developed to remove metadata, none accomplish this deidentification on a systematic, study-wide (or institution-wide) basis (Neuroinformatics Research Group, 2010; MathWorks, 2012).
Any systematic tool for deidentification must be adaptable to the needs of the organization and research studies that it serves. Depending on situational research requirements, different aggregations of PHI/PII in publicly available datasets must be explicitly disallowed. The need for investigators to carefully consider the impact of their data is essential. Once a policy is decided upon, its implementation should involve minimal effort and integrate with ease into the data management workflow. In terms of its accessibility, popularity and open nature, XNAT presents itself as a viable platform for the integration of comprehensive privacy preservation processes for medical images.
The eXtensible Neuroimage Archive Toolkit (XNAT)
XNAT consists of web and desktop tools for quality control, organization and storage of imaging data. It provides workflows to interface scanning hardware with a storage backend (Marcus et al., 2007). XNAT quarantines data sets until they have been verified by a user. This process simplifies the administrative burden associated with data sharing, but it does not provide automated redaction protocols for privacy protection in shared data.
XNAT stores data in a resource hierarchy, with a project as the root type. Subjects in an imaging study are added to projects and possess one or more experiment resources. Scan sessions are created in the corresponding experiment resource, which is linked to the subject.
Currently, investigators must send anonymized images to XNAT and track subjects manually. In addition to DICOM images, XNAT stores subject demographic data, including elements of PHI/PII. This additional storage layer complicates redaction. Investigators must either track subjects in XNAT via their actual PII (making the data useful only within the organization) or use unique identifiers for their subjects tracked through some external resource.
XNAT provides basic support for sanitizing patient data. The DicomBrowser tool allows users to view or edit DICOM metadata both manually and with batch processing (Neuroinformatics Research Group, 2010). It is offered in both a graphical and command-line interface. The flexibility of XNAT allows researchers to upload data in multiple formats with varying study parameters. However, this presents a challenge when standardizing a sanitization process; it requires the end user to tailor the anonymization process to a particular study or session through scripts based on the DICOM tag key-value pairs. Moreover, the current version does not include a tool for defacing.
Medical Image Privacy
Medical images introduce new privacy challenges in addition to those of traditional electronic health records (Kuehler et al., 2011). Textual PHI/PII can be redacted by simply removing or replacing the offending field, but medical images, which can constitute self-identifying data, are not so easily sanitized, and removing this data may destroy useful information. In particular, high resolution structural neuroimages may contain detailed facial features equivalent to a full face image, and thus it is necessary to sanitize both metadata and the neuroimage itself.
There are four categories of primary threats to subject privacy introduced by medical images: (i) direct, (ii) re-linkage, (iii), existential inference, and (iv) inherent identification. The direct threat occurs when the image reveals a condition or diagnosis. This can be mitigated with metadata removal to obscure the subject’s identity. A more common scenario is re-linkage, where the image contains enough metadata to identify the subject. Existential inference exposes a subject by simply suggesting the existence of an image. For example, participation in a study can imply that the subject is part of the case group. Finally, the medical image itself can be inherently identifying by unique features found therein. High resolution structural neuroimages in particular can contain detailed facial features that can be used to reidentify the subject. To mitigate these threats, neuroimages must be deidentified at both the metadata and image level.
Metadata
Many medical data formats (both imaging data and other forms) store embedded information in headers and contain a wealth of PHI/PII. The Digital Imaging and Communications in Medicine (DICOM) standard for medical images is a very rich and robust data format. DICOM implements fields for all relevant medical data associated with a subject, even allowing for complete medical history synopsis (DICOM, 2011). While this information can be helpful in organizing data for a study, much of it must be removed before the data can be safely released to collaborators. Utilities exist to remove data from individual files, but there is a lack of tools to systematically expunge metadata from large studies in order to make data sets immediately sharable. Furthermore, once metadata is expunged, the task of tracking the correspondences between redacted images and data and their original sources is left entirely up to the originating researcher.
Inherent Identifiability
Structural neuroimages can contain facial features that are easily and accurately reconstructed through volume rendering. Several packages for analyzing and viewing neuroimage data provide built-in volume rendering capability. The neuroimage-based likeness can be exploited to identify the subject, and metadata can be used to guide reidentification, filtering the potential subjects using non-PHI fields.
The current limitations of existing automated facial recognition make it tempting to dismiss the feasibility of reidentification based on flawed assumptions: (1) facial recognition will never improve, and (2) only correct identifications are potentially problematic. The latter argument fails to consider the damage caused by an incorrect identification. While a correct identification may reveal sensitive information, challenging a false identity may compel an individual to reveal their records. However, while the challenges of automated facial recognition are not easily confronted, nor are they impossible to solve. Concerns about the feasibility of volume rendering and facial recognition applied to neuroimages are largely the same as the issues with photograph-based recognition. The language of the HIPAA Privacy Rule defines images comparable to photographs as PHI. Therefore, if neuroimage-based recognition can perform with similar accuracy to photographs, such images must be protected as full face images by HIPAA-bound entities.
