Abstract
Members of the genus Bifidobacterium are common inhabitants of the gastrointestinal tracts of humans and other mammals, where they ferment many diet-derived carbohydrates that cannot be digested by their hosts. To extend our understanding of bifidobacterial carbohydrate utilization, we investigated the molecular mechanisms by which 11 strains of Bifidobacterium breve metabolize four distinct α-glucose- and/or α-galactose-containing oligosaccharides, namely, raffinose, stachyose, melibiose, and melezitose. Here we demonstrate that all B. breve strains examined possess the ability to utilize raffinose, stachyose, and melibiose. However, the ability to metabolize melezitose was not common to all B. breve strains tested. Transcriptomic and functional genomic approaches identified a gene cluster dedicated to the metabolism of α-galactose-containing carbohydrates, while an adjacent gene cluster, dedicated to the metabolism of α-glucose-containing melezitose, was identified in strains that are able to use this carbohydrate.
INTRODUCTION
Bifidobacteria are Gram positive, saccharolytic, nonmotile, nonsporulating anaerobic rods with a high G+C genome content that belong to the phylum Actinobacteria and the family Bifidobacteriaceae (1, 2). The first representatives of the genus Bifidobacterium were isolated more than a century ago (3). These bacteria naturally inhabit the gastrointestinal tracts of humans and other mammals and are particularly abundant in breast-fed infants (4, 5). Claims that certain bifidobacterial strains promote and maintain gastrointestinal health have been advanced (6), and these strains are therefore used as health-promoting or probiotic bacterial ingredients in certain functional foods (7). Their reported beneficial effects on the host include the inhibition of bacterial and viral pathogens, alleviation of lactose intolerance, enhancement of natural immunity, and reduction of serum cholesterol levels (8–10).
As saccharolytic microorganisms, bifidobacteria degrade various oligosaccharides and polysaccharides into their monosaccharide constituents, which are then shuttled into a specific hexose fermentation pathway called the fructose-6-phosphate phosphoketolase pathway, or bifid shunt (11). The proliferation of specific species or strains of commensal bifidobacteria is presumed to be stimulated by particular dietary carbohydrates, which for this reason are termed prebiotic substances (12). The term “prebiotics” was first coined in 1995, when it was defined as “nondigestible food ingredients that beneficially affect the host by selectively stimulating the growth and/or activity of one or a limited number of bacterial species already resident in the colon” (13). As much as 8% of the coding capacity of a bifidobacterial genome is dedicated to carbohydrate metabolism, of which half is considered to be responsible for carbohydrate uptake, mainly via ABC transporters, although proton motive force-driven permeases, proton symporters, and phosphoenolpyruvate-dependent phosphotransferase systems (PEP-PTSs) may also be employed for this purpose (14–16). A range of glycosyl hydrolases (GH), enzymes that hydrolyze a specific glycosidic bond between the monosaccharide moieties of certain oligo- and polysaccharides, allow bifidobacteria to grow on dietary and host-derived carbohydrates present in the gastrointestinal tract (15).
Certain bifidobacterial strains have been shown previously to grow on soymilk-derived α-galacto-oligosaccharides, such as raffinose [α-d-Galp-(1→6)-α-d-Glcp-(1→2)-β-d-Fruf], stachyose [α-d-Galp-(1→6)-α-d-Galp-(1→6)-α-d-Glcp-(1→2)-β-d-Fruf], and melibiose [α-d-Galp-(1→6)-α-d-Glcp] (17–19). Stachyose and raffinose (sugars of the so-called raffinose family, which also includes verbascose) are present in a wide variety of plants (20), while the related sugar melibiose (though not a member of the raffinose family) is also found in many plants and is particularly abundant in soybean roots and stems (21). To metabolize such α-galacto-oligosaccharides, bifidobacteria require α-galactosidase enzyme activity, which has been identified and characterized in five bifidobacterial species or strains: Bifidobacterium bifidum JCM 1254 (22), Bifidobacterium adolescentis (23, 24), Bifidobacterium bifidum NCIMB 41171 (25), Bifidobacterium breve 203 (26), and Bifidobacterium longum subsp. longum (27, 28).
The utilization and transcriptional regulation of raffinose have been characterized in more detail in Escherichia coli than in bifidobacteria. In E. coli, raffinose is actively transported into the cell by use of a dedicated raffinose permease (encoded by rafA) and is then hydrolyzed into sucrose and galactose by an α-galactosidase (specified by rafB). The sucrose is then hydrolyzed into glucose and fructose by a sucrose hydrolase, which is encoded by rafD (29).
Various α-glucosidases, such as the enzymes encoded by agl1 and agl2, which were previously identified and characterized in B. breve UCC2003 (30), are produced by Bifidobacterium spp. (30, 31). Agl1 and Agl2, both members of GH family 13, which mainly represents enzymes with α-(1→6)-glucosidase activity (EC 3.2.1.10), have been shown to exhibit hydrolytic activity toward panose, isomaltose, and isomaltotriose, as well as toward four sucrose isomers, isomaltulose (Palatinose), trehalulose, turanose, and maltulose. They have also been shown to partially degrade trehalose and nigerose. The preferred substrates for the Agl1 and Agl2 enzymes have been shown to be panose, isomaltose, and trehalulose, carbohydrates that contain either an α-(1→6)-glucosidic bond (present in panose and isomaltose) or an α-(1→1)-glucosidic bond (present in trehalulose) (30).
Melezitose [α-d-Glcp-(1→3)-β-d-Fruf-(2→1)-α-d-Glcp] is an α-glucose-containing trisaccharide found in honeydew and manna, which are sugar-rich liquid and solid deposits, respectively, associated with the leaves and branches of various trees and shrubs (32). Although it was initially believed that melezitose was an oligosaccharide that was naturally present in various plants (33), it was later concluded that certain insects are responsible for melezitose production, since this sugar is absent from the tree sap used by such insects to form honeydew (32). To the best of our knowledge, no information on how (bifido)bacteria metabolize melezitose is available.
In the current study, we describe the identification of two adjacent gene clusters in the genome of B. breve UCC2003, mel and raf, which are involved in the metabolism of melezitose and raffinose family sugars, respectively.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bifidobacteria were routinely cultured in either de Man Rogosa and Sharpe medium (MRS medium; Difco, BD, Le Pont de Claix, France) supplemented with 0.05% cysteine-HCl or reinforced clostridial medium (RCM; Oxoid Ltd., Basingstoke, England). Carbohydrate utilization by bifidobacterial strains was examined in modified de Man Rogosa and Sharpe (mMRS) medium prepared according to first principles (34), though excluding a carbohydrate source. Prior to inoculation, the mMRS medium was supplemented with cysteine-HCl (0.05%, wt/vol) and a particular carbohydrate source (1%, wt/vol). The carbohydrates used were raffinose, stachyose, melezitose, melibiose, and glucose (all purchased from Sigma-Aldrich, Steinheim, Germany). Bifidobacterial cultures were incubated at 37°C under anaerobic conditions, which were maintained by using an Anaerocult oxygen-depleting system (Merck, Darmstadt, Germany) in an anaerobic chamber. Lactococcus lactis strains were cultivated in M17 broth containing 0.5% glucose (35) at 30°C. Escherichia coli strains were cultured in Luria-Bertani broth (LB) (36) at 37°C with agitation. Where appropriate, growth media contained chloramphenicol (Cm; 5 μg ml−1 for L. lactis, 10 μg ml−1 for E. coli, and 2.5 μg ml−1 for B. breve), erythromycin (Em; 100 μg ml−1 for E. coli), tetracycline (Tet; 10 μg ml−1 for E. coli or B. breve), or kanamycin (Km; 50 μg ml−1 for E. coli).
Table 1.
Strains and plasmids used in this study
| Strain or plasmida | Relevant characteristic(s) | Reference or sourceb |
|---|---|---|
| Strains | ||
| E. coli | ||
| EC101 | Cloning host; repA+ kmr | 44 |
| EC101-pNZ-M.Bbrll+Bbr111 | EC101 harboring a pNZ8048 derivative containing bbrllM and bbrlllM | |
| L. lactis | ||
| NZ9000 | From MG1363, a nisin-inducible overexpression host; pepN::nisRK | 90 |
| NZ9700 | Nisin-producing strain | 90 |
| B. breve | ||
| UCC2003 | Isolate from nursling stool | 14 |
| NCFB 2257 | Isolate from infant intestine | NCFB |
| NCTC 11815 | Isolate from infant intestine | NCTC |
| NCFB 2258 | Isolate from infant intestine | NCFB |
| NCIMB 8815 | Isolate from infant feces | NCIMB |
| JCM 7017 | Isolate from human feces | JCM |
| JCM 7019 | Isolate from infant feces | JCM |
| UCC2005 | Isolate from nursling stool | 30 |
| Yakult | Isolate from nursling stool | 87 |
| Nizo 658 | Isolate from nursling stool | Nizo |
| 461 | Isolate from infant/adult feces | PRL |
| 689 | Isolate from infant/adult feces | PRL |
| 12L | Mother's milk | PRL |
| UCC2003-MelE | pORI19-tet-1856 insertion mutant of UCC2003 | This study |
| UCC2003-MelD | pORI19-tet-1857 insertion mutant of UCC2003 | This study |
| UCC2003-RafB | pORI19-tet-1867 insertion mutant of UCC2003 | This study |
| UCC2003-RafA | pORI19-tet-1869 insertion mutant of UCC2003 | This study |
| UCC2003-MelA | pORI19-tet-1860 insertion mutant of UCC2003 | This study |
| Plasmids | ||
| pORI19 | Emr RepA− ori+ cloning vector | 44 |
| pORI19-tet-MelE | Internal 421-bp fragment of melE and tetW cloned in pORI19 | This study |
| pORI19-tet-MelD | Internal 456-bp fragment of melD and tetW cloned in pORI19 | This study |
| pORI19-tet-MelA | Internal 331-bp fragment of melA and tetW cloned in pORI19 | This study |
| pORI19-tet-RafA | Internal 474-bp fragment of rafA and tetW cloned in pORI19 | This study |
| pORI19-tet-RafB | Internal 394-bp fragment of rafB and tetW cloned in pORI19 | This study |
| pAM5 | pBC1-puC19-Tcr | 45 |
| pNZ8048 | Cmr; nisin-inducible translational fusion vector | 90 |
| pNZ8150 | Cmr; nisin-inducible translational fusion vector | 52 |
| pNZMelE-His | MelE with His tag cloned downstream of nisin-inducible promoter on pNZ8048 | This study |
| pNZMelD-His | MelD with His tag cloned downstream of nisin-inducible promoter on pNZ8048 | This study |
| pNZRafA-His | RafA with His tag cloned downstream of nisin-inducible promoter on pNZ8150 | This study |
NCFB, National Collection of Food Bacteria; NCTC, National Collection of Type Cultures; NCIMB, National Collection of Industrial and Marine Bacteria; JCM: Japanese Collection of Microorganisms; UCC, University College Cork culture collection; Nizo, Nizo food research.