Neuroimage Deidentification
The current approaches to deidentifying neuroimages are categorized as either ‘skull stripping’ or ‘defacing.’ Defacing aims to remove only facial features and leaves behind other nonbrain tissue, whereas skull stripping identifies and removes any extraneous non-brain tissue, including identifiable facial features. Skull stripping is performed as part of a typical neuroimage analysis workflow.
There are several widely used methods for skull stripping, many integrated with neuroimage analysis software. Popular skull stripping tools have been compared and analyzed in detail (Fennema-Notestine et al., 2006).
Because skull stripping is routinely performed, it is a commonly employed deidentification technique. Skull stripping methods can depend highly on fine tuning of parameters, and this may often result into loss of desirable brain tissue. The results may vary widely between methods and the consequential disparities can impact further analysis. Skull stripping may also favor a particular anatomical structure based on the specific study or region of interest. The variation in methods and subjective evaluation can complicate meta-analysis, secondary use, and collaboration by discarding potentially relevant voxels (volumetric pixels).
Unlike skull stripping, defacing preserves non-brain tissue. The MRI Defacer approach removes only non-brain voxels that contain facial features based on a previously constructed and manually labeled face atlas (Bischoff-Grethe et al., 2007). The result appears as though the facial features were eroded, leaving the brain volume intact.
Defacing is an effective method for neuroimage deidentification, and it preserves more potentially relevant brain voxels. As part of the existing analysis workflow, skull stripping is an attractive deidentification approach, but defacing allows for more flexibility in future use. However, MRI Defacer relies on a face atlas to identify features, and because of the potential variation between data sets, a method that does not require prior knowledge may be preferred.
METHODS
Redaction in XNAT
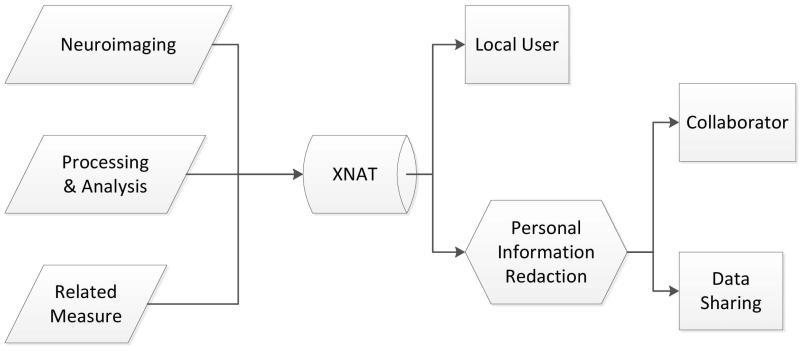
In order to facilitate systematic deidentification, we have implemented an integrated redaction workflow in XNAT. Risk assessment analysis has shown the XNAT platform to be a viable vehicle for implementation of a redaction protocol and data security maintenance (Schimke, 2009). Using the XNAT Redaction Toolkit, a researcher can check out a redacted version of a dataset to any collaborator with confidence that it will comply with the HIPAA Privacy Rule (HIPAA, 2006). Parallel versions of a project (one redacted, and one with the original data) can be maintained automatically by the redaction tool during data collection. The role of redaction in the XNAT data gathering and sharing process is shown in Figure 1.
Figure 1. XNAT and the role of redaction.
Leveraging XNAT’s custom pipeline interface, we have inserted a new redaction workflow into the standard set of XNAT tools. Investigators can invoke this workflow to checkout neuroimage data sets to collaborators with accounts on an accessible XNAT instance. The redaction tool downloads project resources, invokes sanitization routines for each supported resource type, inserts tracking identifiers, and finally uploads this dataset to a new XNAT project accessible by the collaborator. In addition, the redaction tool tracks the PHI and image data which has been redacted, as well as the individuals to whom the data has been released. This information is stored in the Privacy Database.
The architecture follows a three-tiered design pattern consisting of a Privacy Database backend, a Java based application, and several customized XNAT pipelines that provide user interfaces for configuring the redaction process. The Java redaction application is implemented via a set of helper classes for REST queries for data transit, a set of routines for identity tracking across XNAT projects, and an extensible XNATEntity class, which acts as an interface to download, redact, and upload XNAT resources. The XNAT Redaction pipeline leverages the dcm4che2 Java library to read and write from DICOM files (Warnock et al., 2007). It is designed to run in an existing XNAT environment, requiring Java Runtime Environment 1.6, and (optionally) a PHP-enabled Apache web server and HTTPS site.