PRL, Culture collection of probiogenomics, University of Parma.
In order to determine bacterial growth profiles and final optical densities, 5 ml of freshly prepared mMRS medium, including a particular carbohydrate (see above), was inoculated with 50 μl (1%) of a stationary-phase culture of a particular B. breve strain. Uninoculated mMRS medium was used as a negative control. Cultures were incubated anaerobically at 37°C for 16 h, and the optical density at 600 nm (OD600) was determined during this period at 30-min intervals using a PowerWave microplate spectrophotometer (BioTek Instruments, Inc., USA) in conjunction with Gen5 microplate software for Windows.
Nucleotide sequence analysis.
Sequence data were obtained from the Artemis-mediated (37) genome annotations of B. breve UCC2003 (38). Database searches were performed using nonredundant sequences accessible at the National Center for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov) using the basic local alignment search tool (BLAST) (39, 40). Sequences were verified and analyzed using the SeqMan and SeqBuilder programs of the DNAStar software package (version 10.1.2; DNAStar, Madison, WI, USA).
DNA manipulations.
Chromosomal DNA was isolated as described previously (41). Minipreparation of plasmid DNA from E. coli, B. breve, or L. lactis was carried out using the QIAprep Spin plasmid miniprep kit (Qiagen GmBH, Hilden, Germany). For B. breve or L. lactis, an initial lysis step was incorporated into the plasmid isolation procedure by resuspending cells in a lysis buffer supplemented with lysozyme (30 mg ml−1), followed by incubation at 37°C for 30 min. Procedures for DNA manipulations were performed essentially as described previously (36). Restriction enzymes and T4 DNA ligase were used according to the supplier's instructions (Roche Diagnostics, East Sussex, United Kingdom). The synthetic single-stranded oligonucleotide primers used in this study are listed in Fig. S1 in the supplemental material and were synthesized by Eurofins (Ebersberg, Germany). Standard PCRs were performed using Taq PCR master mix (Qiagen) in a Biometra T3000 thermocycler (Biometra, Göttingen, Germany). PCR products were visualized by ethidium bromide (EtBr) staining following agarose gel electrophoresis (1%). B. breve colony PCRs were performed as described previously (42). PCR fragments were purified using the Qiagen PCR purification kit (Qiagen). Plasmid DNA was electroporated into E. coli as described previously (36). B. breve UCC2003 (14) and L. lactis (43) were electrotransformed according to published protocols. The correct orientation and integrity of all plasmid constructs (see also below) were verified by DNA sequencing, performed at Eurofins (Ebersberg, Germany).
Construction of B. breve insertion mutant strains.
Internal fragments of Bbr_1856 (designated melE here) (421 bp, representing codons 242 through 383 of the 620 codons of this gene), Bbr_1857 (designated melD) (456 bp, representing codons 92 to 183 of the 556 codons of this gene), Bbr_1860 (designated melA) (331 bp, representing codons 230 to 341 of the 441 codons of this gene), Bbr_1867 (designated rafB) (394 bp, representing codons 95 to 226 of the 429 codons of this gene), and Bbr_1869 (designated rafA) (474 bp, representing codons 319 to 477 of the 771 codons of this gene) were amplified by PCR using B. breve UCC2003 chromosomal DNA as the template and the oligonucleotide primer combinations 1856fHd3 and 1856rxba1, 1857fHd3 and 1857rxba1, 1860fHd3 and 1860rxba1, 1867fHd3 and 1867rxba1, and 1869Hd3 and 1869Rxba1, respectively. Each of the PCR products generated was ligated to pORI19, an Ori+ RepA− integration plasmid (44), using HindIII and XbaI restriction sites that were incorporated into the primers for the melE, melD, melA, rafB, and rafA fragment-encompassing amplicons, and was introduced into E. coli EC101 by electroporation. Recombinant E. coli EC101 derivatives containing pORI19 constructs were selected on LB agar containing Em and supplemented with X-gal (5-bromo-4-chloro-3-indolyl-d-galactopyranoside) (40 g ml−1) and 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG).
The expected genetic structure of each of the resulting recombinant plasmids, pORI19-melE (pORI19 containing an internal 421-bp fragment of the melE gene), pORI19-melD (pORI19 containing an internal 456-bp fragment of the melD gene), pORI19-melA (pORI19 containing an internal 331-bp fragment of the melA gene), pORI19-rafB (pORI19 containing an internal 394-bp fragment of the rafB gene), and pORI19-rafA (pORI19 containing an internal 474-bp fragment of the rafA gene), was confirmed by restriction mapping and sequencing prior to subcloning of the Tet antibiotic resistance cassette, tetW, from pAM5 (45) as a SacI fragment into the unique SacI site in each of the pORI19 derivatives. The orientation of the tetracycline resistance gene in each of the resulting plasmids, pORI19-tet-melE, pORI19-tet-melD, pORI19-tet-melA, pORI19-tet-rafB, and pORI19-tet-rafA (naming is consistent with the names of their predecessor plasmids [see above], to which the “tet” designation was added), was determined by restriction analysis. The plasmids were subsequently introduced into E. coli EC101 pNZ-MBbrI-MBbrII in order to achieve methylation, and transformants were selected on the basis of Cm and Tet resistance. Methylation of the plasmid complement of the transformants obtained in EC101 pNZ-MBbrI-MBbrII was confirmed by their observed insensitivity to PstI restriction (42). Plasmid preparations of methylated pORI19-tet-melE, pORI19-tet-melD, pORI19-tet-melA, pORI19-tet-rafB, and pORI19-tet-rafA were then introduced into B. breve UCC2003 by electroporation, with subsequent selection for transformants on reinforced clostridial agar (RCA) plates supplemented with Tet. Insertion mutants resulting from site-specific homologous recombination were initially confirmed by colony PCR targeting the tetracycline resistance gene tetW. This was followed by a second PCR, employing a tetW-based primer, either forward or reverse depending on the orientation of tetW, in combination with a primer specific for each targeted gene, to confirm integration at the correct chromosomal position. In this case, a product would be obtained only if the correctly positioned gene disruption had occurred.
Analysis of global gene expression using B. breve DNA microarrays.
Global gene transcription patterns were determined by microarray analysis during the growth of B. breve UCC2003 on raffinose, stachyose, melibiose, melezitose, or sucrose, and these transcriptomes were compared to those obtained from cells that had been grown on ribose as the sole carbohydrate source. All biological replicates were hybridized using a dye swap strategy. DNA microarrays containing oligonucleotide primers representing each of the 1,864 annotated genes on the genome of B. breve UCC2003 were designed by, and obtained from, Agilent Technologies (Palo Alto, CA, USA). Methods for cell disruption, RNA isolation, RNA quality control, cDNA synthesis, and labeling were performed as described previously (46). Labeled cDNA was hybridized using the Agilent Gene Expression hybridization kit (part no. 5188-5242) as described in the Agilent two-color microarray-based gene expression analysis manual, version 4.0 (47). Following hybridization, microarrays were washed in accordance with Agilent standard procedures and were scanned using an Agilent DNA microarray scanner (model G2565A). The scans generated were converted to data files with Agilent's Feature Extraction software (version 9.5). DNA microarray data were processed as described previously (48–50). Differential expression tests were performed with the Cyber-T implementation of a variant of the t test (51). A gene was considered differentially expressed when the P value was <0.001 and the expression ratio was >3 or <0.33 relative to the control.
Expression and purification of RafA, MelE, and MelD.
DNA fragments containing the complete (predicted) α-galactosidase-encoding genes, rafA and melE, or the α-glucosidase-encoding gene, melD, were generated by PCR amplification from chromosomal DNA of B. breve UCC2003 using Taq DNA polymerase and the primer combination 1869EcorVF and 1869Xba1R, 1856EcorVF and 1856Xba1R, or 1857Nco1F and 1857Xba1R, respectively (see Fig. S1 in the supplemental material). An NcoI or EcoRV restriction site and an XbaI restriction site were incorporated at the 5′ ends of each forward and reverse primer, respectively (see Fig. S1). In addition, an in-frame His10-encoding sequence was incorporated into each of the forward primers to facilitate downstream protein purification using the Ni-nitrilotriacetic acid (NTA) affinity system (Qiagen). The three amplicons generated were first digested with either NcoI or EcoRV and XbaI and then ligated into the NcoI- or ScaI- and XbaI-digested nisin-inducible translational fusion plasmid pNZ8048 (which contains an NcoI site) or pNZ8150 (which contains a ScaI site), depending on the restriction sites of the amplified fragment (52). The ligation mixtures were introduced into L. lactis NZ9000 (Table 1) by electrotransformation, and transformants were then selected on the basis of chloramphenicol resistance. The plasmid contents of a number of Cmr transformants were screened by restriction analysis, and the integrity of positively identified clones (carrying pNZMelE-His, containing the melE gene; pNZMelD-His, containing the melD gene; or pNZRafA-His, containing the rafA gene) were verified by sequencing.
In order to (over)express and purify proteins, 400 ml of M17 broth supplemented with 0.5% glucose was inoculated with a 2% inoculum of a particular L. lactis strain, followed by incubation at 30°C until an OD600 of 0.5 was reached. At that point, protein expression was induced by the addition of purified nisin (5 ng ml−1), and incubation was continued at 30°C for 90 min. Cells were harvested by centrifugation, washed, and concentrated 40-fold. Protein was purified using a PrepEase kit specialized for His-tagged protein purification (USB, Germany). Elution fractions were analyzed by SDS-polyacrylamide gel electrophoresis, as described previously (53), on a 12.5% polyacrylamide gel. After electrophoresis, the gels were fixed and stained with Coomassie brilliant blue to identify fractions containing the purified protein. Rainbow prestained low-molecular-weight protein markers (New England BioLabs, Hertfordshire, United Kingdom) were used to estimate the molecular weights of the purified proteins.
Biochemical characterization of MelD, MelE, and RafA.
The putative α-glucosidase activity of MelD and the presumed α-galactosidase activities of RafA and MelE were determined essentially as described previously (54). A 50-μl volume of each purified protein (concentration, 0.5 mg ml−1) was incubated with 20 mM morpholinepropanesulfonic acid (MOPS) (pH 7.0). For RafA and MelE, 0.1 mg ml−1 (wt/vol) of α-(1→4)-galactobiose, α-(1→3)-galactobiose, raffinose, stachyose, melibiose, melezitose, turanose, or sucrose was added to 20 mM MOPS buffer as the enzyme substrate in a final volume of 1 ml at 37°C. For analysis of the catalytic activities of MelD, 0.1 mg ml−1 (wt/vol) of α-(1→4)-galactobiose, α-(1→3)-galactobiose, melezitose, maltulose, isomaltulose, turanose, leucrose, sucrose, raffinose, stachyose, or melibiose was added to 20 mM MOPS buffer at pH 7.0 as the enzyme substrate in a final volume of 1 ml, and the mixture was incubated at 37°C. Following incubation, 200-μl samples were taken at 24-h intervals. Samples were filtered by membrane filtration, using Spin-X centrifuge tube filters (pore size, 0.45 μm; Costar; Corning Inc., NY), and were stored at −20°C prior to high-performance anion-exchange chromatography with pulsed amperometric detection (HPAEC-PAD) analysis (see below).
Kinetic constants for MelD using sucrose or turanose, and for RafA using melibiose, raffinose, or stachyose, were determined by measuring the hydrolysis rates at various substrate concentrations, ranging from 2.5 to 100 mM. Reactions were initiated by the addition of 50 μl of purified protein (concentration, 0.5 mg ml−1) in 20 mM MOPS buffer at the optimum pH and temperature determined for each protein, and the reactions were stopped at different time points (up to 6 min) by heat treatment at 100°C for 15 min. All experiments were performed in duplicate, and the amount of glucose released from each disaccharide substrate, namely, melibiose, sucrose, and turanose, was measured by using the glucose hexokinase assay kit (Sigma) according to the manufacturer's instructions. For raffinose and stachyose hydrolysis, a sucrose assay kit was utilized, in which case enzyme activity is based on the amount of sucrose released as measured according to the manufacturer's instructions (Sigma). Either sucrose, turanose, or melibiose (50 mM) was used as the substrate for the determination of the pH and temperature optima (a pH range of 2.5 to 9.5 and a temperature range of 4°C to 60°C were tested). Reactions were initiated by the addition of 50 μl of purified protein (concentration, 0.5 mg ml−1) in 20 mM MOPS buffer at pH 7.0.
HPAEC-PAD analysis.
For HPAEC-PAD analysis, a Dionex (Sunnyvale, CA) ICS-3000 system was used. Carbohydrate fractions (25-μl aliquots) were separated on a CarboPac PA1 analytical-exchange column (dimensions, 250 mm by 4 mm) with a CarboPac PA1 guard column (dimensions, 50 mm by 4 mm) and a pulsed electrochemical detector (ED40) in the PAD mode (all from Dionex). Elution was performed at a constant flow-rate of 1.0 ml min−1 at 30°C using the following eluents for the analysis: eluent A, 200 mM NaOH; eluent B, 100 mM NaOH plus 550 mM Na acetate; eluent C, Milli-Q water. The following linear gradient of sodium acetate was used with 100 mM NaOH: from 0 to 50 min, 0 mM; from 50 to 51 min, 16 mM; from 51 to 56 min, 100 mM; from 56 to 61 min, 0 mM. Chromatographic profiles of standard carbohydrates were used for comparison of the results of their breakdown by the MelD, MelE, and RafA proteins. Chromeleon software (version 6.70; Dionex Corporation) was used for the integration and evaluation of the chromatograms obtained. A 10-mg ml−1 stock solution of each of the carbohydrates to be used as reference standards was prepared by dissolving the particular sugar in deionized Milli-Q water. The stock solution was then sterilized by membrane filtration using Minisart filters (pore size, 0.45 μm; Sartorius AG, Göttingen, Germany) and was stored at 4°C.
Microarray data accession number.
The microarray data obtained in this study have been deposited in NCBI's Gene Expression Omnibus database and are accessible through GEO Series accession number GSE47448.
RESULTS AND DISCUSSION
Carbohydrate-dependent analysis of the growth of various B. breve strains.
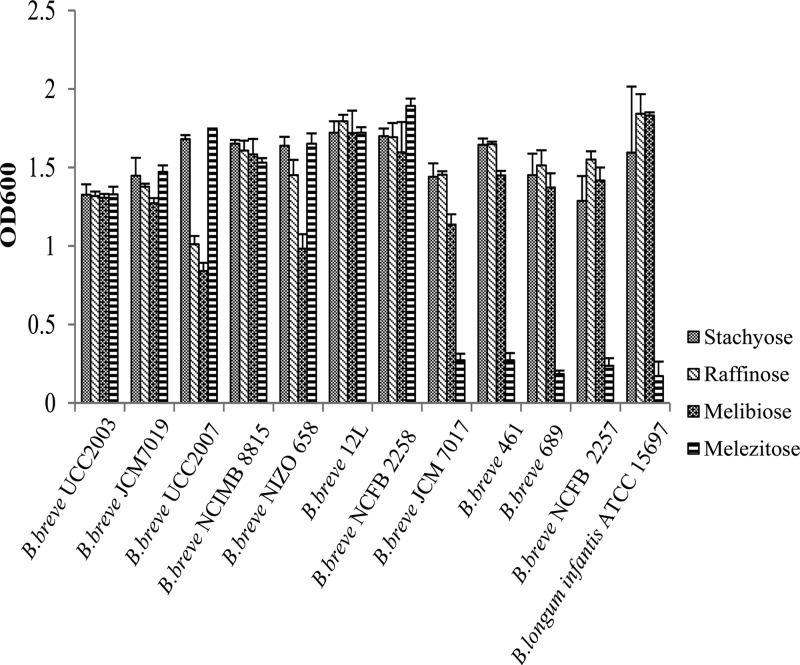
Knowledge of the carbohydrate metabolism of individual bifidobacterial species or strains is important in order to evaluate the prebiotic potential of particular carbohydrates from a dietary perspective. In order to determine whether different strains of B. breve possess the ability to utilize the α-glycosidic bond-containing sugars raffinose, stachyose, melibiose, and melezitose as sole carbohydrate sources (all of approximately 99% purity), growth profiles were determined for 11 B. breve strains. All B. breve strains tested exhibited good growth (final OD600, >1.0) in mMRS medium supplemented with raffinose, stachyose, or melibiose (Fig. 1). Seven strains exhibited good growth in mMRS medium supplemented with melezitose, while five strains, namely, B. breve JCM7017, B. breve 461, B. breve 689, B. breve NCFB 2257, Bifidobacterium longum subsp. infantis ATCC 15697, and B. breve NCFB 2258, were not capable of appreciable growth (final OD600, <0.4) on melezitose as a sole carbohydrate source (Fig. 1).
Fig 1.
Final OD600 values following 16 h of growth of various wild-type B. breve strains on 1% raffinose, 1% stachyose, 1% melibiose, or 1% melezitose. The results are mean values obtained from three separate experiments.
Transcriptome analysis of B. breve UCC2003 grown on raffinose, stachyose, or melibiose.
In order to identify genes involved in the metabolism of raffinose-related carbohydrates, we used DNA microarray analysis (see Materials and Methods) to investigate differences in global gene expression upon the growth of B. breve UCC2003 on raffinose, stachyose, or melibiose, compared with growth on ribose (the metabolic pathway for ribose in B. breve UCC2003 has been characterized previously, and growth on this sugar has been shown to provide a suitable transcriptomic reference) (55). The resulting transcriptomic data showed that transcription of the contiguous genes Bbr_1869, Bbr_1867, Bbr_1866, and Bbr_1865 (here designated rafA, rafB, rafC, and rafD, respectively) was significantly upregulated (fold change, >4.0; P, <0.001) in B. breve UCC2003 cultures grown on raffinose, stachyose, or melibiose, relative to that for cultures grown on the control carbohydrate ribose, thus implicating this gene cluster in raffinose-related sugar metabolism in B. breve UCC2003 (Table 2). Furthermore, transcription of four additional genes was upregulated (relative to the ribose transcriptome) when B. breve UCC2003 was grown on raffinose (and on stachyose, although this transcriptional increase was just below the cutoff value of 4-fold for two of these genes) but not when the strain was grown on melibiose: Bbr_0026, Bbr_0027, Bbr_0030, and Bbr_100, which are predicted to encode two ABC-type permeases, a hypothetical protein, and a putative sucrose phosphorylase, respectively. Since these four genes also exhibit increased transcription when B. breve UCC2003 is grown on sucrose (relative to growth on ribose) (Table 2), we hypothesize that they are involved in the metabolism of this disaccharide, which is released upon removal of the α-galactose moieties from the nonreducing ends of raffinose and stachyose, in contrast to melibiose hydrolysis, which results in galactose and glucose release.
Table 2.