Redaction Process
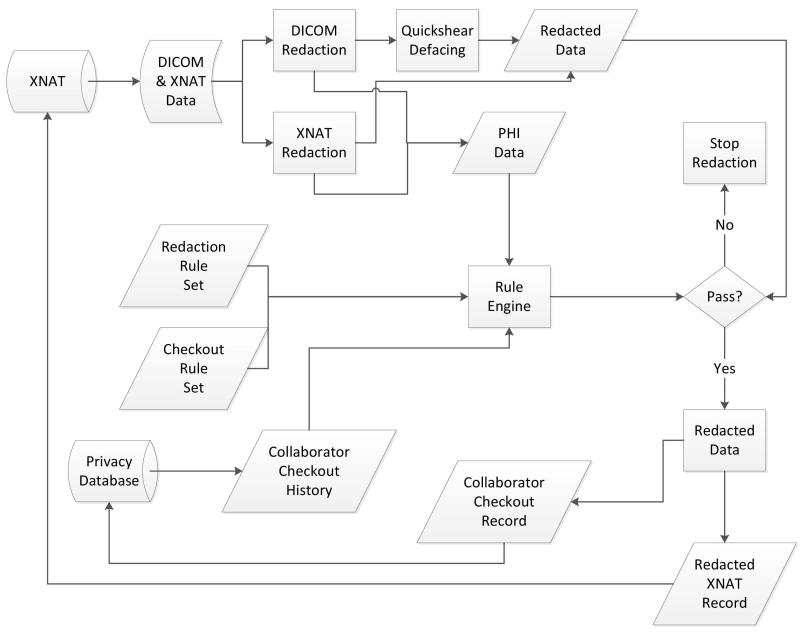
The redaction system in Figure 2 is designed around a previously developed process to examine data at different abstraction layers of the file/media and create a mapping of redacted content to original content (Hale, Manes, Watson, Barclay, & Greer, 2007). Our implementation handles XNAT resources by mimicking the hierarchical resource structure of XNAT. Specifically, the redaction engine keeps an internal data-type for each XNAT resource which must be handled in the redaction process. These internal classes map the PHI/PII fields contained in the various XNAT resources (DICOM, XML) to internal field identifiers. Subsequently, the application’s DICOM redaction and XNAT redaction process elements extract the values of these fields and sanitize the original documents at the bit level by writing zeros over the field contents. This ensures that there is no recoverable private data hidden in the document.
Figure 2. XNAT redaction process.
The PHI/PII data is checked against a database which maps the true identities of subjects onto randomly generated unique identifiers. This is done within the Rule Engine, using the Privacy Database as a storage backend. These identifiers are inserted into the original data types to consistently track individuals in the redacted dataset. Additionally, a subset of PHI may be reinserted into the dataset at the request of an investigator, assuming that the request complies with the policy outlined in the organization’s policy expressed via the policy language. Next, the organizational Privacy Policy is evaluated. The Privacy Database projects a Collaborator Checkout History to assist with the policy validation process executed by the Rule Engine. If the redacted data is approved, then the data is uploaded back to XNAT, and finally information about the redacted data and the execution of the pipeline is stored as part of the Collaborator Checkout record in the Privacy Database.
The pipeline supports the redaction of DICOM datasets (with planned support for automated Quickshear defacing of DICOM images), as well as sanitization of demographic information stored in the XNAT project. Previous research has helped to narrow the scope of our initial redaction efforts to DICOM support, due to its ubiquitous use in medical imaging (Barclay et al., 2009, 2010; Schimke et al., 2010). To ensure support for future neuroimaging resources within the same framework, the redaction pipeline is extensible via the addition of classes mapping internal file format representations to those understood by the redaction pipeline. The presence of any resource type is automatically detected and delegated to the custom resource class by the redaction engine.
Rule Sets
The core of the redaction pipeline is its policy language, which is customizable to enable compliance with any organization’s information privacy policy. The primary function of a variable Privacy Policy rule system is to provide support for the release of PHI necessary to a collaborator’s study and compliant with HIPAA requirements in the particular instance under consideration. In many cases, for example, a data set that includes age but removes all other PHI/PII fields would be useful in a study analysis, and not constitute a breach of privacy.