Carbohydrate-dependent transcriptional upregulation of specific genesa
| Gene ID | Gene name | Function | Fold upregulationb during growth on: |
||||
|---|---|---|---|---|---|---|---|
| Stachyose | Raffinose | Melibiose | Melezitose | Sucrose | |||
| Bbr_0026 | Permease protein of ABC transporter system for sugars | — | 18.55 | — | 4.66 | 23.73 | |
| Bbr_0027 | Permease protein of ABC transporter system for sugars | — | 13.98 | — | 4.55 | 44.95 | |
| Bbr_0030 | Conserved hypothetical protein | 28.53 | 145.75 | — | 86.54 | 216.5 | |
| Bbr_0100 | SPase | Sucrose phosphorylase | 74.26 | 74.26 | — | 128.18 | 41.71 |
| Bbr_1855 | agl1 | Alpha glucosidase | — | 20.43 | 10.74 | 31.95 | — |
| Bbr_1856 | melE | Raffinose synthase or seed inhibition protein | — | — | — | 72.76 | — |
| Bbr_1857 | melD | Alpha glucosidase | — | — | — | 94.33 | — |
| Bbr_1858 | melC | Permease protein of ABC transporter system for sugars | — | — | — | 850.82 | — |
| Bbr_1859 | melB | Permease protein of ABC transporter system for sugars | — | — | — | 679.07 | — |
| Bbr_1860 | melA | Solute binding protein of ABC transporter system for sugars | — | — | — | 724.89 | — |
| Bbr_1865 | rafD | Raffinose transport system permease protein | 21.66 | 98.36 | 105.07 | — | — |
| Bbr_1866 | rafC | Raffinose transport system permease protein | 49.86 | 175.51 | 153.8 | — | — |
| Bbr_1867 | rafB | Raffinose-binding protein | 24.39 | 86.16 | 92.5 | — | — |
| Bbr_1868 | rafR | Transcriptional repressor, ROK family | — | — | — | — | — |
| Bbr_1869 | rafA | α-Galactosidase | 9.23 | 10.53 | 8.07 | — | — |
Based on comparative transcriptome analysis using B. breve UCC2003 grown on 1% raffinose, stachyose, or melibiose compared to growth on ribose. Microarray data were obtained using B. breve UCC2003 grown on 1% raffinose, stachyose, or melibiose and were compared with array data obtained when B. breve UCC2003 was grown on ribose as a control.
The cutoff point is 4-fold, with a P value of <0.001. —, value below the cutoff.
Our data on the raf cluster of B. breve UCC2003 suggest that many bifidobacteria metabolize raffinose and the related sugars stachyose and melibiose, by means of a metabolic route somewhat different from that known for E. coli (56). Raffinose-type sugar uptake in B. breve UCC2003 and other bifidobacteria apparently occurs via an ABC-type transporter system, which is a common way for bifidobacteria to internalize carbohydrates (14, 30, 55, 57–60). Removal of the α-galactose moiety from raffinose and stachyose results in the release of sucrose, which in E. coli is further metabolized via a sucrose hydrolase (56), while in B. breve UCC2003, sucrose utilization appears to occur by a sucrose phosphorylase, since a gene that is predicted to encode such an activity exhibits increased transcription when UCC2003 is grown on sucrose or sucrose-containing sugars (Table 2). Sucrose metabolism by sucrose phosphorylase has been characterized in other bifidobacteria, for example, Bifidobacterium adolescentis DSM20083 (61), B. longum (62, 63), and Bifidobacterium animalis subsp. lactis (64). Based on the ability of all B. breve strains examined to utilize raffinose-type oligosaccharides (Fig. 1), corroborated by the comparative genome analysis presented below (see Fig. 2), and on other reports in the literature regarding raffinose utilization by bifidobacteria, raffinose-type sugar utilization appears to be an ubiquitous property of bifidobacteria, supporting previous publications on the prebiotic potential of (some of) these sugars (65–68).
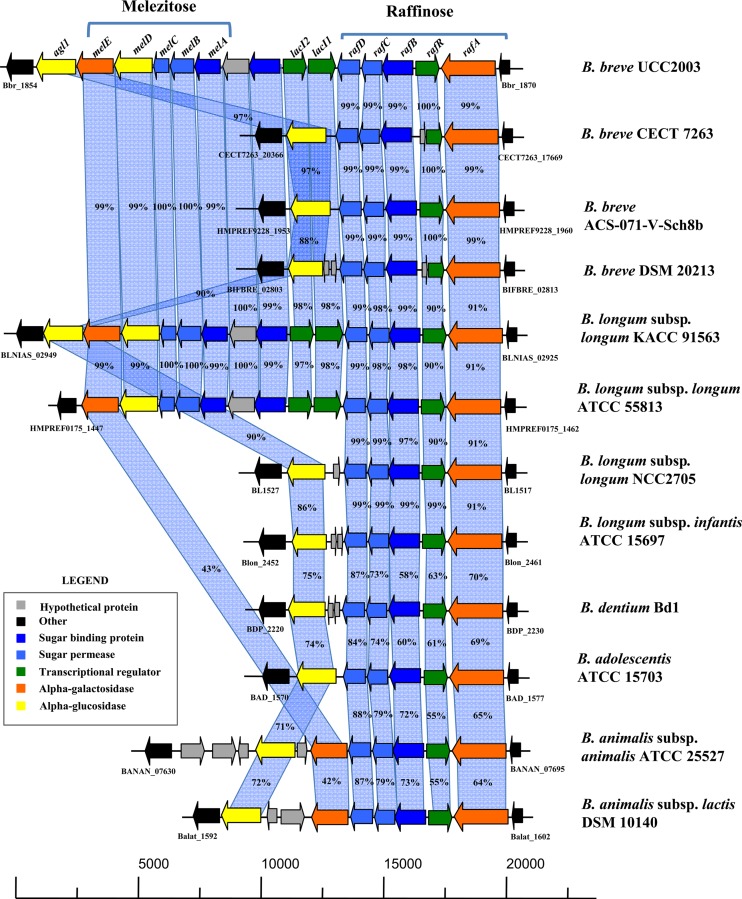
Fig 2.
Comparison of the melezitose, raffinose, and stachyose gene clusters of B. breve UCC2003 with corresponding putative melezitose, raffinose, and stachyose utilization loci of other bifidobacteria. Each solid arrow represents an ORF. The length of each arrow is proportional to the length of the predicted ORF, and the gene locus name, which is indicative of its putative function, is given at the top. Orthologs are shown in the same color. The amino acid identity of each predicted protein to its equivalent protein encoded by B. breve UCC2003, expressed as a percentage, is given above each arrow.
Although transcription of the raf gene cluster of B. breve is induced in the presence of either raffinose, melibiose, or stachyose, it has been shown that the raffinose system in Streptococcus pneumoniae is upregulated in the presence of raffinose but not melibiose, suggesting differential regulatory mechanisms (69).
Transcriptome analysis of B. breve UCC2003 grown on melezitose.
In order to identify genes involved in the utilization of melezitose by B. breve UCC2003, transcriptome analysis was performed for B. breve UCC2003 grown on this trisaccharide, and the results were compared with those obtained when the same organism was grown on ribose. Analysis of transcriptome data obtained from two independent biological replicates revealed that transcription of the contiguous genes Bbr1860 through Bbr_1856, here designated melA, melB, melC, melD, and melE, respectively (see Fig. 2), which are located in close proximity to the raf gene cluster, was significantly upregulated (fold change, >4.0; P, <0.001) in B. breve UCC2003 cultures grown on melezitose relative to that for cultures grown on ribose (Table 2).
These results implicate the melABCDE gene cluster in melezitose metabolism in B. breve UCC2003. Interestingly, on the basis of previously published comparative genome hybridization data (38), we noticed that B. breve strains that did not exhibit growth on melezitose (Fig. 1) lack the melABCDE gene cluster. Furthermore, in findings reminiscent of the results obtained for transcriptome analysis when strain UCC2003 was grown on stachyose and raffinose, the transcription of four additional genes, Bbr_0026, Bbr_0027, Bbr_0030, and Bbr_0100, was upregulated when B. breve UCC2003 was grown on melezitose (Table 2). Since hydrolysis of melezitose produces sucrose if the α-(1→3)-linked glucose is released, the transcriptional induction of these genes is believed to be correlated to sucrose metabolism.
Genetic organization of the raf and mel gene clusters and comparison to other available bifidobacterial genomes.
Our presumption, based on the microarray results, was that the genes of the raf gene cluster, schematically depicted in Fig. 2, are involved in the metabolism of sugars that contain one or more α-(1→6)-linked galactose moieties (e.g., raffinose, stachyose, and melibiose). The rafA gene, which specifies a putative α-galactosidase of GH family 27, is a clear homolog of the B. breve CECT 7263 α-galactosidase-encoding gene, with which it shares 99% sequence identity (70). B. breve CECT 7263 also contains genes with high sequence similarity (99 to 100% identity) to other genes in the B. breve UCC2003 raf gene cluster (Fig. 2). The rafA gene is presumed to be responsible for the breakdown of raffinose, stachyose, and melibiose via hydrolysis of the α-(1→6)-glycosidic bond that is common to these carbohydrates, thereby releasing galactose. The presumptive B. breve UCC2003 α-galactosidase-encoding gene, rafA, is located immediately adjacent to a gene, Bbr_1868 (here designated rafR), that is predicted to encode a ROK-type transcriptional regulator. Members of the ROK protein family include both transcriptional repressors and sugar kinases (71). ROK kinases possess a conserved N-terminal ATP-binding motif, while ROK repressors contain an N-terminal region that includes a canonical helix-turn-helix motif associated with DNA binding (72). BLAST analysis and Pfam searches revealed the presence of such a helix-turn-helix motif, leading to the prediction that the rafR gene product functions as a transcriptional regulator, which may, based on its genomic location, be involved in regulating raffinose (sugar family)-dependent transcription of the other genes of the raf gene cluster. Close homologs of rafA and rafR, and of the neighboring genes rafB, rafC, and rafD, which are predicted to specify a solute binding protein and two permeases, respectively, of a putative ABC-type sugar uptake system, are present in all other publicly available bifidobacterial genomes except for those of Bifidobacterium asteroides PRL2011 and Bifidobacterium bifidum PRL2010 (68, 73) (Fig. 2; also data not shown). Interestingly, the raf gene cluster does not contain a gene with a predicted ATP-binding protein, which is required for providing energy to the ABC-type transport system (74). It is presumed that this activity is encoded by an unconnected gene, whose product can function as an ATP-binding component for multiple ABC transporters, a scenario similar to that suggested for other bifidobacterial ABC-type carbohydrate transport systems (58, 59).
The melABCDE gene cluster, whose transcription is specifically induced by growth on melezitose, is predicted to specify a solute binding protein (melA) and two permease proteins (melB and melC) (again, lacking a gene predicted to encode an ATP-binding protein, representing a genetic configuration similar to that observed for the raf gene cluster [see above]), an α-glucosidase (melD) belonging to GH family 13, and an α-galactosidase/raffinose synthase (melE) of GH family 36 (Table 2 and Fig. 2). The DNA region between the raf and mel gene clusters contains four additional open reading frames (ORFs): Bbr_1863 and Bbr_1864, which encode putative LacI-type transcriptional regulators, here designated lacI2 and lacI1, respectively, and Bbr_1861 and Bbr_1862, which specify a hypothetical protein and a predicted solute binding protein, respectively. The mel and raf genetic loci, including the genes of the intervening region, share high sequence similarity (99 to 100%) at the amino acid level and a conserved gene organization with corresponding regions in the genomes of B. longum subsp. longum KACC 91563 and B. longum subsp. longum ATCC 55813. Interestingly, homologs of the mel gene cluster (including Bbr_1861 to Bbr_1864) are lacking in many bifidobacterial genomes, although such genomes do contain clear homologs of the raf gene cluster and the agl1 gene, which in the UCC2003 genome flank the mel cluster on either end (Fig. 2), suggesting that those bifidobacterial strains lack the (genetic) ability to metabolize melezitose.
Prevalence and genetic organization of raf and mel gene clusters in other bacteria.
The genetic organization of the bifidobacterial raf locus and the raffinose metabolic pathway appears to be quite different from those of various other bacterial species. In E. coli (29), Klebsiella pneumoniae (75), Enterobacter cloacae (76), and Citrobacter freundii (77), among others, raffinose uptake is specified by a single permease-encoding gene, which is cotranscribed with two additional genes that specify an α-galactosidase and a sucrose hydrolase. As mentioned above, uptake of raffinose and related sugars in bifidobacteria appears to be performed by a dedicated ABC-type transport system, whose genes are in close proximity to, though not cotranscribed with, the α-galactosidase-specifying gene, due to their opposing genetic orientation (Fig. 2). Furthermore, in bifidobacteria, the metabolic product of this α-galactosidase activity, i.e., sucrose, is apparently metabolized not by a sucrose hydrolase but by a sucrose phosphorylase, encoded by a gene that is not genetically linked to the raf locus.
Raffinose metabolism has also been investigated in Lactobacillus plantarum ATCC 8014, in which the gene encoding the putative raffinose permease is immediately followed by the α-galactosidase-specifying gene (designated melA) and is preceded by galM, a putative microbial galactose-1-epimerase (78). Interestingly, two genes located downstream of melA, though on the opposite strand, encode the two subunits of a heterodimeric β-galactosidase, which shows that the genetic organization and content of the raffinose utilization cluster of L. plantarum differ substantially from those in bifidobacteria. Recently, transcriptome analyses have shown that B. animalis subsp. lactis Bl-04 possesses a raffinose utilization cluster including three predicted α-glycosidases, exhibiting a transcription profile that is consistent with our observations for B. breve (79).
Very little information is available regarding melezitose metabolism in other microorganisms. Melezitose metabolism has been investigated in the yeast Saccharomyces cerevisiae (80), where a melezitose-metabolizing system was identified, including an α-glucosidase or melezitase that was shown to be capable of hydrolyzing isomaltulose, turanose, maltose, sucrose, and melezitose.
Construction and phenotypes of mutants carrying individual disruptions in the rafA, rafB, melA, melD, and melE genes.
In order to determine whether disruption of a particular gene of the raf gene cluster of B. breve UCC2003 affects the resulting strain's ability to metabolize raffinose, stachyose, and/or melibiose, mutants were made with insertions in rafB, which is predicted to encode a solute binding protein implicated in the internalization of the α-galactose-containing sugars mentioned above, and in rafA, the presumed α-(1→6)-galactosidase-encoding gene. The resulting strains were designated UCC2003-RafB and UCC2003-RafA, respectively (see Materials and Methods). B. breve UCC2003-RafA and B. breve UCC2003-RafB were analyzed for their abilities to grow on mMRS medium supplemented with either raffinose, stachyose, or melibiose as the sole carbon source. As expected, and in contrast to the findings for the wild type, the B. breve insertion mutants were unable to grow on raffinose, stachyose, or melibiose as the sole carbon source. All strains retained their ability to utilize glucose as a sole carbon source (Fig. 3A). These results show not only that the rafA gene is required for growth on raffinose but also that a mutation in rafB causes this growth-deficient phenotype, implying that the rafBCD genes encode an ABC-type transport system responsible for the internalization of α-galactose-containing oligosaccharides. Our results corroborate the findings of a recent study in which a deletion in a homolog of the rafA gene of Bifidobacterium longum 105-A (designated agl) was shown to cause a loss of α-galactosidase activity and a growth deficiency on raffinose or melibiose (28).
Fig 3.
(A) Final OD600 after 16 h of growth of UCC2003 and insertion mutants UCC2003-RafB (raffinose binding protein) and UCC2003-RafA (α-galactosidase) on 1% stachyose, raffinose, melibiose, or glucose. (B) Final OD600 following 16 h of growth of B. breve UCC2003, B. breve UCC2003-MelD [α-(1→3)-glucosidase], and B. breve UCC2003-MelA (solute binding protein) on 1% melezitose or glucose. In both panels, the results are mean values obtained from three separate experiments.
In order to determine whether disruption of a particular gene from the mel cluster results in loss of the ability of B. breve UCC2003 to metabolize melezitose, mutants were made with insertions in melE, which is predicted to encode an α-galactosidase, resulting in strain B. breve UCC2003-MelE; in melD, which is predicted to encode an α-glucosidase, resulting in strain B. breve UCC2003-MelD; and in melA, which encodes a predicted solute binding protein, resulting in strain B. breve UCC2003-MelA (see Materials and Methods). These mutants—B. breve UCC2003-MelE, UCC2003-MelD, and UCC2003-MelA—were then analyzed for their abilities to grow on mMRS medium supplemented with melezitose as the sole carbon source. As expected, and in contrast to the finding for the wild type, the growth of B. breve strain UCC2003-MelA on melezitose was severely reduced from that of the wild-type strain, indicating that melA is indeed required for melezitose catabolism. Mutant strain B. breve UCC2003-MelD showed impaired growth on melezitose compared to that of the wild-type strain UCC2003, though to a lesser extent than strain UCC2003-MelA, and this growth defect is particularly obvious when the corresponding growth profiles of these three strains on melezitose are compared (Fig. 3B). The less severe growth deficiency of the melD insertion mutant than of the melA insertion mutant may be due to the presence of other α-glucosidases produced by B. breve UCC2003 that partially compensate for the melD mutation. Interestingly, the ability of mutant strain UCC2003-MelE to grow on melezitose was not affected, indicating that MelE is not required for growth on this sugar (data not shown). Wild-type UCC2003 and the three mel mutants exhibited no differences in their abilities to utilize glucose as a sole carbon source.
Purification, characterization, and substrate specificity of recombinantly produced MelD, MelE, and RafA.
In order to analyze the glycosyl hydrolase functions of MelD, MelE, and RafA, we purified and biochemically characterized these three predicted sugar-degrading enzymes. All of the overproduced proteins were purified well and in soluble form, and MelE, MelD, and RafA exhibited molecular masses of approximately 68.2 kDa, 62.4 kDa, and 84.1 kDa (inclusive of the His6 tag), respectively, when analyzed by SDS-PAGE (results not shown).
Purified recombinant MelD protein was shown to fully hydrolyze melezitose into glucose and fructose (Fig. 4A, graph I), thereby demonstrating that this protein has both α-(1→2)- and α-(1→3)-glycosyl hydrolase activities. This was further confirmed by our findings that both sucrose [α-d-Glcp-(1→2)-β-d-Fruf] and turanose [α-d-Glcp-(1→3)-β-d-Fruf] are hydrolyzed by MelD to glucose and fructose (Fig. 4A, graphs II and III, respectively). In contrast, MelD was unable, at least under the conditions tested, to hydrolyze leucrose [α-d-Glcp-(1→5)-β-d-Fruf], isomaltulose [α-d-Glcp-(1→6)-β-d-Fruf], or maltulose [α-d-Glcp-(1→4)-β-d-Fruf] (results not shown). These results, therefore, show that MelD is an α-glucosidase with substrate specificities clearly different from those of the two previously characterized α-glucosidases encoded by B. breve UCC2003, Agl1 and Agl2, which cannot hydrolyze leucrose, melezitose, or sucrose but possess hydrolytic activity against turanose, maltulose, and isomaltulose (30). The preferred substrates for Agl1 and Agl2 are thus carbohydrates that contain either an α-(1→6)- or an α-(1→1)-glucosidic bond (30), while MelD is an α-glucosidase with hydrolytic activity against α-(1→2)- and α-(1→3)-glucosidic bonds, which distinguishes it from previously characterized bifidobacterial α-glucosidases (31, 81, 82).
Fig 4.
(A) HPAEC-PAD analysis indicating the breakdown of melezitose and turanose (initial concentration, 0.1 mg ml−1) by the purified recombinant protein MelD in 20 mM MOPS buffer (pH 7.0) over 24 h. The chromatogram shows results for melezitose (graph I), sucrose (graph II), and turanose (graph III) incubated with MelD. The liberation of glucose and fructose is visible as chromatographic peaks eluted at 6.25 and 5.5 min, respectively. Breakdown products are indicated by solid arrows. Chromatographic positions of carbohydrate standards are indicated by dashed arrows above the chromatogram. (B) HPAEC-PAD analysis indicating the breakdown of stachyose, raffinose, and melibiose by the purified recombinant protein RafA in 20 mM MOPS buffer (pH 7.0) over 24 h. (Graphs I and II) Stachyose (graph I) and raffinose (graph II) incubated with RafA. The liberation of galactose and sucrose is visible as chromatographic peaks eluted at 5.75 and 10.5 min, respectively. (Graph III) Melibiose incubated with RafA. The hydrolysis of this substrate to glucose and galactose is visible as a single chromatographic peak eluted at 5.75 min. Arrows are as explained for panel A.