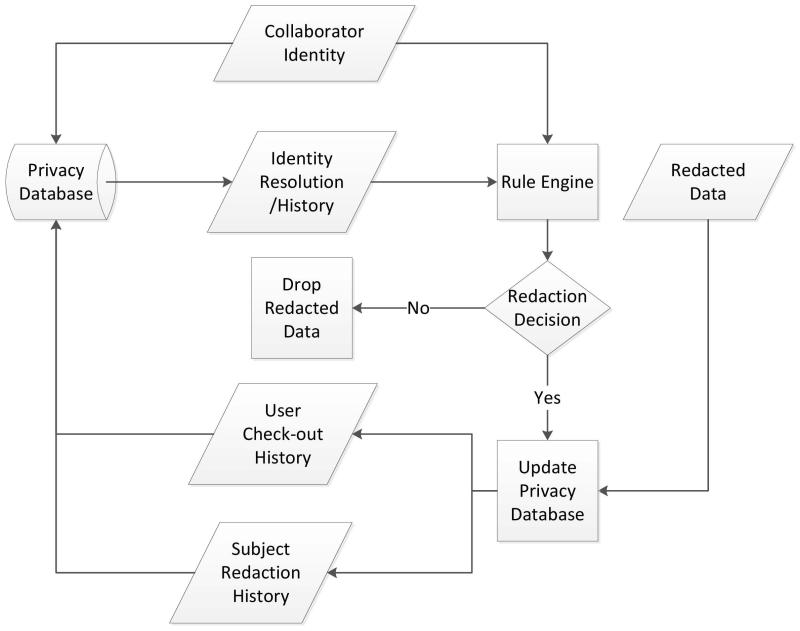
Thus, our tool set allows the preservation of a limited subset of PHI data (subject to the organization’s policy definitions) within the redacted data (for example: Patient Age). Variables which may be useful to collaborators can be released, with the assurance that the status of all released data is tracked and any potential future breaches of privacy can be avoided with no additional administrative effort. This process is detailed in Figure 3.
Figure 3. Detailed view of the privacy and identity management process.
Additional configuration files are implemented for DICOM definitions (mapping DICOM fields to PHI/PII identifiers) and XNAT demographic definitions. Fields slated for redaction can be customized and added to a configurable list using internally recognizable PHI/PII identifiers.
Privacy Management
Retention of specific information from the original DICOM image file may be desirable or necessary for export along with the redacted dataset. In this case, user defined options can keep embedded subject data (such as age, weight) in the resultant DICOM image dataset. In part, the policy language is implemented to prevent breaches of privacy that may occur by allowing the redacted datasets with different retained information to be released. Burdening the investigator with such responsibility has previously led to significant breaches of information privacy (Sweeney, 2002; Ohm, 2009).
To ensure that redacted identities can be tracked automatically, the Redaction Pipeline stores subject PII (names and birth dates) in a relational database. The database correlates this information to a list of identities used to create redacted versions of scan and subject data. Thus, identities can be tracked in both directions. Incoming data to be redacted can be checked to see if the subject has been scanned before, possibly as part of another study. The Redaction Pipeline automatically applies the privacy policy to these situations, and alerts the investigator if a potential privacy violation is detected before data crosses organizational boundaries.
Custom User Interfaces
The XNAT Redaction Pipeline supports execution and task configuration from XNAT’s web interface, using XNAT’s Pipeline Engine to schedule redaction, sanitization, and PHI checkout through simple user interfaces. The Redaction Engine interface is composed of a Velocity screen template, an Apache Turbine screen class, and an Apache Turbine action class (all visible to the user), as well as an XML descriptor. Most pipelines are capable of utilizing the default template, screen class, and action class provided by XNAT without any trouble. The Redaction Pipeline, however, requires a higher degree of integration with XNAT than is provided by these default files. For such scenarios, XNAT supports custom-defined templates, screens, and actions.
The Redaction Pipeline supports automatic as well as manual initiation of redaction for any new scan data uploaded. This is accomplished through a built-in feature which allows pipelines to be launched automatically whenever new images are archived. The flag can be set when a new pipeline is added to a project, or by editing a project’s existing pipelines. In order to initiate redaction, an XNAT project owner or administrator must create a new project to hold the redacted data. The administrator then points the redaction pipeline to the new project. In the case of an existing project, the administrator can specify users to whom the project data will be made available. Users can be specified at redaction time to allow the privacy management system to resolve possible information privacy breeches according to the organization’s privacy policy. Access to redacted projects can also be granted later (used frequently in the case of automated redaction). Granting a user access to any redacted project will automatically result in the notification of the privacy manager, which will then ensure that the access granted complies with the privacy policy.