The putative α-galactosidase MelE failed to exhibit hydrolytic activity toward raffinose, stachyose, or melibiose (results not shown), at least under the conditions tested. However, when two synthetic disaccharides, namely, α-(1→4)- and α-(1→3)-galactobiose, were assayed, MelE was shown to be capable of hydrolyzing both of them (see Fig. S2 in the supplemental material). The finding that the melE gene encodes an α-galactosidase and is present and cotranscribed in a melezitose-induced gene cluster suggests that this locus is also involved in the metabolism of a melezitose-related carbohydrate(s) that contains one or more α-galactose moieties linked through α-(1→4)- and/or α-(1→3)-glycosidic bonds. This sugar, like melezitose, may also be present in honeydew, which is secreted by aphids while they feed on the sugar-rich phloem of their host plants to acquire the amino acids they need for growth and reproduction (83). Such honeydew oligosaccharides, which contain large amounts of melezitose and erlose, have been shown to possess prebiotic potential, since they increase the numbers of bifidobacteria and lactobacilli in an in vitro fermentation system (84).
Purified RafA was shown to hydrolyze stachyose, raffinose, and melibiose (Fig. 4B, graphs I, II, and III) to produce sucrose and galactose, and to cleave melibiose to its monosaccharide constituents glucose and galactose, confirming that RafA functions as an α-galactosidase. RafA was also tested for its abilities to hydrolyze melezitose, sucrose, and a range of sucrose isomers but failed to exhibit hydrolytic activity against any of these carbohydrates (results not shown). However, it did show hydrolytic activity against synthetic α-(1→4)- and α-(1→3)-galactobiose, demonstrating that RafA has rather broad substrate specificity (see Fig. S2 in the supplemental material). Previously, an α-galactosidase from B. bifidum JCM 1254 was shown to be capable of hydrolyzing α-(1→3)-linked galactose in a branched blood group B antigen trisaccharide; however, this α-galactosidase did not possess the ability to hydrolyze α-(1→4)-galactosidic linkages (22). Analysis of the bifidobacterial genomes sequenced to date shows that most of the bifidobacterial strains have at least one gene encoding α-galactosidase (15). Various α-galactosidases, capable of catalyzing the hydrolysis of various α-galacto-oligosaccharides, have been studied in five bifidobacterial species to date (22–27). For example, it was shown that the B. adolescentis α-galactosidase hydrolyzes α-(1→6)-galactosidic bonds from raffinose and stachyose, as well as the α-(1→4) and α-(1→3) bonds of two galactobiose substrates (23, 24), which is consistent with our findings for B. breve UCC2003. Interestingly, the α-galactosidase of B. breve 203 was shown to have the ability to synthesize a trisaccharide [Gal-α-(1→4)-Gal-α-(1→6)-Glc] using melibiose as a substrate (26). Furthermore, the α-galactosidase encoded by B. bifidum JCM 1254 was observed to be capable of hydrolyzing α-(1→3)-linked galactose in a branched blood group B antigen trisaccharide, although this enzyme cannot hydrolyze α-(1→4)-galactosidic linkages (22). The MelD-, MelE-, and RafA-mediated carbohydrate hydrolysis results obtained and the presumed degradation pathways are summarized in Fig. S3 in the supplemental material.
Determination of kinetic parameters of MelD and RafA.
In order to determine the kinetic parameters of the MelD and RafA enzymes, we characterized these two glycosyl hydrolases using the substrates against which they had shown hydrolytic activity [except for α-(1→4)- and α-(1→3)-galactobiose, for which we did not have sufficient amounts to perform such studies]. When sucrose and turanose were used as substrates, the optimum temperature and pH values for MelD activity were determined to be 30°C and pH 7.5, respectively, while for RafA in combination with any of the substrates raffinose, stachyose, and melibiose, the optima were 42°C and pH 6.0, respectively.
Kinetic studies were performed to determine Vmax and Km values, as well as the rate constants (kcat) and catalytic efficiencies (kcat/Km), for MelD using sucrose or turanose as a substrate and for RafA using raffinose, stachyose, or melibiose as a substrate (Table 3). MelD, as shown above, exhibits hydrolytic activity against both α-(1→3)- and α-(1→2)-glucosidic linkages. In order to investigate if MelD exhibited any hydrolytic preference for either of these linkages, we looked at the ability of MelD to hydrolyze sucrose and turanose. The data obtained indicate that the preferred bond cleaved by MelD is the α-(1→2) linkage present in sucrose, since MelD hydrolyzes this bond with an efficacy higher than that for the α-(1→3) linkage present in turanose (Table 3). In order to investigate the preferred substrate of RafA, we determined the kinetic parameters of this enzyme related to its hydrolytic activities toward melibiose, raffinose, and stachyose. The preferred substrate was melibiose, followed by raffinose. However, it should be noted that stachyose contains two α-(1→6) linkages, of which one is present between two galactose moieties, while the other is present between galactose and glucose. The hydrolysis of stachyose by RafA is likely to lead to the generation of galactose and raffinose, the latter again representing a substrate for RafA, as demonstrated by us and as reported previously (17). In our kinetic experiments, we assessed stachyose hydrolysis by measuring the release of sucrose, which would be generated directly by the hydrolysis of stachyose to sucrose and a galacto-disaccharide and indirectly by the initial hydrolysis of stachyose to galactose and raffinose, followed by the hydrolysis of the raffinose to sucrose and galactose. The presence of multiple substrates means that the use of standard kinetic techniques to measure stachyose hydrolysis is not absolutely correct. However, the kinetic values calculated provide a reasonably accurate representation of the substrate preference of the enzyme.
Table 3.
Kinetic study of MelD and RafAa
| Substrate | Protein | Vmax (μmol min−1 mg−1) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|---|
| Melibiose | RafA | 384.305 ± 0.42 | 8.15 ± 1.7 | 542.79 ± 0.63 | 66.6 ± 0.07 |
| Raffinose | RafA | 187.5 ± 2.1 | 14.55 ± 1.9 | 264.815 ± 2.9 | 18.2 ± 2.6 |
| Stachyose | RafA | 11.09±.55 | 3.045 ± 0.51 | 16.665 ± 0.784 | 5.47 ± 1.14 |
| Sucrose | MelD | 7.75 ± 0.07 | 19.3 ± 0.28 | 8.0725 ± 0.07 | 0.4175 ± 0.007 |
| Turanose | MelD | 12.66 ± 1.59 | 91.93 ± 1.07 | 13.16 ± 1.61 | 0.14 ± 0.01 |
kcat, rate constant; kcat/Km, catalytic efficiency. All values are means from two experiments ± standard errors.
A previously published kinetic study of the E. coli K-12 α-galactosidase (85) revealed that this enzyme at an optimal pH of 7.2 exhibits Km values of 3.2 mM for melibiose and 60 mM for raffinose. Since we observed Km values of 8.15 mM for melibiose and 14.55 mM for raffinose at an optimal pH of 6.0 for the B. breve RafA, it is clear that individual α-galactosidases, despite having the same substrate specificities, may still exhibit different kinetic properties.
Concluding remarks.
Bifidobacteria are believed to play an important role in the fermentation of nondigestible carbohydrates in the lower gastrointestinal tract. Consistent with this notion is the prediction that a sizable proportion of the average bifidobacterial genome is dedicated to carbohydrate metabolism (2, 86). More than 50 bifidobacterial carbohydrases have been studied to date (for reviews, see references 42 and 7), and various carbohydrate utilization pathways, such as those dedicated to the metabolism of fructose, galactan, starch, ribose, isomaltulose, cellodextrin, and fructo-oligosaccharides, have been characterized in B. breve UCC2003 (8, 14, 38, 41, 52, 59, 87).
The data assembled in this study provide significant information on the abilities of various B. breve strains to grow on a number of plant-derived α-glucose- and α-galactose-containing oligosaccharides, as well as identifying the genes involved in the metabolism of such sugars. Two adjacent genetic loci dedicated to the utilization of raffinose-containing carbohydrates and melezitose in B. breve UCC2003 were identified, encoding a novel α-glucosidase (specified by melD) and two α-galactosidases (specified by rafD and melE), as well as presumed ABC-type uptake systems for their carbohydrate substrates.
Bifidobacteria appear to encode both common carbohydrate utilization pathways (e.g., for the metabolism of raffinose family sugars), as well as strain- and species-specific pathways (e.g., melezitose metabolism). Such pathways may reflect common elements in the diets of the hosts of such bacteria, while it may also allow certain species/strains the capacity to effectively colonize the gut or reach higher numbers when the host's diet contains more specialized carbohydrates. In a recent study (88), levels of B. animalis subsp. lactis Bl-04 were shown to be selectively increased 10- to 100-fold on melibiose, xylobiose, raffinose, and maltotriose in a model system of the human colon, indicating that these carbohydrates have the potential to serve as prebiotics.
It is possible that the ability of bifidobacteria to lose or acquire a carbohydrate utilization system is similar to that of L. plantarum, an organism that appears to acquire, shuffle, substitute, or delete carbohydrate utilization systems in response to niche requirements, making it a “natural metabolic engineer” (89). Further investigations will be required to determine how and to what extent specific carbohydrate utilization abilities enable certain bifidobacterial species and strains to colonize and persist in the gastrointestinal tracts of their hosts, and the importance of host diet in this regard.
Supplementary Material
ACKNOWLEDGMENTS
This publication is based on research conducted with the financial support of Science Foundation Ireland (SFI) under grants 07/CE/B1368, 08/SRC/B13404, and SFI/12/RC/2273. K.J.O. was supported by a postgraduate fellowship funded through the Tomar trust, while M.O.M. is a recipient of an HRB postdoctoral fellowship (grant PDTM/20011/9).
We sincerely thank J. Thompson (National Institutes of Health, Bethesda, MD) and N. Emphadinhas (Universidade de Coimbra, Coimbra, Portugal) for supplying various sucrose isomers, Francesca Bottacini for bioinformatics input, Aldert Zomer for performing initial studies, and Stephen Cunningham, Marian Keane, and Lokesh Joshi of the Alimentary Glycoscience Research Cluster, NUIG, Ireland, for kindly supplying α-(1→4)- and α-(1→3)-galactobiose.
Footnotes
Published ahead of print 2 August 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.01775-13.