Quickshear Defacing
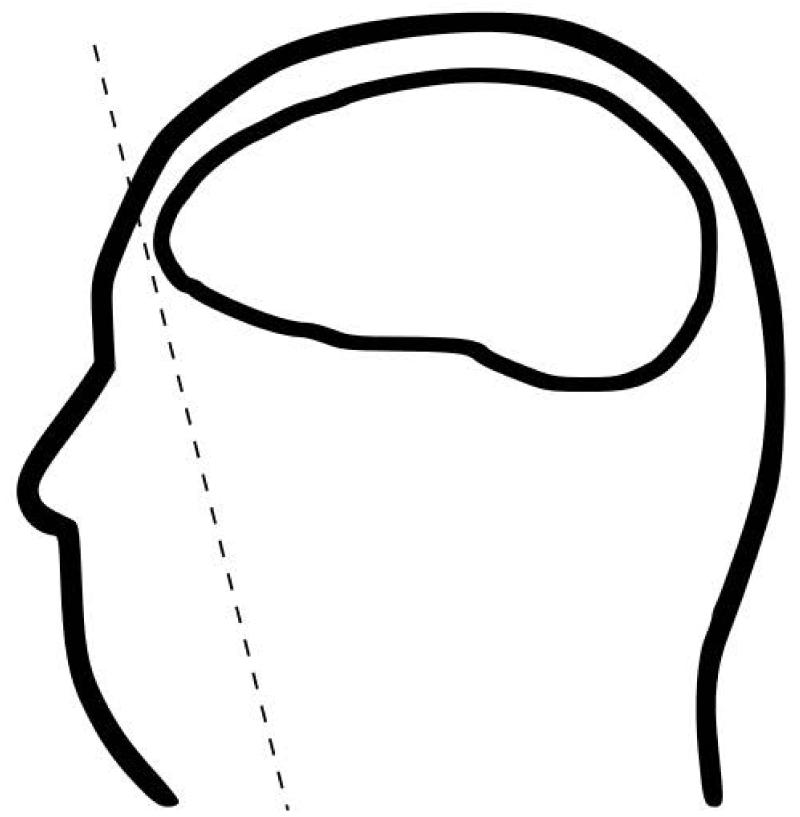
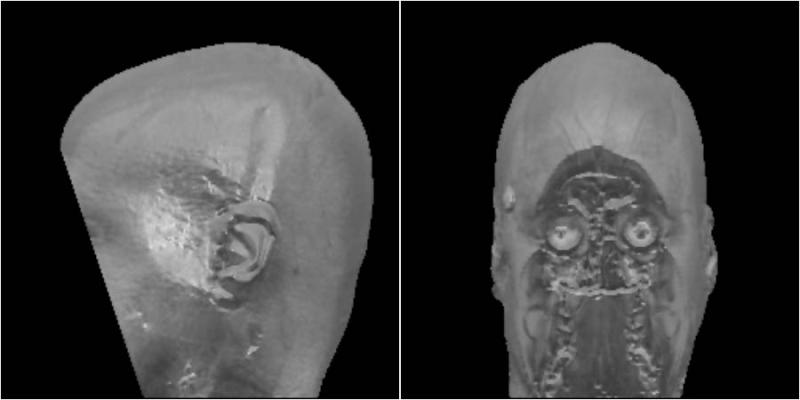
In order to expunge PHI/PII from neuroimages, the redaction process must also remove identifiable facial features data from neuroimages (Schimke et al., 2011a, 2011b) Quickshear defacing eliminates the need for manually labeled face atlases and offers improved performance over other methods while integrating neatly into the neuroimage analysis workflow. The Quickshear technique identifies a plane that divides the neuroimage into two volumes, a face and a brain volume, as illustrated in Figure 4. The voxels of the “face” side are sheared off, removing identifiable facial features and leaving the brain volume intact.
Figure 4. Quickshear defacing illustrated.
The plane is identified using a binary brain mask to determine which voxels contain brain tissue. The mask is created by skull stripping the original volume. While the shortcomings of skull stripping as a deidentification technique have been discussed, it is ideal for creating the binary brain mask. The primary concern with skull stripping is the preservation of brain tissue, and this can be avoided by adding a buffer to the mask.
The goal of Quickshear is to prevent reidentification rather than remove all non-brain tissue, and thus it collapses the original neuroimage and brain mask onto the sagittal plane (profile view) to reduce complexity of identifying the shearing plane. The two-dimensional representation is used to create an edge-of-brain mask and find the convex hull. The line formed by the first point on the lower hull, which is nearest to the forehead, and the next consecutive point is extended into three dimensions to form the shearing plane. By definition of the convex hull, this line is an edge of the polygon, and all brain voxels are contained on the line itself or on the interior of the polygon. All voxels that fall on the “face” side of the plane are set to zero. The result is a defaced volume, as shown in Figure 5.
Figure 5. Volume rendering after Quickshear defacing is applied.
To verify that all brain voxels are intact, the resulting volume is compared to the binary brain mask. Comparison to the original binary mask is a basic sanity check. As another level of verification, a mask generated with an alternate skull stripping technique can be used to check for discarded brain voxels. Facial feature removal can be manually validated by viewing the defaced images directly or automated through face detection techniques such as the OpenCV Face Detector implementation (OpenCV, http://opencv.willowgarage.com/wiki/).
RESULTS
XNAT Redaction Toolkit
The XNAT Redaction Toolkit is a practical tool that addresses a pressing need in the medical imaging community. The integrated workflow provides a means to greatly simplify organizational compliance with data privacy standards. HIPAA and other regulatory measures can place a significant burden on researchers and administrators wishing to share data. This toolkit encourages collaboration with the assurance of privacy policy compliance, thus facilitating data sharing in the scientific community. The specification of flexible privacy rules allows research organizations to implement current privacy policies and adapt as national health privacy policies evolve.
Quickshear Defacing
Quickshear defacing was tested with the Multimodal Reproducibility Study data set from Landman et al. using MPRAGE scans with a 1.0 × 1.0 × 1.2 mm3 resolution (Landman et al., 2010). The data is available from NITRC (http://www.nitrc.org/projects/multimodal) and consists of 42 distinct images. Three skull stripping implementations from popular neuroimaging analysis tools were used to generate binary brain masks for defacing and validation – the Hybrid Watershed Algorithm (HWA) in FreeSurfer (available from http://surfer.nmr.mgh.harvard.edu/), the Brain Extraction Toolkit (BET) in FSL (available from http://www.fmrib.ox.ac.uk/fsl/) and 3dSkullStrip in AFNI (available from http://afni.nimh.nih.gov/afni). The images were also defaced using MRI Defacer, a neuroimage deidentification tool described in Bischoff-Grethe et al. (2007) and available from NITRC (http://www.nitrc.org/projects/mri_deface).
As expected, Quickshear defacing did not discard any voxels when using the same mask as the verification approach. When compared to other masks, on average, Quickshear defacing discarded fewer brain voxels from fewer neuroimages than MRI Defacer. The overall number of voxels discarded is a small percentage of the total brain voxels. An acceptable threshold of discarded voxels should be determined on a situational basis, and flagged volumes manually inspected. Table 1 presents the average number of brain voxels discarded for each defacing mechanism tested.
Table 1. Average voxels discarded in defacing (Number of images with voxels discarded).
| Brain Mask |
||||||
|---|---|---|---|---|---|---|
| Defacing Method | AFNI | BET | HWA | |||
| MRI Defacer | 408.74 | (12) | 75271.93 | (42) | 422.0 | (7) |
| Quickshear AFNI | 0.0 | (0) | 5560.76 | (13) | 0.0 | (0) |
| BET | 0.21 | (1) | 0.0 | (0) | 1.0 | (2) |
| HWA | 0.0 | (0) | 7587.24 | (12) | 0.0 | (0) |
Using the OpenCV face detector, faces were found in 38 of the 42 original, undefaced images. The same detector identified faces in 10 Quickshear defaced images using an AFNI-generated mask, 10 with a BET-generated mask, and 12 with HWA (Table 2). By comparison, 9 were found in the images defaced with MRI Defacer. False positives are potentially due to the presence of eye sockets and nasal cavities that resemble a face. The detection process may benefit from introducing profile-view face detection and combining the results with the frontal face detector. OpenCV provides a profile-view cascade, but it was found unreliable for the test data and only detected faces in 5 of the 42 original images (complete analysis appears in Schimke et al., 2011a). Overall, Quickshear offered comparable defacing efficacy to MRI defacer, while preserving more brain voxels.
Table 2. Number of faces detected under defacing techniques.
| Defacing Method | Faces detected |
|---|---|
| MRI Defacer | 9 |
| Quickshear AFN1 | 10 |
| BET | 10 |
| HWA | 12 |
Table 3 presents the run time performance of the defacing techniques MRI Defacer and Quickshear. For Quickshear defacing, skull stripping and masking times are also shown. The running time is an average per image, over five runs.
Table 3. Runtime performance of defacing techniques.
| Defacing Method | Skull Stripping | Defacing | |
|---|---|---|---|
| Running | Time (s) | Running Time (s) | |
| MRI Defacer | - | 260.17 | |
| Quickshear | AFNI | 205.71 | 4.30 |
| BET | 13.72 | 4.33 | |
| HWA | 29.29 | 4.27 | |
Quickshear defacing for neuroimages is a practical and effective approach for eliminating identifiable facial features from neuroimage data. By leveraging existing skull stripped volumes, it integrates seamlessly into the existing analysis workflow to reduce the administrative burden on the researcher while enhancing subject privacy.
DISCUSSION
Privacy guidelines under HIPAA for the hospital impose extremely strict requirements on controlling PHI/PII data availability. Access to personal information is restricted to individuals who need it to treat the patient. The stipulations for research data allow greater flexibility, but still encumber significant administrative burden. In particular, unless data can be declared immune from reidentification attempts (by an expert biostatistican or a proven deidentification process), it does not qualify for release to the broader research community.
To complicate matters, structural neuroimages can be classified as identifiable data, even absent identifying metadata. Accordingly, practical and effective defacing methods like Quickshear to strip recoverable identity information (full facial images) from a structural image represent a vital component in a comprehensive medical image redaction strategy.