REFERENCES
- 1.Miyake T, Watanabe K, Watanabe T, Oyaizu H. 1998. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol. Immunol. 42:661–667 [DOI] [PubMed] [Google Scholar]
- 2.Ventura M, O'Connell-Motherway M, Leahy S, Moreno-Munoz JA, Fitzgerald GF, van Sinderen D. 2007. From bacterial genome to functionality; case bifidobacteria. Int. J. Food Microbiol. 120:2–12 [DOI] [PubMed] [Google Scholar]
- 3.Tissier H. 1900. Recherches sur la flore intestinale normale pathologique du nourisson. Thesis. University of Paris, Paris, France [Google Scholar]
- 4.Turroni F, Peano C, Pass DA, Foroni E, Severgnini M, Claesson MJ, Kerr C, Hourihane J, Murray D, Fuligni F, Gueimonde M, Margolles A, De Bellis G, O'Toole PW, van Sinderen D, Marchesi JR, Ventura M. 2012. Diversity of bifidobacteria within the infant gut microbiota. PLoS One 7:e36957. 10.1371/journal.pone.0036957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turroni F, Ribbera A, Foroni E, van Sinderen D, Ventura M. 2008. Human gut microbiota and bifidobacteria: from composition to functionality. Antonie Van Leeuwenhoek 94:35–50 [DOI] [PubMed] [Google Scholar]
- 6.Tannock GW. 1997. Probiotic properties of lactic-acid bacteria: plenty of scope for fundamental R & D. Trends Biotechnol. 15:270–274 [DOI] [PubMed] [Google Scholar]
- 7.Stanton C, Ross RP, Fitzgerald GF, van Sinderen D. 2005. Fermented functional foods based on probiotics and their biogenic metabolites. Curr. Opin. Biotechnol. 16:198–203 [DOI] [PubMed] [Google Scholar]
- 8.Leahy SC, Higgins DG, Fitzgerald GF, van Sinderen D. 2005. Getting better with bifidobacteria. J. Appl. Microbiol. 98:1303–1315 [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Jiang Z, Zhou K, Li P, Liu G, Zhang B. 2007. Screening of bifidobacteria with acquired tolerance to human gastrointestinal tract. Anaerobe 13:215–219 [DOI] [PubMed] [Google Scholar]
- 10.Ventura M, Turroni F, Motherway MO, MacSharry J, van Sinderen D. 2012. Host-microbe interactions that facilitate gut colonization by commensal bifidobacteria. Trends Microbiol. 20:467–476 [DOI] [PubMed] [Google Scholar]
- 11.Scardovi V, Trovatelli ID. 1965. The fructose-6-phosphate shunt as a peculiar pattern of hexose degradation in the genus Bifidobacterium. Ann. Microbiol. 15:19–29 [Google Scholar]
- 12.Macfarlane GT, Steed H, Macfarlane S. 2008. Bacterial metabolism and health-related effects of galacto-oligosaccharides and other prebiotics. J. Appl. Microbiol. 104:305–344 [DOI] [PubMed] [Google Scholar]
- 13.Gibson GR, Roberfroid MB. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401–1412 [DOI] [PubMed] [Google Scholar]
- 14.Mazé A, O'Connell-Motherway M, Fitzgerald GF, Deutscher J, van Sinderen D. 2007. Identification and characterization of a fructose phosphotransferase system in Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 73:545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pokusaeva K, Fitzgerald GF, van Sinderen D. 2011. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 6:285–306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, Pridmore RD, Arigoni F. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garro MS, de Valdez GF, Oliver G, de Giori GS. 1999. Hydrolysis of soya milk oligosaccharides by Bifidobacterium longum CRL 849. Z. Lebensmittelunters. Forsch. A 208:57–59 [Google Scholar]
- 18.Minami Y, Yazawa K, Tamura Z, Tanaka T, Yamamoto T. 1983. Selectivity of utilization of galactosyl-oligosaccharides by bifidobacteria. Chem. Pharm. Bull. (Tokyo) 31:1688–1691 [DOI] [PubMed] [Google Scholar]
- 19.Yazawa K, Imai K, Tamura Z. 1978. Oligosaccharides and polysaccharides specifically utilizable by bifidobacteria. Chem. Pharm. Bull. (Tokyo) 26:3306–3311 [DOI] [PubMed] [Google Scholar]
- 20.French D. 1954. The raffinose family of oligosaccharides. Adv. Carbohydr. Chem. 9:149–184 [DOI] [PubMed] [Google Scholar]
- 21.Rehms H, Barz W. 1995. Degradation of stachyose, raffinose, melibiose and sucrose by different tempe-producing Rhizopus fungi. Appl. Microbiol. Biotechnol. 44:47–52 [DOI] [PubMed] [Google Scholar]
- 22.Wakinaka T, Kiyohara M, Kurihara S, Hirata A, Chaiwangsri T, Ohnuma T, Fukamizo T, Katayama T, Ashida H, Yamamoto K. 2013. Bifidobacterial α-galactosidase with unique carbohydrate-binding module specifically acts on blood group B antigen. Glycobiology 23:232–240 [DOI] [PubMed] [Google Scholar]
- 23.Leder S, Hartmeier W, Marx SP. 1999. α-Galactosidase of Bifidobacterium adolescentis DSM 20083. Curr. Microbiol. 38:101–106 [DOI] [PubMed] [Google Scholar]
- 24.Van Laere KM, Hartemink R, Beldman G, Pitson S, Dijkema C, Schols HA, Voragen AG. 1999. Transglycosidase activity of Bifidobacterium adolescentis DSM 20083 α-galactosidase. Appl. Microbiol. Biotechnol. 52:681–688 [DOI] [PubMed] [Google Scholar]
- 25.Goulas T, Goulas A, Tzortzis G, Gibson GR. 2009. A novel α-galactosidase from Bifidobacterium bifidum with transgalactosylating properties: gene molecular cloning and heterologous expression. Appl. Microbiol. Biotechnol. 82:471–477 [DOI] [PubMed] [Google Scholar]
- 26.Zhao H, Lu L, Xiao M, Wang Q, Lu Y, Liu C, Wang P, Kumagai H, Yamamoto K. 2008. Cloning and characterization of a novel α-galactosidase from Bifidobacterium breve 203 capable of synthesizing Gal-α-1,4 linkage. FEMS Microbiol. Lett. 285:278–283 [DOI] [PubMed] [Google Scholar]
- 27.Garro MS, de Giori GS, de Valdez GF, Oliver G. 1994. α-d-Galactosidase (EC 3.2.1.22) from Bifidobacterium longum. Lett. Appl. Microbiol. 19:16–19 [Google Scholar]
- 28.Hirayama Y, Sakanaka M, Fukuma H, Murayama H, Kano Y, Fukiya S, Yokota A. 2012. Development of a double-crossover markerless gene deletion system in Bifidobacterium longum: functional analysis of the α-galactosidase gene for raffinose assimilation. Appl. Environ. Microbiol. 78:4984–4994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aslanidis C, Schmid K, Schmitt R. 1989. Nucleotide sequences and operon structure of plasmid-borne genes mediating uptake and utilization of raffinose in Escherichia coli. J. Bacteriol. 171:6753–6763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pokusaeva K, O'Connell-Motherway M, Zomer A, Fitzgerald GF, van Sinderen D. 2009. Characterization of two novel α-glucosidases from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 75:1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van den Broek LA, Struijs K, Verdoes JC, Beldman G, Voragen AG. 2003. Cloning and characterization of two α-glucosidases from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 61:55–60 [DOI] [PubMed] [Google Scholar]
- 32.Bacon JS, Dickinson B. 1957. The origin of melezitose: a biochemical relationship between the lime tree (Tilia spp.) and an aphis (Eucallipterus tiliae L.). Biochem. J. 66:289–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hudson CS. 1946. Melezitose and turanose. Adv. Carbohydr. Chem. 2:1–36 [DOI] [PubMed] [Google Scholar]
- 34.De Man JC, Rogosa M, Sharpe ME. 1960. A medium for the cultivation of lactobacilli. J. Appl. Microbiol. 23:130–135 [Google Scholar]
- 35.Terzaghi BE, Sandine WE. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 29:807–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. 2000. Artemis: sequence visualization and annotation. Bioinformatics 16:944–945 [DOI] [PubMed] [Google Scholar]
- 38.O'Connell Motherway M, Zomer A, Leahy SC, Reunanen J, Bottacini F, Claesson MJ, O'Brien F, Flynn K, Casey PG, Munoz JA, Kearney B, Houston AM, O'Mahony C, Higgins DG, Shanahan F, Palva A, de Vos WM, Fitzgerald GF, Ventura M, O'Toole PW, van Sinderen D. 2011. Functional genome analysis of Bifidobacterium breve UCC2003 reveals type IVb tight adherence (Tad) pili as an essential and conserved host-colonization factor. Proc. Natl. Acad. Sci. U. S. A. 108:11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403–410 [DOI] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Riordan K, Fitzgerald GF. 1999. Molecular characterisation of a 5.75-kb cryptic plasmid from Bifidobacterium breve NCFB 2258 and determination of mode of replication. FEMS Microbiol. Lett. 174:285–294 [DOI] [PubMed] [Google Scholar]
- 42.O'Connell Motherway M, O'Driscoll J, Fitzgerald GF, van Sinderen D. 2009. Overcoming the restriction barrier to plasmid transformation and targeted mutagenesis in Bifidobacterium breve UCC2003. Microb. Biotechnol. 2:321–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wells JM, Wilson PW, Le Page RW. 1993. Improved cloning vectors and transformation procedure for Lactococcus lactis. J. Appl. Bacteriol. 74:629–636 [DOI] [PubMed] [Google Scholar]
- 44.Law J, Buist G, Haandrikman A, Kok J, Venema G, Leenhouts K. 1995. A system to generate chromosomal mutations in Lactococcus lactis which allows fast analysis of targeted genes. J. Bacteriol. 177:7011–7018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alvarez-Martin P, O'Connell-Motherway M, van Sinderen D, Mayo B. 2007. Functional analysis of the pBC1 replicon from Bifidobacterium catenulatum L48. Appl. Microbiol. Biotechnol. 76:1395–1402 [DOI] [PubMed] [Google Scholar]
- 46.Zomer A, Fernandez M, Kearney B, Fitzgerald GF, Ventura M, van Sinderen D. 2009. An interactive regulatory network controls stress response in Bifidobacterium breve UCC2003. J. Bacteriol. 191:7039–7049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.AgilentTechnologies 2007 Two-color microarray-based gene expression analysis, version 4.0. G4140-90050. Agilent Technologies, Palo Alto, CA [Google Scholar]
- 48.Garcia de la Nava J, Santaella DF, Alba JC, Carazo JM, Trelles O, Pascual-Montano A. 2003. Engene: the processing and exploratory analysis of gene expression data. Bioinformatics 19:657–658 [DOI] [PubMed] [Google Scholar]