Moreover, internal review boards commonly require that researchers submit a plan to protect identifiers from improper use or disclosure, and to destroy identifiers unless retention is required by law. The XNAT Redaction Tool establishes a standardized and proven workflow for the deidentification of PHI, greatly simplifying the planning process. Thus, in no small measure it alleviates considerable administrative burden from the researcher.
Large scale neuroimage studies confront researchers with a number of challenges. Notably, access to data sets is hindered by concerns over managing privacy issues. The XNAT Redaction Tool described here offers an integrated strategy that transparently applies deidentification solutions over metadata and structural data within the normal archival workflow. This approach is key to fostering collaboration and data sharing among researchers employing medical imaging in their studies.
ACKNOWLEDGMENT
We gratefully acknowledge support from the William K. Warren Foundation and the National Institutes of Health.
Biographies
Matthew Matlock is a research assistant and scientific programmer in the Center for Biological Systems Engineering and the Department of Pathology and Immunology of the Washington University School of Medicine. He received his Bachelor of Science degree in 2009 and Master of Science in 2011, both from the University of Tulsa. His research interests include neuroinformatics, artificial intelligence, and computational techniques for drug discovery.
Nakeisha Schimke is a postdoctoral researcher in the Institute of Bioinformatics and Computational Biology at The University of Tulsa. She received her PhD in Computer Science from The University of Tulsa in 2011. Her research interests are in the areas of neuroinformatics, information security and privacy.
Liang Kong is a doctoral student in Computer Science at The University of Tulsa. He received Bachelor of Science in 2009 from Lanzhou University and Master’s of Science in 2011 from University of Tulsa. He works in the Human Computer Interaction (HCI) Laboratory at the University of Tulsa. His research interests are spatial access control and digital technologies for collaborative workflow.
Stephen Macke is an undergraduate researcher in the Institute of Bioinformatics and Computational Biology at The University of Tulsa. A 2011 Barry M. Goldwater Scholar, his research interests include bioinformatics, machine learning and medical informatics.
John Hale is a Professor in the Tandy School of Computer Science and a faculty researcher in the Institute for Information Security at The University of Tulsa. He received his Bachelor’s of Science in 1990, Master’s of Science in 1992 and doctorate degree in 1997, all in computer science from the University of Tulsa. Dr. Hale has overseen the development of one of the premier information assurance curricula in the nation while at iSec. In 2000, he earned a prestigious National Science Foundation CAREER award for his education and research initiatives at iSec. His research interests include cyber attack modeling, analysis and visualization, enterprise security management, secure operating systems, distributed system verification and policy coordination.
Contributor Information
Matt Matlock, Center for Biological Systems Engineering, Department of Pathology and Immunology, Washington University School of Medicine.
Nakeisha Schimke, Institute of Bioinformatics and Computational Biology, Tandy School of Computer Science, University of Tulsa, USA.
Liang Kong, Institute of Bioinformatics and Computational Biology, Tandy School of Computer Science, University of Tulsa, USA.
Stephen Macke, Institute of Bioinformatics and Computational Biology, Tandy School of Computer Science, University of Tulsa, USA.
John Hale, Institute of Bioinformatics and Computational Biology, Tandy School of Computer Science, University of Tulsa, USA.
REFERENCES
- Barclay A, Schimke N, Hale J. Comprehensive neuroimage redaction; Poster presented at the USENIX Security Symposium; Montreal, QC, Canada. Aug, 2009. [Google Scholar]
- Barclay A, Schimke N, Hale J. Redacting PHI in neurological images using XNAT. Comprehensive neuroimage redaction; Poster presented at the USENIX Security Symposium; Washington, DC. Aug, 2010. [Google Scholar]
- Bischoff-Grethe A, Ozyurt IB, Busa E, Quinn BT, Fennema-Notestine C, Clark CP. A technique for the deidentification of structural brain MR images. Human Brain Mapping. 2007;28:892–903. doi: 10.1002/hbm.20312. doi:10.1002/hbm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costafreda SG. Pooling fMRI data: Metaanalysis, mega-analysis and multi-center studies. Frontiers in Neuroinformatics. 2009;3(33) doi: 10.3389/neuro.11.033.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fennema-Notestine C, Ozyurt IB, Clark CP, Morris S, Bischoff-Grethe A, Bondi MW. Quantitative evaluation of automated skull-stripping methods applied to contemporary and legacy images: Effects of diagnosis, bias correction, and slice location. Human Brain Mapping. 2006;27:99–113. doi: 10.1002/hbm.20161. doi:10.1002/hbm.20161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale J, Manes G, Watson L, Barclay A, Greer D. Redaction digital information from electronic devices. In: Craiger P, Shenoi S, editors. Advances in digital forensics III. Springer; New York, NY: 2007. pp. 205–214. [Google Scholar]
- Kolata G. Rare sharing of data leads to progress on Alzheimer’s. The New York Times. 2010 Aug 12; Retrieved March 19, 2012, from http://www.nytimes.com/2010/08/13/health/research/13alzheimer.html. [Google Scholar]
- Kuehler M, Schimke N, Hale J. Privacy considerations for electronic health records. In: Yee G, editor. Privacy protection measures and technologies in business organizations: Aspects and standards. IGI Global; Hershey, PA: 2011. pp. 210–226. doi:10.4018/978-1-61350-501-4.ch008. [Google Scholar]
- Landman BA, Huang AJ, Gifford A, Vikram DS, Lim IA, Farrell JA. Multi-parametric neuroimaging reproducibility: A 3T resource study. NeuroImage. 2011;54:2854–2866. doi: 10.1016/j.neuroimage.2010.11.047. doi:10.1016/j.neuroimage.2010.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The extensible neuroimaging archive toolkit: An informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- MathWorks Dicomanon. 2012 Retrieved from http://www.mathworks.com/help/toolbox/images/ref/dicomanon.html. [Google Scholar]
- Mueller SG, Weiner MW, Thal LJ, Peterson RC, Jack C, Jagust W. The Alzheimer’s disease neuroimaging initiative. Neuroimaging Clinics of North America. 2005;15:869–877. doi: 10.1016/j.nic.2005.09.008. doi:10.1016/j.nic.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Electrical Manufacturer’s Association Digital imaging and communications in medicine. 2011 Retrieved from http://medical.nema.org/standard.html. [Google Scholar]
- Neuroinformatics Research Group DicomBrowser. 2010 Retrieved from http://nrg.wustl.edu/projects/DICOM/DicomBrowser. [Google Scholar]
- Ohm P. UCLA Law Review. University of California. Vol. 57. School of Law; Los Angeles: 2010. Broken promises of privacy: Responding to the surprising failure of anonymization; pp. 1701–1777. [Google Scholar]
- Schimke N, Barclay A, Gehres P, Hale J. PHI redaction for neuroimagery; Poster presented at the Sixth International Imaging Genetics Conference; Irvine, CA. Jan, 2010. [Google Scholar]
- Schimke N, Kuehler M, Hale J. Li Y, editor. Preserving privacy in structural neuroimages. Proceedings of the 25th Annual IFIP TC Conference on Data and Applications Security and Privacy. 2011a:301–308. LNCS 6818. [Google Scholar]
- Schimke N, Kuehler M, Hale J. Proceedings of the Second USENIX Workshop on Health Security and Privacy. San Francisco, CA: 2011b. Quickshear defacing for neuroimages; p. 11. [Google Scholar]
- Schimke N, Singleton N, Hale J. XNAT security assessment (Tech. Rep.) University of Tulsa; Tulsa, OK: 2009. [Google Scholar]
- Sweeney L. k-anonymity: A model for protecting privacy. International Journal of Uncertainty Fuzziness and Knowledge Based Systems. 2002;10:557–570. doi:10.1142/S0218488502001648. [Google Scholar]
- Toga AW. Neuroimage databases: The good, the bad, and the ugly. Nature Neuroscience. 2002;3:302–309. doi: 10.1038/nrn782. doi:10.1038/nrn782. [DOI] [PubMed] [Google Scholar]
- Toga AW, Crawford KL, Alzheimer’s Disease Neuroimaging Initiative The informatics core of the Alzheimer’s disease neuroimaging. Alzheimer’s & Dementia. 2010;6(3):247–256. doi: 10.1016/j.jalz.2010.03.001. doi:10.1016/j.jalz.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- U.S. Department of Health and Human Services. Retrieved from http://www.hhs.gov/ocr/privacy/hipaa/administrative/privacyrule/adminsimpregtext.pdf HIPAA administrative simplification: Regulation text. 2006 http://www.hhs.gov/ocr/privacy/hipaa/administrative/privacyrule/adminsimpregtext.pdf [Google Scholar]
- Van Horn JD, Toga AW. Is it time to reprioritize neuroimaging databases and digital repositories? NeuroImage. 2009;47:1720–1734. doi: 10.1016/j.neuroimage.2009.03.086. doi:10.1016/j.neuroimage.2009.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnock MJ, Toland C, Evans D, Wallace B, Nagy P. Benefits of using the DCM4CHE DICOM archive. Journal of Digital Imaging. 2007;20(1):125–129. doi: 10.1007/s10278-007-9064-1. doi:10.1007/s10278-007-9064-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC. The Alzheimer’s disease neuroimaging initiative: A review of papers published since its inception. Alzheimer’s & Dementia. 2012;1(1):S1–S68. doi: 10.1016/j.jalz.2011.09.172. doi:10.1016/j.jalz.2011.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]