- 49.van Hijum SAFT, De JA, Baerends RJ, Karsens HA, Kramer NE, Larsen R, den Hengst CD, Albers CJ, Kok J, Kuipers OP. 2005. A generally applicable validation scheme for the assessment of factors involved in reproducibility and quality of DNA-microarray data. BMC Genomics 6:77. 10.1186/1471-2164-6-77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Hijum SA, Garcia de la Nava J, Trelles O, Kok J, Kuipers OP. 2003. MicroPreP: a cDNA microarray data pre-processing framework. Appl. Bioinformatics 2:241–244 [PubMed] [Google Scholar]
- 51.Long AD, Mangalam HJ, Chan BY, Tolleri L, Hatfield GW, Baldi P. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276:19937–19944 [DOI] [PubMed] [Google Scholar]
- 52.Mierau I, Leij P, van Swam I, Blommestein B, Floris E, Mond J, Smid E. 2005. Industrial-scale production and purification of a heterologous protein in Lactococcus lactis using the nisin-controlled gene expression system NICE: the case of lysostaphin. Microb. Cell Fact. 4:15. 10.1186/1475-2859-4-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laemmli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685 [DOI] [PubMed] [Google Scholar]
- 54.Börnke F, Hajirezaei M, Sonnewald U. 2001. Cloning and characterization of the gene cluster for palatinose metabolism from the phytopathogenic bacterium Erwinia rhapontici. J. Bacteriol. 183:2425–2430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pokusaeva K, Neves AR, Zomer A, O'Connell-Motherway M, MacSharry J, Curley P, Fitzgerald GF, van Sinderen D. 2010. Ribose utilization by the human commensal Bifidobacterium breve UCC2003. Microb. Biotechnol. 3:311–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Aslanidis C, Schmitt R. 1990. Regulatory elements of the raffinose operon: nucleotide sequences of operator and repressor genes. J. Bacteriol. 172:2178–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Connell Motherway M, Fitzgerald GF, Neirynck S, Ryan S, Steidler L, van Sinderen D. 2008. Characterization of ApuB, an extracellular type II amylopullulanase from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 74:6271–6279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.O'Connell Motherway M, Fitzgerald GF, van Sinderen D. 2011. Metabolism of a plant derived galactose-containing polysaccharide by Bifidobacterium breve UCC2003. Microb. Biotechnol. 4:403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pokusaeva K, O'Connell-Motherway M, Zomer A, MacSharry J, Fitzgerald GF, van Sinderen D. 2011. Cellodextrin utilization by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 77:1681–1690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ryan SM, Fitzgerald GF, van Sinderen D. 2005. Transcriptional regulation and characterization of a novel β-fructofuranosidase-encoding gene from Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 71:3475–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van den Broek LA, van Boxtel EL, Kievit RP, Verhoef R, Beldman G, Voragen AG. 2004. Physico-chemical and transglucosylation properties of recombinant sucrose phosphorylase from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 65:219–227 [DOI] [PubMed] [Google Scholar]
- 62.Kim M, Kwon T, Lee HJ, Kim KH, Chung DK, Ji GE, Byeon E-S, Lee J-H. 2003. Cloning and expression of sucrose phosphorylase gene from Bifidobacterium longum in E. coli and characterization of the recombinant enzyme. Biotechnol. Lett. 25:1211–1217 [DOI] [PubMed] [Google Scholar]
- 63.Kwon T, Kim CT, Lee J-H. 2007. Transglucosylation of ascorbic acid to ascorbic acid 2-glucoside by a recombinant sucrose phosphorylase from Bifidobacterium longum. Biotechnol. Lett. 29:611–615 [DOI] [PubMed] [Google Scholar]
- 64.Trindade MI, Abratt VR, Reid SJ. 2003. Induction of sucrose utilization genes from Bifidobacterium lactis by sucrose and raffinose. Appl. Environ. Microbiol. 69:24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amaretti A, Tamburini E, Bernardi T, Pompei A, Zanoni S, Vaccari G, Matteuzzi D, Rossi M. 2006. Substrate preference of Bifidobacterium adolescentis MB 239: compared growth on single and mixed carbohydrates. Appl. Microbiol. Biotechnol. 73:654–662 [DOI] [PubMed] [Google Scholar]
- 66.Dinoto A, Suksomcheep A, Ishizuka S, Kimura H, Hanada S, Kamagata Y, Asano K, Tomita F, Yokota A. 2006. Modulation of rat cecal microbiota by administration of raffinose and encapsulated Bifidobacterium breve. Appl. Environ. Microbiol. 72:784–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rada V, Bartonova J, Vlkova E. 2002. Specific growth rate of bifidobacteria cultured on different sugars. Folia Microbiol. (Praha) 47:477–480 [DOI] [PubMed] [Google Scholar]
- 68.Turroni F, Strati F, Foroni E, Serafini F, Duranti S, van Sinderen D, Ventura M. 2012. Analysis of predicted carbohydrate transport systems encoded by Bifidobacterium bifidum PRL2010. Appl. Environ. Microbiol. 78:5002–5012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rosenow C, Maniar M, Trias J. 1999. Regulation of the α-galactosidase activity in Streptococcus pneumoniae: characterization of the raffinose utilization system. Genome Res. 9:1189–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jimenez E, Villar-Tajadura MA, Marin M, Fontecha J, Requena T, Arroyo R, Fernandez L, Rodriguez JM. 2012. Complete genome sequence of Bifidobacterium breve CECT 7263, a strain isolated from human milk. J. Bacteriol. 194:3762–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Titgemeyer F, Reizer J, Reizer A, Saier MH. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349–2354 [DOI] [PubMed] [Google Scholar]
- 72.Conejo MS, Thompson SM, Miller BG. 2010. Evolutionary bases of carbohydrate recognition and substrate discrimination in the ROK protein family. J. Mol. Evol. 70:545–556 [DOI] [PubMed] [Google Scholar]
- 73.Bottacini F, Milani C, Turroni F, Sanchez B, Foroni E, Duranti S, Serafini F, Viappiani A, Strati F, Ferrarini A, Delledonne M, Henrissat B, Coutinho P, Fitzgerald GF, Margolles A, van Sinderen D, Ventura M. 2012. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7:e44229. 10.1371/journal.pone.0044229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schneider E, Hunke S. 1998. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol. Rev. 22:1–20 [DOI] [PubMed] [Google Scholar]
- 75.Hama H, Wilson TH. 1992. Primary structure and characteristics of the melibiose carrier of Klebsiella pneumoniae. J. Biol. Chem. 267:18371–18376 [PubMed] [Google Scholar]
- 76.Okazaki N, Jue XX, Miyake H, Kuroda M, Shimamoto T, Tsuchiya T. 1997. A melibiose transporter and an operon containing its gene in Enterobacter cloacae. J. Bacteriol. 179:4443–4445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shimamoto T, Shimamoto T, Xu XJ, Okazaki N, Kawakami H, Tsuchiya T. 2001. A cryptic melibiose transporter gene possessing a frameshift from Citrobacter freundii. J. Biochem. 129:607–613 [DOI] [PubMed] [Google Scholar]
- 78.Silvestroni A, Connes C, Sesma F, De Giori GS, Piard JC. 2002. Characterization of the melA locus for α-galactosidase in Lactobacillus plantarum. Appl. Environ. Microbiol. 68:5464–5471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Andersen J, Barrangou R, Hachem M, Lahtinen S, Goh Y, Svensson B, Klaenhammer T. 2013. Transcriptional analysis of oligosaccharide utilization by Bifidobacterium lactis Bl-04. BMC Genomics 14:312. 10.1186/1471-2164-14-312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hwang DS, Lindegren CC. 1964. Palatinose element of the receptor of the melezitose locus in Saccharomyces. Nature 203:791–792 [DOI] [PubMed] [Google Scholar]
- 81.Degnan BA, Macfarlane GT. 1994. Synthesis and activity of α-glucosidase produced by Bifidobacterium pseudolongum. Curr. Microbiol. 29:43–47 [Google Scholar]
- 82.Igaue I, Watabe H, Oda T, Oyamada K. 1983. Enzymes of human intestinal bacteria active in degrading maltitol. 2. Some characteristics of maltitol-hydrolyzing enzyme (α-glucosidase) from a strain of Bifidobacterium adolescentis. J. Agric. Chem. Soc. Jpn. 57:995–999 [Google Scholar]
- 83.Wool D, Hendrix DL, Shukry O. 2006. Seasonal variation in honeydew sugar content of galling aphids (Aphidoidea: Pemphigidae: Fordinae) feeding on Pistacia: host ecology and aphid physiology. Basic Appl. Ecol. 7:141–151 [Google Scholar]
- 84.Sanz ML, Polemis N, Morales V, Corzo N, Drakoularakou A, Gibson GR, Rastall RA. 2005. In vitro investigation into the potential prebiotic activity of honey oligosaccharides. J. Agric. Food Chem. 53:2914–2921 [DOI] [PubMed] [Google Scholar]
- 85.Schmid K, Schmitt R. 1976. Raffinose metabolism in Escherichia coli K12. Eur. J. Biochem. 67:95–104 [DOI] [PubMed] [Google Scholar]
- 86.Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. 2007. Genomics of actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Oishi K, Sato T, Yokoi W, Yoshida Y, Ito M, Sawada H. 2008. Effect of probiotics, Bifidobacterium breve and Lactobacillus casei, on bisphenol A exposure in rats. Biosci. Biotechnol. Biochem. 72:1409–1415 [DOI] [PubMed] [Google Scholar]
- 88.van Zanten GC, Knudsen A, Roytio H, Forssten S, Lawther M, Blennow A, Lahtinen SJ, Jakobsen M, Svensson B, Jespersen L. 2012. The effect of selected synbiotics on microbial composition and short-chain fatty acid production in a model system of the human colon. PLoS One 7:e47212. 10.1371/journal.pone.0047212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Siezen R, van Hylckama Vlieg J. 2011. Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microb. Cell Fact. 10(Suppl 1):S3. 10.1186/1475-2859-10-S1-S3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.de Ruyter PG, Kuipers OP, Beerthuyzen MM, van Alen-Boerrigter I, de Vos WM. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434–3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.